Abstract
The Tol2 element is a naturally occurring active transposable element found in vertebrate genomes. The Tol2 transposon system has been shown to be active from fish to mammals and considered to be a useful gene transfer vector in vertebrates. However, cis-sequences essential for transposition have not been characterized. Here we report the characterization of the minimal cis-sequence of the Tol2 element. We constructed Tol2 vectors containing various lengths of DNA from both the left (5′) and the right (3′) ends and tested their transpositional activities both by the transient excision assay using zebrafish embryos and by analyzing chromosomal transposition in the zebrafish germ lineage. We demonstrated that Tol2 vectors with 200 bp from the left end and 150 bp from the right end were capable of transposition without reducing the transpositional efficiency and found that these sequences, including the terminal inverted repeats (TIRs) and the subterminal regions, are sufficient and required for transposition. The left and right ends were not interchangeable. The Tol2 vector carrying an insert of >11 kb could transpose, but a certain length of spacer, <276 but >18 bp, between the left and right ends was necessary for excision. Furthermore, we found that a 5-bp sequence, 5′-(A/G)AGTA-3′, is repeated 33 times in the essential subterminal region. Mutations in the repeat sequence at 13 different sites in the subterminal region, as well as mutations in TIRs, severely reduced the excision activity, indicating that they play important roles in transposition. The identification of the minimal cis-sequence of the Tol2 element and the construction of mini-Tol2 vectors will facilitate development of useful transposon tools in vertebrates. Also, our study established a basis for further biochemical and molecular biological studies for understanding roles of the repetitive sequence in the subterminal region in transposition.
TRANSPOSONS are genetic elements that move from one locus to another locus and reside in the genome as repetitive sequences. These transposon sequences are grouped into two classes: (1) autonomous members, such as Ac in maize (Fedoroff et al. 1983; Müller-Neumann et al. 1984; Pohlman et al. 1984), that encode a functional transposase and can transpose by itself; (2) nonautonomous members, such as Ds in maize (Döring et al. 1984; Sutton et al. 1984), that have mutations or deletions in the transposase gene, but retain essential cis-sequences, and can transpose in the presence of the transposase activity. The Tol2 element was found from the genome of a small teleost, the medaka fish. Tol2 belongs to the hAT family of transposons (Koga et al. 1996). An autonomous member of the Tol2 element was identified (Kawakami et al. 1998). The autonomous Tol2 contains a gene encoding a fully functional transposase that is capable of catalyzing transposition (Kawakami and Shima 1999; Kawakami et al. 2000). Thus, to date, Tol2 is the only active autonomous transposable element found in a vertebrate genome.
We have identified mRNA transcribed from the transposase gene, cloned its cDNA (Kawakami and Shima 1999), and developed two-component transposition systems in vertebrate animals. In zebrafish, the transposition system is composed of a transposon-donor plasmid containing a nonautonomous Tol2 construct and mRNA synthesized in vitro by using the transposase cDNA as a template (Kawakami et al. 2000, 2004b). The nonautonomous Tol2 construct cannot transpose by itself since part of the transposase coding region is replaced by a foreign gene, but can transpose when the transposase activity is supplied in trans. These are introduced into zebrafish cells by micro-injection into fertilized eggs. Then, in developing embryos, the transposase protein produced from the mRNA catalyzes excision of the Tol2 construct from the plasmid and integration of the excised construct into the chromosomal DNA. Since transposition can occur in the zebrafish germ lineage very efficiently, the Tol2 transposon system has been successfully applied to transgenesis and gene trapping (Kawakami et al. 2000, 2004b; Kawakami 2005). In mouse, a transposon-donor plasmid containing the Tol2 construct with the neomycin resistance gene and the helper plasmid containing the transposase cDNA placed under the control of a strong promoter are introduced in embryonic stem (ES) cells by electroporation. In the ES cells, the transposase produced from the helper plasmid catalyzes transposition of the Tol2 construct from the plasmid to the mouse genome (Kawakami and Noda 2004). The transposition frequency was increased as the transposase activity was increased (Kawakami and Noda 2004), suggesting that the Tol2 transposon system may not suffer from a phenomenon termed “overproduction inhibition,” which has been observed in the case of the Sleeping Beauty transposon system (Miskey et al. 2005). In Xenopus, the transposon-donor plasmid and the transposase mRNA are injected into fertilized eggs. The transposase produced in the injected embryos catalyzes excision of the transposon construct from the plasmid (Kawakami et al. 2004a). Recently, we have shown that the excised transposon was integrated in the Xenopus genome and transmitted to the next generation through the germ lineage (Johnson Hamlet et al. 2006). Thus, Tol2 can be used as a gene transfer vector in vertebrates. The current Tol2 vectors contain ∼2.8 kb of DNA from the original Tol2 element (Kawakami and Noda 2004; Kawakami et al. 2004b). To increase the usefulness of Tol2 as a genetic tool, it is important to remove the unnecessary part from the vector. First, manipulation of the transposon vector will become easier. Second, possible effects of the Tol2 sequence on expression of a foreign gene cloned in the transposon vector and/or on expression of chromosomal genes located near the transposon integration site will be minimized. To this end, it is important to analyze cis-sequences that are sufficient and required for transposition.
cis-Elements of other transposons belonging to the hAT family have been studied. In the case of Ac from maize, 238 bp from the 5′-end and 209 bp from the 3′-end are sufficient and required for transposition (Coupland et al. 1989). These subterminal sequences contain 13 AAACGG sequences (Chatterjee and Starlinger 1995), which are the binding motif for the Ac transposase (Kunze and Starlinger 1989). While mutations in some of these motifs reduced or abolished the cis-activity required for excision (Chatterjee and Starlinger 1995), some binding motifs were dispensable, and roles of these sequences in transposition have not been elucidated (Chatterjee and Starlinger 1995). In the case of Tag1 from Arabidopsis, 98 bp from the 5′-end and 109 bp from the 3′ end are sufficient for transposition. Tag1 is unique since the 5′ and 3′ subterminal sequences contain repeats of different sequences (Liu et al. 2001). Mutagenesis of these repetitive sequences, however, has not been carried out for Tag1. Thus, although there have been evidences that the subterminal regions are important for transposition of transposons of the hAT family, their roles in transposition have not been well understood. Therefore, it is important to know the structure–function relationship in the subterminal region of Tol2, a vertebrate member of the hAT family.
In this study, we aimed to characterize cis-elements of the Tol2 transposable element essential for transposition. First, to determine minimal sequences sufficient and required for transposition, Tol2 vectors carrying various sizes of the Tol2 end sequences were created and tested for their activities both in excision in zebrafish embryos and in transposition in the zebrafish germ lineage. Second, to further characterize sequences required for transposition, base-substitution mutations were introduced in the terminal inverted repeats (TIRs) and in the subterminal regions and their effects on the excision activity were analyzed. These studies led to identification of the minimal cis-sequence of Tol2 and the functional repetitive sequence in the subterminal region.
MATERIALS AND METHODS
Transposon constructs:
The structures of the Tol2 constructs created and used in this study are shown in Figure 1A. To construct pT2KXIGΔin, pT2KXIG (Kawakami et al. 2004b) was digested with NruI, ligated to a BglII linker and then digested with BglII and self-ligated. To construct T2AL200R200G (accession no. AB262448), T2AL175R200G (AB262449), T2AL150R200G (AB262450), T2AL200R175G (AB262451), T2AL200R150G (AB262452), T2AL200R100G (AB262453), and T2AL50R50G (AB262454), DNA fragments containing various lengths of the left and right end sequences plus the backbone plasmid DNA were amplified by PCR and ligated with the GFP expression cassette, which contains the Xenopus EF1α enhancer–promoter, the rabbit β-globin intron, the EGFP gene, and the SV40 poly(A) signal at XhoI and BglII sites. The structures of all of these plasmids were confirmed by DNA sequencing. T2AL200L200G (AB262455) and T2AR200R200G (AB262456) were constructed by replacing the right end and the left end of T2AL200R200G with the PCR-amplified BglII–KpnI fragment of the left end and the PCR-amplified SpeI–XhoI fragment of the right end, respectively. The primers used for the PCR amplification were designed to retain the 8-bp target duplication sequence. The structures of the plasmids were confirmed by DNA sequencing. T2AL200R150-1139, T2AL200R150-502, T2AL200r150-276, and T2AL200R150-18 were constructed by deleting, respectively, the ApaI–ApaI, BglII–HindIII, ClaI–XhoI, and BglII–XhoI fragmentsfrom T2AL200R150G. The structures of Tol2 constructs (T2AL200R150GmL series and T2AL200R150GmR series) carrying substitution mutations in the left- and right-end sequences, respectively, are shown in Figure 4 and Table 4. To construct these plasmids, the backbone DNA fragments were amplified by PCR using pT2AL200R150G as a template and primers containing an MluI site at the 5′-end. The amplified DNA was digested with MluI and self-ligated. The structures of the plasmids were confirmed by DNA sequencing.
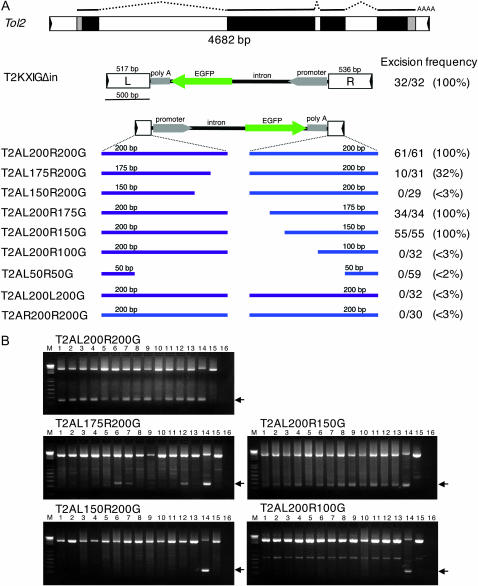
Figure 1.—
The structures and activities of mini-Tol2 constructs. (A) The structures of the full-length Tol2, T2KXIGΔin, and mini-Tol2 constructs created in this study and their activities in the excision assay. The full-length Tol2 (4682 bp) encodes the transposase gene. Exons are shown by gray (untranslated region) and black (translated region) boxes. Lines (exons), dotted lines (intron), and AAAA (polyadenylation) above Tol2 indicate mRNA for the transposase. The left end (5′-end) and the right end (3′-end) are designated with respect to orientation of the transcript. The mini-Tol2 constructs contain various lengths of DNA from the left-end (purple) and the right-end (blue) sequences. All of these constructs contain the GFP expression cassette in the middle. The excision frequency represents “the number of DNA samples from which the excision product was amplified” per “the number of DNA samples analyzed.” (B) The examples of the excision assay performed in this study. DNA samples prepared from embryos injected with the transposase mRNA and plasmids containing T2AL200R200G, T2AL175R200G, T2AL150R200G, T2AL200R150G, and T2AL200R100G were analyzed by PCR. Top bands represent PCR products from unexcised transposon constructs and bottom bands represent excision products. Lanes 1–13: PCR products amplified from DNA samples prepared from each injected embryo. Lane 14: positive control (a DNA sample was prepared from an embryo injected with T2KXIGΔin). Lane 15: negative control (no transposase mRNA). Lane 16: negative control (a DNA sample was prepared from an uninjected embryo). The weak bands are seen in T2AL175R200G (lanes 10, 11, 15) and T2AL200R100G (lanes 1–13, 15) artifacts, which tend to be amplified when the amount of the excision product is too low to be amplified.
Figure 4.—
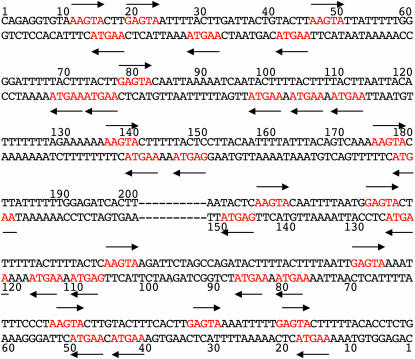
The left- and right-end sequences of Tol2. The 200-bp left end (5′-end) and the 150-bp right end (3′-end) sequences sufficient and required for transposition. The numbers above the sequence show bases from the left end and the numbers below the sequence show bases from the right end. The 5-bp repeats, 5′-(A/G)AGTA-3′, are in red on the sequence and also indicated by arrows.
TABLE 4.
Positions and sequences of substitution mutations in the subterminal region of Tol2 by MluI mutagenesis
| Mutant namea | Wild-type sequence (positionb) | Mutant sequence |
|---|---|---|
| mTIRL1-6 | CAGAGGTGTAAAGTA (L1-15) | ACGCGTTGTAAAGTA |
| mTIRR1-6 | CAGAGGTGTAAAAAG (R1-15) | ACGCGTTGTAAAAAG |
| mL13-16 | TGTAAAGTACTTGAG (L7-21) | TGTAAACGCGTTGAG |
| mL26-30 | GTAATTTTACTTGAT (L21-35) | GTAATACGCGTTGAT |
| mL48-51 | ACTTAAGTATTATTT (L42-56) | ACTTAACGCGTATTT |
| mL66-70 | GGATTTTTACTTTAC (L61-75) | GGATTACGCGTTTAC |
| mL106-108 | CTTTTACTTTTACTT (L99-113) | CTTTTACGCGTACTT |
| mL136-148 | AAAGTACTTTTTACT (L135-149) | ATTTTACGCGTTTTT |
| mL169-181 | AGTCAAAAAGTACTT (L168-182) | AAACACGCGTTAAAT |
| mR15-18 | TTTTGAGTACTTTTT (R24-10) | TTTTGACGCGTTTTT |
| mR27-31 | ACTTGAGTAAAATTT (R37-23) | ACTTGACGCGTATTT |
| mR48-51 | CCCTAAGTACTTGTA (R57-43) | CCCTAACGCGTTGTA |
| mR63-67 | AATTGAGTAAAATTT (R73-59) | AATTGACGCGTATTT |
| mR103-108 | TTTTACTCAAGTAAG (R113-99) | TTTTAACGCGTTAAG |
| mR114-126 | GAGTACTTTTTTACT (R127-113) | GTTTAACGCGTTTTT |
| mL88-92 | CAATTAAAAATCAAT (L83-97) | CAATTACGCGTCAAT |
| mL158-162 | TACAATTTTATTTAC (L153-167) | TACAAACGCGTTTAC |
| mR89-94 | GATTCTAGCCAGATA (R99-85) | GATTCACGCGTGATA |
The bases substituted in the mutants are underlined. TIR, terminal inverted repeat.
L and R indicate mutations in the left end and the right end, respectively. Numbers indicate positions of the bases substituted in the mutants.
The positions of the first and the last base of the sequence shown here are indicated in parentheses as base-pair distances from the left end (L) or the right end (R).
Micro-injection and excision assay:
Transposase mRNA was synthesized as described previously (Kawakami 2004; Kawakami et al. 2004b). Approximately 1 nanoliter of a DNA–RNA solution containing 25 ng/μl circular DNA of a transposon-donor plasmid and 25 ng/μl transposase mRNA were injected into fertilized eggs. Approximately 10 hr after the injection, DNA samples were prepared from the injected embryos and the transient excision assay was performed as described (Kawakami and Shima 1999; Kawakami 2004) with some modifications. In the experiments described in Figure 1 and Figure 5, the excision products were amplified by using Takara Ex Taq (Takara Bio, Shiga, Japan) and the primers T2AexL (5′-CGC AAT TAA CCC TCA CTA AAG G-3′) and T2AexR (5′-ACC CAA CTG ATC TTC AGC ATC T-3′). In the experiments described in Figure 3, the DNA samples were digested with ScaI prior to PCR to cut five ScaI sites in the transposon constructs on the plasmid DNA, which sometimes hampered the amplification of the excision products. PCR was carried out by using the Expand high fidelity PCR system (Roche Diagnostics GmbH, Mannheim, Germany) and the primers T2AexL-2 (5′-ACC CTC ACT AAA GGG AAC AAA AG-3′) and T2AexR-2 (5′-GTG CGG GCC TCT TCG CTA TTA C-3′).
Figure 5.—
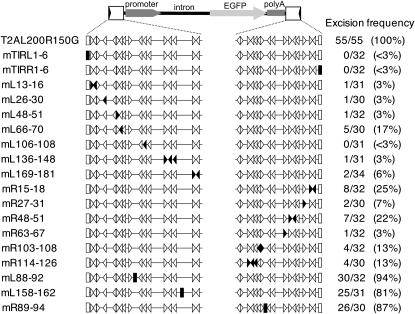
The structures and activities of Tol2 constructs with substitution mutations created by the MluI mutagenesis. Boxes at the both ends represent the TIRs and the TIR mutations (mTIRL1-6 and mTIRR1-6) are indicated as solid boxes. Open arrowheads indicate positions and directions of the repeat 5′-(A/G)AGTA-3′, and solid arrowheads indicate repeats mutated by base substitutions shown in Table 4. In mL88-92, mL158-162, and mR89-94, the substitution mutations in the sequences outside of the repeats (Table 4) are shown as solid boxes. Excision frequency represents “the number of DNA samples from which the excision product was amplified” per “the number of DNA samples analyzed.”
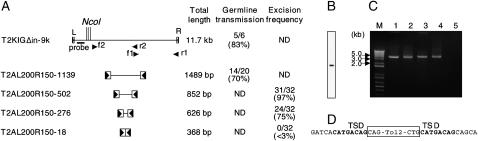
Figure 3.—
Spacing capacity and requirements. (A) The structures of transposon constructs carrying various lengths of inserts between the left and right ends, their total lengths, and activities determined by germline transmission and the transient excision assay. NcoI sites in T2KIGΔin-9k were used for the Southern blot analysis in B. f1, r1, f2, and r2: positions and directions of primers used for PCR in C. ND, not determined. (B) Southern blot hybridization analysis of NcoI-digested genomic DNA from transgenic fish. The result indicates an insertion of a single copy of T2KIGΔin-9k. (C) The genomic DNA from the transgenic fish carrying the T2KIGΔin-9k insertion (lanes 1 and 3) and the injected plasmid DNA (lanes 2 and 4) were used for PCR. PCR was carried out by using f1 and f2 (lanes 1 and 2) and f2 and r2 (lanes 3 and 4). M, size marker; lane 5, no DNA control. (D) The genomic DNA surrounding the T2KIGΔin-9k integration site was determined by adaptor ligation PCR. The target-site duplication (TSD) is in boldface type. The T2KIGΔin-9k insertion was mapped within an intron of zgc:65966 (Pho GDP dissociation inhibitor).
GFP expression in embryos:
GFP expression in embryos was analyzed by using a fluorescence stereomicroscope MZ16 FA (Leica Microsystems GmbH, Wetzlar, Germany), and photos were taken by using a digital color camera DFC300 FX (Leica Microsystems GmbH).
Southern blot hybridization, inverse PCR, and adaptor ligation PCR:
Southern blot hybridization was performed as described previously (Kawakami 2004; Kawakami et al. 2004b). The genomic DNA samples were digested either with BglII (Figure 2) or with NcoI (Figure 3), which gave rise to one hybridization band from one insertion. The junction fragments containing the Tol2 ends and the genomic DNA were cloned either by inverse PCR (Kawakami 2004; Kawakami et al. 2004b) or by adaptor ligation PCR (Kotani et al. 2006). The adaptor ligation PCR method described previously (Siebert et al. 1995; Spertini et al. 1999) was applied to our system as follows. The genomic DNA from transgenic fish is digested with MboI, BglII, BamHI, SpeI, XbaI, or NheI. MboI, BglII, and BamHI generate a 5′-GATC-3′ cohesive end, and SpeI, XbaI, and NheI generate a 5′-CTAG-3′ cohesive end. The adaptors are prepared by annealing AL (5′-CTA ATA CGA CTC ACT ATA GGG CTC GAG CGG CCG CGG GGG CAG GT-3′) with phosphorylated GATC-AS (5′-pGAT CAC CTG CCC CCG CTT-3′) or phosphoylated CTAG-AS (5′-pCTA GAC CTG CCC CCG CTT-3′) at 95° for 1 min. These adaptors are ligated to the digested genomic DNA with appropriate cohesive ends. The ligated samples are diluted to 10-fold and used for PCR. To clone 5′ junctions, the first PCR is performed by using the primers Ap1 (5′-GGA TCC TAA TAC GAC TCA CTA TAG GG-3′) and 175L-out (5′-TTT TTG ACT GTA AAT AAA ATT G-3′), and the second PCR is performed by using the primers Ap2 (5′-CAC TAT AGG GCT CGA GCG G-3′) and 150L-out (5′-GAG TAA AAA GTA CTT TTT TTT CT-3′). To clone 3′ junctions, the first PCR is performed by using the primers Ap1 and 175R-out (5′-TTC TTG CTT TTA CTT TTA CTT CC-3′), and the second PCR is performed by using the primers Ap2 and 150R-out (5′-AAT ACT CAA GTA CAA TTT TA-3′). The PCR products for the 5′ and 3′ junctions are gel extracted and sequenced by using 100L-out (5′-AGT ATT GAT TTT TAA TTG TA-3′) and 100R-out (5′-AGA TTC TAG CCA GAT ACT-3′), respectively.
Figure 2.—
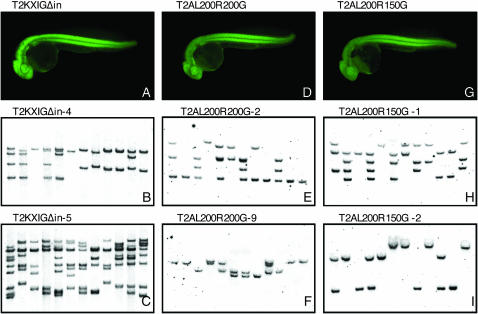
Germline transmission using mini-Tol2 constructs. (A, D, F) GFP expression in transgenic embryos carrying a single insertion of the mini-Tol2 constructs T2KXIGΔin (A), T2AL200R200G (D), and T2AL200R150G (F). (B, C, E, F, H, I) Southern blot hybridization analysis of transposon insertions in F1 fish. F1 fish from founder fish injected with the transposase mRNA and a plasmid containing T2KXIGΔin (B and C), T2AL200R200G (E and F), or T2AL200R150G (H and I) were analyzed by Southern blot hybridization. The genomic DNA was prepared from tail fins of 12 F1 transgenic fish from each founder fish (T2KXIGΔin-4, T2KXIGΔin-5, T2AL200R200G-2, T2AL200R200G-9, T2AL200R150G-1, and T2AL200R150G-2: see Table 1) and were used for the Southern blot analysis.
RESULTS
The minimal sequence sufficient and required for excision:
To characterize the cis-sequence essential for transposition of the Tol2 transposable element, we created Tol2 constructs that contained various lengths of DNA from the 5′-and 3′-ends (Figure 1A). The 5′- and 3′-ends were designated with respect to the orientation of the transposase gene (Kawakami and Shima 1999) and are also referred to as left (L) and right (R), respectively, in this article (Figure 1A). The activities of these transposon constructs were analyzed by the transient excision assay using zebrafish embryos (Figure 1, A and B). First, the excision product was detected from all embryos injected with the transposase mRNA and a transposon-donor plasmid containing T2KXIGΔin (100%: 32/32), which carried 517 bp of DNA from the 5′-end and 536 bp of DNA from the 3′-end. It should be noted that a Tol2 construct containing the end sequences of similar sizes was not excised in the initial excision assay (Kawakami and Shima 1999). We have found that the transposase activity in the initial condition was much lower than that in the present condition (Kawakami et al. 2000, 2004b). We hypothesize that there may be sequences that are essential for excision under lower transposase activities, but are dispensable for excision under higher transposase activities. While such sequences remain to be determined, by the transient excision assay and using the present condition, we further analyzed a smaller Tol2 construct to determine minimal cis-sequences. The T2AL200R200G construct, containing 200 bp of DNA from both ends, fully retained the excision activity (100%: 61/61). To determine the minimal DNA sequence from the left end, T2AL175R200G and T2AL150R200G were constructed and analyzed. The excision activity was reduced in T2AL175R200G (32%: 10/31) and abolished nearly completely in T2AL150R200G (<3%: 0/29), indicating that 200 bp of DNA from the left end is sufficient and >175 bp of DNA is required for the full excision activity. Then, to determine the minimal DNA sequence from the right end, T2AL200R175G, T2AL200R150G, and T2AL200R100G were constructed. T2AL200R175G and T2AL200R150G retained the excision activity (34/34 and 55/55, respectively). However, in T2AL200R100G the excision activity was drastically decreased (<3%: 0/32), indicating that 150 bp of DNA from the right end is sufficient and >100 bp of DNA is required for the excision activity.
The left and right ends are not interchangeable:
To determine whether the left and right ends of the Tol2 element are functionally equivalent, we constructed T2AL200L200G containing the 200-bp left-end sequence at both ends and T2AR200R200G containing the 200-bp right-end sequence at both ends and analyzed their activities by the transient excision assay (Figure 1A). Excision products were detected from neither of these plasmids (0/32 and 0/30, respectively), indicating that the left and right ends of Tol2 are functionally different and not interchangeable.
Germline transmission using mini-Tol2 vectors:
We have developed a highly efficient transgenesis method in zebrafish using the Tol2 transposon system (Kawakami et al. 2004b; Kawakami 2005). To determine whether the smaller “mini-Tol2” constructs are active not only for excision but also for integration in new loci, and whether they can transpose in the zebrafish germ lineage as efficiently as the authentic vector, we injected transposase mRNA and transposon-donor plasmids containing T2KXIGΔin, T2AL200R200G, T2AL200R150G, and T2AL50R50G into fertilized eggs. T2AL50R50G was used as a negative control since no excision products were detected from embryos injected with this construct (Figure 1A). The injected fish were raised to adulthood and crossed with uninjected wild-type fish, and F1 embryos were analyzed for GFP expression under a fluorescent microscope. In the progeny from 7 of 11 fish (63%) injected with pT2KXIGΔin, 7 of 10 fish (70%) injected with pT2AL200R200G, and 6 of 10 fish (60%) injected with pT2AL200R150G, embryos expressing GFP were identified (Figure 2, A, D, and G), indicating that the injected DNA was integrated into the genome and transmitted to the F1 generation very efficiently (Table 1). None of the F1 embryos from 10 fish injected with pT2AL50R50G expressed GFP, indicating that the frequency of integration of this construct in the genome was considerably low (Table 1). The injected fish that transmit the transposon insertions to the F1 generation are hereafter referred to as founder fish. These results are consistent with the results obtained from the transient excision assay using the same transposon constructs. Thus, 200 bp of DNA from the left end and 150 bp of DNA from the right end are sufficient for highly efficient transposition in the zebrafish germ lineage.
TABLE 1.
Germline transmission frequency using different transposon vectors
| T2KXIGa | T2KXIGΔin | T2AL200R200G | T2AL200R150G | T2AL50R50G | |
|---|---|---|---|---|---|
| Injected fish | 10 | 11 | 10 | 10 | 10 |
| Founder fishb | 5 | 7 | 7 | 6 | 0 |
| No. of insertionsc | 5.6 | 6.9 | 6.9 | 5.5 | NA |
The result with T2KXIG was described previously (Kawakami et al. 2004b).
The injected fish that transmit the transposon insertions to the F1 generation.
The average numbers of insertions transmitted per founder fish detected by Southern blot hybridization (Table 2).
We then characterized the chromosomal insertions of the transposon constructs. First, the GFP-positive F1 embryos were raised to adulthood and the genomic DNA prepared from their tail fins were analyzed by Southern blot hybridization. In total, 68 transgenic F1 fish from seven founder fish injected with T2KXIGΔin (Figure 2, B and C), 84 transgenic F1 fish from seven founder fish injected with T2AL200R200G (Figure 2, E and F), and 72 transgenic F1 fish from six founder fish injected with T2AL200R150G (Figure 2, H and I) were subjected to Southern blot analysis. The germline of these founder fish was highly mosaic, and GFP-positive F1 fish contained different numbers of insertions in the genome, from 1 to >10 (Figure 2, B, C, E, F, H, and I). The total number of insertions transmitted by a single founder fish also varied, from 1 to 15 (Table 2). The number of bands detected by Southern blot analysis was counted and summarized in Table 2. The average number of transposon insertions transmitted by one founder fish was 6.9, 6.9, and 5.5 for T2KXIGΔin, T2AL200R200G, and T2AL200R150G, respectively (Table 1). Transgenic fish carrying a single copy of the T2KXIGΔin, T2AL200R200G, or T2AL200R150G insertion, which was identified by Southern blot analysis, expressed GFP in a quite similar manner (Figure 2, A, D, and G), indicating that those vectors have a similar ability to express a foreign gene in transgenic fish. Second, to determine whether these insertions were created by transposition, we established more transgenic fish lines with single insertions, cloned junction fragments from those fish by the inverse PCR and adaptor ligation PCR methods, and analyzed them by DNA sequencing. Both 5′ and 3′ flanking genomic sequences were identified for 4 T2KXIGΔin insertions, 6 T2AL200R200G insertions, and 6 T2AL200R150G insertions (Table 3). Duplication of the 8-bp target sequence, which is characteristic of the integration site of transposons of the hAT family, was found in all of these 16 insertions, indicating that these insertions were indeed created by transposition. The genomic sequences surrounding the insertions were analyzed by BLAST search against the ensembl database Zv6. Of the 16 insertions, 11 were mapped within either an intron or an exon of known and predicted genes (Table 3).
TABLE 2.
GFP-positive fish in F1 offspring and insertions transmitted by single founder fish
| Founder IDa | GFP+/total F1 (%)b | GFP+ F1 fish analyzedc | No. of insertionsd |
|---|---|---|---|
| T2KXIGΔin-1 | 15/136 (11) | 7 | 6 |
| T2KXIGΔin-2 | 41/185 (22) | 7 | 3 |
| T2KXIGΔin-3 | 30/99 (30) | 11 | 9 |
| T2KXIGΔin-4 | 30/55 (55) | 12 | 6 |
| T2KXIGΔin-5 | 102/165 (62) | 12 | 14 |
| T2KXIGΔin-6 | 81/137 (59) | 11 | 5 |
| T2KXIGΔin-7 | 34/114 (30) | 8 | 5 |
| T2AL200R200G-1 | 130/247 (53) | 12 | 7 |
| T2AL200R200G-2 | 79/182 (43) | 12 | 6 |
| T2AL200R200G-4 | 98/150 (65) | 12 | 7 |
| T2AL200R200G-5 | 21/141 (15) | 12 | 1 |
| T2AL200R200G-7 | 255/300 (85) | 12 | 12 |
| T2AL200R200G-8 | 141/302 (47) | 12 | 11 |
| T2AL200R200G-9 | 36/219 (16) | 12 | 4 |
| T2AL200R150G-1 | 14/88 (16) | 12 | 6 |
| T2AL200R150G-2 | 24/136 (18) | 12 | 3 |
| T2AL200R150G-4 | 59/332 (18) | 12 | 15 |
| T2AL200R150G-6 | 78/189 (41) | 12 | 6 |
| T2AL200R150G-8 | 33/188 (18) | 12 | 2 |
| T2AL200R150G-9 | 15/144 (10) | 12 | 1 |
The ID number was given to each injected fish (founder fish) that transmited insertions to the F1 fish.
The number of GFP-expressing embryos per the total number of F1 embryos observed.
The number of GFP-positive F1 fish from each founder fish analyzed by Southern blot hybridization.
The total number of different insertions detected by the Southern blot analysis among F1 fish descended from each founder fish.
TABLE 3.
Sequences of the transposon integration sites and genes located near the insertions
| Insertion name | Integration-site sequencea | Chromosome | Insertion site |
|---|---|---|---|
| T2KXIGΔin-1A | GTTACTAATT GTCGTGCG ATTGCATTGC | 24 | ∼6.5 kb downstream of GENSCAN00000029096 |
| T2KXIGΔin-1B | AGATTTTCGT ACTGACGG TCACGGATTT | 1 | ∼800-bp downstream of zgc:92601 |
| T2KXIGΔin-1C | GTTTCTGTGA GACAGACA TGCCTCAAAC | 7 | Intron of GENSCAN00000033024 |
| T2KXIGΔin-3A | TTAATACTCA ATAATGAA CCTTTTATTT | 20 | ∼200-bp downstream of zgc:85792 (sodium-glucose cotransporter) |
| T2AL200R200G-1A | CCAATTCGAT CATAACTC ACAGCTCAAC | 24 | Intron of GENSCAN00000020107 |
| T2AL200R200G-1B | CCCCGGGTCC CCCAGGAG CCCCCTGCTT | 11 | Exon of GENSCAN00000020963 (collagen α-chain precursor) |
| T2AL200R200G-1C | CAAATACTGT GGGATAAG AGTAGTCAAA | 22 | Intron of GENSCAN00000003589 |
| T2AL200R200G-9A | AATGTAATAA ATTTTATA TACACTGGCG | 20 | intron of GENSCAN00000003761 (osteopetrosis-associated transmembrane protein 1) |
| T2AL200R200G-9B | AGGTACATCA GCGCTCTT ACCAGTGCAG | 22 | Intron of ENSDARG00000057016 (CDC14 homolog) |
| T2AL200R200G-9C | TTCTGTGGTC CTTTTATG TTTAGCAAAA | 17 | Intron of si:dkey-177f17.1 (kinase D-interacting substance) |
| T2AL200R150G-2A | TCACCATCTG CTCGGCAC TGATGTGAAC | 15 | Exon of GENSCAN00000022253 |
| T2AL200R150G-2B | AGTCTGATGA ACCTACTA ATACATTTCT | 5 | Intron of GENSCAN00000029052 |
| T2AL200R150G-8A | TCATACTATA CCTGTAGT TAACACTCAG | ND | |
| T2AL200R150G-8B | AGATTGGTGA CCCCTGAG TTAGAATGTT | ND | |
| T2AL200R150G-10A | ACAGCTCAAC TGCAGAGT GTGTTACAGC | ND | Intron of GENSCAN00000010677 |
| T2AL200R150G-10B | TTGTTATCGC AGAGTAAC TCTGAGCTAT | 13 | Intron of zgc:101599 (CDK-activating kinase assembly factor MAT1) |
ND, the chromosomal location of the integration site was not determined by BLAST against the ensembl database Zv6.
Eight-base-pair duplications of the target sequence are underlined.
Spacing capacity and requirements:
To test whether the transposon vector can carry a large DNA fragment, we replaced the EF1α promoter sequence of T2KXIGΔin with an ∼9-kb fragment of the zebrafish genomic DNA. The total length between the left and right ends of the resulting Tol2 construct became ∼11.7 kb (T2KIGΔin-9k; Figure 3). Transposase mRNA and a plasmid DNA containing this construct were injected into fertilized eggs and the germline transmission frequency was analyzed. Five of six injected fish (83%) transmitted the transposon insertion to the F1 generation (Figure 3A). We identified transgenic fish carrying a single insertion by Southern blot hybridization (Figure 3B) and tested whether any structural changes, such as deletions or rearrangements, occurred with integration of the large transposon construct. We performed PCR using two sets of primers (Figure 3A: f1 and r1; f2 and r2) within the construct and compared the sizes of DNA bands amplified from the genomic DNA of the transgenic fish with those amplified from the plasmid DNA used for injection. No significant difference was detected (Figure 3C). Further, we cloned the junction fragments and identified 8-bp direct repeats at both ends of the insertion (Figure 3D), indicating that the insertion was indeed created by transposition. The insertion was mapped within an intron of a known gene (zgc:65966). From these results, we concluded that the Tol2 transposon construct of 11.7 kb can transpose without reducing its transpositional activity and without causing anomalies in its structure.
We also tested whether the distance between the left and right ends is important for transposition. We showed that T2AL200R150G, which is 2552 bp in length, could transpose efficiently (Figures 1 and 2, Table 1). We created T2AL200R150-based constructs, which contained various lengths of DNA inserts between the 200-bp left end and the 150-bp right end and analyzed their activities either by germline transmission or by the transient excision assay. Of 20 fish injected with the transposase mRNA and a transposon-donor plasmid containing T2AL200R150-1139, which carried a 1139-bp DNA insert between the left and right ends, 14 fish transmitted transposon insertions to the F1 generation, and the excision products were detected from 97% (31/32) of embryos injected with the transposase mRNA and a transposon-donor plasmid containing T2AL200R150-502, which contained a 502-bp DNA insert (Figure 3A). These frequencies were comparable to those of T2AL200R150G (Figure 1A). However, the excision activity was slightly decreased in the excision assay using T2AL200R150-276 (75%: 24/32), which contained a 276-bp DNA insert and was not detected in the experiment using T2AL200R150-18 (<3%: 0/32), which contained a DNA insert of only 18 bp (Figure 3A), indicating that a certain length of a spacer between the left and the right ends is required for transposition.
Identification of a repetitive sequence in the subterminal region:
In this study, we found that 200 bp of DNA from the 5′-end and 150 bp of DNA from the 3′-end are sufficient and required for efficient transposition of Tol2. Tol2 has perfect TIRs of 12 bp at both ends. Which sequence(s) in these regions is indeed required for transposition? To answer the question, we first created the mTIRL1-6 and mTIRR1-6 mutations in the left and right TIR, respectively (Table 4). The mutations in TIRs resulted in a loss of the excision activity (<3%; Figure 4), indicating that these sequences are essential for transposition. Then we carefully analyzed the sequence of the subterminal region and found that 5-bp sequences, 5′-AAGTA-3′ and 5′-GAGTA-3′, are repeated 24 and 9 times, respectively, in these regions. We tentatively considered 5′-(A/G)AGTA-3′ to be a consensus sequence. The 200-bp left- and the 150-bp right-subterminal region contain 17 and 16 copies of this repeat, respectively (Figure 5). Overlapping of the repeat in an opposite direction creates ScaI sites (5′-AGTACT-3′). Indeed, five ScaI sites are found in the subterminal regions.
To determine whether the repeats in the subterminal regions are functionally important, we created mutations in these sequences by “MluI mutagenesis.” In the mL13-16, mL26-30, mL48-51, mL66-70, mL106-108, mL136-148, and mL169-181 mutations, wild-type sequences containing one, two, or three repeats at seven different sites in the left subterminal region were substituted for mutant sequences containing one MluI site (Table 4). Also, in the mR15-18, mR27-31, mR48-51, mR63-67, mR103-108, and mR114-126 mutations, wild-type sequences containing one, two, or three repeats at seven different sites in the right subterminal region were substituted for mutant sequences containing one MluI site (Table 4). The activities of the T2AL200R150G constructs carrying these mutations were analyzed by the transient excision assay. The excision products were detected in 3% (1/31), 3% (1/30), 3% (1/32), 17% (5/30), <3% (0/31), 3% (1/31), 6% (2/34), 25% (8/32), 7% (2/30), 22% (7/32), 3% (1/32), 13% (4/32), and 13% (4/30) of embryos injected with the T2AL200R150G construct carrying the mL13-16, mL26-30, mL48-51, mL66-70, mL106-108, mL136-148, mL169-181, mR15-18, mR27-31, mR48-51, mR63-67, mR103-108, and mR114-126 mutations, respectively (Figure 4). Although the transient excision assay is based on micro-injection and PCR techniques, which are not easy to be controlled quantitatively, and factors that make the difference between 25% (8/32) and 3% (1/30) are unknown, it is obvious that all of these mutations drastically decreased the excision activity in comparison with the wild-type T2AL200R150G. One may argue that any substitutions created by the MluI mutagenesis in the subterminal region may cause such decreases in the excision frequency. To exclude this possibility, we created three mutations, mL88-92, mL158-162, and mR88-94, in which sequences outside of the repeats were substituted for one MluI site (Table 4), and analyzed their activities. The Tol2 constructs carrying these mutations, mL88-92, mL158-162, and mR88-94, exhibited the excision activities of 94% (30/32), 81% (25/31), and 87% (26/30), respectively (Figure 4), indicating that these mutant constructs retained the excision activity comparable to that of T2AL200R150G. From these results, we concluded that the repeats that we identified in the subterminal region should play important roles in transposition.
DISCUSSION
Minimal cis-sequences of Tol2:
In this study, we demonstrated that 200 bp of DNA from the left end and 150 bp of DNA from the right end are sufficient for excision, the first step of transposition. Although the lengths may be shortened slightly, most of those sequences are required for excision since T2AL175R200G, carrying 175-bp of DNA from the left end, showed a reduced excision activity, and T2AL200R100G, carrying 100 bp of DNA from the right end, could not be excised. The previous Tol2 construct with the GFP expression cassette T2KXIG contains 2229 bp from the left and 536 bp from the right end (Kawakami et al. 2004b). The 2229-bp left-side sequence contains ∼300-bp inverted repeats that may form a large secondary structure (Izsvák et al. 1999). We have been sometimes told by others that they encountered difficulties in manipulating the previous transposon vector. We infer that this may have happened because of the large secondary structure; i.e., a decreased copy number of the plasmid in Escherichia coli or reduced E. coli transformation efficiencies, etc. Such a problem will not happen when the smaller Tol2 vectors such as T2KXIGΔin and T2AL200R150, which lack the inverted repeats, are used. Furthermore, Tol2 contains the promoter and the poly(A) signal for the transposase gene near the 5′-end and the 3′-end, respectively (Kawakami and Shima 1999). The minimal Tol2 vector T2AL200R150 also lacks these regulatory elements. Therefore, it can be expected that the minimal vector should have minimal effects both on expression of a foreign gene cloned in the vector and on expression of chromosomal genes located near the integration site of the vector. Thus, identification of minimal cis-sequences should increase the usefulness of Tol2 as a gene transfer vector.
Transposition of mini-Tol2 vectors:
The Tol2 transposon system can create chromosomal integration in the zebrafish germ lineage very efficiently (Kawakami et al. 2004b; Kawakami 2005). Two aspects of the transpositional activities can be examined. One is the frequency of obtaining germline-transmitting founder fish and the other is the number of different transposon insertions transmitted by a single founder fish. First, in our previous study, 50% (5/10) of the fish injected with the transposase mRNA and a transposon-donor plasmid containing T2KXIG could transmit transposon insertions to the next generation (Kawakami et al. 2004b). In this study, we demonstrated that 64, 70, and 60% of fish injected with the transposase mRNA and transposon-donor plasmids containing T2KXIGΔin, T2AL200R200G, and T2AL200R150G, respectively, transmitted transposon insertions to their offspring (Table 1). Thus, the germline transmission frequencies using these mini-Tol2 vectors are comparable to, or even higher than, that observed in transgenesis using T2KXIG. Second, in the previous study, the number of insertions transmitted by a single founder fish injected with T2KXIG was 5.6 on average (Kawakami et al. 2004b). In this study, the average number of insertions transmitted by fish injected with T2KXIGΔin, T2AL200R200G, and T2AL200R150G were 6.9, 6.9, and 5.5, respectively (Table 1). Although the number with T2AL200R150G was slightly lower than those with the other mini-Tol2 constructs, the numbers of insertions transmitted by fish injected with T2KXIGΔin, T2AL200R200G, and T2AL200R150G are still comparable to that with T2KXIG. Therefore, we concluded that 200 bp from the left end and 150 bp from the right end contain cis-sequences sufficient not only for excision but also for efficient transposition.
In this study, we characterized a total of 17 integration sites of Tol2 at the sequence level. The 8-bp duplication of the target site in these 17 cases was created as has been described (Kawakami et al. 2000, 2004b). Although we did not find any obvious consensus sequences at the integration sites, we found that 12 of 17 target loci (71%) were mapped within either an intron or an exon of known or predicted genes. Although further analyses are needed to determine whether all of these predicted genes are indeed transcribed, this result may suggest that Tol2 is preferentially integrated within transcriptional units.
Insert-size capacity and requirements:
We demonstrated that the Tol2 construct of 11.7 kb could transpose without reducing the germline transmission frequency in zebrafish. This construct contained a DNA insert of 10.6 kb, including the zebrafish genomic DNA, the rabbit β-globin intron, the EGFP gene, and the poly(A) signal, between the 517-bp left and 536-bp right ends of the Tol2 sequence. Therefore, if the mini-Tol2, T2AL200R150, was used as a vector, one can clone ∼11.3-kb of DNA into the vector and perform transgenesis with high efficiencies in zebrafish and presumably in other vertebrates. This may be a great advantage for Tol2 to be used as a gene transfer vector in comparison to transposons belonging to the Tc1/mariner family since, in the case of the Tc1/mariner-type transposon Himar1, an ∼38% decrease in the transposition frequency has been observed for each 1-kb increase in transposon size within a range of 2–8 kb (Lampe et al. 1998). The transgenic fish carrying the T2KIGΔin-9k insertion in the genome expressed GFP in the kidney as expected from the nature of the zebrafish genomic DNA placed upstream of the EGFP gene (K. Hoshijima, personal communication), indicating that a large insert can be put in the transposon vector to achieve tissue- or organ-specific expression of a gene of interest.
We found that the T2AL200R150 vector carrying 18 bp of DNA between the left and the right ends could not be excised even though it contained all of the cis-sequences required for excision. The same vector carrying 276 or 502 bp of DNA was active in the transient excision assay, indicating that Tol2 has a minimal size requirement for transposition. The spacer between two ends may be essential for the left and right ends in forming a synaptic complex with oligomers of the transposase proteins.
Essential repeats in the subterminal region:
The 200-bp left-end and the 150-bp right-end sequences essential for transposition contain 12-bp perfect TIRs and the subterminal regions. Mutations in the TIRs drastically reduced the excision activity, indicating that TIRs have a critical role in transposition. In the subterminal regions, we found that a 5-bp sequence, 5′-(A/G)AGTA-3′, is repeated 33 times. Mutations in these repeats reduced the excision activity to various levels, suggesting that the repeats are functionally important and the activity of each repeat is somewhat different. In the case of Ac, 238 bp from the 5′-end and 209 bp from the 3′-end, containing 11-bp TIRs and the subterminal regions, are required for the excision activity (Coupland et al. 1989). The TIRs of Ac are also essential for excision (Chatterjee and Starlinger 1995). The Ac transposase binds to a 6-bp sequence, AAACGG (Kunze and Starlinger 1989), and the 5′ and 3′ subterminal sequences contain 10 and 3 copies of this motif, respectively (Chatterjee and Starlinger 1995). While mutations in these motifs reduce the excision activity, none of them by itself is indispensable and only combinations of mutations in more than six motifs abolish the excision activity nearly completely (Chatterjee and Starlinger 1995). The mechanism of how these multiple binding sequences regulate transposition is unknown. These observations are similar to what we observed in the case of Tol2. Therefore, one possible function of the subterminal repeat of Tol2 may be interaction with the transposase protein. Although similar numbers of repeats are observed in the left end (17 repeats) and the right end (16 repeats) (Figure 5), these ends are not functionally interchangeable. Such dissimilar ends are observed also in Ac (Coupland et al. 1989) and Tag1 (Liu et al. 2001). It will be interesting to learn the mechanism that determines how the left and right subterminal regions play different roles in transposition, which may be common for transposons belonging to the hAT family.
In summary, our study provides useful information for Tol2 to be used as a gene transfer vector and also establishes a basis for further biochemical and molecular biological studies of the roles of the subterminal sequence in transposition.
Acknowledgments
We thank S. Miyabayashi, N. Mouri, T. Uematsu, and S. Takeda for fish room maintenance, and K. Hoshijima for personal communication. G. Morvan was supported by a fellowship from the Japan Society for the Promotion of Science. This work was supported by National Institutes of Health/National Institute of General Medical Sciences grant R01GM069382 and by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Chatterjee, S., and P. Starlinger, 1995. The role of subterminal sites of transposable element Ds of Zea mays in excision. Mol. Gen. Genet. 249: 281–288. [DOI] [PubMed] [Google Scholar]
- Coupland, G., C. Plum, S. Chatterjee, A. Post and P. Starlinger, 1989. Sequences near the termini are required for transposition of the maize transposon Ac in transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 86: 9385–9388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring, H. P., E. Tillmann and P. Starlinger, 1984. DNA sequence of the maize transposable element Dissociation. Nature 307: 127–130. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N., S. Wessler and M. Shure, 1983. Isolation of the transposable maize controlling elements Ac and Ds. Cell 35: 235–242. [DOI] [PubMed] [Google Scholar]
- Izsvák, Z., Z. Ivics, N. Shimoda, D. Mohn, H. Okamoto et al., 1999. Short inverted-repeat transposable elements in teleost fish and implications for a mechanism of their amplification. J. Mol. Evol. 48: 13–21. [DOI] [PubMed] [Google Scholar]
- Johnson Hamlet, M. R., M. Takeda, M. Taira, K. Kawakami and P. E. Mead, 2006. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis 44: 438–445. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., 2004. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 77: 201–222. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., 2005. Transposon tools and methods in zebrafish. Dev. Dyn. 234: 244–254. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., and T. Noda, 2004. Transposition of the Tol2 element, an Ac-like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics 166: 895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K., and A. Shima, 1999. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene 240: 239–244. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., A. Koga, H. Hori and A. Shima, 1998. Excision of the Tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene 225: 17–22. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., A. Shima and N. Kawakami, 2000. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 97: 11403–11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, K., K. Imanaka, M. Itoh and M. Taira, 2004. a Excision of the Tol2 transposable element of the medaka fish Oryzias latipes in Xenopus laevis and Xenopus tropicalis. Gene 338: 93–98. [DOI] [PubMed] [Google Scholar]
- Kawakami, K., H. Takeda, N. Kawakami, M. Kobayashi, N. Matsuda et al., 2004. b A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev. Cell 7: 133–144. [DOI] [PubMed] [Google Scholar]
- Koga, A., M. Suzuki, H. Inagaki, Y. Bessho and H. Hori, 1996. Transposable element in fish. Nature 383: 30. [DOI] [PubMed] [Google Scholar]
- Kotani, T., S. Nagayoshi, A. Urasaki and K. Kawakami, 2006. Transposon-mediated gene trapping in zebrafish. Methods 39: 199–206. [DOI] [PubMed] [Google Scholar]
- Kunze, R., and P. Starlinger, 1989. The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J. 8: 3177–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe, D. J., T. E. Grant and H. M. Robertson, 1998. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149: 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., A. Mack, R. Wang, M. Galli, J. Belk et al., 2001. Functional dissection of the cis-acting sequences of the Arabidopsis transposable element Tag1 reveals dissimilar subterminal sequence and minimal spacing requirements for transposition. Genetics 157: 817–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskey, C., Z. Izsvak, K. Kawakami and Z. Ivics, 2005. DNA transposons in vertebrate functional genomics. Cell. Mol. Life Sci. 62: 629–641. [DOI] [PubMed] [Google Scholar]
- Müller-Neumann, M., J. I. Yoder and P. Starlinger, 1984. The DNA sequence of the transposable element Ac of Zea mays L. Mol. Gen. Genet. 198: 19–24. [Google Scholar]
- Pohlman, R. F., N. V. Fedoroff and J. Messing, 1984. The nucleotide sequence of the maize controlling element Activator. Cell 37: 635–643. [DOI] [PubMed] [Google Scholar]
- Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov and S. A. Lukyanov, 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23: 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spertini, D., C. Beliveau and G. Bellemare, 1999. Screening of transgenic plants by amplification of unknown genomic DNA flanking T-DNA. Biotechniques 27: 308–314. [DOI] [PubMed] [Google Scholar]
- Sutton, W. D., W. L. Gerlach, D. Schwartz and W. J. Peacock, 1984. Molecular analysis of Ds controlling element mutations at the Adh1 locus of maize. Science 223: 1265–1268. [DOI] [PubMed] [Google Scholar]