Abstract
There has been very little undisputed evidence for recombination in animal mitochondrial DNA (mtDNA) provided so far. Previous unpublished results suggestive of mtDNA recombination in the scorpion family Buthidae, together with cytological evidence for a unique mechanism of mitochondrial fusion in that family, prompted us to investigate this group in more details. First, we sequenced the complete mtDNA genome of Mesobuthus gibbosus, and chose two genes opposing each other (16S and coxI). We then sequenced 150 individuals from the natural populations of four species of Buthidae (Old World genera Buthus and Mesobuthus). We observed strong evidence for widespread recombination through highly significant negative correlations between linkage disequilibrium and physical distance in three out of four species. The evidence is further confirmed when using five other tests for recombination and by the presence of a high amount of homoplasy in phylogenetic trees.
Keywords: cytochrome oxidase subunit I, linkage disequilibrium, mitofusion, mitochondrial DNA, scorpions, Buthidae
1. Introduction
Mitochondrial DNA (mtDNA) is widely used for phylogeographic and phylogenetic studies for two principal reasons. First, mtDNA is a compact double-stranded circular DNA molecule (approximately 15–16 kb) and second, recombination (the exchange of homologous DNA sequences between different chromosomes) is thought to be absent in animals owing to the clonal inheritance of mtDNA. Numerous phylogenetic studies rely on mtDNA genes (both protein-coding and ribosomal RNA genes) amplified through conserved polymerase chain reaction (PCR) primers (Simon et al. 1994). Most models of genetic divergence and phylogenetic methods assume absence of recombination. Thus, major conclusions drawn from these analyses may be inaccurate since tree-building algorithms are misled by recombination (Avise 1994; Schierup & Hein 2000; Posada & Crandall 2002).
Despite the fact that animal mitochondria undoubtedly have the necessary toolkit of enzymes to recombine (Thyagarajan et al. 1996; Eyre-Walker et al. 1999; Yaffe 1999; Rokas et al. 2003), evidence for recombination in animal mtDNA is scarce (Rokas et al. 2003). Evidence based on different indirect methods was found in humans, mice (Mus musculus), flatfish (Platichthys spp.), frogs (Rana spp.) and bivalve mussels (Mytilus spp.) However, only in the last case is there a direct experimental confirmation for homologous recombination owing to biparental inheritance (Awadalla et al. 1999; Ladoukakis & Zouros 2001). Whether human mtDNA is really recombining has been questioned since Awadalla et al. (1999) used a potentially uncertain measure of linkage disequilibrium (LD) in their test for recombination (Ingman et al. 2000; Kivisild & Villems 2000; Kumar et al. 2000) and there is a possibility of sequencing errors in their data (Macaulay et al. 1999; Wiuf 2001; Innan & Nordborg 2002).
While working on mtDNA phylogeographies of Old World buthid scorpions (genera Buthus and Mesobuthus; Gantenbein & Largiadèr 2002, 2003; Gantenbein et al. 2003), results were repeatedly obtained that led to the suspicion that there might be recombination in the mtDNA of some of these species. These suspicions were further fuelled by cytological evidence, reporting the fusion of mitochondria into a ring (mitofusion) in the Buthidae scorpion family. The unique cytological phenomenon of mitofusion together with the preliminary statistical evidence for recombination prompted the investigation of this issue in more detail.
Here, we present strong statistical evidence for recombination in newly sequenced populations of west and Central Asian scorpions of the genus Mesobuthus and in previously published scorpion populations (Gantenbein & Largiadèr 2002, 2003; Gantenbein et al. 2001, 2003), sequenced at two mitochondrial loci, the large ribosomal rRNA unit (16S) and a fragment of the cytochrome oxidase subunit I (coxI). In order to precisely estimate the physical distance between the two sequenced loci, which is required for the correlation between genetic LD and distance, we sequenced the complete mitochondrial genome of the scorpion Mesobuthus gibbosus, which will be published elsewhere (Gantenbein in progress).
2. Material and methods
(a) Samples
The DNA sequences from the GenBank of three published datasets were extracted (Gantenbein & Largiadèr 2002, 2003; Gantenbein et al. 2003), including four different buthid species, Buthus mardochei (Simon 1878; Morocco, North Africa), Mesobuthus caucasicus (Nordmann 1840; Central Asia), M. eupeus (C. L. Koch 1839; Anatolia and Central Asia) and M. gibbosus (Brullé 1832; Anatolia and Greece), for which several haplotypes have been sequenced. For the three Mesobuthus species, we added more localities and sample sizes and genotyped specimens for two mitochondrial (mt) fragments (large ribosomal rRNA subunit 16S, 344 bp, and cytochrome c oxidase subunit I, coxI, 461 bp). The samples of M. caucasicus and M. eupeus from Central Asia (Turkmenistan, Kazakhstan, Uzbekistan and Tajikistan) were collected between June 2000 and May 2002. New sampling sites and a detailed list of identified haplotypes can be downloaded at http://www.science.marshall.edu/fet/euscorpius/mt_buthid.htm.
(b) DNA methods
DNA was extracted according to a standard phenol–chloroform/ethanol precipitation method of 100% ethanol-stored tissue samples (Gantenbein & Largiadèr 2002). Approximately 200–400 ng total genomic DNA was used for 20 μl PCR reactions as previously reported. We used the same PCR profiles as in Gantenbein et al. (2003) and Gantenbein & Largiadèr (2003). Amplicons were checked for clean single band amplification on 1–2% agarose gel, and subsequently purified as previously described (Gantenbein et al. 2003). Sequencing reactions were carried out using one of the PCR primers (Gantenbein & Largiadèr 2003) and were run on an automated sequencer (either ABI377XL or on a capillary sequencer ABI3730). All sequence traces were inspected manually for sequencing errors. Haplotypes with singletons were reamplified and resequenced from both directions. In order to determine the distance between the two gene fragments (16S and coxI), the complete mitochondrial genome of M. gibbosus from the Greek mainland was sequenced by amplifying three overlapping fragments and using the primer walking technique (B. Gantenbein, unpublished data). The sequences reported in this paper have been deposited in the EMBL nucleotide sequence database (accession numbers AJ783454–AJ783614).
(c) Statistical tests for recombination
All sequences were aligned using ClustalW1.5 (Thompson et al. 1997) and by eye. The coxI data could be unambiguously aligned because of the presence of an open reading frame. Following Posada & Crandall (2001) and Wiuf et al. (2001), the five following recombination tests were applied: (i) Recombination Detection Program (RDP; Martin & Rybicki 2000), (ii) Geneconv (Sawyer 1989), (iii) Maxchi (Maynard Smith 1992), (iv) Chimaera (Posada & Crandall 2001; a slight modification of Maxchi) and (v) the homoplasy test (Maynard Smith & Smith 1998). The first four tests were run using RDP (Martin & Rybicki 2000). In RDP, we set the highest acceptable p value to 0.05 (=the probability that sequences could share high identities in potentially recombinant regions by chance alone) and we corrected for multiple comparisons using the standard Bonferroni method for Geneconv, and sequential Bonferroni for RDP, Maxchi and Chimaera (Sokal & Rohlf 1995; Martin & Rybicki 2000). In RDP p values are binomial and are of the Karlin–Altschul-type in Geneconv, and χ2-distributed in Maxchi and Chimaera. The significance of χ2 peaks in Maxchi and Chimaera were determined with 1000 permutations. Tests were run on the concatenated 16S and coxI data or on the protein-coding coxI exclusively. The homoplasy test (Maynard Smith & Smith 1998) is based on an estimation of expected homoplasies under clonality and compares this estimate with the true number of homoplasies in a dataset. When computing the homoplasy test, we only considered third coding positions of the coding region (coxI) since non-coding regions characterized by high mutation rate biases can lead to erroneous results when applying this test (Piganeau & Eyre-Walker 2004). As an out-group for the estimation of the effective number of sites, the species Androctonus australis (L., 1758; Buthidae) was included. These tests were run using the Qbasic program available at http://www.biols.susx.ac.uk/home/John_Maynard_Smith/.
Additionally, LD between polymorphic sites using two different measures of LD, |D′| (Lewontin 1964) and R2 (Hill & Robertson 1968) was estimated, the latter being more robust to variation in mutation rates (Awadalla et al. 1999; Kivisild & Villems 2000; Kumar et al. 2000; Meunier & Eyre-Walker 2001; Wiuf 2001; Innan & Nordborg 2002; Piganeau & Eyre-Walker 2004). LD was estimated using the LDhat computer package (available at http://www.stats.ox.ac.uk/~mcvean/LDhat/LDhat.html). |D′| and R2 versus physical distance was then correlated. A spacer fragment, was included corresponding to 5038 bp in M. gibbosus, thus, the spacer fragment was approximated to ∼5 kb for all correlations in all four species (16S-5 kb-coxI). The correlation was also repeated with maximum power going the longer way around the mitochondrial genome with a ∼10 kb spacer fragment (coxI-10 kb-16S). Significance of the Pearson correlation coefficient was assessed by using the one-tailed Mantel test computed over 10 000 permutations.
3. Results
The DNA sequence datasets for the four species contain considerable polymorphism in terms of the total number of segregating sites (table 1). The significant decline of LD versus distance is regarded as strong evidence for recombination (Awadalla & Charlesworth 1999). When using R2 highly significant correlation was obtained in all four species (p<0.000 1; table 2). Since R2 is dependent on allele frequencies, the analysis was additionally restricted to sites at which both alleles were present at a frequency greater than 10%. It has been argued that this restriction limits the analysis to alleles, which tend to be older and are more likely to show evidence for past recombination events (Awadalla et al. 1999). Using this subset of polymorphisms, significant correlations were still observed for three out of four species, with the correlation in M. gibbosus becoming non-significant (table 2). Performing the correlation with |D′|, the correlations remain highly significant only for B. mardochei (both when considering all polymorphisms or only those above 10%), and marginally so only for polymorphisms in frequencies above 10% in M. gibbosus (table 2). Conversely, the homoplasy test finds strong evidence for recombination only in M. caucasicus (table 3). The best evidence for recombination is thus found in the two species B. mardochei and M. caucasicus (tables 2 and 3), whereas in the two other species (M. eupeus and M. gibbosus) the evidence is somewhat conflicting. In M. eupeus in particular, only the correlation between R2 and physical distance is significant.
Table 1.
The total number of sites and the number of segregating sites.
| species | n | total number of sites | segregating sites (S) | ||
|---|---|---|---|---|---|
| 16S | coxI | 16S | coxI | ||
| B. occitanus | |||||
| B. o. mardochei | 34 | 360 | 464 | 111 | 181 |
| Mesobuthus | |||||
| M. caucasicus | 36 | 349 | 461 | 151 | 97 |
| M. eupeus | 38 | 354 | 461 | 151 | 125 |
| M. gibbosus | 42 | 341 | 461 | 77 | 96 |
Table 2.
Correlation between linkage disequilibrium and physical distance in four scorpion species. (Estimates of R2 and |D′| were carried out using the computer package ‘LDhat’. p values were calculated from a one-sided Mantel test with 10 000 permutations. Genes were tested in the order 16S-5 kb-coxI using the shorter distance. Estimates do not change remarkably if they are tested using the longer connection of approximately 10 kb. ***p values <0.000 1, **p values >0.000 1 and <0.01, and *p values >0.01 and <0.05.)
| species | length (bp) | number of comparisons | R2 | |D′| | |||
|---|---|---|---|---|---|---|---|
| correlation | p-value | correlation | p value | ||||
| B. mardochei | 5824 | 29 976 | all sites | −0.038 35 | <0.000 1*** | −0.042 14 | <0.000 1*** |
| 4465 | allele frequency>10% | −0.041 6 | 0.008 6** | −0.062 17 | <0.000 1*** | ||
| M. caucasicus | 5810 | 17 020 | all sites | −0.210 34 | <0.000 1*** | −0.011 58 | 0.093 4 |
| 861 | allele frequency>10% | −0.075 13 | 0.021 9* | −0.023 2 | 0.225 6 | ||
| M. eupeus | 5815 | 16 653 | all sites | −0.220 51 | <0.000 1*** | 0.007 8 | 0.810 7 |
| 1326 | allele frequency>10% | −0.158 99 | <0.000 1*** | 0.019 98 | 0.800 2 | ||
| M. gibbosus | 5802 | 11 325 | all sites | −0.094 76 | <0.000 1*** | 0.014 02 | 0.899 |
| 1711 | allele frequency>10% | −0.037 02 | 0.200 8 | −0.056 58 | 0.075 5 | ||
Table 3.
Homoplasy test in four scorpion species. (***p values <0.000 1, **p values >0.000 1 and <0.01, and *p values >0.01 and <0.05.)
| polymorphic sitesa | informative sites | effective number of sites (Se) | observed homoplasies | expected homoplasies | p value | homoplasy index from MPT | |
|---|---|---|---|---|---|---|---|
| B. mardochei | 123 | 99 | 136 | 198 | 192.633 | 0.364 | 0.349 |
| M. caucasicus | 105 | 79 | 146 | 136 | 93.512 | 0.000*** | 0.331 |
| M. eupeus | 117 | 95 | 124 | 159 | 230.82 | 1 | 0.356 |
| M. gibbosus | 88 | 78 | 126 | 69 | 61.653 | 0.229 | 0.349 |
The number of polymorphic sites differs from table 1, owing to the inclusion of the out-group A. australis and only third codon positions are included for analysis.
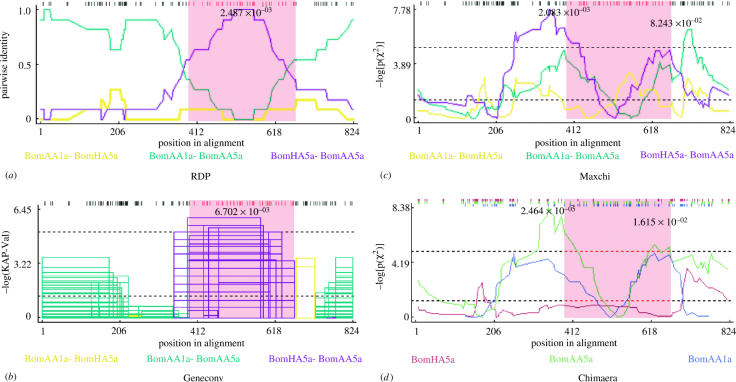
Testing for recombination using the general RDP, Geneconv, Maxchi and Chimaera, on a concatenated ‘supergene’ of the 16S and coxI data recovered many putative recombinant ‘daughter’ haplotypes between ‘parental’ haplotypes in the B. mardochei sequence alignment. An example is given in figure 1, where the RDP and the Geneconv test provide strong support for two ‘breakage points’ at sequence positions 319 and 685: The pairwise identity in the regions 1–319 and 686–823 bp is relatively high between BomAA1a and BomAA5a (haplotypes sampled from two populations from the Moroccan Anti-Atlas) relative to a third sequence (BomHA5a, a sequence from the Moroccan Haut-Atlas; figure 1a). In the middle piece (320–685 bp), however, the two haplotypes BomAA5a and BomHA5a share many polymorphic sites, thus being more similar than the two haplotypes BomAA1a and BomAA5a. ‘Breaking points’ similar to those shown in figure 1 are found for the three haplotypes BomAS1a (As Saghir region, Morocco), BomHA1a and BomHA1b. When only the protein-coding region was tested using the same four recombination tests, no potential recombination events were found using RDP and Geneconv, whereas the algorithms Maxchi and Chimaera found 12 and 8 putative recombination events in B. mardochei, 18 and 1 in M. caucasicus, 2 and 1 in M. eupeus, and finally, no recombination events in M. gibbosus.
Figure 1.
Test results for recombination in B. mardochei scorpion mtDNA of a concatenated 16S and coxI ‘supergene’ using (a) the general recombination method (RDP), (b) the Geneconv method, (c) the Maxchi and (d) the Chimaera test statistics. The x-axis gives the nucleotide positions of the alignment, whereas the y-axis represents the particular test statistics for each test. Peaks in the log P of χ2 values in the Maxchi and Chimaera tests mark potential points of recombination. Dashed lines represent p value cut-offs: uncorrected (lower line) and corrected for multiple comparisons (upper line) at the 0.05 level.
4. Discussion
(a) Correlation of LD versus distance
Our mtDNA data show a clear decline of LD versus physical distance, a pattern that is typical for nuclear recombining genes (Awadalla & Charlesworth 1999). It has been shown that decline of LD versus distance can also be reached with unusual models of mutation rates such as ‘mutational hot spots’ or adaptive substitution (McVean 2001; Innan & Nordborg 2002). However, we do not believe that the observed decline is owing to mutational hot spots or to adaptive substitution since the decline of LD versus distance is also observed but less statistically supported if only protein-coding sequences or synonymous third codon sites are compared (correlation coefficients are significant for R2 in B. mardochei: −0.070 41, p=<0.001, including all sites and −0.053 2, p=0.043 9, including only alleles with a frequency greater than 10%, and in M. eupeus: −0.139 49, p<0.001, including all sites, all other tests are non-significant at the 0.05 level). Moreover, mutational hot spots are less likely to occur in protein-coding regions owing to codon usage bias (Piganeau & Eyre-Walker 2004). Comparing evolutionary rates of these two genes, we estimated an approximate 1% net sequence divergence per Myr for the 16S gene in the species M. gibbosus (Gantenbein & Largiadèr 2002). Using the identical protocol to calibrate a clock for the coxI gene, we estimate here a rate of ∼1.4% per Myr for the coxI gene. Thus, the mutation rates of these two genes might be assumed to be relatively equal.
The choice of the measure of LD (|D′| or R2) had a strong impact on the correlations of the scorpion data: correlations using R2 were significant in all four species analysed (table 2), whereas correlations using |D′| were only significant in B. mardochei (table 2). This observation is in agreement with criticisms on the evidence for recombination in human mtDNA (Awadalla et al. 1999; Kivisild & Villems 2000; Kumar et al. 2000), where strong correlations disappeared if |D′| was used instead of R2. Following Meunier & Eyre-Walker's (2001) recommendation to limit the analysis to alleles with a frequency greater than 10% weakened the correlation in two cases and turned the correlation to non-significant in one species. All four tests using the intra-subspecific mtDNA data of B. mardochei, however, were significant, independently of the number of alleles considered or on how LD was estimated (table 2).
The homoplasy test was significant only in the species M. caucasicus (table 3). However, the assumptions of this test are less likely to be met since it assumes a specific model of sequence divergence, whereas the LD versus distance test is more robust to such violations (McVean 2001; Piganeau & Eyre-Walker 2004). Furthermore, the mean sequence divergence was greater than 5% in these datasets, which is higher than recommended (Maynard Smith & Smith 1998). Thus, the results of the homoplasy test should be interpreted with caution.
(b) Remark on the ‘Buthus occitanus complex’
The fact that evidence for recombination is most strongly supported in the species B. mardochei may not be random. It fits well with morphological and phylogeographic observations. This species was, until recently, treated as a subspecies of the Buthus occitanus complex before these were split very recently into different species based on new morphological evidence (Lourenço 2003). Vachon (1952) summarized five subspecies and a number of ‘variations’ under the taxon B. occitanus, thus underlining their close phylogenetic relationship. A recent phylogeographic study on this species on four out of five subspecies using the same two mitochondrial makers as in this study revealed a total of 11 highly divergent clades (with approximately 12–15% sequence divergence between these clades; Gantenbein & Largiadèr 2003). Furthermore, the most parsimonious tree in this study suffered from a low consistency index (CI=0.44) and a relatively high number of homoplasies (table 3). Eight out of these 11 Moroccan mtDNA clades belong to the recently elevated species B. mardochei. Nuclear allozyme data and nuclear ITS-1 sequence data completely disagreed with mtDNA sequence data with respect to the Moroccan populations (Gantenbein & Largiadèr 2003; Gantenbein 2004): nuclear data indicated a common gene pool for all the Moroccan samples, suggesting high gene flow among these population groups. We interpret this strong discrepancy between nuclear and mitochondrial data in the Moroccan samples (all other European and Tunisian clades being congruent in phylogenetic analysis) as evidence that the genetic population structure of Buthus prevents the extinction of these phylogenetically old and diverse mtDNA lineages, and that these Moroccan population groups ‘hybridize’. While hybridization between subspecies might itself increase mtDNA recombination, the higher divergence between sequences will obviously ease its detection through statistical means.
(c) Mitofusion and heteroplasmy in buthid scorpions
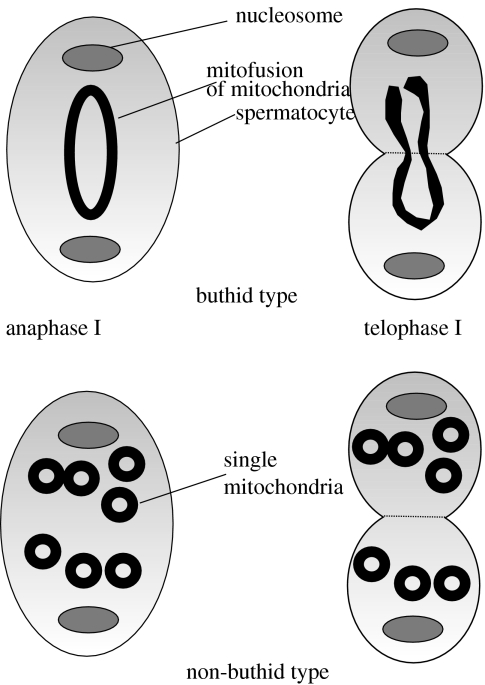
Although these results show indirect evidence for recombination, here, a clear link is seen to early cytological studies on the spermatogenesis of buthid scorpions. The studies by Wilson et al. (Wilson 1916, 1931; Wilson & Pollister 1937) were the first to observe two very different mechanisms of mitochondrial segregation in scorpions during early meiosis—fusion of mitochondria into a ring in the primary spermatocytes in the family Buthidae (genus Centruroides) as opposed to a non-buthid species (genus Opisthacanthus; family Liochelidae), which does not have such fusion. This ring is pulled apart during the first meiotic division, with a half-ring moving to each pole (figure 2). The half-ring straightens into a rod which divides so that each secondary spermatocyte receives two mitochondrial rods. During the second meiotic division, these rods divide again, and each spermatid receives two rods, which later join to form a new fused structure posterior to the nucleus called Nebenkern (German for ‘next to the nucleus’). This phenomenon has also been observed in the European buthid species, B. occitanus (Tuzet 1938) and in the Chinese buthid, Mesobuthus martensii (Sato 1940). However, such mitochondrial fusion before the first meiotic division was not observed in other scorpion families which appear to have random distribution of separate mitochondria: recorded evidence includes species of Hadrurus (Caraboctonidae), Euscorpius (Euscorpiidae), Opisthacanthus (Liochelidae), Vaejovis (Vaejovidae) and Heterometrus (Scorpionidae; Sokolow 1913; Bhattacharya & Gatenby 1924; Gatenby 1925). Electron microscopy supports the existence of a unique pattern limited to Buthidae. There are two elongated mitochondria in the middle piece of the buthid spermatozoon (Hood et al. 1972; Da Cruz Landim & Ferreira 1973; Alberti 1983) as opposed to four to nine mitochondria in Euscorpiidae and Vaejovidae (Andre 1959; Hood et al. 1972; Jespersen & Hartwick 1973).
Figure 2.
Schematic drawing of buthid versus non-buthid scorpion spermatogenesis (after Wilson 1916; Sato 1940).
Formation of the fused mitochondrial structure called Nebenkern during late spermatogenesis is known in many invertebrates and is well-studied in Drosophila (Fuller 1993). During flagellum elongation, the Nebenkern unfolds and results in two mitochondrial derivatives, later producing ATP to power the sperm's flagellum (Yaffe 1999). Mitochondrial fusion and subsequent elongation may allow coordinated ATP delivery to all regions of the flagellar axoneme. The Nebenkern in Drosophila forms during spermatid differentiation, i.e. after the meiosis is completed. On the contrary, the scorpion family Buthidae represents a unique case when mitochondria fuse much earlier (at the very beginning of meiosis) into a ring-shaped Nebenkern, which undergoes two regular divisions (Wilson 1916; Sato 1940). To the best of our knowledge, this phenomenon has not been reported in any other animal.
It is very likely that the fusion of mitochondria during the early stages of spermatogenesis provides increased opportunities for the mtDNA molecules to recombine, either through homologous recombination (intermolecular) or non-homologous recombination (intramolecular). The first case of non-homologous recombination of mtDNA has been reported in the nematode Meloidogyne javanica (Lunt & Hyman 1997). It was later shown in vitro that both types of recombination events can occur in human mtDNA (Lunt & Hyman 1997; Kraytsberg et al. 2004). Conditions for homologous recombination are met if at least two types of mtDNA molecules (haplotypes) are found in the same cell (heteroplasmy). In mammals, it is known that there are different mechanisms acting to prevent recombination of mtDNA (Rokas et al. 2003). Up to 100 paternal mitochondria enter the egg but are destroyed during the first few hours following fertilization. Several mechanisms ensure that the zygote contains only maternal mitochondria. However, it has been reported that the maternal inheritance of mitochondria in mammals seems to be merely a quantitative phenomenon, and situations of paternal leakage have been reported in mice (Gyllensten & Wilson 1987; Gyllensten et al. 1991). Failure of the recognition mechanisms for paternal mitochondria seems to be more common in hybrid populations, in which the specificity in the recognition process is relaxed, as was shown in drosophilids (Kondo et al. 1990).
Two recent papers provide strong support for actual recombination in human mtDNA. A recent reevaluation of lately added/corrected data found clear indirect evidence for recombination in human mtDNA using LD versus distance correlations (Piganeau & Eyre-Walker 2004). Moreover, a patient having paternal and maternal mtDNA (=heteroplasmic) in his muscle tissue was recently screened for recombined haplotypes using single template PCR (Kraytsberg et al. 2004). A frequency of approximately 0.7% of recombinant genotypes was reported, providing direct proof that human mtDNA is able to recombine. A recent compilation on published mtDNA sequence data from various vertebrates and invertebrates also showed indirect evidence for recombination in primates, insects and crustaceans (Piganeau et al. 2004).
The finding of strong negative correlation between LD and distance, together with the unique cytological evidence for mitofusion in early spermatogenesis, provides a strong suggestion for recombination in buthid scorpions. A combination of cytological methods to document scorpion spermatogenesis with controlled crossing experiments of populations of known haplotypes with subsequent genotype screening of offspring would provide a powerful direct approach to address recombination in buthid scorpion mtDNA (Ladoukakis & Zouros 2001; Rokas et al. 2003). If mtDNA recombination were directly linked to the phenomenon of mitofusion, non-buthids (e.g. Euscorpiidae, Scorpionidae) should not show any indirect evidence for recombination. Such a major difference between buthid and non-buthid scorpions would not be entirely unexpected as Buthidae represent a separate, ancient lineage within the scorpion phylogeny (Soleglad & Fet 2003) and possess a number of other unique, derived evolutionary mechanisms (such as mammal-specific neurotoxins).
Acknowledgments
B.G. was supported by an SNF-IHP grant (83-EU-065528), The Royal Society (R-36579) and the ‘Basler Stiftung für experimentelle Zoologie’ (c/o Eidgenössisches Tropeninstitut). Field trips of B.G. were supported by the commission for travel grants of the Swiss Academy of Natural Science (SANW). The field trip to Central Asia in March–May 2002 was supported by the National Geographic Society Research and Exploration grant 7001-01 to V.F. We thank Alexander V. Gromov for his help in the field and additional specimens of Mesobuthus. We also thank Gwenaël Piganeau, Michael Gardner and Adam Eyre-Walker for their helpful correspondence and for granting access to their unpublished manuscript that included evidence for recombination in scorpions based on a subset of M. eupeus. We are particularly grateful to Bill Amos who let us use his sequencer free of charge to finish the project when our finances had run out. Lily Jin ran three gels for us on the ABI377. B.G. would like to acknowledge Matt E. Braunwalder for his skilled guidance on scorpion biology and for leading him into the field of ‘scorpiology’.
One anonymous reviewer and Adam Eyre-Walker significantly improved an earlier version of the manuscript.
Footnotes
As this paper exceeds the maximum length normally permitted, the authors have agreed to contribute to production costs.
Present address: AO Research Institute (ARI), Clavadelerstrasse, CH-7270 Davos Platz, Switzerland.
References
- Alberti G. Fine structure of scorpion spermatozoa (Buthus occitanus; Buthidae, Scorpiones) J. Morphol. 1983;177:205–212. doi: 10.1002/jmor.1051770207. [DOI] [PubMed] [Google Scholar]
- Andre J. Etude au microscope électronique de l'évolution du chondriome pendant la spermatogénèse du scorpion Euscorpius flavicaudis. J. Ultrastruct. Res. 1959;2:208–308. doi: 10.1016/s0022-5320(59)80003-4. [DOI] [PubMed] [Google Scholar]
- Avise J. Chapman & Hall; New York: 1994. Molecular markers, natural history and evolution. [Google Scholar]
- Awadalla P, Charlesworth D. Recombination and selection at Brassica self-incompatibility loci. Genetics. 1999;152:413–425. doi: 10.1093/genetics/152.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla P, Eyre-Walker A, Maynard Smith J. Linkage disequilibrium and recombination in hominid mitochondrial DNA. Nature. 1999;286 doi: 10.1126/science.286.5449.2524. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D.R, Gatenby J.B. Spermatogenesis in an Indian scorpion. Nature. 1924;113:858. [Google Scholar]
- Da Cruz Landim C, Ferreira A. The spermatozoa of Tityus bahiensis (Perty) (Scorpiones, Buthidae) Cytologia. 1973;38:187–194. doi: 10.1508/cytologia.38.187. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Smith N.H, Maynard Smith J. Reply to Macaulay et al. (1999): mitochondrial DNA recombination—reasons to panic. Proc. R. Soc. B. 1999;266:2041–2042. (doi:10.1098/rspb.1999.0884) [Google Scholar]
- Fuller M.T. Spermatogenesis. In: Bate M, Arias A.M, editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; New York: 1993. pp. 71–147. [Google Scholar]
- Gantenbein B. The genetic population structure of Buthus occitanus (Scorpiones: Buthidae) across the Strait of Gibraltar—calibrating a molecular clock using nuclear allozyme variation. Biol. J. Linn. Soc. 2004;81:519–534. [Google Scholar]
- Gantenbein B, Largiadèr C.R. Mesobuthus gibbosus (Scorpiones: Buthidae) on the island of Rhodes—hybridisation between Ulysses' stowaways and native scorpions? Mol. Ecol. 2002;11:925–938. doi: 10.1046/j.1365-294x.2002.01494.x. [DOI] [PubMed] [Google Scholar]
- Gantenbein B, Largiadèr C.R. The phylogeographic importance of the Strait of Gibraltar as a gene flow barrier in terrestrial arthropods: a case study with the scorpion Buthus occitanus as model organism. Mol. Phylogenet. Evol. 2003;28:119–130. doi: 10.1016/s1055-7903(03)00031-9. [DOI] [PubMed] [Google Scholar]
- Gantenbein B, Fet V, Barker M.D. Mitochondrial DNA reveals a deep, divergent phylogeny in Centruroides exilicauda (Wood, 1863) (Scorpiones: Buthidae) In: Fet V, Selden P.A, editors. Scorpions 2001: Memoriam Gary A. Polis. The British Arachnological Society; Burnham Beeches, Bucks: 2001. pp. 235–244. [Google Scholar]
- Gantenbein B, Fet V, Gromov A.V. The first DNA phylogeny of four species of Mesobuthus Vachon, 1950 (Scorpiones, Buthidae) from Eurasia. J. Arachnol. 2003;31:412–420. [Google Scholar]
- Gatenby J.B. The scorpion spermateleosis. Nature. 1925;114:380–381. [Google Scholar]
- Gyllensten U, Wilson A.C. Interspecific mitochondrial DNA transfer and the colonization of Scandinavia by mice. Gen. Res. Camb. 1987;49:25–29. doi: 10.1017/s0016672300026690. [DOI] [PubMed] [Google Scholar]
- Gyllensten U, Wharton D, Josefsson A, Wilson A.C. Paternal inheritance of mitochondrial DNA in mice. Nature. 1991;352:255–257. doi: 10.1038/352255a0. [DOI] [PubMed] [Google Scholar]
- Hill W.G, Robertson A. The effects of inbreeding at loci with heterozygote advantage. Genetics. 1968;60:615–628. doi: 10.1093/genetics/60.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood R.D, Watson O.F, Deason T.R, Benton C.L.J. Ultrastructure of scorpion spermatozoa with atypical axonemes. Cytobios. 1972;5:167–177. [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Innan H, Nordborg M. Recombination or mutational hot spots in human mtDNA? Mol. Biol. Evol. 2002;19:1122–1127. doi: 10.1093/oxfordjournals.molbev.a004170. [DOI] [PubMed] [Google Scholar]
- Jespersen A, Hartwick R. Fine structure of spermiogenesis in scorpions from the family Vaejovidae. Ultrastruct. Res. 1973;45:366–383. doi: 10.1016/s0022-5320(73)80068-1. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Villems R. Questioning evidence for recombination in human mitochondrial DNA. Science. 2000;288:1931. [PubMed] [Google Scholar]
- Kondo R, Satta Y, Matsura E.T, Ishiwa H, Takahata N, Chigousa S.I. Incomplete maternal transmission of mitochondrial DNA in Drosophila. Genetics. 1990;126:657–663. doi: 10.1093/genetics/126.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraytsberg Y, Schwartz M, Brown T.A, Ebralidse K, Kunz W.S, Clayton D.A, Vissing J, Khrapko K. Recombination of human mitochondrial DNA. Science. 2004;304:981. doi: 10.1126/science.1096342. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedrick P, Dowling T.E. Questioning evidence for recombination in human mitochondrial DNA. Science. 2000;288:1931a. [PubMed] [Google Scholar]
- Ladoukakis E.D, Zouros E. Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol. 2001;18:1168–1175. doi: 10.1093/oxfordjournals.molbev.a003904. [DOI] [PubMed] [Google Scholar]
- Lewontin R.C. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço W.R. Compléments à la faune de scorpions (Arachnida) de l'Afrique du Nord, avec des considérations sur le genre Buthus Leach, 1815. Rev. Suisse Zool. 2003;110:875–912. [Google Scholar]
- Lunt D.H, Hyman B.C. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Sykes B.C. Mitochondrial DNA recombination—no need to panic. Proc. R. Soc. B. 1999;266:2037–2039. doi: 10.1098/rspb.1999.0883. (doi:10.1098/rspb.1999.0883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992;35:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Smith N.H. Detecting recombination from gene trees. Mol. Biol. Evol. 1998;15:590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- McVean G.A.T. What do patterns of genetic variability reveal about mitochondrial recombination? Heredity. 2001;87:613–620. doi: 10.1046/j.1365-2540.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- Meunier J, Eyre-Walker A. The correlation between linkage disequilibrium and distance: implications for recombination in hominid mitochondria. Mol. Biol. Evol. 2001;18:2132–2135. doi: 10.1093/oxfordjournals.molbev.a003756. [DOI] [PubMed] [Google Scholar]
- Piganeau G, Eyre-Walker A. A reanalysis of the indirect evidence for recombination in human mitochondrial DNA. Heredity. 2004;92:282–288. doi: 10.1038/sj.hdy.6800413. [DOI] [PubMed] [Google Scholar]
- Piganeau G, Gardner M, Eyre-Walker A. A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 2004;21:2319–2325. doi: 10.1093/molbev/msh244. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl Acad. Sci. USA. 2001;98:13 757–13 762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. The effect of recombination on the accuracy of phylogeny estimation. J. Mol. Biol. 2002;54:396–402. doi: 10.1007/s00239-001-0034-9. [DOI] [PubMed] [Google Scholar]
- Rokas A, Ladoukakis E.D, Zouros E. Animal mitochondrial DNA recombination revisited. Trends Ecol. Evol. 2003;18:411–417. [Google Scholar]
- Sato I. Studies on the cytoplasmic phenomena in the spermatogenesis of the oriental scorpion Buthus martensii, with special reference to the structure of the chondriosome ring and the dichyokinesis. J. Sci. Hiroshima Univ. Zool. 1940;8:1–116. [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Schierup M.H, Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–891. doi: 10.1093/genetics/156.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Entomol. Soc. Am. 1994;87:651–701. [Google Scholar]
- Sokal R.R, Rohlf F.J. W. H. Freeman & Company; New York: 1995. Biometry. [Google Scholar]
- Sokolow I. Untersuchungen über die Spermatogenese bei den Arachniden. I. Über die Spermatogenese der Skorpione. Arch. Zellforsch. 1913;9:399–432. [Google Scholar]
- Soleglad M.E, Fet V. High-level systematics and phylogeny of the extant scorpions (Scorpiones: Orthosterni) Euscorpius. 2003;11:1–175. [Google Scholar]
- Thompson J.D, Gibson T.J, Plewniak F, Jeanmougin F, Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B, Padua R.A, Campbell C. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 1996;271:27 536–27 543. doi: 10.1074/jbc.271.44.27536. [DOI] [PubMed] [Google Scholar]
- Tuzet O. Sur la spermatogénèse de Buthus occitanus (Amor.) Arch. Zool. Exp. Gén. 1938;80:335–351. [Google Scholar]
- Vachon M. Institut Pasteur d'Algérie; Algérie: 1952. Études sur les scorpions. [PubMed] [Google Scholar]
- Wilson E.B. The distribution of the chondriosomes to the spermatozoa in scorpions. Proc. Natl Acad. Sci. USA. 1916;2:321–324. doi: 10.1073/pnas.2.6.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E.B. The distribution of sperm-forming materials in scorpions. J. Morphol. 1931;52:429–464. [Google Scholar]
- Wilson E.B, Pollister A.W. Observations on sperm formation in the Centrurid scorpions with special reference to the Golgi material. J. Morphol. 1937;60:407–443. [Google Scholar]
- Wiuf C. Recombination in human mitochondrial DNA? Genetics. 2001;159:749–756. doi: 10.1093/genetics/159.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf C, Christensen T, Hein J. A simulation study of the reliability of recombination detection methods. Mol. Biol. Evol. 2001;18:1929–1939. doi: 10.1093/oxfordjournals.molbev.a003733. [DOI] [PubMed] [Google Scholar]
- Yaffe M.P. The machinery of mitochondrial inheritance and behavior. Science. 1999;283:1493–1497. doi: 10.1126/science.283.5407.1493. [DOI] [PubMed] [Google Scholar]