Abstract
Sexual reproduction and recombination are important for maintaining a stable copy number of transposable elements (TEs). In sexual populations, elements can be contained by purifying selection against host carriers with higher element copy numbers; however, in the absence of sex and recombination, asexual populations could be driven to extinction by an unchecked proliferation of TEs. Here we provide a theoretical framework for analyzing TE dynamics under asexual reproduction. Analytic results show that, in an infinite asexual population, an equilibrium in copy number is achieved if no element excision is possible, but that all TEs are eliminated if there is some excision. In a finite population, computer simulations demonstrate that small populations are driven to extinction by a Muller's ratchet-like process of element accumulation, but that large populations can be cured of vertically transmitted TEs, even with excision rates well below transposition rates. These results may have important consequences for newly arisen asexual lineages and may account for the lack of deleterious retrotransposons in the putatively ancient asexual bdelloid rotifers.
TRANSPOSABLE elements (TEs) are mobile DNA sequences that are abundant in the genomes of nearly all living organisms, including bacteria, protists, fungi, plants, and animals (Craig et al. 2002). Although there are cases in which mobile genetic elements may be coopted to serve host regulatory or structural functions (Kidwell and Lisch 2001), like most other mutator mechanisms, TEs are known to reduce the fitness of their host organism. However, unlike other classes of mutation, TEs are capable of autonomous self-replication. When elements transpose, they replicate faster than their host genome, with rates of transposition above rates of spontaneous deletion (Charlesworth and Langley 1989; Nuzhdin and Mackay 1995; Maside et al. 2000). This permits TE persistence despite their deleterious effects, as postulated by the “selfish DNA” hypothesis (Doolittle and Sapienza 1980; Orgel and Crick 1980).
Experimental and theoretical studies suggest that the ability of TEs to propagate can be balanced by natural selection against individuals with high element copy number (Charlesworth and Charlesworth 1983; Kaplan and Brookfield 1983). Three main sources of deleterious effects on fitness of segregating TEs have been postulated (see review by Nuzhdin 1999): insertions disrupting gene function (Finnegan 1992), chromosomal rearrangements generated by ectopic exchange (Montgomery et al. 1987), and a selective cost of transposition itself (Brookfield 1991). Host regulatory mechanisms may play a role as well, and there may be rare beneficial insertions, but it is likely that nearly all TE insertions are deleterious. There is substantial evidence from natural populations and experimental studies for such deleterious effects (Charlesworth et al. 1994).
Coevolution between TEs and their hosts depends critically on the presence of sexual reproduction. Outcrossing provides a means for TEs to spread to all individuals in a population. Despite being deleterious, TEs can persist in a sexual population while inflicting a severe fitness penalty on their hosts because they can increase in number faster than the host genome (Hickey 1982). This would not be true in an asexual population. In the absence of horizontal transmission, there is no between-lineage transmission, and TEs cannot initially spread in an asexual population (Hickey 1982). However, asexual lineages generally arise from sexual progenitors whose genomes are riddled with TEs. Upon the abandonment of sex, elements are coupled entirely to their hosts. The proliferation of TEs would be detrimental to the host to which they are confined. In the long run, TEs would be expected to become inactive and domesticated, as lineages with inactive elements should have a selective advantage and outcompete other lineages (Doolittle et al. 1984). However, theoretical studies show that TEs in outcrossing populations will generally evolve maximum transposition rates (Charlesworth and Langley 1986). Asexual lineages that arise from sexual populations will contain actively transposing TEs, selected to multiply without considering the fitness of the ancestral sexual host. By doing so, they would seal the fates of their asexual hosts. Accordingly, a long-term advantage of sex may result from the early extinction of asexual lineages, due to the unchecked proliferation of TEs upon the abandonment of sex (Arkhipova and Meselson 2005a).
Despite the fact that sex facilitates the spread of TEs (Hickey 1982), it may thus also be necessary to contain their proliferation. In this article, we examine this idea by modeling the population dynamics of transposable elements under asexual reproduction. While the abandonment of sex in small populations will probably lead to element buildup and eventual host extinction in small populations, large populations with some level of element excision or deletion may be able to eliminate vertically transmitted deleterious TEs, providing a potential benefit of asexuality.
METHODS
Assumptions of the models:
We consider a diploid asexual lineage arising from an ancestral sexual population that contains deleterious TEs. With offspring genotypes identical to those of their parents, we assume that a clonal lineage originates from a single ancestor, such that initially all individuals have a TE copy number x. Elements are transmitted only vertically from parent to offspring. The copy number may increase due to transposition, at a rate u per element, or decrease due to excision at a rate v per element. We first consider analytic models of TE dynamics in infinite asexual populations. We then employ a stochastic simulation method to investigate the effects of finite population sizes.
Infinite populations:
To examine TE dynamics with infinite population size, we adapt the exact recurrence relations of Kimura and Maruyama (1966; Equation 3.1), originally devised to calculate the mutation load in asexual populations. We assume an infinite number of insertion sites and Poisson distributions of the numbers of transposition and excision events as an approximation for binomial distributions. We calculate the transposition load in the presence and absence of excision, without specifying the relation between fitness and copy numbers. We then consider specific fitness functions and determine the mean and variance element of copy number using iterations and approximations based on the method of Charlesworth (1990).
Finite populations:
Computer simulations were used to examine the effects of excision in finite asexual populations. The population size N is constant, with offspring genomes identical to those of their parents except for transposition and excision events. The genome is made up of two diploid chromosomes, with each chromosome able to carry 200 elements, although since the population is asexual and nonrecombining, this is equivalent to any number of chromosomes of any ploidy level. A simulation run is initiated with a single individual carrying x elements, which founds the clonal population, such that all N individuals are initially identical. TE dynamics are then monitored over many generations, where each generation reproduces asexually, followed by transposition and excision. Reproduction involves randomly sampling individuals to produce a new offspring population of size N, where the probability of an individual being selected as a parent is proportional to its fitness. As in the case of the infinite population size model, the numbers of transposition and excision events were drawn from Poisson distributions, with constant probabilities per element of u and v, respectively. New insertions were placed randomly in the genome at unoccupied sites, and the exact locations of elements were maintained between generations unless they had been excised.
Simulations were run until all elements had been lost from the population or until the mean TE copy number had accumulated substantially (>150). Above this number, a runaway process of element buildup is observed (results not shown). No stable equilibrium with an intermediate TE copy number was ever observed, consistent with results from an asexual model of TE sequence evolution for simulations in which TE inactivation through mutation was not included (Docking et al. 2006). We calculated the proportion of simulation runs where TEs were eliminated, to examine the conditions under which asexual populations are expected to cure themselves of deleterious TEs or be driven to early extinction. We also monitored the variance in copy number and the minimum TE copy number in the population. C++ files of this simulation program are available upon request. The random numbers were implemented using the Mersenne Twister pseudorandom number generator (Matsumoto and Nishimura 1998), adapted for C++ by J. Bedaux (http://www.bedaux.net/mtrand/).
Selection:
Fitness was assumed to be a decreasing function of TE abundance, as would be expected in the ancestral population. In sexual populations, a stable equilibrium copy number occurs only when there are synergistic fitness interactions between elements (Charlesworth and Charlesworth 1983). Thus, the fitness of an individual with n elements was represented by an exponential quadratic, decreasing function of the copy number,
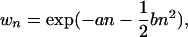 |
(1) |
where a and b are constant selection coefficients (Charlesworth 1990).
Initial copy number of clonal lineage:
We set the initial copy number, x, to be equal to the equilibrium mean copy number of the ancestral sexual population. In outcrossing populations, with low frequencies of elements at each occupied site, copy number dynamics are described by
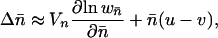 |
(2) |
where 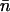 is the mean population copy number, Vn is the variance in copy number, and
is the mean population copy number, Vn is the variance in copy number, and 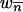 is the fitness of an individual carrying the mean number of elements (Charlesworth 1985). Solving Equation 2 for
is the fitness of an individual carrying the mean number of elements (Charlesworth 1985). Solving Equation 2 for 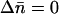 , assuming a Poisson distribution of elements, and using the quadratic fitness formula, the equilibrium mean element copy number of the ancestral sexual population is
, assuming a Poisson distribution of elements, and using the quadratic fitness formula, the equilibrium mean element copy number of the ancestral sexual population is
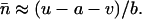 |
(3) |
The transposition load of a sexual population is measured as
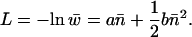 |
(4) |
However, due to the synergism in fitness effects, there is a departure from independence among elements, resulting in the equilibrium load due to TEs being approximately half that of Equation 4 when 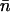 is given by Equation 3 (Kimura and Maruyama 1966; Crow 1970; Charlesworth 1990; Charlesworth and Barton 1996). We thus set the equilibrium load,
is given by Equation 3 (Kimura and Maruyama 1966; Crow 1970; Charlesworth 1990; Charlesworth and Barton 1996). We thus set the equilibrium load, 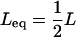 , and solve for the equilibrium mean copy number under synergistic fitness, such that
, and solve for the equilibrium mean copy number under synergistic fitness, such that
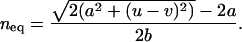 |
(5) |
Model parameters:
We set the parameters in our simulations by using estimates from studies of Drosophila populations. Average transposition rates per element per generation are ∼10−4, regardless of the class of element, with excision rates at least one order of magnitude smaller (Nuzhdin and Mackay 1995; Vieira and Biémont 1997; Pasyukova et al. 1998; Maside et al. 2000). We set 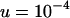 and investigate excision rates of
and investigate excision rates of 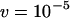 and
and 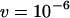 . The strength of selection on segregating elements in natural populations should be of the order 10−5–10−4 per copy, if no insertions are completely neutral (Charlesworth et al. 1994). With synergism between elements, the strength of selection will depend on both a and b. We set
. The strength of selection on segregating elements in natural populations should be of the order 10−5–10−4 per copy, if no insertions are completely neutral (Charlesworth et al. 1994). With synergism between elements, the strength of selection will depend on both a and b. We set 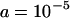 and let b vary according to Equation 5 for a given
and let b vary according to Equation 5 for a given 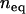 . We set the initial copy number, x, of the founding clone to
. We set the initial copy number, x, of the founding clone to 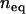 and assume that asexual lineages will experience the same parameters as the sexual ancestor. We use these values of transposition and selection to investigate the impact of population size on different excision rates.
and assume that asexual lineages will experience the same parameters as the sexual ancestor. We use these values of transposition and selection to investigate the impact of population size on different excision rates.
RESULTS
Infinite populations without excision:
We modify the exact recurrence relations of Kimura and Maruyama (1966; Equation 3.1) to analyze TE dynamics in an infinite asexual population without element excision. We write fi for the frequency of individuals with i elements, whose fitness is wi. We assume a Poisson distribution of transposition insertions with mean ui in a genome with i elements, where each transposition event leads to a new insertion. With no excision (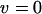 ), the frequency of individuals with i elements in the next generation is
), the frequency of individuals with i elements in the next generation is
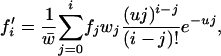 |
(6) |
where 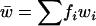 is the mean fitness. Since there is no excision to reduce the TE copy number below that of the initial clone, the frequency of individuals with the lowest copy number is fx. Following the derivation of Kimura and Maruyama (1966),
is the mean fitness. Since there is no excision to reduce the TE copy number below that of the initial clone, the frequency of individuals with the lowest copy number is fx. Following the derivation of Kimura and Maruyama (1966),
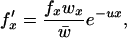 |
(7) |
and the lineage reaches an equilibrium with mean fitness
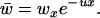 |
(8) |
Since individuals with the lowest element copy number have the highest fitness, all else being equal, a stable equilibrium is achieved with a transposition load,
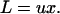 |
(9) |
This is equivalent to the mutational load of an asexual population (Kimura and Maruyama 1966), where the genomic mutation rate, U, is replaced by the product of the transposition rate and initial copy number. Thus, the load depends critically on the initial number of TEs in the founding clone. This result is quite general and independent of the fitness function.
The equilibrium mean and variance in element copy number can be calculated by iteration, given a specific fitness function, transposition rate, and initial copy number. Using Equation 1, a normal-distribution approximation can be used to estimate the equilibrium solutions. We adapt Equations 2–4 of Charlesworth (1990), originally developed to calculate the equilibrium number of mutations in an asexual population under mutation–selection balance, to determine the equilibrium properties of transposable elements under asexuality without excision. Using his method of assuming an approximately normal distribution of copy number among individuals in the population, at equilibrium we have
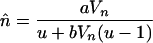 |
(10) |
and
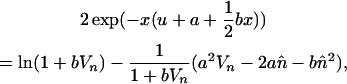 |
(11) |
where 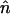 is the mean asexual population TE copy number and Vn is the variance in copy number.
is the mean asexual population TE copy number and Vn is the variance in copy number.
These equations allow the mean and variance in TE copy number at equilibrium to be calculated by iteration (Charlesworth 1990). We set 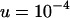 ,
, 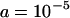 ,
, 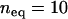 and evaluate b using Equation 5, giving
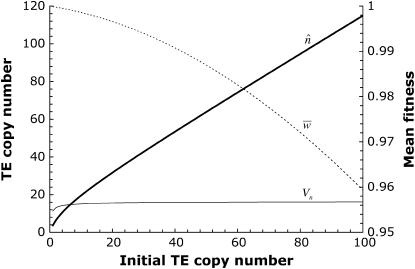
and evaluate b using Equation 5, giving 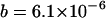 . We then calculate the equilibrium solutions for an asexual population, with TE copy numbers in the initial clone from 1–100. Figure 1 shows the equilibrium properties of an infinite asexual population for the given set of parameters solved, using the normal approximation and confirmed by iteration of Equation 6. A stable equilibrium is always achieved, with a mean copy that depends nearly linearly on x. The mean fitness declines with larger x, while the variance in copy number is nearly constant.
. We then calculate the equilibrium solutions for an asexual population, with TE copy numbers in the initial clone from 1–100. Figure 1 shows the equilibrium properties of an infinite asexual population for the given set of parameters solved, using the normal approximation and confirmed by iteration of Equation 6. A stable equilibrium is always achieved, with a mean copy that depends nearly linearly on x. The mean fitness declines with larger x, while the variance in copy number is nearly constant.
Figure 1.—
Equilibrium mean properties of an asexual population in the absence of excision, at transposition–selection balance with a quadratic fitness function. The primary y-axis shows how the mean copy number (thick line) and variance in copy number (thin line) vary with the initial copy number at equilibrium. The dashed line uses the secondary y-axis to denote the mean fitness of the equilibrium population. The parameter values used are: rate of transposition per element u = 10−4, selection coefficient a = 10−5, and synergism coefficient b = 6.1 × 10−6.
Unlike sexual populations, where the logarithm of fitness must decline faster than linearly with TE copy number (Charlesworth and Charlesworth 1983), a stable equilibrium can be achieved in an asexual population with an arbitrary fitness function. We now consider selection in the absence of synergism (b = 0), as may be the case if these interactions are dependent on the presence of sexual reproduction and recombination (e.g., with meiotic ectopic exchange; Langley et al. 1988). Without synergism, the mean and variance in TE copy number simplify to
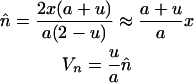 |
(12) |
These equations were confirmed by comparison with the exact equilibrium solutions obtained by iteration of Equation 6. It can be seen that the mean and variance scale linearly with the initial TE copy number, x. If the rate of transposition is much greater than the strength of selection against segregating TEs, the mean copy number and variance can be exceedingly high in the absence of synergism. For example, with the parameters used previously (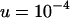 ,
, 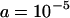 ),
), 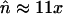 and
and 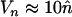 . Thus, while a stable equilibrium is theoretically achieved in the absence of synergism between elements, such fitness interactions may be necessary to hold copy number at a biologically relevant equilibrium.
. Thus, while a stable equilibrium is theoretically achieved in the absence of synergism between elements, such fitness interactions may be necessary to hold copy number at a biologically relevant equilibrium.
Infinite populations with excision:
With element excision, the previous analysis no longer holds. The lowest copy number can be reduced below that of the initial clone, and the individuals with fewer TEs should have a selective advantage, leading to a decline in the mean number of elements in the population. Regardless of the TE count of the initial clone, in an infinite asexual population, it is eventually possible for all TEs to be excised, creating a class of individuals without any elements. Unlike the situation with recurrent mutations, once such a zero class has been generated, it is immune to further transposition, assuming that there is no horizontal transfer. Due to its fitness advantage, this class will spread to fixation, effectively curing the population of deleterious TEs. This was confirmed by modifying Equation 6 to include excision and solving by iteration. The mean and variance in TE copy number always decline to zero irrespective of the rates of transposition and excision, the fitness function, or the initial copy number.
Parameter scaling of finite populations:
To further investigate whether asexual populations achieve the equilibrium expectations without excision and whether populations with excision can eliminate all their deleterious TEs, computer simulations were used to examine the effects of finite population sizes. Running simulations using the biologically realistic parameters derived from Drosophila populations is very time-consuming, because of the large population sizes and small transposition rates, excision rates, and selection coefficients. According to population genetics theory, if evolutionary forces are weak, so that diffusion approximations can be used, the properties of the system are determined by the values of the products of the deterministic forces and the effective population size (Ewens 1979, Chap. 4). If these values are maintained constant, we should expect the same evolutionary outcomes.
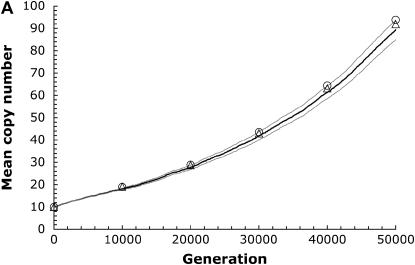
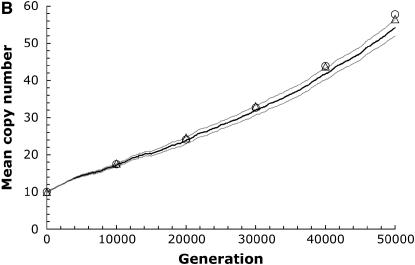
We confirmed this expectation by comparing simulations with the parameters from Drosophila and an adjusted version of these parameters for N = 104, such that 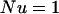 , with the other ratios kept constant as well. To achieve this, we scale up u, v, a, and b one and two orders of magnitude while scaling down N and the numbers of generations by the same amount. The accuracy of this parameter adjustment is demonstrated in Figure 2. By reducing the population size and the time of the simulation accordingly, this simplification seems to be reasonably accurate, although perhaps somewhat overestimating the mean copy number, especially with higher excision rates. However, this allows us to perform a greater number of simulation runs in a fraction of the computation time and should be a conservative correction, since it potentially biases our results against finding situations when complete TE elimination is expected. We continued by using 100-fold greater transposition, excision, and selection values than those estimated from natural populations. This implies that the population sizes and times used in our simulations need to be scaled upward two orders of magnitude to reflect biologically relevant values. The results presented below show the uncorrected parameter values used in the computer simulations. However, since the magnitude of the population size is so critical to our results, we also include the scaled population sizes in parentheses.
, with the other ratios kept constant as well. To achieve this, we scale up u, v, a, and b one and two orders of magnitude while scaling down N and the numbers of generations by the same amount. The accuracy of this parameter adjustment is demonstrated in Figure 2. By reducing the population size and the time of the simulation accordingly, this simplification seems to be reasonably accurate, although perhaps somewhat overestimating the mean copy number, especially with higher excision rates. However, this allows us to perform a greater number of simulation runs in a fraction of the computation time and should be a conservative correction, since it potentially biases our results against finding situations when complete TE elimination is expected. We continued by using 100-fold greater transposition, excision, and selection values than those estimated from natural populations. This implies that the population sizes and times used in our simulations need to be scaled upward two orders of magnitude to reflect biologically relevant values. The results presented below show the uncorrected parameter values used in the computer simulations. However, since the magnitude of the population size is so critical to our results, we also include the scaled population sizes in parentheses.
Figure 2.—
Demonstrating the accuracy of scaling parameters. The thick line shows the mean copy number from computer simulations run over 50,000 generations for a population size N = 104, with initial copy number x = 10, rate of transposition per element u = 10−4, selection coefficient a = 10−5, and synergism coefficient b. The equilibrium is solved for a given rate of excision per element according to Equation 5. (A) An excision rate two orders of magnitude lower than transposition (v = 10−6; b = 6.0 × 10−6; number of simulation runs, r = 25). (B) An excision rate one order of magnitude lower than transposition ( v = 10−5; b = 5.4 × 10−6; 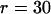 ). The thin lines denote the standard error of the mean between runs of the simulation. The triangles show the mean copy number from simulations scaled 10-fold (r = 100). The circles show the mean copy number from simulations scaled 100-fold (r = 1000). The generation time is also scaled upward to compare with the unadjusted simulation runs.
). The thin lines denote the standard error of the mean between runs of the simulation. The triangles show the mean copy number from simulations scaled 10-fold (r = 100). The circles show the mean copy number from simulations scaled 100-fold (r = 1000). The generation time is also scaled upward to compare with the unadjusted simulation runs.
Finite populations without excision:
Unlike the case of infinite populations without excision, in simulations with 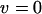 , a large but finite population size,
, a large but finite population size, 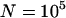 (equivalent to 107 when rescaled), and using a scaled set of the parameters presented in Figure 1 (
(equivalent to 107 when rescaled), and using a scaled set of the parameters presented in Figure 1 (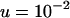 ,
, 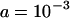 , and
, and 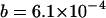 ), no equilibrium was ever observed for moderately low initial copy numbers,
), no equilibrium was ever observed for moderately low initial copy numbers, 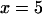 and
and 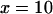 . Elements always accumulated substantially within several thousand generations (up to millions of generations, when rescaled), far above the infinite population equilibrium values of
. Elements always accumulated substantially within several thousand generations (up to millions of generations, when rescaled), far above the infinite population equilibrium values of 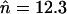 and
and 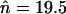 , respectively. This process of element accumulation is accelerated in smaller populations, with larger initial copy numbers, and with weaker selection. This is consistent with the results for TE sequence evolution in simulations with no excision, where no stable equilibrium in copy number was ever reached (Docking et al. 2006). This suggests that asexual populations with no excision of TEs are likely to accumulate large numbers of copies within a time frame of millions of generations, even if the effective population size is in the millions. Of course, with a sufficiently large population size, the population will spend a very long time near the equilibrium derived in the previous section, so that it will appear as though no accumulation is happening, but ultimately the population is expected to experience an unbounded proliferation of TEs.
, respectively. This process of element accumulation is accelerated in smaller populations, with larger initial copy numbers, and with weaker selection. This is consistent with the results for TE sequence evolution in simulations with no excision, where no stable equilibrium in copy number was ever reached (Docking et al. 2006). This suggests that asexual populations with no excision of TEs are likely to accumulate large numbers of copies within a time frame of millions of generations, even if the effective population size is in the millions. Of course, with a sufficiently large population size, the population will spend a very long time near the equilibrium derived in the previous section, so that it will appear as though no accumulation is happening, but ultimately the population is expected to experience an unbounded proliferation of TEs.
Finite populations with excision:
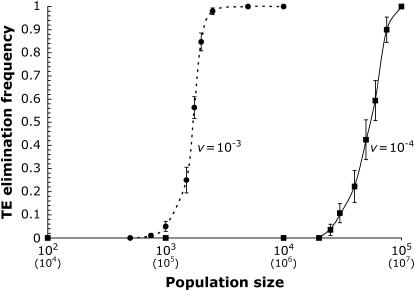
As the results of infinite populations with element excision showed, deleterious TEs can be completely purged from asexual populations when 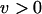 . The simulation results illustrate the interaction of population size and excision rate on the probability of TE elimination (Figure 3). In contrast to the expectation of complete element elimination in an infinite asexual population, TEs tend to accumulate substantially in small populations. Even with element excision, deleterious TEs accumulate in small populations well above the copy number expected at equilibrium in the absence of excision (see Figure 1). On the other hand, in large populations, the efficacy of selection and excision is enhanced. Consistent with the findings for the infinite population model, a sufficiently large asexual lineage can effectively purge itself of vertically transmitting deleterious TEs upon the abandonment of sex.
. The simulation results illustrate the interaction of population size and excision rate on the probability of TE elimination (Figure 3). In contrast to the expectation of complete element elimination in an infinite asexual population, TEs tend to accumulate substantially in small populations. Even with element excision, deleterious TEs accumulate in small populations well above the copy number expected at equilibrium in the absence of excision (see Figure 1). On the other hand, in large populations, the efficacy of selection and excision is enhanced. Consistent with the findings for the infinite population model, a sufficiently large asexual lineage can effectively purge itself of vertically transmitting deleterious TEs upon the abandonment of sex.
Figure 3.—
TE elimination frequency and population size. The initial copy number was x = 10, u = 10−2, and a = 10−3; the synergism coefficient, b, was calculated for a given rate of excision per element using Equation 5. The solid line represents excision two orders of magnitude below transposition (v = 10−4; b = 6.0 × 10−4). The dashed line represents excision one order of magnitude below transposition (v = 10−3; b = 5.4 × 10−4). With v = 10−4, all points with a TE elimination frequency of 0 or 1 are based on at least 10 simulation runs, and all points with an intermediate frequency include at least 30 simulation runs. With v = 10−3, all points are based on at least 60 simulation runs. Error bars denote one standard error. To reflect plausible parameters for natural populations, all transposition and selection terms must be scaled down 100-fold. Scaled population sizes are shown in parentheses below the x-axis.
Two main factors contribute to the proliferation of TEs in small populations. As with sexual populations, this is partly due to the reduction in the variance in copy number between individuals in a finite population, thereby limiting the power of natural selection (Brookfield and Badge 1997). While we present results only on the final state of the populations, we also monitored the variance in copy number between generations over the course of the simulations. In comparisons among populations that accumulate TEs, smaller populations indeed have a lower variance (results not shown). In addition, random genetic drift may lead to the loss of the class of individuals with the lowest copy number. In the absence of excision, the loss of this class would be irreversible, and TEs would continue to build up, leading to a decline in the mean fitness of the population. This process would be similar to that described for mutations, known as Muller's ratchet (Muller 1964; Stephan and Kim 2002), except that the genomic rate of transposition would increase with the copy number, leading to accelerated rates of fitness deterioration. With element excision, the process is reversible, but the rate at which the least-loaded class is lost may greatly exceed the rate of excision to generate a new best class. We kept track of the minimum TE copy number in the population during our simulations and observed the copy number of the least-loaded class to increase gradually above x as a population accumulated TEs (results not shown). Both of these factors probably contributed to the increased rate of TE proliferation in smaller populations.
In addition to the rate of excision relative to transposition, the ability of an asexual population to cure itself of deleterious TEs depends critically on the amount of synergism between elements, the magnitude of both transposition and excision, and the initial copy number to be eliminated. Synergism affects the ability of selection to limit TE proliferation in asexual populations. Higher rates of transposition result in faster TE proliferation, even if the ratio of transposition to excision is kept constant. The total number of initial TEs reduces the population mean fitness and affects the total genomewide rate of transposition. All of these parameters influence the ability of selection and excision to limit copy number.
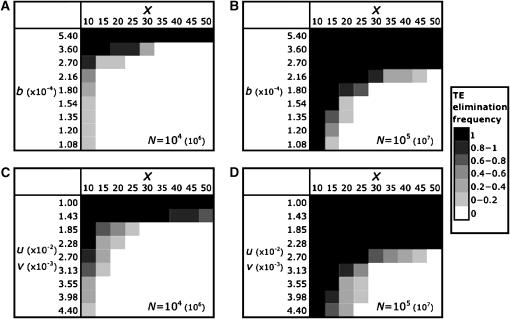
To explore these factors, we looked at the effect of varying the degree of synergism, b, and changing the magnitude of transposition and excision, while maintaining the ratio of u:v constant at 10. None of these parameters are strictly independent, because they influence the expected equilibrium distribution of elements in the ancestral sexual population. We varied the initial copy number, x, from 10 to 50 and used Equation 5 to solve for the corresponding equilibrium value of b and the equilibrium values of u and v. In both cases, for the parameters that were held constant, we used the values employed previously. Figure 4 shows the effects of synergism, magnitude of transposition–excision, and initial copy number on the probability of TE elimination at two large population sizes.
Figure 4.—
The effect of varying the degree of synergism, magnitude of transposition and excision, and initial copy number on the frequency of TE elimination for two large population sizes. (A and B) The effect of changing b. (C and D) The effect of changing the magnitude of u and v while keeping the ratio of u:v constant at 10, while allowing x to vary. The ancestral sexual equilibrium solutions according to Equation 5 are located along the diagonal. The parameters that do not change are u = 10−2, v = 10−3, a = 10−3, and b = 5.4 × 10−4. All points are based on at least 10 simulation runs. To reflect plausible parameters for natural populations, all transposition and selection terms must be scaled down 100-fold. Scaled population sizes are shown in parentheses next to the values used in the simulations.
All else being equal, increased synergism and lower transposition–excision rates reduce the population size necessary for TE elimination. Increased synergism strengthens selection against large copy numbers, and lower rates of transposition–excision slow the rate of TE accumulation, thereby improving the efficacy of selection. With strong synergism or low rates of transposition–excision, TEs are quite effectively removed from large populations regardless of the initial copy number. In contrast, with low initial copy numbers, TEs are likely to be eliminated, even with weak synergism or strong transposition–excision. Using parameters where intermediate frequencies of TE elimination are observed, a fine balance exists between selection and genetic drift. Slight differences in any of the parameters or the population size can shift this balance and greatly affect whether an asexual population increasingly accumulates or cures itself of deleterious TEs. Weaker synergism or higher transposition–excision rates also correspond to larger equilibrium mean copy numbers in an ancestral sexual population. Thus, the expected initial copy number will be greater, further hindering the ability of an asexual lineage to purge TEs. However, our approach may be viewed as conservative, since for all analyses other than that in Figure 4 we initiated the asexual population with the TE copy number, x, set to the sexual equilibrium. Given the variance in copy number in the ancestral population, however, an asexual lineage may arise from lower copy number individuals, thereby reducing x and increasing the probability of TE elimination. The asexual populations that survive are more likely to have arisen from low copy number founding populations.
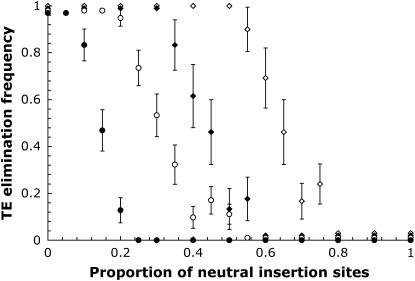
Our model has so far supposed that all TE insertions are deleterious, but TEs may vary in their selective effects. In the absence of meiotic ectopic exchange, TE insertions in intergenic regions of the genome may not affect fitness if these regions have little or no functional role. We consider a model with two classes of insertion sites: those in which TEs affect fitness according to Equation 1 and those in which TEs are selectively neutral (Charlesworth 1991). We assume that new TE insertions are neutral with a constant probability p and are deleterious with a probability 1 − p. Figure 5 shows the effect of varying p on the TE elimination frequency. With a high proportion of neutral sites, asexual populations are far less likely to be able to eliminate TEs, since elements tend to build up at neutral sites and continue to transpose. This effect is magnified if TEs inherited from the ancestral sexual population are also present at neutral sites, as would be the case if TEs in this population were primarily contained by ectopic exchange, and hence found in sites where insertions have little or no direct fitness effects in the absence of recombination (Montgomery et al. 1987; Charlesworth et al. 1994).
Figure 5.—
TE elimination frequencies incorporating neutral sites. The open symbols show when all x sites containing the initial TEs in the founding clone are under selection; the solid symbols show when these are neutral. The circles show simulations done with 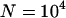 , diamonds show simulations done with
, diamonds show simulations done with 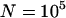 , and the remaining parameters are as before: x = 10, u = 10−2, v = 10−3, a = 10−3, and b = 5.4 × 10−4. All symbols are based on at least 30 simulation runs for
, and the remaining parameters are as before: x = 10, u = 10−2, v = 10−3, a = 10−3, and b = 5.4 × 10−4. All symbols are based on at least 30 simulation runs for 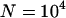 and at least 10 simulation runs for
and at least 10 simulation runs for 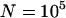 . Error bars denote one standard error. To reflect plausible parameters for natural populations, all transposition and selection terms must be scaled down 100-fold, and the population size must be scaled up 100-fold.
. Error bars denote one standard error. To reflect plausible parameters for natural populations, all transposition and selection terms must be scaled down 100-fold, and the population size must be scaled up 100-fold.
DISCUSSION
Whether an asexual population can cure itself of deleterious TEs depends critically on the ability of selection and excision to limit the propagation of elements. The probability of a population achieving a TE-free state is influenced by the parameters controlling TE dynamics, including the scale of transposition and excision, and the degree of synergism between elements. However, all else being equal, the major factor affecting TE elimination is the population size of the asexual lineage. Since the strength of selection on segregating elements is expected to be relatively low, selection will be ineffective in removing TEs unless the population size is large, and TEs will continue to accumulate at an ever-increasing rate.
Our results are in accordance with the hypothesis that deleterious TEs can drive the extinction of asexuals (Arkhipova and Meselson 2005a). However, we also provide a means by which an asexual lineage may be able to rid itself of the transposition load that it inherits upon the abandonment of sex. This possibility would provide a long-term benefit to asexual rather than sexual reproduction. Small populations may suffer from a reduced variance in TE copy number and a random loss of the class of individuals with low TE copy numbers. In large populations, however, selection is more effective and the Muller's ratchet-like process of element accumulation can be reversed because of excision. Even rates of TE excision well below that of transposition can improve overall fitness in large populations. Excision events are doubly advantageous, by both raising fitness and reducing the genomewide transposition rate because of reduced copy number. This allows mean fitness to improve faster than the stochastic loss of the least-loaded class, leading to the eventual elimination of all TEs. Once a class of individuals free of all TEs has been achieved, they are immune to further transposition and can spread through the population, arresting the process of TE-induced fitness deterioration. But retrotranposons notoriously show little or no excision (Craig et al. 2002), which poses a problem for this model that we consider below.
Most theoretical models of transposable elements are based on the biology of sexual eukaryotes, as selfish DNA can invade a population only if there is sexual exchange between individuals (Hickey 1982). The widespread presence of TEs in bacteria has prompted some theoretical analyses of TE dynamics in asexual populations. These models have shown that TE maintenance can be explained by frequent horizontal transmission (Sawyer et al. 1987; Hartl and Sawyer 1988), by potential positive impacts of TEs, including beneficial TE insertions causing the fixation of other neutral or slightly deleterious TEs in the genome by hitchhiking (Martiel and Blot 2002), or by selective advantages of TE insertions in fluctuating environments with environment-dependent selection (Edwards and Brookfield 2003). Basten and Moody (1991) obtained analytic expressions for equilibrium distributions of TEs in prokaryotic populations, using branching process theory and a number of selection models. In the absence of beneficial insertions, maintenance of TEs requires horizontal transfer, and the net rate of element change due to transposition and excision must be a decreasing function of TE copy number.
Here we focus exclusively on vertically transmitted deleterious TEs in eukaryotic asexual populations. This may be more representative of retroelements, for which horizontal transfer is extremely rare (Jordan et al. 1999; Malik et al. 1999), than of DNA-based transposons, for which there is frequent evidence for horizontal transfer (Silva et al. 2004). Previous authors have proposed that, with negligible transmission rates between individuals, TEs should either accumulate in asexual populations to the point of lineage extinction or be completely eliminated (Arkhipova and Meselson 2000, 2005a; Wright and Finnegan 2001; Nuzhdin and Petrov 2003). However, it remained unclear how asexual lineages could escape long-term degeneration and completely eliminate all the TEs in the genome. TEs can decay after accumulating mutations, but it seems likely that they will continue to multiply upon the abandonment of sex, given that the rate of inactivating mutations in TEs is probably similar to the rate of transposition (Nuzhdin et al. 1998). Docking et al. (2006) carried out simulations of TE sequence evolution after the abandonment of sex, including the effects of inactivating mutations, They found that, while all elements were eventually lost from asexual populations, it took many thousands of generations for selection on TE sequences to be relaxed, even with higher mutation and transposition rates than expected in nature. This suggests that new asexual lineages should indeed harbor many active TEs, perhaps for millions of generations assuming a scaling of parameters similar to that done in our model.
Nuzhdin and Petrov (2003) suggested that suppressor alleles in a lineage abandoning sex could result in the instantaneous inactivation of TEs. Indeed, our results for infinite populations imply that a lineage with a trans-acting repressor (located in either the host or the TE genomes) that reduces the rate of transposition, u, for all elements would have a fitness advantage, by reducing the load due to TEs, 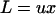 (Equation 9). Nontransposing elements would eventually decay under mutation pressure. Our results provide, however, a simpler explanation for complete TE elimination, based on straightforward population genetic processes and biologically plausible parameters and requiring only large population sizes to enhance the efficacy of selection and excision.
(Equation 9). Nontransposing elements would eventually decay under mutation pressure. Our results provide, however, a simpler explanation for complete TE elimination, based on straightforward population genetic processes and biologically plausible parameters and requiring only large population sizes to enhance the efficacy of selection and excision.
The model presented here generally assumes that TEs are uniformly deleterious on a neutral background. The potential for TE elimination may be confounded by the presence of beneficial mutations, whether derived from TEs or otherwise, which may fix linked deleterious TEs by selective sweeps (Charlesworth et al. 1992). However, this requires selection coefficients for beneficial mutations to be substantially greater than those for deleterious TEs (Johnson and Barton 2002; Bachtrog and Gordo 2004). If excision occurs, this would only reduce the rate of TE elimination, by fixing a clonal lineage with a new initial copy number and lower mean fitness. A further limitation on TE elimination arises if a fraction of sites allow neutral TE insertions (Charlesworth 1991). The continual insertion of TEs into such sites causes a buildup of elements and reduces the ability of selection to contain them at other sites in the genome.
Our model has investigated the eventual fate of an asexual lineage, either complete TE elimination or runaway element accumulation, but has not generally considered the time frame involved. In our simulations, the time to reach either final state generally ranges from several hundred generations to tens of thousands of generations, depending on the parameters involved (results not shown). However, the number of generations generally scales with the population size (see Figure 2). This implies that, with the parameter adjustment employed in our model, the time required to drive an asexual lineage to the point of extinction could be as high as millions of generations for larger populations. Similarly, complete TE elimination could take a very long time, especially with high initial copy numbers or weaker selection and excision. Thus, asexual lineages could persist with active deleterious TEs for a substantial period of time before arriving at either final state; however, on a long-term evolutionary scale (i.e., tens of millions of years), it may be the case that only asexual populations that have purged all their deleterious TEs will persist (Arkhipova and Meselson 2005a).
Asexual lineages generally compose only a single “species” and rarely embrace a taxonomic group of higher rank. While asexual lineages that arise from sexual populations may prosper in the short term, they almost invariably suffer early extinction (see review by Judson and Normark 1996). Our results have important implications for the existence of ancient asexual taxa: those eukaryotic taxa that have evolved and reproduced for a long stretch of evolutionary time without sex or recombination and have been labeled as “evolutionary scandals” (Maynard Smith 1986). While many claims of ancient asexuality have been refuted upon further investigation, the most compelling evidence for ancient asexuality exists for the bdelloid rotifers, which are not known to have males, hermaphrodites, or meiosis (Arkhipova and Meselson 2005a). Strikingly, bdelloid rotifers appear to lack deleterious vertically transmitted retrotransposons. Unlike all sexually reproducing eukaryotic species that have been examined, including the monogonont rotifers, reverse transcriptases of two superfamilies of retrotransposons, gypsy-like and LINE-like retrotransposons, were not detectable in any of five bdelloid species (Arkhipova and Meselson 2000). This contrasts sharply with the patchy distribution of various DNA transposons found in bdelloid rotifers (Arkhipova and Meselson 2000, 2005b), in agreement with their often horizontal mode of transmission, and the presence of a domesticated retrotransposon (Arkhipova et al. 2003).
Presumably, the common sexual ancestor of the bdelloid and monogont rotifers harbored active deleterious TEs, including retrotransposons. It is not certain what mechanism has allowed bdelloids to become free of these TEs. Our models provide a plausible scenario for complete elimination of vertically transmitting TEs, assuming that bdelloids have fairly large population sizes and that excision takes place. It is not known what the effective population size, Ne, of bdelloids might be. This can be estimated from measures of sequence diversity, which is a function of Neμ, where μ is the mutation rate. While the mutation rate of bdelloid rotifers is unknown, there is no evidence that bdelloids have an unusual mutation rate, as relative rate tests between the nucleotide mutation rates of bdelloid and monogonont rotifers show no significant difference at either synonymous or replacement sites (Mark Welch and Meselson 2001). Mitochondrial nucleotide diversity in independently evolving bdelloid clades appears to be similar to that of sexual organisms and averages ∼1% (Birky et al. 2005; C. W. Birky, personal communication). Mutation rates of mitochondrial DNA are generally higher than those for nuclear DNA, although this varies between taxa. If we suppose a mutation rate of 10−9–10−7, we can estimate the effective population size to be ∼105–107. This is in the range of being large enough to facilitate complete TE elimination under biologically realistic transposition parameters, even with rates of excision well below that of transposition (see Figures 3 and 4). Thus, unusually high excision rates or attenuated transposition rates are not necessary to explain the existence of TE-free ancient asexuals.
The transposition, excision, and selection parameters that applied to bdelloid rotifers upon the abandonment of sex are also not known. However, it has been suggested that the unusual ecology of bdelloid rotifers, by causing frequent DNA damage and repair, may have facilitated their loss of TEs (M. Meselson, personal communication). Bdelloids are capable of anhydrobiosis, a form of dormancy triggered by desiccation (Ricci 1998). However, unlike most organisms capable of anhydrobiosis, including the monogonont rotifers, bdelloids do not produce the water-replacement substance trehalose (Lapinski and Tunnacliffe 2003). Bdelloids tolerate high levels of ionizing radiation, causing DNA double-strand breaks (DSBs) that are repaired within a few hours (E. Gladyshev and M. Meselson, unpublished results). The bacterium Deinococcus radiodurans is well known for its ability to repair DNA damage following radiation or dehydration by efficient repair mechanisms (Battista 1997), suggesting that the response to irradiation in bdelloids may be an outcome of selection for desiccation resistance.
DSB repair processes in bdelloids might either lead to direct elimination of TEs or cause synergistic selection between deleterious TEs, as a result of ectopic exchange between them (M. Meselson, personal communication). Although bdelloids do not undergo meiosis, ectopic pairing of TEs during DSB repair could lead to deleterious rearrangements or unrepaired breaks, with a frequency proportional to the square of the copy number (Montgomery et al. 1987; Charlesworth et al. 1994). Such ectopic pairing may lead to the excision of heterozygous TEs, which is thought to be extremely rare for retrotransposons, and would prevent any TE insertion from being selectively neutral, just as in sexual species, enhancing the possibility of complete TE elimination (see Figure 5).
We have shown that newly arisen asexual lineages may be able to avoid early extinction due to TEs. The ability to do so depends critically on large population sizes, with some level of excision. The unique ecology and stress-resistance mechanism of the bdelloid rotifers, which probably evolved as a result of adaptation to their transient aquatic habitats, may have provided the means under which to eliminate deleterious TEs upon the abandonment of sex. Perhaps other asexual lineages were not as fortunate (Arkhipova and Meselson 2005a).
Acknowledgments
We thank M. Meselson for helpful discussions and L. Loewe for assistance with the simulation modeling. This research was supported by postgraduate scholarships to E.S.D. from the Natural Sciences and Engineering Research Council (Canada) and the University of Edinburgh School of Biological Sciences. B.C. is supported by the Royal Society (United Kingdom).
References
- Arkhipova, I., and M. Meselson, 2000. Transposable elements in sexual and ancient asexual taxa. Proc. Natl. Acad. Sci. USA 97: 14473–14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova, I., and M. Meselson, 2005. a Deleterious transposable elements and the extinction of asexuals. BioEssays 27: 76–85. [DOI] [PubMed] [Google Scholar]
- Arkhipova, I., and M. Meselson, 2005. b Diverse DNA transposons in rotifers of the class Bdelloidea. Proc. Natl. Acad. Sci. USA 102: 11781–11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova, I. R., K. I. Pyatkov, M. Meselson and M. B. Evgen'ev, 2003. Retroelements containing introns in diverse invertebrate taxa. Nat. Genet. 33: 123–124. [DOI] [PubMed] [Google Scholar]
- Bachtrog, D., and I. Gordo, 2004. Adaptive evolution of asexual populations under Muller's ratchet. Evolution 58: 1403–1413. [DOI] [PubMed] [Google Scholar]
- Basten, C. J., and M. E. Moody, 1991. A branching-process model for the evolution of transposable elements incorporating selection. J. Math. Biol. 29: 743–761. [DOI] [PubMed] [Google Scholar]
- Battista, J. R., 1997. Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51: 203–224. [DOI] [PubMed] [Google Scholar]
- Birky, Jr., C. W., C. Wolf, H. Maughan, L. Herbertson and E. Henry, 2005. Speciation and selection without sex. Hydrobiologia 546: 29–45. [Google Scholar]
- Brookfield, J. F. Y., 1991. Models of repression of transposition in P-M hybrid dysgenesis by P cytotype and by zygotically encoded repressor proteins. Genetics 128: 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield, J. F. Y., and R. M. Badge, 1997. Population genetic models of transposable elements. Genetica 100: 281–294. [PubMed] [Google Scholar]
- Charlesworth, B., 1985. The population genetics of transposable elements, pp. 213–232 in Population Genetics and Molecular Evolution, edited by T. Ohta and K. Aoki. Springer-Verlag, Berlin.
- Charlesworth, B., 1990. Mutation-selection balance and the evolutionary advantage of sex and recombination. Genet. Res. 55: 199–221. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1991. Transposable elements in natural populations with a mixture of selected and neutral insertion sites. Genet. Res. 57: 127–135. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., and N. H. Barton, 1996. Recombination load associated with selection for increased recombination. Genet. Res. 67: 27–41. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., and D. Charlesworth, 1983. The population dynamics of transposable elements. Genet. Res. 42: 1–27. [Google Scholar]
- Charlesworth, B., and C. H. Langley, 1986. The evolution of self-regulated transposition of transposable elements. Genetics 112: 359–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., and C. H. Langley, 1989. The population genetics of Drosophila transposable elements. Annu. Rev. Genet. 23: 251–287. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., A. Lapid and D. Canada, 1992. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. II. Inferences on the nature of selection against element. Genet. Res. 60: 115–130. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., P. Sniegowski and W. Stephan, 1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. [DOI] [PubMed] [Google Scholar]
- Craig, N. L., R. Craigie, M. Gellert and A. M. Lambowitz (Editors), 2002. Mobile DNA II. ASM Press, Washington, DC.
- Crow, J. F., 1970. Genetic loads and the cost of natural selection, pp. 128–177 in Mathematical Topics in Population Genetics, edited by K. Kojima. Springer-Verlag, Berlin.
- Docking, T. R., F. E. Saadé, M. C. Elliott and D. J. Schoen, 2006. Retrotransposon sequence variation in four asexual plant species. J. Mol. Evol. 62: 375–387. [DOI] [PubMed] [Google Scholar]
- Doolittle, W. F., and C. Sapienza, 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601–603. [DOI] [PubMed] [Google Scholar]
- Doolittle, W. F., T. B. L. Kirkwood and M. A. H. Dempster, 1984. Selfish DNA and self-restraint. Nature 307: 501–502. [DOI] [PubMed] [Google Scholar]
- Edwards, R. J., and J. F. Y. Brookfield, 2003. Transiently beneficial insertions could maintain mobile DNA sequences in variable environments. Mol. Biol. Evol. 20: 30–37. [DOI] [PubMed] [Google Scholar]
- Ewens, W. J., 1979. Mathematical Population Genetics. Springer-Verlag, Berlin.
- Finnegan, D. J., 1992. Transposable elements, pp. 1096–1107 in The Genome of Drosophila melanogaster, edited by D. L. Lindsley and G. Zimm. Academic Press, New York.
- Hartl, D. L., and S. A. Sawyer, 1988. Why do unrelated insertion sequences occur together in the genome of Escherichia coli? Genetics 118: 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey, D. A., 1982. Selfish DNA: a sexually-transmitted nuclear parasite. Genetics 101: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, T., and N. H. Barton, 2002. The effect of deleterious alleles on adaptation in asexual populations. Genetics 162: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, I. K., L. V. Matyunina and J. F. McDonald, 1999. Evidence for recent horizontal transfer of long terminal repeat retrotransposons. Proc. Natl. Acad. Sci. USA 96: 12621–12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson, O. P., and B. B. Normark, 1996. Ancient asexual scandals. Trends Ecol. Evol. 11: 41–46. [DOI] [PubMed] [Google Scholar]
- Kaplan, N. L., and J. F. Y. Brookfield, 1983. Transposable elements in Mendelian populations. III. Statistical results. Genetics 104: 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, M. G., and D. R. Lisch, 2001. Transposable elements, parasitic DNA, and genome evolution. Evolution 55: 1–24. [DOI] [PubMed] [Google Scholar]
- Kimura, M., and T. Maruyama, 1966. The mutational load with epistatic gene interactions in fitness. Genetics 54: 1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley, C. H., E. A. Montgomery, R. Hudson, N. Kaplan and B. Charlesworth, 1988. On the role of unequal exchange in the containment of transposable element copy number. Genet. Res. 52: 223–236. [DOI] [PubMed] [Google Scholar]
- Lapinski, J., and A. Tunnacliffe, 2003. Anhydrobiosis without trehalose in bdelloid rotifers. FEBS Lett. 553: 387–390. [DOI] [PubMed] [Google Scholar]
- Malik, H. S., W. D. Burke and T. H. Eickbush, 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16: 793–805. [DOI] [PubMed] [Google Scholar]
- Mark Welch, D. B., and M. Meselson, 2001. Rates of nucleotide substitution in sexual and anciently asexual rotifers. Proc. Natl. Acad. Sci. USA 98: 6720–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiel, J.-L., and M. Blot, 2002. Transposable elements and fitness of bacteria. Theor. Popul. Biol. 61: 509–518. [DOI] [PubMed] [Google Scholar]
- Maside, X., S. Assimacopoulos and B. Charlesworth, 2000. Rates of movement of transposable elements on the second chromosome of Drosophila melanogaster. Genet. Res. 75: 275–284. [DOI] [PubMed] [Google Scholar]
- Matsumoto, M., and T. Nishimura, 1998. Mersenne twister: a 623-dimensionally equidistributed uniform pseudo-random number generator. ACM Trans. Mod. Comput. Simul. 8: 3–30. [Google Scholar]
- Maynard Smith, J., 1986. Contemplating life without sex. Nature 324: 300–301. [DOI] [PubMed] [Google Scholar]
- Montgomery, E. A., B. Charlesworth and C. H. Langley, 1987. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet. Res. 49: 31–41. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1964. The relation of recombination to mutational advance. Mutat. Res. 1: 2–9. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., 1999. Sure facts, speculations, and open questions about evolution of transposable elements. Genetica 107: 129–137. [PubMed] [Google Scholar]
- Nuzhdin, S. V., and T. F. C. Mackay, 1995. The genomic rate of transposable element movement in Drosophila melanogaster. Mol. Biol. Evol. 12: 180–181. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., and D. A. Petrov, 2003. Transposable elements in clonal lineages: lethal hangover from sex. Biol. J. Linn. Soc. 79: 33–41. [Google Scholar]
- Nuzhdin, S. V., E. G. Pasyukova, T. V. Morozova and A. J. Flavell, 1998. Quantitative genetic analysis of copia retrotransposon activity in inbred Drosophila melanogaster lines. Genetics 150: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel, L. E., and F. H. Crick, 1980. Selfish DNA: the ultimate parasite. Nature 284: 1517–1523. [DOI] [PubMed] [Google Scholar]
- Pasyukova, E. G., S. V. Nuhzdin and D. A. Filatov, 1998. The relationship between the rate of transposition and transposable element copy number for copia and Doc retrotransposons of Drosophila melanogaster. Genet. Res. 72: 1–11. [DOI] [PubMed] [Google Scholar]
- Ricci, C., 1998. Anhydrobiotic capabilities of bdelloid rotifers. Hydrobiologia 388: 321–326. [Google Scholar]
- Sawyer, S. A., D. E. Dykhuizen, R. F. DuBose, L. Green, T. Mutangadura-Mhlanga et al., 1987. Distribution and abundance of insertions sequences among natural isolates of Escherichia coli. Genetics 115: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, J. C., E. L. Loreto and J. B. Clark, 2004. Factors that affect the horizontal transfer of transposable elements. Curr. Issues Mol. Biol. 6: 57–71. [PubMed] [Google Scholar]
- Stephan, W., and Y. Kim, 2002. Recent applications of diffusion theory to population genetics, pp. 72–93 in Modern Developments in Theoretical Population Genetics, edited by M. Slatkin and M. Veuille. Oxford University Press, Oxford.
- Vieira, C., and C. Biémont, 1997. Transposition rate of the 412 retrotransposable element is independent of copy number in natural populations of Drosophila simulans. Mol. Biol. Evol. 14: 185–188. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., and D. J. Finnegan, 2001. Genome evolution: sex and the transposable element. Curr. Biol. 11: R296–R299. [DOI] [PubMed] [Google Scholar]