Abstract
The transforming growth factorβ (TGFβ) superfamily regulates a broad spectrum of biological responses throughout embryonic development and adult life, including cell proliferation and differentiation, epithelial-to-mesenchymal transition, apoptosis, and angiogenesis. TGFβ members initiate signaling by bringing together a complex of serine/threonine kinase receptors that transmit signals through intracellular Smad proteins. Genetic alterations in numerous components of the TGFβ signaling pathway have been associated with several human cancers. In addition, tight regulation of TGFβ signaling is pivotal to the maintenance of homeostasis and the prevention of carcinogenesis. The ubiquitin/proteosome system is one mechanism by which cells regulate the expression and activity of effectors of the TGFβ signaling cascade. Mounting evidence also suggests that disruption of the ubiquitin-dependent degradation of components of the TGFβ pathway leads to the development and progression of cancer. Therefore, understanding how these two pathways intertwine will contribute to the advancement of our knowledge of cancer development.
Keywords: TGFβ, Smad, Smurf, ubiquitination, cancer
Introduction
The transforming growth factor β (TGFβ) superfamily is a large family of multifunctional cytokines involved in a number of biological responses during embryonic development and adult tissue homeostasis [1,2]. Because it promotes cell growth inhibition, apoptosis, and differentiation, TGFβ has been described as a potent tumor suppressor [3,4]. Supporting this notion, mutations in the components of the TGFβ signaling cascade have been identified in a number of human cancers, including hereditary nonpolyposis colon cancer, hepatocellular carcinoma (HCC), and pancreatic and ovarian cancers [5]. TGFβ also functions as a tumor promoter by stimulating angiogenesis, immunosuppression, and epithelial-to-mesenchymal transition (EMT) in later stages of the disease [4,6]. In recent years, ubiquitin-dependent proteosomal degradation has proven to be an important mechanism by which cells control TGFβ signaling output. Therefore, it is likely that disruptions in the proteosomal degradation of TGFβ pathway components may promote the development and progression of tumors. This review will focus on how the TGFβ signaling cascade is regulated by the ubiquitin/proteosome pathway and how deregulation of this may contribute to cancer.
The TGFβ Signaling Pathway
TGFβ is the prototypic member of the TGFβ superfamily, which also includes activins, nodals, bone morphogenetic proteins (BMPs), and anti-Mullerian factor. The cytokines signal through a heteromeric complex of type I and type II serine/threonine kinase receptors. Activation of the receptor complex through ligand binding results in the phosphorylation of the type I receptor by the type II receptor kinase [1–3,7,8]. Subsequently, active type I receptors transiently interact with and phosphorylate receptor-regulated Smads (R-Smads), which are intracellular transducers of TGFβ signals. The specificity of TGFβ or BMP responses is dictated by the ability of BMP type I receptors to phosphorylate and activate the R-Smads, Smad1, Smad5, and Smad8, whereas TGFβ or activin type I receptors phosphorylate the R-Smads, Smad2 and Smad3. Phosphorylated R-Smads then associate with Smad4, the common Smad (co-Smad), and shuttle to the nucleus [1,2,7–9]. By interacting with a large repertoire of transcription factors such as FoxH1, Mixer, LEF-1/TCF, OAZ, GATA-4, or Runx-related proteins, and cofactors such as CBP/p300, c-ski, SnoN, and histone deacetylases (HDACs), Smads either positively or negatively regulate specific transcriptional responses to TGFβ and BMP signaling [1,2,7–9]. A third class of Smads—the inhibitory Smads (I-Smads), which include Smad6 and Smad7—has been identified as negative regulators of TGFβ and BMP signaling. By interacting with type I receptors, I-Smads block the access of R-Smads to their specific receptors and inhibit signaling. In addition, I-Smads can downregulate signaling by targeting cell surface receptors for ubiquitin-dependent proteosomal degradation [1,2,7–9].
Smads contain two well-conserved globular domains known as MH1 and MH2 domains, which are coupled to each other by a divergent proline-rich linker region [1,2,7–9]. Although the C-terminal MH2 domain is highly conserved across all Smads, the amino-terminal domain of I-Smads shows only a weak sequence similarity to the N-terminal MH1 domain of other Smads. Both the MH1 and MH2 domains interact with transcription factors, but only the MH1 domain is able to directly interact with DNA [1,2,7–9]. Furthermore, the MH1 domain contains nuclear localization signals and plays a pivotal role in the nuclear shuttling of Smads [10]. In addition to mediating association with DNA-binding partners, the MH2 is crucial for Smad oligomerization and receptor interaction. It has also been shown to mediate the interaction between Smad2 and Smad3 with the Smad anchor for receptor activation (SARA). This FYVE domain-containing protein, which is mainly localized into early endosomes, enhances the recruitment of R-Smads to TGFβ receptors and facilitates TGFβ signaling [11]. Although the linker region is less conserved among Smads, this region comprises a PY motif that mediates the recruitment of E3 ubiquitin ligases and a number of phosphorylation sites that are important for crosstalk with other signaling pathways, such as receptor tyrosine kinase-mediated pathways [8,12].
Smads are the classic intracellular effectors of TGFβ signaling; however, mounting evidence shows that biological responses can also be elicited through Smad-independent pathways [12]. Furthermore, there is evidence demonstrating that TGFβs and BMPs can signal through MAP kinases (such as ERK, JNK, and p38), PKB/Akt, and LIM kinase 1 [12]. More recently, TGFβ signaling has been shown to regulate EMTand cell migration through PAR6, an important component of the epithelial polarity complex and a regulator of tight junction assembly [13].
Ubiquitin-Dependent Regulation of R-Smads
R-Smads play a pivotal role in the transmission of TGFβ/BMP signaling, and their degradation through the ubiquitin-dependent proteosomal pathway is an important mechanism by which cells tightly control Smad steady-state levels and activity. [14]. Ubiquitination occurs through a three-step process involving ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin ligase (E3) enzymes [15]. E3 ubiquitin ligases are generally divided into three classes: HECT (homologous to the E6-associated protein C-terminus) type, RING (really interesting gene) type, and U-box type [15]. Although HECT domain-containing E3 ligases directly catalyze the transfer of ubiquitin to the substrate, RING and U-box domain E3 ligases act as molecular scaffolds that facilitate the ubiquitination of target proteins [15]. Although structurally related, the U-box domain differs from the RING finger domain, as it uses hydrogen bonds, instead of zinc binding, to stabilize its structure [16].
Smad ubiquitination-related factor 1 (Smurf1) was the first E3 ubiquitin ligase of the C2/WW/HECT domain class to be identified as a regulator of TGFβ/BMP signaling (Figure 1, Table 1) [17]. It was shown to target noninduced BMP-specific Smad1 and Smad5 for degradation through a specific interaction between the Smurf1 WW domain and the PY motif of BMP-regulated R-Smads [17]. The cytoplasmic pool of Smad1 and Smad2 is also regulated by Smurf2 [18,19], a Smurf1-related E3 HECT-containing ubiquitin ligase shown to be expressed in response to TGFβ signaling [20]. For its part, the U-box E3 ligase, CHIP (carboxyl terminus of Hsc70-interacting protein), has also been shown to downregulate Smad1 and Smad4steady-state levels (Figure 1, Table 1) [21].
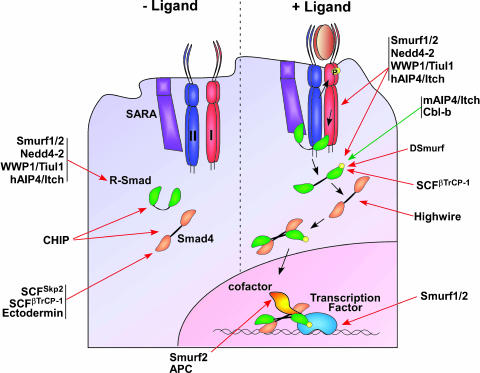
Figure 1.
E3 ubiquitin ligases regulating the TGFβ signaling pathway. HECT domain, RING type, and U-box E3 ubiquitin ligases regulate both basal levels and activated components of the TGFβ signaling pathway. Black arrows illustrate TGFβ signal transduction. Positive (green arrows) and negative (red arrows) regulation of TGFβ signaling components by the ubiquitin/proteosome pathway are also indicated.
Table 1.
E3 Ubiquitin Ligases Targeting Components of the TGFβ Signaling Pathway.
| E3 Ub Ligase | Target | Signaling Dependence | Adaptor | Modulator | Effect | Reference |
| HECT domain | ||||||
| Smurf1 | TβRII/I | A | Smad6/7 | [49] | ||
| BMPRII/I | A | Smad6/7 | Inhibition of BMP signaling in Xenopus and mammalian cells | [52] | ||
| Smad1, Smad5 | B | Downregulation of Smad1 target genes and secondary axis formation | [17] | |||
| B | LMP-1 | Interference with Smurf/Smad interaction; enhancement of BMP responsiveness | [78] | |||
| Smad4 | A (TGFβ) | R/I-Smad | [32] | |||
| Smad7 | B | p300 | Acetylation decreases ubiquitination and enhances the stability of Smad7 | [75] | ||
| B | HDAC1 | Deacetylation enhances ubiquitination and decreases the stability of Smad7 | [43,76] | |||
| Enhancement of Smad7 Ub and promotion of renal fibrosis by Smurf1/2 | [91] | |||||
| RhoA | B | Cdc42/PAR6/aPKC | Cell migration, neurite outgrowth | [106,107] | ||
| A (TGFβ) | PAR6/aPKC | EMT | [104] | |||
| B | Synaptodin | Interference with Smurf1/RhoA interaction; inhibition of podocyte cell migration | [108] | |||
| MEKK2 | A (BMP2) | Regulation of osteoblast activity | [87] | |||
| Runx2 | B | Smad6 | Inhibition of BMP-dependent Runx2-induced transcription | [83] | ||
| A (BMP2) | p300 and HDAC4/5 | Regulation of Runx2 ubiquitin-dependent degradation and bone formation by dynamic acetylation | [85] | |||
| TNF | Induction of Smurf1/Smurf2 expression (which mediates Runx2 degradation) by TNF | [86] | ||||
| Runx3 | A (TGFβ) | p300 and HDAC5 | Regulation of Smurf1/2-mediated Runx3 degradation by dynamic acetylation | [84] | ||
| Smurf2 | TβRII/I | A | Smad7 | Downregulation of TGFβ transcriptional activity | [39] | |
| Alk4 (ActRIB) | A | Smad7 | FKBP12 | Regulation of the duration of activin signaling | [53] | |
| Smad1, Smad2 | B | Robust decrease of Smad1 levels but modest decrease of Smad2 levels; specific inhibition of Smad1 function | [18,19] | |||
| Smad2 | A | Downregulation of Smad2 levels; inhibition of TGFβ transcriptional activity | [18,19] | |||
| Smad4 | B | I-Smad | [32] | |||
| Smad4 | A (TGFβ) | R-Smad | [32] | |||
| SnoN | A (TGFβ) | Smad2 | [23] | |||
| RNF11 | B | Blocks Smurf2-dependent inhibibion of TGFb signaling | [99] | |||
| DSmurf | Mad | A | Morphologic defects in larvae cuticle | [88] | ||
| Nedd4-2 | TβRII/I | A | Smad6/7 | Downregulation of TGFβ-dependent transcriptional activity | [24] | |
| Smad2 | A | Downregulation of TGFβ-dependent transcriptional activity | [24] | |||
| Smad4 | B | [32] | ||||
| WWP1/Tiul1 | TβRII/I | A | Smad7 | Downregulation of TGFβR complex and Smad signaling; inhibition of TGFβ growth arrest | [25,26] | |
| Smad2 | A | TGIF | Enhancement of Smad2 turnover; inhibition of TGFβ growth arrest | [26] | ||
| Smad4 | B | R/I-Smad | [32] | |||
| hAIP4/Itch | Smad7 | B | Stabilization of activated TβRI/Smad7 complex; inhibition of TGBβ signaling in a Ub-independent manner | [54] | ||
| HEF1 | B | Smad3 | Induction of HEF1 degradation through Smad3 by TGFβ | [58,59] | ||
| mAIP4/Itch | Smad2 | A | Increase in Smad2 phosphorylation and TGFβ growth arrest | [112] | ||
| RING type | ||||||
| Single subunit | ||||||
| Highwire | BMP-like signaling Medea | A | Regulation of synaptic boutons | [36] | ||
| Ectodermin | Smad4 | B | Restriction of excessive BMP signaling in vegetal hemisphere of Xenopus blastula; restriction of TGFβ/nodal signaling to animal pole; TGFβ growth inhibition in HepG2 cells | [37] | ||
| Cbl-b | Smad2 | A | Increase in Smad2 phosphorylation; in vivo resistance of Cbl-b-/- T cells to TGFβ | [31] | ||
| Multisubunit | ||||||
| APC | SnoN | A (TGFβ) | Smad3 | CDH1 | Regulation of cell cycle | [56,57] |
| F-box type | ||||||
| SCFSkp2 | Smad4 R100T | B | Accelerated degradation | [65] | ||
| Smad4 G65V | B | Accelerated degradation | [65] | |||
| ROC-1/SCFβTrCP1 | Smad3 | A | p300 | [27] | ||
| Smad4 | A (TGFβ) | Jab1 | Inhibition of TGFβ growth arrest | [33,34] | ||
| Smad4 R100T | B | Accelerated degradation | [65] | |||
| Smad4 G65V | B | Accelerated degradation | [65] | |||
| MFB1 | DAF-7 pathway | Negative regulation of Dauer formation | [28] | |||
| U-box type | ||||||
| CHIP | Smad1, Smad4 | B | [21] |
A, interaction enhanced by signaling; B, interaction observed in basal state.
Ubiquitin-mediated proteosomal degradation is important not only for controlling spurious activation of TGFβ/BMP signaling cascades but also for turning off signaling output once the biological response has occurred. The growing number of E3 ubiquitin ligases that are able to downregulate activated R-Smads and their ubiquitous expression in adult tissues reflect the importance of the proper regulation of these signaling molecules for the maintenance of tissue homeostasis. Degradation of phosphorylated Smad2 was first observed in human keratinocytes, and although the E2-conjugating enzymes UbcH5b/c and Ubc3 were implicated in the transfer of ubiquitin moieties onto phospho-Smad2, a candidate E3 ubiquitin ligase responsible for substrate specificity had not been identified at the time [22]. In recent years, a number of E3 ubiquitin ligases have been shown to target activated Smad2 for proteosomal degradation (Figure 1, Table 1). Although Smurf2 constitutively regulates R-Smads, the association between Smurf2 and Smad2/3 is enhanced on TGFβ stimulation, suggesting a role for Smurf2 in the regulation of activated R-Smads [18,23]. Two other members of the E3 HECT domain ubiquitin ligase class, Nedd4-2 and WWP1/Tiul1, constitutively bind Smad2, and as with Smurf2/Smad2 interaction, these constitutive interactions are enhanced in response to an activated TGFβ type I receptor (TβRI) (Figure 1, Table 1) [24–26]. Interestingly, although Nedd4-2 enhanced the polyubiquitination and degradation of Smad2 in the presence of an activated TβRI, the ability of WWP1/Tiul1 to target activated Smad2 for ubiquitin-dependent proteosomal degradation is not as clear. Although Komuro et al. [25] demonstrated that the interaction between WWP1/Tiul1 and Smad2 was enhanced in the presence of activated TβRI, WWP1/Tiul1 does not appear to promote the polyubiquitination and degradation of activated Smad2. However, Seo et al. [26] found that WWP1/Tiul1 was able to induce the ubiquitin-dependent degradation of Smad2 in the presence of the transcriptional corepressor, TGIF. Therefore, it is possible that the ability of WWP1/Tiul1 to mediate ubiquitin-dependent proteosomal degradation of Smad2 relies on the presence of additional protein partners such as TGIF. Interestingly, Smurfs, Nedd4-2, and WWP1/Tiul1 show a distinct pattern of expression in human tissues and human carcinoma cell lines [24,25]. Although WWP1/Tiul1 protein expression levels were found to be moderate to high in the heart, liver, skeletal muscles, and kidneys, only low levels of Smurf1 expression were observed in these tissues [25]. Similarly, Nedd4-2 and Smurf2 also have distinct distribution patterns in certain tissues such the kidneys, prostate, and testes [24].
The regulation of activated R-Smads is not exclusive to HECT domain-containing E3 ubiquiting ligases, but may also occur through the multisubunit RING E3 ligase, Skp-1/Cul/F-Box (SCF) complex (Figure 1, Table 1). Roc-1, a component of SCFFbw1a/βTrcP1, interacts with Smad3 and promotes the SCFFbw1a/βTrcP1-dependent ubiquitination and degradation of phosphorylated Smad3 in the cytoplasm [27]. As the interaction between Roc1 and Smad3 is enhanced in the presence of the transcriptional coactivator p300, it is thought that SCF/Roc1-mediated proteosomal degradation is necessary to terminate Smad3 transcriptional activity [27]. Although molecular targets have not been identified, MFB-1, a novel F-box-type ubiquitin ligase, negatively regulates Dauer formation in Caenorhabditis elegans by modulating the DAF-7/TGFβ-like signaling pathway [28]. Proteosomal degradation of activated Smad1 has also been reported to occur through a complex comprising the ornithine decarboxylase antizyme (Az) and the 20S proteosome β subunit, HsN3 [29]. The targeting of R-Smads for degradation by all three classes of E3 ubiquitin ligases (Figure 1, Table 1) suggests that ubiquitin-dependent proteosomal degradation is an important mechanism by which a cell controls its ability to respond to both TGFβ and BMP signaling and that this occurs only when appropriate, thereby preventing aberrant activation of cascades.
Although the polyubiquitination of R-Smads by a variety of E3 ligases appears to negatively regulate TGFβ/BMP signaling, there is also emerging evidence that suggests a role for this posttranslational modification in the enhancement of TGFβ signaling (Figure 1, Table 1). Loss of the AIP4/Itch E3 ligase in mouse embryonic fibroblasts results in resistance to TGFβ-induced cell growth inhibition [30]. Although the turnover rate of TβRIs and Smad2 remained unchanged, phosphorylation of Smad2 was decreased in AIP4/Itch-/- cells when compared to AIP4/Itch+/- cells [30]. Biochemical studies demonstrated that wild-type, but not catalytically inactive, AIP4/Itch mediated the TGFβ-dependent ubiquitination of Smad2, as well as enhanced the interaction between Smad2 and activated TβRI [30]. Recently, the E3 ubiquitin ligase Cbl-b has also been shown to enhance TGFβ-dependent Smad2 phosphorylation in T cells [31]. Therefore, by promoting Smad2 phosphorylation, E3 ubiquitin ligases may also function as positive regulators of TGFβ signaling.
Regulation of Smad4 through the Ubiquitin-Dependent Proteosomal Pathway
Being a common intracellular effector of both the TGFβ and BMP signaling pathways, Smad4 is a critical point at which both cascades can be modulated to maintain homeostasis. Like R-Smads, Smad4 levels are also regulated by HECT domain E3 ubiquitin ligases such as Smurf1, Smurf2, Nedd4-2, and WWP1/Tiul1 (Figure 1, Table 1) [32]. However, because Smad4 lacks a PY motif, it cannot directly associate with HECT-containing E3 ligases, but rather recruits the enzymes through adaptors such as I-Smads and R-Smads [32]. Consequently, mutations disrupting the interaction between adaptors and Smad4 also interfere with the ubiquitin-dependent degradation of Smad4 [32]. Overexpression of the Jun-activating domain binding protein 1 (Jab1), a subunit of COP9 signalosome, promotes the interaction between Smad4 and the Roc1/SCFβTrCP1 complex, resulting in the ubiquitination and proteosomal degradation of Smad4 [33,34]. Ectopic expression of oncogenic Ras has also been shown to enhance Smad4 proteosomal degradation. However, the molecular mechanism regulating this process has yet to be defined [35]. Drosophila Highwire (Hiw), a RING-H2-type E3 ligase, was shown to bind the Smad4-like protein Medea (Med) in yeast-two hybrid and in vitro binding assays [36]. Although ubiquitination and proteosomal degradation of Med were not directly demonstrated, complete genetic removal of Med in hiw mutants suppresses excessive synaptic growth displayed in hiw single mutants. The hiw phenotype is also suppressed in wit (a BMP type II receptor) mutants [36]. Although the neuronal overexpression of yeast UBP2 or Drosophila Fat Facet (Faf) deubiquitinases resulted in synaptic overgrowth in wild-type larvae, ectopic expression of UBP2 and Faf did not cause synaptic overexpansion in med or wit mutants [36]. Taken together, these observations suggest that Hiw regulates BMP signaling through Med in a ubiquitin-dependent mechanism [36]. Recently, Ectodermin, a single-subunit RING-type E3 ligase, was shown to prevent excessive BMP signaling in the animal pole of Xenopus blastula, allowing for the proper development of ectodermal and neuronal tissues, as well as the restriction of TGFβ/nodal-mediated mesodermal induction to the vegetal hemisphere of the embryo [37]. Furthermore, human Ectodermin was also shown to restrict TGFβ-induced growth arrest in HepG2 cells [37]. Ectodermin appears to mediate these biological responses by targeting Smad4 for ubiquitination and proteosomal degradation, which result in the downregulation of both TGFβ and BMP signaling cascades [37].
In addition to being an important posttranslational modification by which Smad4 protein levels are controlled, ubiquitination is also a mechanism by which Smad4 activity is modulated [38]. Monoubiquitination of lysine 507 in the MH2 domain of Smad4 enhances the association of co-Smad with R-Smads and promotes Smad4 transcriptional activity [38]. Therefore, as for Smad2, ubiquitination of Smad4 can act to both positively and negatively regulate Smad4 function.
Role of Smads as Adaptor for E3 Ligases
Although Smads have clearly been shown to be substrates for E3 ubiquitin ligases, they can also function as adaptors to recruit ubiquitin ligases to other target proteins. This novel role for Smads was first described by Kavsak et al. [39]. It was shown that, upon TGFβ stimulation, the I-Smad, Smad7 interacts with Smurf2 and promotes the export of the complex from the nucleus to the cell surface, where Smad7 acts as a bridge to target Smurf2 to the TGFβ receptor complex (Figure 1, Table 1) [39]. Although the Smad7 MH2 domain interacts with activated TβRI, its PY motif associates with the WW domains of Smurf2 [17,39–41]. Furthermore, the amino-terminal domain (NTD) of Smad7, through a leucine-rich motif, recruits the E2-conjugating enzyme UbcH7 to the HECT domain of Smurf2 and stimulates Smurf2 catalytic activity [42]. Once recruited to the receptor complex, Smurf2 ubiquitinates Smad7 and promotes the degradation of both Smad7 and the receptors [39], which occurs in the lipid-raft/caveolar-dependent endocytic pathway [43]. Alternatively, TGFβ receptors also internalize through the clathrin-dependent endocytic route where they associate with SARA and cPML and promote Smad-dependent signaling [11,43–47]. Because TGFβ ligand does not seem to preferentially target the receptors to one compartment over another, it is not known what causes receptors to partition into two different internalization compartments [43]. However, it is likely that proper partitioning is required for the fine-tuning of TGFβ superfamily signaling. In fact, a recent study demonstrated that memory of activin exposure relied on the time spent by the activin-activin receptor signaling complex in the clathrin-dependent endocytic pathway and was abolished by Smad7/Smurf2 [48].
I-Smads can recruit HECT-E3 ligases other than Smurf2 to receptor complexes. Smad7 was also shown to associate with Smurf1 and to recruit it to the TGFβ receptor complex (Table 1) [49]. The nuclear export of the Smad7/Smurf1 complex is mediated by chromosomal region maintenance 1 (CRM1), an importin β-related nuclear transport receptor, and the nuclear export signal located in the HECT domain of Smurf1 [50,51]. As with TGFβ receptor complexes, Smad6 and Smad7 are capable of targeting Smurf1 to cell surface activin and BMP receptors, and of promoting their ubiquitination and turnover [52,53]. Interestingly, the recruitment of Smad7/Smurf1 to the activin type I receptor, ALK4 (ActRIB), is enhanced by FKBP12, an intracellular inhibitor of TGFβ signaling [53]. Furthermore, Smad6 and Smad7 also recruit non-Smurf HECT E3 ligases such as Nedd4-2, WWP1/Tiul1, and human AIP4/Itch [24–26,54] to TGFβ receptor complexes (Figure 1, Table 1). Nedd4-2 and WWP1/Tiul1 promote the ubiquitin-dependent degradation of TGFβ receptor complexes, which results in the downregulation of TGFβ-dependent transcription and growth arrest [24–26]. Although the mechanism by which human AIP4/Itch inhibits TGFβ signaling has not been described, it appears to be independent of the ubiquitin-dependent proteosomal degradation of receptors and Smads [54]. Interestingly, although human AIP4/Itch inhibits TGFβ signaling, the mouse homolog promotes the phosphorylation of Smad2 and the induction of TGFβ signaling [30]. This difference may be due to tissue or cell type-specific effects. Recent evidence also suggests an important role for deubiquitinases such as UCH37 in the regulation of TGFβ receptor complexes [55]. A balanced recruitment of both deubiquitinases and E3 ubiquitin ligases is most likely required to assure proper TGFβ and BMP responses.
In addition to I-Smads, R-Smads play an important role in recruiting E3 ubiquitin ligases to specific substrates. Smad2 is known to recruit Smurf2 and to promote the ubiquitination and proteosomal degradation of the transcriptional corepressor SnoN (Figure 1, Table 1) [23]. Likewise, the anaphase-promoting complex (APC) requires Smad3 as an adaptor for the efficient ubiquitination and degradation of SnoN (Table 1) [56,57]. The TGFβ-dependent degradation of SnoN, either through Smurf2 or APC, is thought to be a mechanism through which the amplitude of TGFβ signals is modulated as SnoN is itself a negative regulator of TGFβ target genes. Smad3 has also been shown to bind human AIP4/Itch and HEF1, and to promote ubiquitin-dependent proteosomal degradation of this Cas family member (Figure 1, Table 1) [58,59].
Ubiquitination and Degradation of Oncogenic Smad Mutants
A number of inactivating mutations have been identified in Smad2 and Smad4 in a wide range of human carcinomas, including colorectal, pancreatic, and lung carcinomas [3–5]. In most cases, missense and nonsense mutations cluster in the MH2 domain of Smads and have been shown to interfere with Smad homo-oligomerization, hetero-oligomerization, DNA binding, and nuclear translocation [5]. However, several mutations also affect Smad protein stability. The HCC-derived mutation glutamine 407 to arginine (Q407R), as well as the colorectal cancer-associated mutation leucine 369 to arginine (L369R), in the MH2 domain of Smad2 is highly unstable and, in the case of Q407R, rapidly targets Smad2 for ubiquitin-dependent proteosomal degradation [60,61]. A nonsense mutation of Smad4 identified in pancreatic adenocarcinomas, which results in the deletion of the last 38 amino acids of the MH2 domain, not only inhibits Smad2 recruitment and DNA binding but also targets Smad4 for degradation [62].
Although most Smad mutations localize to the MH2 domain, several mutations have also been described in the MH1 domain [38,63,64]. An arginine-to-cysteine mutation at residue 133 of the MH1 domain of Smad2 leads to the increased ubiquitination and degradation of Smad2 [63]. Likewise, tumor-derived Smad4 L43S, G65V, R100T, and P130S mutants all exhibit accelerated polyubiquitination and proteosomal degradation when compared to wild-type Smad4 [38,63,64]. A recent study shows that the SCF complex, comprising either βTrCP-1 or Skp2 as the F-box component, exhibits stronger binding to cancer-derived Smad4 mutants (R1 00Tand G65V) and catalyzes a more rapid degradation of these mutants when compared to wild-type Smad4 (Table 1) [65]. In summary, a number of inactivating mutations in Smad2 and Smad4 cause accelerated ubiquitin-dependent proteosomal degradation and likely result in aberrant TGFβ signaling, thereby promoting cancer development.
Regulation of Smads by other Posttranslational Modifications
Protein stability and function are regulated by not only ubiquitination but also a number of other ubiquitin-like modifications, such as SUMOylation, NEDDylation, and ISGylation [66–69]. Of these three posttranslational modifications, SUMOylation is the only one to date to be implicated in the regulation of TGFβ pathway components. SUMOylation of target substrates appears to play an important role in the modulation of subcellular localization, protein-protein, and protein-DNA interactions, as well as enzyme activity and ubiquitin-dependent degradation [66–69]. Several studies have demonstrated that the SUMO E3 ligase PIASγ (protein inhibitor of activated STATγ) interacts with Smad4 and promotes its SUMOylation, which results in enhanced nuclear accumulation, protein stability, and transcriptional activity [70–72]. However, a recent report also demonstrates that SUMOylation decreases the ability of Smad4 to transactivate an artificial GAL4 promoter, suggesting that SUMOylation may affect Smad4 transcriptional activity either positively or negatively on different promoters [73]. PIASγ has also been shown to modify Smad3 [74]; therefore, the contradictory effects resulting from Smad4 SUMOylation may be, in part, explained by the simultaneous SUMOylation of Smad3. Smad3 modification may inhibit complex formation with Smad4 or regulate Smad3 binding to DNA, which could both result in the downregulation of Smad4 transcriptional activity. Alternatively, SUMOylation of Smad4 may lead to the recruitment of cofactors, and the specificity of this recruitment may be cell type-specific, which could also explain the different effects observed on Smad4 transcriptional activity.
A number of proteins, including transcription factors and other nuclear proteins, have been found to be modified by the addition of an acetyl group on the q amino group of lysine residues. Like SUMOylation, the functional consequences of acetylation are as diverse as increasing protein stability, regulating protein-protein and protein-DNA interactions, and inhibiting nuclear export. The histone acetyltransferase p300 was shown to interact and acetylate Smad7 on two lysine residues located in the amino-terminus of the I-Smad. Although acetylation neither interfered with Smad7/Smurf1 complex formation nor prevented nuclear export or recruitment of the complex to cell surface receptors, it did appear to protect Smad7 from polyubiquitination [75]. Furthermore, deacetylation of Smad7 by HDACs enhances both Smad7 polyubiquitination and turnover [76]. Taken together, these observations suggest that a balance between acetylation and deacetylation controls Smad7 protein stability. Acetylation of Smad7 may protect it from premature Smurf1-mediated degradation, allowing the recruitment of the Smad7/Smurf1 complex to cell surface receptors. However, once Smad7/Smurf1 is recruited to the receptors, deacetylation may be induced to promote the ubiquitination and degradation of the TGFβ receptor/Smad7/Smurf1 complex.
Biological Role of Smurfs
As negative regulators of TGFβ and BMP signaling, Smurfs have been proven to have key functions during both normal biological responses (such as EMT, cellular migration, and bone formation) and pathogenic processes (such as fibrosis and cancer).
Recently, a number of in vitro and in vivo studies have provided insight on the physiological role of the ubiquitin/proteosome pathway in the downregulation of BMP signaling during bone development. Ectopic expression of Smurf1 induces the proteosomal degradation of Smad5 and thereby blocks BMP-induced osteogenic conversion of pluripotent C2Cl2 myoblasts [77]. Recent studies have also demonstrated that LMP-1, a LIM domain protein, inhibits Smad1 and Smad5 recruitment to Smurf1 and subsequent degradation, resulting in enhanced BMP signaling and bone nodule mineralization [78]. Gain-of-function studies in mice have demonstrated that overexpression of Smurf1, under the control of the Col1a1 promoter, leads to inhibition of osteoblast differentiation and reduced bone formation [79]. In contrast, a subsequent study showed that mice in which ectopic expression of Smurf1 was driven by the Col1a2 promoter exhibited no appreciable phenotype [80]. However, mating Smurf1 and Smad6 transgenic animals produced double-transgenic pups with a similar but more severe phenotype than that of the Smad6 transgenic mice, which included delayed chondrocyte hypertrophy and postnatal dwarf ism with osteopenia [80]. This phenotype was due to an impairment of BMP signaling, as decreased phospho-Smad1, Smad5, and Smad8 were observed in trabecular bone sections [80]. Although these studies show different phenotypes for Smurf1 transgenic mice, likely due to the use of different promoters to drive the overexpression of the transgene, both studies show that Smurf1 plays a specific role in bone formation in vivo, even if only a supportive role to Smad6. Interestingly, Smurf1 has also been shown to regulate BMP-induced embryonic lung growth by down regulating Smad1 and Smad5, suggesting that the requirement of Smurf1 for proper BMP signaling is important for homeostasis in a number of tissues [81].
The Runx family of transcription factors plays critical functions during development and disease, and all three Runx proteins have been shown to interact with R-Smads and to regulate several TGFβ/BMP target genes [82]. In addition, Runx proteins are themselves regulated by components of the TGFβ/BMP pathway [82]. Overexpression of Smurf1 in 2T3 osteoblasts downregulates both Smad1 and Runx2 protein levels and inhibits terminal osteoblast differentiation [79]. Moreover, Smad6 recruits Smurf1, Smurf2, and WWP1/Tiul1 to downregulate Runx2 protein levels (Table 1) [83]. Recent evidence also suggests that, although TGFβ/BMP-dependent acetylation of Runx2 and Runx3 by p300 counteracts Smurf1-dependent ubiquitination and degradation, HDAC4- and HDAC5-mediated deacetylation of Runx proteins appears to promote their turnover [84,85]. Interestingly, tumor necrosis factor promotes Runx2 ubiquitin-dependent proteosomal degradation by upregulating Smurf1 and Smurf2 protein expression in osteoblasts [86]. Therefore, regulation of Runx2 by Smurfs may also occur independently of BMP signaling (Table 1).
Although overexpression studies in cell culture or gain-of-function studies in mice have confirmed the importance of Smurfs in Smad-dependent TGFβ/BMP signaling, loss-of-function studies have demonstrated that disruption of the mouse Smurf1 gene does not alter Smad-dependent signaling but rather affects BMP-induced osteoblast activity by promoting the ubiquitination and destruction of MEKK2, an activator of JNK signaling (Table 1) [87]. In Drosophila, disruption of DSmurf activity leads to both spatial and temporal expansions of phosphorylated MAD, an R-Smad-like protein [88]. Morphologically, expansion of phospho-MAD results in the appearance of a posterior hole in the cuticle, as well as hindgut defects, in mutant embryos (Figure 1, Table 1) [88]. Overlapping the expression and activity of numerous E3 ubiquitin ligases may explain why loss of Smurf1 activity does not significantly affect TGFβ or BMP signaling in mice. However, in Drosophila, where only two E3 ubiquitin ligases (DSmurf and Hiw) have been described to negatively regulate BMP-like signaling, disruption of DSmurf activity has more severe effects on Smad-dependent signaling, thereby highlighting their pivotal role in regulating Smad function in vivo.
Although TGFβs have been shown to inhibit the proliferation of most cell types, their principal effect on mesenchymal cells is to stimulate the proliferation and production of the extracellular matrix, and to mediate fibrogenesis [89]. Fibrotic diseases such as scleroderma, pulmonary fibrosis, liver cirrhosis, and a variety of nephropathies have been linked to aberrant TGFβ signaling [89]. Moreover, evidence suggests that disruption of the ubiquitin/proteosome-dependent regulation of TGFβ signaling promotes fibrosis. A recent study of glomeruli isolated from rats with antithymocyte serum nephritis demonstrated down regulation of Smad2 protein levels that are inversely correlated with increased Smurf2 levels [90]. Similarly, progressive fibrosis and enhanced TGFβ signaling in kidneys from a mouse model with progressive tubulointerstitial fibrosis were associated with increased Smurf1/2 protein levels and a concomitant decreased in Smad7 protein levels (Table 1) [91]. Interestingly, gene expression profiling of scleroderma-associated lung fibroblasts revealed increased Smad7 and Smurf2 expression in response to TGFβ stimulation [92]. Furthermore, two studies have shown that deregulated Smad7 expression is associated with impaired TGFβ signaling in scleroderma (Ssc) fibroblasts [93,94]. Dong et al. [93] showed that decreased Smad7 expression in Ssc fibroblasts was associated with increased phospho-Smad2/3 levels and enhanced TGFβ-dependent PAI-1 gene expression, suggesting that decreased Smad7 expression resulted in enhanced TGFβ signaling. In contrast, Asano et al. [94] reported that Ssc fibroblasts exhibited a marked increase in Smad7 expression and enhanced phospho-Smad2 and TβRI proteins levels, and hypothesized that impaired Smad7-dependent degradation of TβRI could be due to mutations in either Smad7 or Smurfs. However, overexpression of wild-type Smad7 or Smurfs in Ssc fibroblasts did not affect TβRI levels, suggesting that other components of the ubiquitin/proteosome pathway may be disrupted in Ssc fibroblasts [94]. UbcH7, which is recruited by Smad7 to Smurf2 [42], may be mutated in Ssc fibroblasts, and this may affect TβRI turnover. Alternatively, Caveolin-1, which was shown to regulate Smad7/Smurf2-mediated receptor degradation [43], is downregulated in lung fibroblasts of scleroderma patients [95], and this may be responsible for impaired receptor degradation.
Disruption of TGFβ signaling is commonly observed in human cancers, and genetic alterations of different components of the TGFβ signaling cascade, such as the receptors Smad2, Smad4, and Smad7, have been described in a number of pancreatic, lung, breast, gastrointestinal, and gynecologic cancers [2,3,5]. Being important regulators of various components of the TGFβ signaling cascade, misregulated expression or aberrant function of E3 ubiquitin ligases, such as Smurfs, Nedd4-2, WWP1/Tiul1, AIP4/Itch, Ectodermin, and the SCF complex, would gravely affect TGFβ signal transmission and potentially result in human cancer development and progression. cDNA microarray-based comparative genomic hybridization analysis of a set of pancreatic carcinoma cell lines has identified Smurf1 in DNA amplifications [96]. Likewise, reverse transcription-polymerase chain reaction studies have shown that human carcinoma cell lines such as colon HT-29, breast MDA-MB-231, gastric MKN-1, and ovarian OVCAR-5 all display high levels of one or more E3 ligases, including Smurf2, Ectodermin, Nedd4-2, and WWP1/Tiul1 [24,25,37]. In addition, PRAJA, a RING-H2 E3 ubiquitin ligase that targets ELF (a positive regulator of Smad4) for degradation is overexpressed in two gastric cancer cell lines (NCI-187 and SNU-1) and likely blocks TGFβ signaling by downregulating Smad4 activity through ELF [97]. Because HT-29 and MKN-1 cells have also been described as being resistant to TGFβ growth inhibition [24], overexpression of E3 ubiquitin ligases in proliferating cells likely results in downregulation of TGFβ signaling, and, consequently, allows these cells to escape TGFβ-induced growth inhibition and to participate in tumor development.
Although evidence of aberrant TGFβ signaling resulting from altered E3 ubiquitin ligase activity is still scarce in human cancers, high expression levels of Smurf2, associated with low levels of Smad2 phosphorylation, have been detected in esophageal squamous carcinoma [98]. This suggests that downregulation of TGFβ signaling by Smurf2 is not limited to cell lines maintained in culture indefinitely but actually promotes tumor development in humans. In contrast, upregulation of TGFβ signaling through downregulation of E3 ubiquitin ligase activity is also likely to enhance the tumor promoter activity of TGFβ in later stages of the disease. RNF11, a RING-H2 finger protein highly expressed in prostate and invasive breast cancers, has been shown to block Smurf2-dependent activity and to promote TGFβ signaling in human tumors [99,100]. RNF11 has also been shown to interact with Smurf1, AIP4/Itch, and WWP1/Tiul1; thus, it may be a novel common adaptor for E3 ubiquitin ligases that regulate TGFβ signaling [99,100]. Interestingly, recent studies have also shown that Smurf2, upregulated by telomere attrition, uses the p53 and Rb pathways to induce replicative senescence through E3 ubiquitin ligase-independent activity [101]. These data also suggest a novel function for Smurf2 in tumor development, which is independent of its role in the TGFβ signaling cascade.
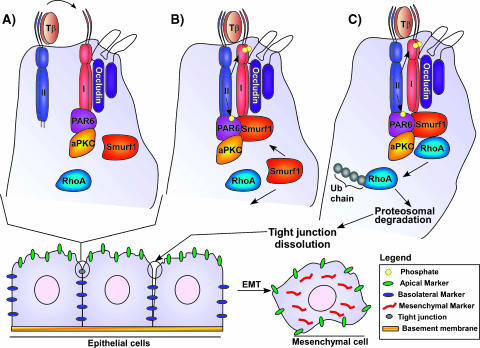
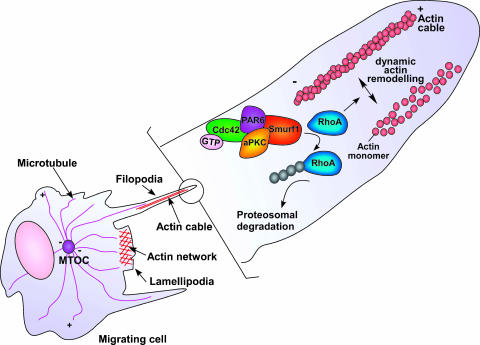
The importance of TGFβ signaling in cancer is not limited to its capacity to promote growth inhibition and apoptosis in early cancer development, but also includes its ability to induce angiogenesis, immunosuppression, and epithelial-to-mesenchymal (EMT) transition (Fig. 2) in later stages of the disease [2]. EMT is a multistep process involving dissolution of epithelial tight junctions; disruption of adherens junctions; cytoskeletal reorganization; loss of cell polarity; repression of epithelial markers such as E-cadherin, ZO-1, and b4 integrin; and upregulation of mesenchymal proteins such as vimentin [102]. Regulation of EMT by TGFβ is commonly thought to occur through the induction of a mesenchymal gene expression profile in either a Smad-dependent or a Smad-independent mechanism [103]. However, recent evidence reveals a new mechanism by which TGFβ regulates EMT in a Smad-independent and transcription-independent manner (Figure 2) [104]. In polarized epithelial cells, TβRIs and TGFβ type II receptors (TβRII) have distinct localization patterns [104]. Although TβRII is localized to puncta distributed over the cell surface, occludin restricts TβRI to tight junctions where it recruits PAR6 (Figure 2A) [104,105]. In response to TGFβ stimulation, TGFβRII is recruited to tight junctions where it interacts with TβRI and directly phosphorylates PAR6 (Figure 2B) [104]. TGFβ-dependent phosphorylation of PAR6 allows the recruitment of Smurf1, which in turn mediates the localized ubiquitination and degradation of RhoA, resulting in tight junction dissolution and EMT (Figure 2C) [104]. Smurf1, through the Cdc42/PAR6/PKCη complex, also regulates dynamic actin cytoskeletal remodeling by mediating localized RhoA degradation in filopodia and lamellipodia (Figure 3) [106]. Altogether, the Smurf1-mediated degradation of RhoA appears to be involved in multiple steps during the progression of cancer. By contributing to the dissolution of tight junctions, Smurf1 supports the transdifferentiation of epithelial cells to a fibroblastoid phenotype and, subsequently, by regulating cytoskeletal remodeling, promotes cell migration. Smurf1-dependent down regulation of RhoA has also been reported to regulate neurite outgrowth in Neuro2A neuroblastoma cells, as well as cell migration in kidney podocytes [107,108]. Synaptopodin, a proline-rich actin-associated protein, regulates podocyte cell migration through RhoA ubiquitin-dependent degradation by competing with Smurf1 for RhoA binding [108]. Taken together, these data suggest that both normal and transformed cells regulate dynamic actin cytoskeletal remodeling through localized Smurf1-mediated ubiquitination and degradation of RhoA (Figure 3).
Figure 2.
Smurf1-regulated RhoA degradation mediates EMT. (A) TβRI is restricted to tight junctions by occludin. In tight junctions, TβRI interacts with PAR6. (B) In response to TGFβ, TβRII is recruited to tight junctions and forms a complex with TβRI and PAR6. TβRII phosphorylates PAR6, thereby stimulating the recruitment of Smurf1 to tight junctions. (C) Smurf1 promotes the ubiquitination and degradation of RhoA, resulting in tight junction dissolution and EMT.
Figure 3.
Smurf1-dependent RhoA degradation mediates cell migration. The Cdc42/PAR6/aPKC complex recruits Smurf1 to filopodia and lamellipodia where it locally degrades RhoA and promotes cell migration.
Conclusions
Like many other signaling cascades, the TGFβ and BMP pathways are tightly regulated at different levels by ubiquitin-dependent proteosomal degradation. The complexity by which the ubiquitin/proteosome pathway regulates what appears, at first, to be a simple linear TGFβ signaling pathway is astounding. By regulating unactivated cytoplasmic pools of R-Smads, the ubiquitin/proteosome pathway prevents spurious activation of the TGFβ/BMP cascade and assures that cells remain competent to receive incoming signaling cues. In addition, the targeted ubiquitination and degradation of receptors, Smads, and transcription factors, in response to TGFβ or BMP stimulation, are a means by which signaling is turned off. A growing list of HECT domain, RING type, and U-box E3 ubiquitin ligases, both directly or through adaptors, targets components of the signaling pathway for degradation and thus assures proper signaling over a wide variety of tissues and organs. Exciting new evidence shows that E3 ubiquitin ligases not only act as negative regulators but also enhance TGFβ signaling by promoting R-Smad phosphorylation. By controlling the turnover of many tumor suppressors and oncoproteins, the ubiquitin/proteosome pathway plays a pivotal role in the development and progression of cancer [15,109–111]. Alteration of ubiquitin-dependent proteosomal degradation of TGFβ signaling pathway components is also associated with cancer development. Overexpression of E3 ubiquitin ligases, as described in a number of human carcinomas and cancer cell lines, likely contributes to cancer development by downregulating TGFβ pathway components, resulting in decreased TGFβ-dependent expression of genes involved in growth inhibition and apoptosis. Adventitious expression of Smurf1 may promote cancer invasion and metastasis by potentiating EMT and cell migration [104]. Undeniably, the pivotal role held by E3 ubiquitin ligases in the regulation of TGFβ-dependent biological responses makes it a worthy target for the development of small-molecule or peptide-based inhibitors for use in future therapeutic treatments.
Acknowledgement
We thank Christine LeRoy for insightful comments.
Footnotes
Research in the laboratory was supported by grants from the Canadian Institute for Health Research and the National Cancer Institute of Canada with funds from the Cancer Research Society. L.A. is a Canada Research Chair and L.I. holds a Heart and Stroke Doctoral Award.
References
- 1.Feng XH, Derynck R. Specificity and versatility in TGF-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 2.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3(11):807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 3.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-β signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 4.Derynck R, Akhurst RJ, Balmain A. TGF-β signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 5.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 6.Wakefield LM, Roberts AB. TGF-β signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22–29. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 7.Moustakas A, Souchelnytskyi S, Heldin C-H. Smad regulation in TGF-β signal transduction. J Cell Sci. 2001;114:4359–4369. doi: 10.1242/jcs.114.24.4359. [DOI] [PubMed] [Google Scholar]
- 8.Attisano L, Wrana JL. Signal transduction by the TGF-b superfamily. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 9.ten Dijke P, Miyazono K, Heldin C-H. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem Sci. 2000;25:64–70. doi: 10.1016/s0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- 10.Reguly T, Wrana JL. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends Cell Biol. 2003;13(5):216–220. doi: 10.1016/s0962-8924(03)00075-8. [DOI] [PubMed] [Google Scholar]
- 11.Le Roy C, Wrana JL. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6(2):112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 12.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118(Pt 16):3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 13.Bose R, Wrana JL. Regulation of Par6 by extracellular signals. Curr Opin Cell Biol. 2006;18(2):206–212. doi: 10.1016/j.ceb.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Izzi L, Attisano L. Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene. 2004;23(11):2071–2078. doi: 10.1038/sj.onc.1207412. [DOI] [PubMed] [Google Scholar]
- 15.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 16.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27(7):368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 18.Lin X, Liang M, Feng X-H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohashi N, Yamamoto T, Uchida C, Togawa A, Fukasawa H, Fujigaki Y, Suzuki S, Kitagawa K, Hattori T, Oda T, et al. Transcriptional induction of Smurf2 ubiquitin ligase by TGF-beta. FEBS Lett. 2005;579(12):2557–2563. doi: 10.1016/j.febslet.2005.03.069. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Xin H, Xu X, Huang M, Zhang X, Chen Y, Zhang S, Fu XY, Chang Z. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol Cell Biol. 2004;24(2):856–864. doi: 10.1128/MCB.24.2.856-864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo RS, Massague J. Ubiquitin-dependent degradation of TGF-β-activated Smad2. Nat Cell Biol. 1999;1:472–478. doi: 10.1038/70258. [DOI] [PubMed] [Google Scholar]
- 23.Bonni S, Wang H-R, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL. TGF-β induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- 24.Kuratomi G, Komuro A, Goto K, Shinozaki M, Miyazawa K, Miyazono K, Imamura T. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor. Biochem J. 2005;386(Pt 3):461–470. doi: 10.1042/BJ20040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, Miyazawa K. Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1) Oncogene. 2004;23(41):6914–6923. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- 26.Seo SR, Lallemand F, Ferrand N, Pessah M, L'Hoste S, Camonis J, Atfi A. The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J. 2004;23(19):3780–3792. doi: 10.1038/sj.emboj.7600398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuchi M, Imamura T, Chiba T, Ebisawa T, Kawabata M, Tanaka K, Miyazono K. Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of Roc1 and associated proteins. Mol Biol Cell. 2001;12:1430–1443. doi: 10.1091/mbc.12.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aoyama Y, Urushiyama S, Yamada M, Kato C, Ide H, Higuchi S, Akiyama T, Shibuya H. MFB-1, an F-box-type ubiquitin ligase, regulates TGF-beta signalling. Genes Cells. 2004;9(11):1093–1101. doi: 10.1111/j.1365-2443.2004.00792.x. [DOI] [PubMed] [Google Scholar]
- 29.Gruendler C, Lin Y, Farley J, Wang T. Proteasomal degradation of Smad1 induced by bone morphogenetic proteins. J Biol Chem. 2001;276(49):46533–46543. doi: 10.1074/jbc.M105500200. [DOI] [PubMed] [Google Scholar]
- 30.Bai Y, Yang C, Hu K, Elly C, Liu YC. Itch E3 ligase-mediated regulation of TGF-beta signaling by modulating smad2 phosphorylation. Mol Cell. 2004;15(5):825–831. doi: 10.1016/j.molcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Wohlfert EA, Gorelik L, Mittler R, Flavell RA, Clark RB. Cutting edge: deficiency in the E3 ubiquitin ligase Cbl-b results in a multifunctional defect in T cell TGF-beta sensitivity in vitro and in vivo. J Immunol. 2006;176(3):1316–1320. doi: 10.4049/jimmunol.176.3.1316. [DOI] [PubMed] [Google Scholar]
- 32.Moren A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280(23):22115–22123. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- 33.Wan M, Cao X, Wu Y, Bai S, Wu L, Shi X, Wang N, Cao X. Jab1 antagonizes TGF-beta signaling by inducing Smad4 degradation. EMBO Rep. 2002;3:171–176. doi: 10.1093/embo-reports/kvf024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wan M, Tang Y, Tytler EM, Lu C, Jin B, Vickers SM, Yang L, Shi X, Cao X. Smad4 protein stability is regulated by ubiquitin ligase SCF beta-TrCP1. J Biol Chem. 2004;279(15):14484–14487. doi: 10.1074/jbc.C400005200. [DOI] [PubMed] [Google Scholar]
- 35.Saha D, Datta PK, Beauchamp RD. Oncogenic ras represses transforming growth factor-beta/Smad signaling by degrading tumor suppressor Smad4. J Biol Chem. 2001;276(31):29531–29537. doi: 10.1074/jbc.M100069200. [DOI] [PubMed] [Google Scholar]
- 36.McCabe BD, Hom S, Aberle H, Fetter RD, Marques G, Haerry TE, Wan H, O'Connor MB, Goodman CS, Haghighi AP. Highwire regulates presynaptic BMP signaling essential for synaptic growth. Neuron. 2004;41(6):891–905. doi: 10.1016/s0896-6273(04)00073-x. [DOI] [PubMed] [Google Scholar]
- 37.Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121(1):87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 38.Moren A, Hellman U, Inada Y, Imamura T, Heldin CH, Moustakas A. Differential ubiquitination defines the functional status of the tumor suppressor smad4. J Biol Chem. 2003;278(35):33571–33582. doi: 10.1074/jbc.M300159200. [DOI] [PubMed] [Google Scholar]
- 39.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets TGFβ receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y-Y, Grinnell BW, Richardson MA, Topper JN, Gimbrone M, Jr, Wrana JL, et al. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 41.Chong PA, Lin H, Wrana JL, Forman-Kay JD. An expanded WW domain recognition motif revealed by the interaction between Smad7 and the E3 ubiquitin ligase Smurf2. J Biol Chem. 2006;281(25):17069–17075. doi: 10.1074/jbc.M601493200. [DOI] [PubMed] [Google Scholar]
- 42.Ogunjimi AA, Briant DJ, Pece-Barbara N, Le Roy C, Di Guglielmo GM, Kavsak P, Rasmussen RK, Seet BT, Sicheri F, Wrana JL. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19(3):297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 43.Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5(5):410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 44.Ehrlich M, Shmuely A, Henis YI. A single internalization signal from the di-leucine family is critical for constitutive endocytosis of the type II TGF-beta receptor. J Cell Sci. 2001;114(Pt 9):1777–1786. doi: 10.1242/jcs.114.9.1777. [DOI] [PubMed] [Google Scholar]
- 45.Hayes S, Chawla A, Corvera S. TGF beta receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J Cell Biol. 2002;158(7):1239–1249. doi: 10.1083/jcb.200204088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penheiter SG, Mitchell H, Garamszegi N, Edens M, Dore JJ, Jr, Leof EB. Internalization-dependent and -independent requirements for transforming growth factor beta receptor signaling via the Smad pathway. Mol Cell Biol. 2002;22(13):4750–4759. doi: 10.1128/MCB.22.13.4750-4759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431(7005):205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- 48.Jullien J, Gurdon J. Morphogen gradient interpretation by a regulated trafficking step during ligand-receptor transduction. Genes Dev. 2005;19(22):2682–2694. doi: 10.1101/gad.341605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki C, Murakami G, Fukuchi M, Shimanuki T, Shikauchi Y, Imamura T, Miyazono K. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J Biol Chem. 2002;277(42):39919–39925. doi: 10.1074/jbc.M201901200. [DOI] [PubMed] [Google Scholar]
- 51.Tajima Y, Goto K, Yoshida M, Shinomiya K, Sekimoto T, Yoneda Y, Miyazono K, Imamura T. Chromosomal region maintenance 1 (CRM1)-dependent nuclear export of Smad ubiquitin regulatory factor 1 (Smurf1) is essential for negative regulation of transforming growth factor-beta signaling by Smad7. J Biol Chem. 2003;278(12):10716–10721. doi: 10.1074/jbc.M212663200. [DOI] [PubMed] [Google Scholar]
- 52.Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14(7):2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamaguchi T, Kurisaki A, Yamakawa N, Minakuchi K, Sugino H. FKBP12 functions as an adaptor of the Smad7-Smurf1 complex on activin type I receptor. J Mol Endocrinol. 2006;36(3):569–579. doi: 10.1677/jme.1.01966. [DOI] [PubMed] [Google Scholar]
- 54.Lallemand F, Seo SR, Ferrand N, Pessah M, L'Hoste S, Rawadi G, Roman-Roman S, Camonis J, Atfi A. AIP4 restricts transforming growth factor-beta signaling through a ubiquitination-independent mechanism. J Biol Chem. 2005;280(30):27645–27653. doi: 10.1074/jbc.M500188200. [DOI] [PubMed] [Google Scholar]
- 55.Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Dijke PT, Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24(54):8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- 56.Stroschein SL, Bonni S, Wrana JL, Luo K. Smad3 recruits the anaphase promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 2001;15:2822–2836. doi: 10.1101/gad.912901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wan Y, Liu X, Kirschner MW. The anaphase-promoting complex mediates TGF-β signaling by targeting SnoN for destruction. Mol Cell. 2001;8:1027–1039. doi: 10.1016/s1097-2765(01)00382-3. [DOI] [PubMed] [Google Scholar]
- 58.Feng L, Guedes S, Wang T. Atrophin-1-interacting protein 4/human Itch is a ubiquitin E3 ligase for human enhancer of filamentation 1 in transforming growth factor-beta signaling pathways. J Biol Chem. 2004;279(28):29681–29690. doi: 10.1074/jbc.M403221200. [DOI] [PubMed] [Google Scholar]
- 59.Liu X, Elia AE, Law SF, Golemis EA, Farley J, Wang T. A novel ability of Smad3 to regulate proteasomal degradation of a Cas family member HEF1. EMBO J. 2000;19:6759–6769. doi: 10.1093/emboj/19.24.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L-C, Bapat B, Gallinger S, Andrulis I, et al. MADR2 maps to 18q21 and encodes a TGFβ MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 61.Dumont E, Lallemand F, Prunier C, Ferrand N, Guillouzo A, Clement B, Atfi A, Theret N. Evidence for a role of Smad3 and Smad2 in stabilization of the tumor-derived mutant Smad2. 407R. J Biol Chem. 2003;278(27):24881–24887. doi: 10.1074/jbc.M212496200. [DOI] [PubMed] [Google Scholar]
- 62.Maurice D, Pierreux CE, Howell M, Wilentz RE, Owen MJ, Hill CS. Loss of Smad4 function in pancreatic tumors: C-terminal truncation leads to decreased stability. J Biol Chem. 2001;276(46):43175–43181. doi: 10.1074/jbc.M105895200. [DOI] [PubMed] [Google Scholar]
- 63.Xu J, Attisano L. Mutations in the tumour suppressors Smad2 and Smad4 inactivate TGFβ signalling by targeting Smads to the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 2000;97:4820–4825. doi: 10.1073/pnas.97.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moren A, Itoh S, Moustakas A, Dijke P, Heldin CH. Functional consequences of tumorigenic missense mutations in the amino-terminal domain of Smad4. Oncogene. 2000;19(38):4396–4404. doi: 10.1038/sj.onc.1203798. [DOI] [PubMed] [Google Scholar]
- 65.Liang M, Liang YY, Wrighton K, Ungermannova D, Wang XP, Brunicardi FC, Liu X, Feng XH, Lin X. Ubiquitination and proteolysis of cancer-derived Smad4 mutants by SCFSkp2. Mol Cell Biol. 2004;24(17):7524–7537. doi: 10.1128/MCB.24.17.7524-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desterro JM, Rodriguez MS, Hay RT. Regulation of transcription factors by protein degradation. Cell Mol Life Sci. 2000;57(8–9):1207–1219. doi: 10.1007/PL00000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Melchior F. SUMO—nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 68.Muller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2(3):202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 69.Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28(6):321–328. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- 70.Lin X, Liang M, Liang YY, Brunicardi FC, Feng XH. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J Biol Chem. 2003;278(33):31043–31048. doi: 10.1074/jbc.C300112200. [DOI] [PubMed] [Google Scholar]
- 71.Lee PS, Chang C, Liu D, Derynck R. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J Biol Chem. 2003;278(30):27853–27863. doi: 10.1074/jbc.M301755200. [DOI] [PubMed] [Google Scholar]
- 72.Lin X, Liang M, Liang YY, Brunicardi FC, Melchior F, Feng XH. Activation of transforming growth factor-beta signaling by SUMO-1 modification of tumor suppressor Smad4/DPC4. J Biol Chem. 2003;278(21):18714–18719. doi: 10.1074/jbc.M302243200. [DOI] [PubMed] [Google Scholar]
- 73.Long J, Wang G, He D, Liu F. Repression of Smad4 transcriptional activity by SUMO modification. Biochem J. 2004;379(Pt 1):23–29. doi: 10.1042/BJ20031867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Imoto S, Sugiyama K, Muromoto R, Sato N, Yamamoto T, Matsuda T. Regulation of transforming growth factor-β signaling by protein inhibitor of activated STAT, PIASy through Smad3. J Biol Chem. 2003;278(36):34253–34258. doi: 10.1074/jbc.M304961200. [DOI] [PubMed] [Google Scholar]
- 75.Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10(3):483–493. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- 76.Simonsson M, Heldin CH, Ericsson J, Gronroos E. The balance between acetylation and deacetylation controls Smad7 stability. J Biol Chem. 2005;280(23):21797–21803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- 77.Ying SX, Hussain ZJ, Zhang YE. Smurf1 facilitates myogenic differentiation and antagonizes the bone morphogenetic protein-2-induced osteoblast conversion by targeting Smad5 for degradation. J Biol Chem. 2003;278(40):39029–39036. doi: 10.1074/jbc.M301193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sangadala S, Boden SD, Viggeswarapu M, Liu Y, Titus L. LMP-1 potentiates BMP responsiveness via a novel interaction with Smurf1 resulting in decreased ubiquitination of Smads. J Biol Chem. 2006;281(25):17212–17219. doi: 10.1074/jbc.M511013200. [DOI] [PubMed] [Google Scholar]
- 79.Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR, Chen D. Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J Biol Chem. 2004;279(13):12854–12859. doi: 10.1074/jbc.M313294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horiki M, Imamura T, Okamoto M, Hayashi M, Murai J, Myoui A, Ochi T, Miyazono K, Yoshikawa H, Tsumaki N. Smad6/Smurf1 overexpression in cartilage delays chondrocyte hypertrophy and causes dwarfism with osteopenia. J Cell Biol. 2004;165(3):433–445. doi: 10.1083/jcb.200311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi W, Chen H, Sun J, Chen C, Zhao J, Wang YL, Anderson KD, Warburton D. Overexpression of Smurf1 negatively regulates mouse embryonic lung branching morphogenesis by specifically reducing Smad1 and Smad5 proteins. Am J Physiol Lung Cell Mol Physiol. 2004;286(2):L293–L300. doi: 10.1152/ajplung.00228.2003. [DOI] [PubMed] [Google Scholar]
- 82.Bae SC, Lee YH. Phosphorylation, acetylation and ubiquitination: the molecular basis of RUNX regulation. Gene. 2006;366(1):58–66. doi: 10.1016/j.gene.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 83.Shen R, Chen M, Wang YJ, Kaneki H, Xing L, O'Keefe RJ, Chen D. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J Biol Chem. 2006;281(6):3569–3576. doi: 10.1074/jbc.M506761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jin YH, Jeon EJ, Li QL, Lee YH, Choi JK, Kim WJ, Lee KY, Bae SC. Transforming growth factor-beta stimulates p300-dependent RUNX3 acetylation, which inhibits ubiquitination-mediated degradation. J Biol Chem. 2004;279(28):29409–29417. doi: 10.1074/jbc.M313120200. [DOI] [PubMed] [Google Scholar]
- 85.Jeon EJ, Lee KY, Choi NS, Lee MH, Kim HN, Jin YH, Ryoo HM, Choi JY, Yoshida M, Nishino N, et al. Bone morphogenetic protein-2 stimulates RUNX2 acetylation. J Biol Chem. 2006;281(24):16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 86.Kaneki H, Guo R, Chen D, Yao Z, Schwarz EM, Zhang YE, Boyce BF, Xing L. Tumor necrosis factor promotes Runx2 degradation through upregulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem. 2006;281(7):4326–4333. doi: 10.1074/jbc.M509430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121(1):101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Podos SD, Hanson KK, Wang YC, Ferguson EL. The DSmurf ubiquitin-protein ligase restricts BMP signaling spatially and temporally during Drosophila embryogenesis. Dev Cell. 2001;1(4):567–578. doi: 10.1016/s1534-5807(01)00057-0. [DOI] [PubMed] [Google Scholar]
- 89.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J. 2004;18(7):816–827. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- 90.Togawa A, Yamamoto T, Suzuki H, Fukasawa H, Ohashi N, Fujigaki Y, Kitagawa K, Hattori T, Kitagawa M, Hishida A. Ubiquitin-dependent degradation of Smad2 is increased in the glomeruli of rats with anti-thymocyte serum nephritis. Am J Pathol. 2003;163(4):1645–1652. doi: 10.1016/S0002-9440(10)63521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fukasawa H, Yamamoto T, Togawa A, Ohashi N, Fujigaki Y, Oda T, Uchida C, Kitagawa K, Hattori T, Suzuki S, et al. Downregulation of Smad7 expression by ubiquitin-dependent degradation contributes to renal fibrosis in obstructive nephropathy in mice. Proc Natl Acad Sci USA. 2004;101(23):8687–8692. doi: 10.1073/pnas.0400035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Renzoni EA, Abraham DJ, Howat S, Shi-Wen X, Sestini P, Bou-Gharios G, Wells AU, Veeraraghavan S, Nicholson AG, Denton CP, et al. Gene expression profiling reveals novel TGFbeta targets in adult lung fibroblasts. Respir Res. 2004;5(1):24. doi: 10.1186/1465-9921-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dong C, Zhu S, Wang T, Yoon W, Li Z, Alvarez RJ, ten Dijke P, White B, Wigley FM, Goldschmidt-Clermont PJ. Deficient Smad7 expression: a putative molecular defect in scleroderma. Proc Natl Acad Sci USA. 2002;99(6):3908–3913. doi: 10.1073/pnas.062010399. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Asano Y, Ihn H, Yamane K, Kubo M, Tamaki M. Impaired Smad7-Smurf-mediated negative regulation of TGF-beta signaling in scleroderma fibroblasts. J Clin Invest. 2004;113(2):253–264. doi: 10.1172/JCI16269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tourkina E, Gooz P, Pannu J, Bonner M, Scholz D, Hacker S, Silver RM, Trojanowska M, Hoffman S. Opposing effects of protein kinase C alpha and protein kinase C epsilon on collagen expression by human lung fibroblasts are mediated via MEK/ERK and caveolin-1 signaling. J Biol Chem. 2005;280(14):13879–13887. doi: 10.1074/jbc.M412551200. [DOI] [PubMed] [Google Scholar]
- 96.Bashyam MD, Bair R, Kim YH, Wang P, Hernandez-Boussard T, Karikari CA, Tibshirani R, Maitra A, Pollack JR. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia. 2005;7(6):556–562. doi: 10.1593/neo.04586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Saha T, Vardhini D, Tang Y, Katuri V, Jogunoori W, Volpe EA, Haines D, Sidawy A, Zhou X, Gallicano I, et al. RING finger-dependent ubiquitination by PRAJA is dependent on TGF-beta and potentially defines the functional status of the tumor suppressor ELF. Oncogene. 2006;25(5):693–705. doi: 10.1038/sj.onc.1209123. [DOI] [PubMed] [Google Scholar]
- 98.Fukuchi M, Fukai Y, Masuda N, Miyazaki T, Nakajima M, Sohda M, Manda R, Tsukada K, Kato H, Kuwano H. High-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Res. 2002;62(24):7162–7165. [PubMed] [Google Scholar]
- 99.Subramaniam V, Li HX, Wong M, Kitching R, Attisano L, Wrana JL, Zubovits J, Burger A, Seth A. The RING-H2 protein RNF11 is overexpressed in breast cancer and is a target of Smurf2 E3 ligase. Br J Cancer. 2003;89(8):1538–1544. doi: 10.1038/sj.bjc.6601301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kitching R, Wong MJ, Koehler D, Burger AM, Landberg G, Gish G, Seth A. The RING-H2 protein RNF11 is differentially expressed in breast tumours and interacts with HECT-type E3 ligases. Biochim Biophys Acta. 2003;1639(2):104–112. doi: 10.1016/j.bbadis.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 101.Zhang H, Cohen SN. Smurf2 up-regulation activates telomere-dependent senescence. Genes Dev. 2004;18(24):3028–3040. doi: 10.1101/gad.1253004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4(8):657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 103.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 104.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGF-beta receptors controls epithelial cell plasticity. Science. 2005;307(5715):1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 105.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307(5715):1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 106.Wang H-R, Zhang Y, Ozdamar B, Ogunjimi AO, Alexandrova E, Thomsen GH, Wrana JL. The ubiquitin ligase Smurf1 targets the Small GTPase RhoA for degradation and regulates cell polarity and protrusion formation. Science. 2003;302(5651):1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- 107.Bryan B, Cai Y, Wrighton K, Wu G, Feng XH, Liu M. Ubiquitination of RhoA by Smurf1 promotes neurite outgrowth. FEBS Lett. 2005;579(5):1015–1019. doi: 10.1016/j.febslet.2004.12.074. [DOI] [PubMed] [Google Scholar]
- 108.Asanuma K, Yanagida-Asanuma E, Faul C, Tomino Y, Kim K, Mundel P. Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat Cell Biol. 2006;8(5):485–491. doi: 10.1038/ncb1400. [DOI] [PubMed] [Google Scholar]
- 109.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 110.Pray TR, Parlati F, Huang J, Wong BR, Payan DG, Bennett MK, Issakani SD, Molineaux S, Demo SD. Cell cycle regulatory E3 ubiquitin ligases as anticancer targets. Drug Resist Updat. 2002;5(6):249–258. doi: 10.1016/s1368-7646(02)00121-8. [DOI] [PubMed] [Google Scholar]
- 111.Sakamoto KM. Ubiquitin-dependent proteolysis: its role in human diseases and the design of therapeutic strategies. Mol Genet Metab. 2002;77(1–2):44–56. doi: 10.1016/s1096-7192(02)00146-4. [DOI] [PubMed] [Google Scholar]
- 112.Bai S, Shi X, Yang X, Cao X. Smad6 as a transcriptional corepressor. J Biol Chem. 2000;275:8267–8270. doi: 10.1074/jbc.275.12.8267. [DOI] [PubMed] [Google Scholar]