Abstract
The interaction between genotype and environment is recognized as an important source of experimental variation when complex traits are measured in the mouse, but the magnitude of that interaction has not often been measured. From a study of 2448 genetically heterogeneous mice, we report the heritability of 88 complex traits that include models of human disease (asthma, type 2 diabetes mellitus, obesity, and anxiety) as well as immunological, biochemical, and hematological phenotypes. We show that environmental and physiological covariates are involved in an unexpectedly large number of significant interactions with genetic background. The 15 covariates we examined have a significant effect on behavioral and physiological tests, although they rarely explain >10% of the variation. We found that interaction effects are more frequent and larger than the main effects: half of the interactions explained >20% of the variance and in nine cases exceeded 50%. Our results indicate that assays of gene function using mouse models should take into account interactions between gene and environment.
IT is widely recognized that environmental variables, such as who carries out the experiment and when, and physiological variables, such as sex and weight, are confounds that need to be accounted for during the collection of mouse phenotypes. Many articles attest to the effect of these variables on phenotypic values (e.g., Chesler et al. 2002a; Champy et al. 2004) and point out the need for rigorous standardization of laboratory practice (Henderson 1970; Crabbe et al. 1999; Brown et al. 2005). It is also acknowledged that the size and even direction of environmental effects on a phenotype can vary with genotype, a phenomenon known as gene-by-environment interaction, and this has been documented in studies of rodents over the past 50 years (e.g., Cooper and Zubek 1958).
Following a report on the importance of laboratory-by-strain interaction (Crabbe et al. 1999), recent interest has focused on the prevalence and size of such interactions, as well as their ability to increase power in genetic mapping experiments (Wang et al. 2006). Table 1 summarizes the available data and shows that the picture of how much genetic and environmental factors interact is piecemeal: our knowledge of the relative size of interaction and main effects is limited to a handful of phenotype–covariate combinations.
TABLE 1.
Recent reports of gene-by-environment interactions in mouse
| Covariate | Phenotype | QTL (i.e., single locus) or polygenic (e.g., strain) effect | Main-effect variancea (%) | Interaction-effect variancea (%) | Reference |
|---|---|---|---|---|---|
| Laboratory | Elevated plus maze | Polygenic | 32.7b | 21b | Crabbe et al. (1999) |
| Body weight | Polygenic | 20.4b | 7.1b | ||
| Cocaine-induced activity | Polygenic | 5.3b | 8.6b | ||
| Sex | Body weight | Polygenic | 63.7b | 7b | |
| Open field test | Polygenic | — | 4.5b | ||
| Diet | Obesity | QTL | — | — | York et al. (1999) |
| Diet (food shortage) | Amphetamine-induced activity | — | — | Cabib et al. (2000) | |
| Maternal lactational environment | Plasma glucose | Polygenic | — | — | Reifsnyder et al. (2000) |
| Experimenter | Tail-withdrawal latency | Polygenic | 42 | 18 | Chesler et al. (2002) |
| Sex | |||||
| Testing order | |||||
| Time of day | |||||
| Laboratory | Locomotion | Polygenic | 11.9–28.4b | 10.9–16.5b | Wahlsten et al. (2003) |
| Elevated plus maze | Polygenic | 25.2–30b | 13–14.3b | ||
| Diet | Agressiveness | Polygenic | — | — | Nyberg et al. (2004) |
| Diet | Liver weight | QTL | — | — | Ehrich et al. (2005) |
| Serum insulin | QTL | — | — | ||
| Fat pad | QTL | — | — | ||
| Diet | Liver weight | Polygenic | — | — | Biddinger et al. (2005) |
| Leptin | Polygenic | — | — | ||
| Glucose tolerance test | Polygenic | — | — | ||
| Laboratory | Open field test | Polygenic | 0–20.3 | 0.1–8.7 | Kafkafi et al. (2005) |
| Sex | Gonadal fat mass | QTL | — | — | Wang et al. (2006) |
The proportion of variance attributable to the main or interaction effect of the covariate, with “—” representing cases where this figure was not reported.
The proportion of variance is given as the partial ω2-statistic.
During an investigation of the genetic basis of complex traits in 2448 genetically heterogeneous stock (HS) mice (1220 female, 1228 male) (Solberg et al. 2006), we collected environmental and physiological covariates. The mice we used were descended from eight inbred strains (A/J, AKR/J, BALBc/J, CBA/J, C3H/HeJ, C57BL/6J, DBA/2J, and LP/J) (Demarest et al. 2001), incorporating more genetic variation from a single cross than has hitherto been assessed in mice. The generality of our findings is enhanced by our use of a battery of tests that includes both behavioral and a broad range of physiological phenotypes (Solberg et al. 2006), summarized in Table 2 (the names of all phenotypes are given in Table 3).
TABLE 2.
Summary of phenotypes analyzed, number of animals, and mean age (in days) at which the animals were analyzed
| Phenotype | Description | No. of animals | Mean age (days) |
|---|---|---|---|
| Weight, 6 wk | Body weight at the beginning of testing. | 2516 | 42 |
| Immunology | CD4, CD3, CD8, and B220 antibody staining. | 1872 | 42 |
| OFT | Open field arena: distance in the perimeter, the center, and total distance in 5 min. | 2504 | 45 |
| EPM | Elevated plus maze: distance traveled, time spent, and entries into closed and open arms. | 2452 | 46 |
| FN | Food hyponeophagia: time taken to sample a novel foodstuff (overnight food deprivation). | 2474 | 47 |
| Burrowing | No. of pellets removed from burrow in 1.5 hr. | 2455 | 48 |
| Activity | Activity measured in a home cage in 30 min. | 2445 | 48 |
| Startle | Startle to a loud noise. | 1948 | 52 |
| Context freezing | Freezing to the context in which a tone is associated with a foot shock. | 2070 | 55 |
| Cue freezing | Freezing to a tone after association with a foot shock. | 2110 | 56 |
| Plethysmography | Animals sensitized by injection with ovalbumin inhale metacholine and changes in lung function are measured by plethysmography (a model of asthma). Respiratory rate, tidal volume, minute volume, expiratory time, inspiratory time, and enhanced pause are recorded with and without exposure to metacholine. | 2304 | 63 |
| IPGTT | Glucose and insulin values taken at 0, 15, 30, and 75 min after i.p. glucose injection (a model of type 2 diabetes mellitus). | 2334 | 68 |
| Weight, 10 wk | Body weight at the end of testing. | 2319 | 70 |
| FBC | Full blood count (hematocrit, Hb concentration, mean cellular volume, mean cellular Hb concentration, white cell count, platelet count). | 1892 | 71 |
| Tissue harvest | Adrenal weight. | 2309 | 71 |
| Wound healing | Reduction in size of a 2-mm ear punch hole. | 2273 | 71 |
| Biochemistry | Albumin, alkaline phosphatase, alanine transaminase, aspartate transaminase, calcium, chloride, creatinine, high-density lipoprotein, low-density lipoprotein, phosphorous, sodium, total cholesterol, total protein, triglycerides. | 1890 | 71 |
TABLE 3.
Phenotypes assessed in the project
| Test | Measure |
|---|---|
| Open field arena | Total activity |
| Fecal boli | |
| Elevated plus maze | Closed-arm distance |
| Open-arm distance | |
| Closed-arm time | |
| Open-arm time | |
| Closed-arm entries | |
| Open-arm entries | |
| New home-cage activity | Total beam breaks (30 min) |
| Total beam breaks (first 5 min) | |
| Total beam breaks (last 5 min) | |
| Fine movement | |
| Context freezing | Time freezing to context (sec) |
| Cue conditioning | Time freezing during cue (sec) |
| Time freezing after cue (sec) | |
| Fecal boli | |
| Fear-potentiated startle | Startle response |
| Change in startle after training | |
| Plethysmography | Enhanced pause (baseline) |
| Enhanced pause (metacholine) | |
| Expiratory time (baseline) | |
| Expiratory time (metacholine) | |
| Inspiratory time (baseline) | |
| Inspiratory time (metacholine) | |
| PenH difference | |
| Respiratory rate (baseline) | |
| Respiratory rate (metacholine) | |
| Tidal minute volume (baseline) | |
| Tidal minute volume (metacholine) | |
| Tidal volume (baseline) | |
| Tidal volume (metacholine) | |
| IPGTT | AUC-G (mg/dl) |
| AUC-IRI (ng/ml) | |
| AUC-IRI/AUC-G | |
| DG (mg/dl) | |
| DIRI (ng/ml) | |
| DIRI/DG | |
| Glucose 0 (mg/dl) | |
| Glucose 15 (mg/dl) | |
| Glucose 30 (mg/dl) | |
| Glucose 75 (mg/dl) | |
| Insulin 0 (ng/ml) | |
| Insulin 15 (ng/ml) | |
| Insulin 30 (ng/ml) | |
| Insulin 75 (ng/ml) | |
| Insulin slope | |
| K (glucose slope) | |
| Immunology | %B220+ |
| %CD3+ | |
| %CD4+ | |
| %CD4+/CD3+ | |
| %CD8+ | |
| %CD8+/CD3+ | |
| %NK cells | |
| Hematology | Hematocrit (%) |
| Hemoglobin (g/dl) | |
| Mean cellular volume (fl) | |
| Platelets (n/μl) | |
| Red blood cell count (n/μl) | |
| White blood cell count (n/μl) | |
| Mean cellular Hb concentration (%) | |
| Red cell distribution width | |
| Mean corpuscular hemoglobin (pg) | |
| Lymphocytes | |
| Plateletcrit (%) | |
| Biochemistry | Alkaline phosphatase (units/liter) |
| Alanine transaminase (units/liter) | |
| Aspartate transaminase (units/liter) | |
| Albumin (g/liter) | |
| Calcium (mmol) | |
| Chloride (mmol) | |
| High-density lipoproteins (mmol) | |
| Low-density lipoproteins (mmol) | |
| Phosphorous (mmol) | |
| Sodium (mmol) | |
| Total cholesterol (mmol) | |
| Total protein (g/liter) | |
| Triglycerides (mmol) | |
| Urea (mmol) | |
| Weight, length, and growth | Body length |
| Body mass index | |
| Growth slope | |
| Weight, 10 wk | |
| Weight, 6 wk | |
| Weight, 7 wk | |
| Weight, 8 wk | |
| Adrenal weight | Adrenal weight (g) |
| Wound healing | Ear hole area (from ear punch) (mm2) |
METHODS
Animals:
Original Northport HS mice were obtained from Robert Hitzemann at the Oregon Health Sciences Unit, Portland, Oregon. At the time the animals arrived they had passed 50 generations of pseudorandom breeding (Demarest et al. 2001). A breeding colony in open cages was established at Oxford University to generate animals for phenotyping. The animals' pedigree comprising the parents and grandparents of the phenotyped animals was recorded.
Phenotypes and covariates:
The phenotypes used in this study and the protocol used to collect them are fully described in Solberg et al. (2006) and summarized in Table 2. We collected 15 covariates (Table 4). Seven are mouse-specific covariates (short names quoted in brackets where needed): sex, age, cage identifier (i.e., a unit of shared environment), weight at 9 weeks (“weight”), number of animals in a cage (“cage density”), sibship (“family”), and which litter the mouse came from (“litter”; e.g., “3” means the animal came from his parents' third litter); three are test-specific covariates: experimenter, test order, and apparatus (if more than one was used); and five covariates are for the time of the experiment: year, season (the group of three months), month, hour (time rounded to the nearest hour), and “study day,” defined as the number of days from start of the study on January 20, 2003.
TABLE 4.
Covariates used in the study
| Covariate | Encoding | Description | Summary |
|---|---|---|---|
| Age | Integer | Age in days | Mean = 61, SD = 4, 31–85 |
| Apparatus | Categorical | Experimental unit used | Groups = 4, size = 348–526 |
| Cage | Categorical | Cage in which animal was housed | Groups = 435–549, size = 1–7 |
| Cage density | Integer | No. of animals in a cage | Mean = 4.7, SD = 1.1, 2–7 |
| Experimenter | Categorical | Who performed the test | Groups = 2–12, size = 7–457 |
| Family | Categorical | Sibship of animal | Groups = 160–180, size = 1–52 |
| Hour | Categorical | Hour of the day test was performed | Groups = 1–11, size = 1–2307 |
| Litter | Integer | No. litter the animal came from | Mean = 2.2, SD = 1.3, 1–8 |
| Month | Categorical | Month test was performed | Groups = 12, size = 32–314 |
| Season | Categorical | Season test was performed | Groups = 4, size = 284–788 |
| Sex | Categorical | Sex of the animal | Groups = 2, size = 806–1293 |
| Study day | Integer | Day into study that test was performed (day 1 is Jan. 20, 2003) | Mean = 306, SD = 160, 1–621 |
| Test order | Integer | Order in which animal was tested that day | Mean = 2.8, SD = 1.4, 1–7 |
| Weight | Real no. | Body weight (g) at 9 wk | Mean = 23.9, SD = 4.2, 12–39.1 |
| Year | Categorical | Year of test | Groups = 2, size = 711–1517 |
“Encoding” refers to how a covariate was modeled statistically. For numerical covariates, the column headed “Summary” gives the grand mean and standard deviation over all phenotypes, followed by the minimum and maximum values observed for any given phenotype. For categorical covariates Summary gives the number and size of categories seen for a typical phenotype. For example, for phenotypes in which the experimenter covariate was present, there were between 2 and 12 experimenters who each recorded data for between 7 and 457 mice.
In the analysis, we fitted statistical models for each phenotype, first testing the significance of each covariate as a main effect and then its interaction with genetic background. Covariates were either treated as continuous variables [age, cage density, litter, study day (continuous), weight] or encoded as categorical factors taking discrete levels (apparatus, cage, experimenter, sex, hour, month, season, year, and family). Note that although hour could have been treated as continuous, that would have allowed detection of only linear trends between time and phenotype, whereas as a factor it can be used to detect nonlinear relationships.
Statistical analysis:
All analysis was carried out using the R statistical package (R Development Core Team 2004), along with the add-on packages lme4 (Pinheiro and Bates 2000), MASS (Venables and Ripley 2002), and regress (Clifford and McCullagh 2005).
We applied normalizing transformations to each phenotype, guided by the Box–Cox procedure (Venables and Ripley 2002), and in most cases this comprised a simple exponentiation or log transform to correct skewness (see Table 5). Phenotypes with symmetrical but highly long-tailed distributions were corrected with a simplified Blom transformation (Blom 1958), in which the value is replaced by the probit of its empirical distribution function probability. Asymmetric highly skewed long-tailed distributions best modeled as exponential or gamma distributions were excluded from the analysis, as were categorical phenotypes and latency phenotypes that require survival analysis. After transformation, each phenotype was trimmed by removing values more than 3 standard deviations from the mean to moderate the effects of outliers.
TABLE 5.
Transformations, heritabilities, and common environment effects for 88 phenotypes listed in order of heritability
| Phenotype | Transformation | Category | % variance due to additive genetic variation (i.e., heritability) | % variance due to common environment |
|---|---|---|---|---|
| %CD8+ | x | Physiological | 88.91 | 11.09 |
| CD4+/CD8+ | x−(1/3) | Physiological | 80.48 | 14.59 |
| Weight, 7 wk (g) | x1/3 | Physiological | 79.36 | 20.64 |
| Weight, 6 wk (g) | x1/3 | Physiological | 74.48 | 25.52 |
| %CD4+/CD3+ | x2 | Physiological | 72.73 | 18.48 |
| Weight, 8 wk (g) | x1/3 | Physiological | 71.99 | 18.29 |
| High density lipoproteins (mmol) | x | Physiological | 69.11 | 17.01 |
| Alkaline phosphatase (units/liter) |  |
Physiological | 62.83 | 20.47 |
| Weight, 10 wk (g) | x1/3 | Physiological | 62.35 | 18.02 |
| %B220+ |  |
Physiological | 59.86 | 24.66 |
| Glucose 0 (mg/dl) |  |
Physiological | 55.33 | 32.08 |
| Red cell distribution width | x−2 | Physiological | 55.29 | 12.98 |
| Mean cellular Hb conc. (%) | x | Physiological | 52.16 | 39.97 |
| %CD3+ | x2 | Physiological | 51.30 | 22.51 |
| Ear hole area (mm2) |  |
Physiological | 51.02 | 14.46 |
| Mean cellular volume (fl) | x | Physiological | 50.89 | 20.60 |
| Calcium (mmol) | x | Physiological | 48.89 | 31.39 |
| Lymphocytes |  |
Physiological | 48.29 | 17.85 |
| Mean corpuscular hemoglobin (pg) | x | Physiological | 47.94 | 20.24 |
| Inspiratory time (metacholine) | x−1 | Physiological | 44.96 | 10.81 |
| Chloride (mmol) | x | Physiological | 44.78 | 38.43 |
| Open-arm distance | x1/3 | Behavioral | 42.06 | 6.19 |
| %CD4+ | x | Physiological | 40.70 | 26.46 |
| Startle response | x1/3 | Behavioral | 40.67 | 4.20 |
| White blood cell count (n/μl) | log10(x + 1) | Physiological | 40.65 | 23.15 |
| Sodium (mmol) | x | Physiological | 39.34 | 37.83 |
| Closed-arm distance | x | Behavioral | 38.81 | 7.95 |
| Open-arm entries |  |
Behavioral | 38.57 | 5.46 |
| Open-arm time |  |
Behavioral | 37.92 | 6.08 |
| Enhanced pause (baseline) | log10(x) | Physiological | 37.81 | 26.70 |
| Total cholesterol (mmol) | x | Physiological | 37.50 | 17.62 |
| Total beam breaks (30 min) |  |
Behavioral | 37.17 | 11.14 |
| Respiratory rate (metacholine) | log10(x) | Physiological | 36.13 | 12.81 |
| Expiratory time (metacholine) | log10(x) | Physiological | 35.00 | 13.94 |
| Glucose 15 (mg/dl) |  |
Physiological | 34.83 | 29.86 |
| Total activity | x | Behavioral | 33.86 | 5.81 |
| Inspiratory time (baseline) | x−2 | Physiological | 32.76 | 16.55 |
| Alanine transaminase (units/liter) | log10(x + 3) | Physiological | 32.20 | 29.18 |
| Respiratory rate (baseline) | x | Physiological | 31.57 | 18.95 |
| Tidal volume (metacholine) | x1/3 | Physiological | 30.95 | 21.69 |
| Low density lipoproteins (mmol) | log10(x) | Physiological | 30.70 | 18.25 |
| Urea (mmol) | log10(x + 1) | Physiological | 30.59 | 21.54 |
| Growth slope | x | Physiological | 30.52 | 37.39 |
| Time freezing during cue (sec) | x | Behavioral | 30.51 | 0.00 |
| Expiratory time (baseline) | log10(x) | Physiological | 29.52 | 21.37 |
| Fine movement | x2 | Behavioral | 29.45 | 10.04 |
| Total beam breaks (first 5 min) |  |
Behavioral | 29.27 | 12.26 |
| Enhanced pause (metacholine) | log10(x) | Physiological | 27.30 | 28.26 |
| Adrenal weight | log10(x) | Physiological | 27.00 | 36.09 |
| Closed-arm time |  |
Behavioral | 26.65 | 7.47 |
| Tidal minute volume (metacholine) | x1/3 | Physiological | 26.59 | 20.01 |
| Albumin (g/liter) | x | Physiological | 26.42 | 22.42 |
| Insulin 30 (ng/ml) | x1/4 | Physiological | 26.34 | 21.62 |
| Glucose 75 (mg/dl) | 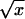 |
Physiological | 26.28 | 22.94 |
| Insulin 15 (ng/ml) | log10(x) | Physiological | 25.85 | 22.24 |
| Time freezing to context (sec) |  |
Behavioral | 25.23 | 12.09 |
| PenH difference | x1/3 | Physiological | 25.20 | 28.85 |
| Platelets (n/μl) | x | Physiological | 25.07 | 19.94 |
| DIRI/DG | x1/4 | Physiological | 24.61 | 23.89 |
| Triglycerides (mmol) | log10(x) | Physiological | 22.55 | 21.94 |
| AUC-IRI/AUC-G | x1/4 | Physiological | 22.48 | 22.86 |
| Total beam breaks (last 5 min) |  |
Behavioral | 22.39 | 7.58 |
| Glucose 30 (mg/dl) |  |
Physiological | 22.18 | 27.00 |
| % NK cells | x−(1/2) | Physiological | 21.88 | 30.24 |
| DG (mg/dl) | x | Physiological | 21.82 | 24.96 |
| Body length (cm) | x | Physiological | 21.34 | 19.92 |
| AUC-G (mg/dl) | x | Physiological | 21.24 | 24.88 |
| DIRI (ng/ml) | x1/3 | Physiological | 21.02 | 19.04 |
| Aspartate transaminase (units/liter) | x−(1/2) | Physiological | 20.96 | 18.47 |
| AUC-IRI (ng/ml) | x1/2 | Physiological | 19.24 | 18.87 |
| Closed-arm entries | x | Behavioral | 19.20 | 7.00 |
| Tidal volume (baseline) | x1/3 | Physiological | 18.56 | 25.07 |
| Insulin 0 (ng/ml) | log10(x) | Physiological | 17.83 | 26.01 |
| Tidal minute volume (baseline) | x1/3 | Physiological | 16.51 | 22.21 |
| Phosphorous (mmol) | log10(x + 1) | Physiological | 16.10 | 28.41 |
| Insulin slope | x1/3 | Physiological | 15.21 | 6.49 |
| Red blood cell count (n/μl) | x3 | Physiological | 15.14 | 18.00 |
| Hemoglobin (g/dl) | x3 | Physiological | 15.12 | 17.83 |
| Time freezing after cue (sec) | x | Behavioral | 13.81 | 0.00 |
| Change in startle after training | Blom(x) | Behavioral | 13.61 | 4.48 |
| Fecal boli |  |
Behavioral | 13.38 | 13.02 |
| Body mass index | x | Physiological | 13.21 | 14.75 |
| Insulin 75 (ng/ml) | x1/3 | Physiological | 13.11 | 26.72 |
| Plateletcrit (%) |  |
Physiological | 12.91 | 20.24 |
| Hematocrit (%) | x3 | Physiological | 10.86 | 18.98 |
| Fecal boli after cue |  |
Behavioral | 9.91 | 6.97 |
| Total protein (g/liter) | x2 | Physiological | 8.51 | 28.59 |
| K (glucose slope) | x1/2 | Physiological | 7.60 | 10.28 |
Transformations use the following conventions: x = phenotype; log10, log to base 10; Blom, replace each point with the probit of its relative cumulative frequency.
Modeling the heritability and the effect of common environment:
We used a variance-components approach to model the effect of genetic background. Here the genetic effect on an animal's phenotype is a value drawn from a normal distribution constrained such that the genetic effects of different animals correlate with their relatedness. First we fitted a standard additive genetic, common environmental error, unique environmental error (ACE) model to obtain estimates of the proportion of phenotypic variance attributable to additive genetic effects (i.e., the heritability) and to shared environmental effects. Second, we used an approximation to the ACE model that could be extended to test for the effect of individual environmental covariates.
We formulated the ACE model as follows. Let n be the total number of animals,  be the number of cages, μ be the grand mean,
be the number of cages, μ be the grand mean, 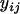 be the phenotype of the ith animal in the jth cage,
be the phenotype of the ith animal in the jth cage, 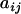 be that animal's additive genetic random effect,
be that animal's additive genetic random effect,  be its value for covariate c,
be its value for covariate c,  be the fixed effect associated with covariate c, C be the set of fixed-effect covariates,
be the fixed effect associated with covariate c, C be the set of fixed-effect covariates,  be the random effect of cage j, and
be the random effect of cage j, and  be the random effect of uncorrelated environmental noise. Then
be the random effect of uncorrelated environmental noise. Then
 |
(1) |
where, the n-vector  , the
, the  -vector
-vector  , and the n-vector
, and the n-vector  , where
, where  is the
is the  additive genetic relationship matrix (e.g., see Lynch and Walsh 1998) computed from the pedigree. We estimated the heritability of each phenotype, i.e., the proportion of variance attributable to additive genetic variation, as
additive genetic relationship matrix (e.g., see Lynch and Walsh 1998) computed from the pedigree. We estimated the heritability of each phenotype, i.e., the proportion of variance attributable to additive genetic variation, as  and the size of the common environmental effect as
and the size of the common environmental effect as 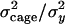 , where
, where  is the phenotypic variance. The set of covariates chosen for C was sex, litter, and, for phenotypes not directly related to body mass, weight. Fitting was done by restricted estimate maximum likelihood (REML), using the R package regress.
is the phenotypic variance. The set of covariates chosen for C was sex, litter, and, for phenotypes not directly related to body mass, weight. Fitting was done by restricted estimate maximum likelihood (REML), using the R package regress.
Testing main effects of covariates:
For each phenotype we tested the significance of individual covariates using an approximation to the ACE model above. We employed a random family effect as a surrogate for the genetic effect, replacing the random effect 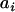 , specific to individual i, with a random effect
, specific to individual i, with a random effect  , specific to family q. As explained below, this substitution amounts to a reparameterization that affects in a predictable fashion only the estimated variance of random terms. Also, because we wish to examine the effects of individual environmental covariates, we excluded a catch-all random effect for cage, which would otherwise be heavily confounded with any individual environmental covariate. Using notation similar to that above, the model for testing the significance of covariate
, specific to family q. As explained below, this substitution amounts to a reparameterization that affects in a predictable fashion only the estimated variance of random terms. Also, because we wish to examine the effects of individual environmental covariates, we excluded a catch-all random effect for cage, which would otherwise be heavily confounded with any individual environmental covariate. Using notation similar to that above, the model for testing the significance of covariate  was
was
 |
(2) |
where  are the fixed effects associated with covariate c,
are the fixed effects associated with covariate c, 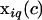 is the component of the design matrix representing the ith animal's value for covariate c,
is the component of the design matrix representing the ith animal's value for covariate c, 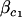 and
and  are defined similarly for
are defined similarly for  , and
, and 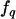 is such that if there are
is such that if there are  nuclear families then the
nuclear families then the 
 . We measured the significance of the covariate
. We measured the significance of the covariate  as the improvement in fit conferred by covariate
as the improvement in fit conferred by covariate 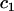 after certain basic covariates (C) had already been included. The set C usually comprised sex and, for phenotypes not directly related to body mass, weight. When
after certain basic covariates (C) had already been included. The set C usually comprised sex and, for phenotypes not directly related to body mass, weight. When  was weight, C comprised only sex; when
was weight, C comprised only sex; when  was sex, C was empty. The significance of the fixed effect
was sex, C was empty. The significance of the fixed effect  was assessed using an approximation to the sequential F-test based on the Wald test (Pinheiro and Bates 2000). We fit all models by REML using the lmer function from the R package lme4 (Pinheiro and Bates 2000).
was assessed using an approximation to the sequential F-test based on the Wald test (Pinheiro and Bates 2000). We fit all models by REML using the lmer function from the R package lme4 (Pinheiro and Bates 2000).
Testing interaction effects between covariates and family:
We define the “interaction model” for the covariate 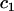 and family by adding a term to the main-effects model in Equation 3 to allow each family to have its own effect for that covariate. For factor covariates, the interaction model included a random intercept nested within family, i.e.,
and family by adding a term to the main-effects model in Equation 3 to allow each family to have its own effect for that covariate. For factor covariates, the interaction model included a random intercept nested within family, i.e.,
 |
(3) |
where  is the fixed effect associated with category k of covariate
is the fixed effect associated with category k of covariate 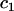 , and
, and 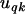 is the random effect for category k in family q, such that if there are
is the random effect for category k in family q, such that if there are  unique combinations of category and family then the
unique combinations of category and family then the  -vector
-vector  . For continuous covariates, the interaction model included a random slope for
. For continuous covariates, the interaction model included a random slope for  conditioned on family, i.e.,
conditioned on family, i.e.,
 |
(4) |
where 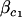 is the fixed coefficient of covariate
is the fixed coefficient of covariate 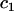 ,
, 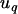 is the random deviation from that coefficient in family q, and the correlation between the random intercept f and slope u is unrestricted. We assessed the significance of the interaction model (Equation 3 or Equation 4) by a likelihood-ratio test (LRT) with the corresponding main-effects model. Note that by using the change in the number of degrees of freedom to parameterize the chi-square distribution used for the LRT, our P-values for interaction effects are slightly conservative (Self and Liang 1987). We used the Dunn–Šidák correction, an exact form of the Bonferroni correction (Sahai and Ageel 2000), to take account of the number of tests performed. For N tests, the corrected 5% threshold is
is the random deviation from that coefficient in family q, and the correlation between the random intercept f and slope u is unrestricted. We assessed the significance of the interaction model (Equation 3 or Equation 4) by a likelihood-ratio test (LRT) with the corresponding main-effects model. Note that by using the change in the number of degrees of freedom to parameterize the chi-square distribution used for the LRT, our P-values for interaction effects are slightly conservative (Self and Liang 1987). We used the Dunn–Šidák correction, an exact form of the Bonferroni correction (Sahai and Ageel 2000), to take account of the number of tests performed. For N tests, the corrected 5% threshold is  .
.
The magnitude of a covariate's effect is defined as the percentage of phenotypic variance it explains, estimated in the model used to test its significance. For fixed effects, this is the percentage of the total sum of squares attributable to the effect in a sequential ANOVA table after fitting the other covariates (known in some literature as η2; Olejnik and Algina 2003). For random effects, it is the estimated variance of the effect expressed as a percentage of the total phenotypic variance. Where the random effect is based on an interaction with family, we report the percentage variance as twice that of the estimated amount, in accordance with the reparameterization formulas described below.
Our use of family as a surrogate for the genetic effect means we underestimate the effect size of interactions by a factor of two. However, this difference is entirely superficial. Suppose the n animals are sorted in order of their  nuclear families. When fitting the family effect, the n-vector of random effects is distributed as
nuclear families. When fitting the family effect, the n-vector of random effects is distributed as  , where the matrix F is block diagonal such that
, where the matrix F is block diagonal such that 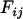 is 1 if i and j are in the same sibship and 0 otherwise (note that parents are not included in the analysis because phenotypes were collected only on the offspring). The covariance matrix for all random effects is therefore
is 1 if i and j are in the same sibship and 0 otherwise (note that parents are not included in the analysis because phenotypes were collected only on the offspring). The covariance matrix for all random effects is therefore
 |
(5) |
where 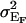 is the environmental variance when using family for the genetic effect. This models all animals within a sibship as if they were genetically identical and all sibships as nuclear. Treating sibships as nuclear is reasonable in our case since the sparsity of our additive genetic relationship matrix means that
is the environmental variance when using family for the genetic effect. This models all animals within a sibship as if they were genetically identical and all sibships as nuclear. Treating sibships as nuclear is reasonable in our case since the sparsity of our additive genetic relationship matrix means that  , where
, where  when i = j, 0.5 when i and j are sibs, and 0 otherwise, and we found empirically that in this data set the likelihood ratios using the full pedigree A matrix were very close to those obtained using the nuclear approximation S. Using the approximation S for A, our heritability models a covariance matrix
when i = j, 0.5 when i and j are sibs, and 0 otherwise, and we found empirically that in this data set the likelihood ratios using the full pedigree A matrix were very close to those obtained using the nuclear approximation S. Using the approximation S for A, our heritability models a covariance matrix
 |
(6) |
Substituting the equality  and equating the coefficients of F and I, it follows that
and equating the coefficients of F and I, it follows that  such that when estimated,
such that when estimated,  , which agrees with our observed discrepancy between family-effect size and heritability. Similarly
, which agrees with our observed discrepancy between family-effect size and heritability. Similarly  . Thus the two models are reparameterizations of each other. When fitted, they have identical likelihood ratios, and hence
. Thus the two models are reparameterizations of each other. When fitted, they have identical likelihood ratios, and hence  is an estimate of the true additive genetic variance.
is an estimate of the true additive genetic variance.
Our estimates of the variance attributable to gene-by-environment effects also rely on the use of the family surrogate. Applying a similar argument to that above we can show that those variance estimates are also half what they would be if we used the S matrix. The variance of the interaction model for categorical covariates (Equation 4) is
 |
(7) |
where 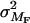 is the variance of the interaction and
is the variance of the interaction and  is its correlation matrix, which is simply F but with
is its correlation matrix, which is simply F but with  when animals i and j are in different categories. If we were to use S (an approximation for A) in place of F we would have
when animals i and j are in different categories. If we were to use S (an approximation for A) in place of F we would have
 |
(8) |
with  being the interaction between the categorical covariate and the additive genetic effect. However, since
being the interaction between the categorical covariate and the additive genetic effect. However, since  and
and  , it follows that
, it follows that 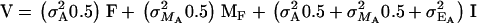 and therefore
and therefore  . For interactions between a continuous covariate x and family (Equation 5) the variance is
. For interactions between a continuous covariate x and family (Equation 5) the variance is
 |
(9) |
where  when x is the n-vector of x for the n animals. If we were to use S-approximation for A the variance would be
when x is the n-vector of x for the n animals. If we were to use S-approximation for A the variance would be
 |
(10) |
Substituting  as before,
as before, 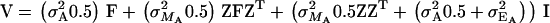 , which implies
, which implies  . Hence, in all cases the estimated variance of an additive genetic component is simply twice that of the corresponding family component.
. Hence, in all cases the estimated variance of an additive genetic component is simply twice that of the corresponding family component.
RESULTS
Of the 102 phenotypes available for analysis (Solberg et al. 2006), 88 could be accommodated in our linear mixed modeling framework (see methods). We obtained data for 15 covariates (Table 4): age, apparatus (for those tests where multiple units were used), cage (a variable indicating animals that were housed in the same cage), cage density (the number of animals in a cage), experimenter, family (defined as the offspring of two parents), sex, hour, litter (a number representing the birth order of each litter for a given sire and dam), month, season, study day, test order, weight, and year. An average of 10.3 covariates were recorded per phenotype (since not all phenotype–covariate combinations were available), leading to an average of 69.4 phenotypes measured per covariate. In total, we performed 1804 statistical tests. The significance of results is reported as the negative base 10 logarithm of the P-value (log P) of the relevant test. We took account of multiple testing by using the Dunn–Šidák correction, which for α = 5% comparisonwise error rate yielded a significance threshold of log P = 4.55.
We assessed initially the importance of three physiological covariates (sex, weight, and age). We fitted the covariates sequentially in the order sex, then weight, then age, so that, for instance, our reported significance for weight refers to how much it improved the fit of a model that already included sex. We included family in all models to ensure tested covariates were significant over and above genetic effects. Family, modeled as a random effect, is highly correlated with heritability (correlation of 0.89) and so acts a surrogate for the effect of additive genetic variation (see methods). We report estimates of heritability for all phenotypes in Table 5.
The effects of sex, weight, and age were relatively small (Figure 1b, “main effect” rows): sex effects explained >10% of the variance for 14 phenotypes; in more than half of the cases the effect was <5%; weight accounted for >10% of the variance for three phenotypes; all age effects were <2% (see appendix).
Figure 1.—
Main effects and interactions. (a) The log P (i.e., the −log10 of the P-value) for main and interaction effects of 12 covariates. Each box shows significance scores for one covariate on all applicable phenotypes. The shaded bar marks the corrected 5% threshold for significance (log P = 4.55). For example, Apparatus has significant main effects for a few phenotypes but significant interactions for none, whereas Hour has few significant main effects but has significant interaction effect for a number of phenotypes. (b) The estimated percentage of variance significant effects contributed to the phenotype. Note that log P's are capped at 20 for display purposes and that results for test order, which had no significant effects, are not shown.
Appendix.
Significant main effects and interactions of 13 covariates in 88 phenotypes
| Main effects
|
Interactions
|
||||||
|---|---|---|---|---|---|---|---|
| Test | Phenotype | Covariate | No. observed | log P | % variance explained | log P | % variance explained |
| Adrenal weight | Adrenal weight | Age | 2256 | — | — | 7.15 | 0.03 |
| Adrenal weight | Adrenal weight | Cage density | 2256 | 9.18 | 0.71 | 10.65 | 7.33 |
| Adrenal weight | Adrenal weight | Experimenter | 2257 | 11.79 | 1.02 | 7.11 | 21.28 |
| Adrenal weight | Adrenal weight | Litter | 2244 | — | — | 19.45 | 13.99 |
| Adrenal weight | Adrenal weight | Month | 2256 | 12.46 | 1.53 | 38.39 | 28.19 |
| Adrenal weight | Adrenal weight | Season | 2256 | — | — | 27.85 | 28.39 |
| Adrenal weight | Adrenal weight | Sex | 2257 | 306.75 | 36.47 | 31.10 | 29.83 |
| Adrenal weight | Adrenal weight | Study day | 2256 | — | — | 20.21 | 0.03 |
| Adrenal weight | Adrenal weight | Weight | 2257 | 18.91 | 1.57 | 19.96 | 0.68 |
| Adrenal weight | Adrenal weight | Year | 2256 | 5.75 | 0.43 | 7.37 | 26.54 |
| Biochemistry | Alanine transaminase (units/liter) | Cage density | 1897 | — | — | 6.90 | 11.83 |
| Biochemistry | Alanine transaminase (units/liter) | Litter | 1887 | — | — | 21.79 | 18.17 |
| Biochemistry | Alanine transaminase (units/liter) | Month | 1589 | 24.03 | 6.70 | 25.21 | 48.49 |
| Biochemistry | Alanine transaminase (units/liter) | Season | 1589 | 14.47 | 3.36 | 18.73 | 45.51 |
| Biochemistry | Alanine transaminase (units/liter) | Sex | 1897 | 9.11 | 1.53 | 6.29 | 19.04 |
| Biochemistry | Alanine transaminase (units/liter) | Study day | 1589 | 26.68 | 5.81 | — | — |
| Biochemistry | Alanine transaminase (units/liter) | Weight | 1897 | — | — | 6.55 | 0.51 |
| Biochemistry | Alanine transaminase (units/liter) | Year | 1589 | 8.30 | 1.70 | 4.97 | 31.62 |
| Biochemistry | Albumin (g/liter) | Cage density | 1999 | — | — | 6.52 | 11.16 |
| Biochemistry | Albumin (g/liter) | Litter | 1990 | — | — | 9.08 | 12.27 |
| Biochemistry | Albumin (g/liter) | Month | 1680 | 20.84 | 5.74 | 11.81 | 31.16 |
| Biochemistry | Albumin (g/liter) | Season | 1680 | 11.81 | 2.66 | 9.37 | 31.15 |
| Biochemistry | Albumin (g/liter) | Sex | 1999 | 25.05 | 4.44 | 4.93 | 16.30 |
| Biochemistry | Albumin (g/liter) | Study day | 1680 | 15.31 | 3.12 | — | — |
| Biochemistry | Albumin (g/liter) | Year | 1680 | 11.51 | 2.31 | 4.87 | 24.13 |
| Biochemistry | Alkaline phosphatase (units/liter) | Age | 1701 | 7.71 | 1.16 | 11.99 | 0.93 |
| Biochemistry | Alkaline phosphatase (units/liter) | Litter | 2011 | — | — | 10.65 | 10.17 |
| Biochemistry | Alkaline phosphatase (units/liter) | Month | 1701 | 8.93 | 2.35 | 17.78 | 31.72 |
| Biochemistry | Alkaline phosphatase (units/liter) | Season | 1701 | — | — | 10.64 | 26.09 |
| Biochemistry | Alkaline phosphatase (units/liter) | Sex | 2021 | 11.28 | 1.49 | 11.77 | 23.24 |
| Biochemistry | Alkaline phosphatase (units/liter) | Study day | 1701 | — | — | 6.20 | 0.02 |
| Biochemistry | Alkaline phosphatase (units/liter) | Year | 1701 | — | — | 4.95 | 24.45 |
| Biochemistry | Aspartate transaminase (units/liter) | Litter | 1933 | — | — | 9.83 | 10.96 |
| Biochemistry | Aspartate transaminase (units/liter) | Month | 1629 | 9.03 | 3.02 | 8.89 | 25.09 |
| Biochemistry | Aspartate transaminase (units/liter) | Season | 1629 | 5.73 | 1.35 | 10.02 | 27.87 |
| Biochemistry | Aspartate transaminase (units/liter) | Sex | 1942 | 27.03 | 4.91 | — | — |
| Biochemistry | Aspartate transaminase (units/liter) | Study day | 1629 | 25.52 | 5.27 | — | — |
| Biochemistry | Aspartate transaminase (units/liter) | Weight | 1942 | 10.20 | 1.72 | — | — |
| Biochemistry | Aspartate transaminase (units/liter) | Year | 1629 | 7.03 | 1.35 | 5.49 | 27.42 |
| Biochemistry | Calcium (mmol) | Age | 1688 | — | — | 4.70 | 0.77 |
| Biochemistry | Calcium (mmol) | Cage density | 2004 | — | — | 4.72 | 8.84 |
| Biochemistry | Calcium (mmol) | Litter | 1994 | — | — | 13.69 | 15.89 |
| Biochemistry | Calcium (mmol) | Month | 1688 | 12.22 | 3.66 | 14.95 | 35.51 |
| Biochemistry | Calcium (mmol) | Season | 1688 | 6.95 | 1.59 | 13.60 | 38.72 |
| Biochemistry | Calcium (mmol) | Sex | 2004 | 11.48 | 1.65 | 15.18 | 32.18 |
| Biochemistry | Calcium (mmol) | Study day | 1688 | 13.11 | 2.55 | 10.28 | 0.02 |
| Biochemistry | Calcium (mmol) | Weight | 2004 | 10.36 | 1.48 | 11.42 | 0.83 |
| Biochemistry | Calcium (mmol) | Year | 1688 | 11.79 | 2.26 | — | — |
| Biochemistry | Chloride (mmol) | Age | 1744 | — | — | 13.36 | 2.21 |
| Biochemistry | Chloride (mmol) | Cage density | 2068 | — | — | 7.82 | 11.10 |
| Biochemistry | Chloride (mmol) | Litter | 2058 | — | — | 26.60 | 38.71 |
| Biochemistry | Chloride (mmol) | Month | 1744 | 7.69 | 2.39 | 43.88 | 69.06 |
| Biochemistry | Chloride (mmol) | Season | 1744 | — | — | 30.28 | 70.86 |
| Biochemistry | Chloride (mmol) | Sex | 2068 | 23.74 | 3.32 | 20.74 | 35.48 |
| Biochemistry | Chloride (mmol) | Study day | 1744 | 8.33 | 1.42 | 14.12 | 0.02 |
| Biochemistry | Chloride (mmol) | Weight | 2068 | 10.63 | 1.40 | 14.04 | 0.78 |
| Biochemistry | High-density lipoproteins (mmol) | Age | 1612 | — | — | 5.36 | 0.45 |
| Biochemistry | High-density lipoproteins (mmol) | Month | 1612 | — | — | 5.86 | 12.67 |
| Biochemistry | High-density lipoproteins (mmol) | Sex | 1912 | 173.55 | 22.74 | 14.96 | 21.61 |
| Biochemistry | High-density lipoproteins (mmol) | Study day | 1612 | 4.58 | 0.49 | — | — |
| Biochemistry | High-density lipoproteins (mmol) | Weight | 1912 | 19.46 | 2.01 | 14.86 | 0.71 |
| Biochemistry | High-density lipoproteins (mmol) | Year | 1612 | 5.34 | 0.58 | — | — |
| Biochemistry | Low-density lipoproteins (mmol) | Cage density | 1947 | — | — | 4.77 | 8.53 |
| Biochemistry | Low-density lipoproteins (mmol) | Month | 1646 | 9.20 | 3.05 | 5.54 | 18.53 |
| Biochemistry | Low-density lipoproteins (mmol) | Sex | 1947 | 13.44 | 2.32 | 7.73 | 19.19 |
| Biochemistry | Phosphorous (mmol) | Age | 1495 | 6.27 | 1.41 | — | — |
| Biochemistry | Phosphorous (mmol) | Month | 1495 | — | — | 12.77 | 40.25 |
| Biochemistry | Phosphorous (mmol) | Season | 1495 | — | — | 5.77 | 26.54 |
| Biochemistry | Phosphorous (mmol) | Sex | 1783 | — | — | 5.21 | 24.81 |
| Biochemistry | Phosphorous (mmol) | Study day | 1495 | 4.86 | 1.05 | — | — |
| Biochemistry | Phosphorous (mmol) | Year | 1495 | 8.05 | 1.84 | — | — |
| Biochemistry | Sodium (mmol) | Age | 1734 | — | — | 9.72 | 1.85 |
| Biochemistry | Sodium (mmol) | Litter | 2048 | — | — | 28.54 | 33.14 |
| Biochemistry | Sodium (mmol) | Month | 1734 | 6.63 | 2.10 | 37.62 | 62.02 |
| Biochemistry | Sodium (mmol) | Season | 1734 | — | — | 21.62 | 50.65 |
| Biochemistry | Sodium (mmol) | Sex | 2058 | 34.14 | 5.01 | 17.45 | 31.96 |
| Biochemistry | Sodium (mmol) | Study day | 1734 | 7.04 | 1.17 | 11.85 | 0.02 |
| Biochemistry | Sodium (mmol) | Weight | 2058 | 14.14 | 1.96 | 12.64 | 0.76 |
| Biochemistry | Sodium (mmol) | Year | 1734 | 7.08 | 1.17 | — | — |
| Biochemistry | Total cholesterol (mmol) | Month | 1704 | 15.12 | 3.48 | 6.68 | 16.02 |
| Biochemistry | Total cholesterol (mmol) | Season | 1704 | 11.76 | 2.09 | — | — |
| Biochemistry | Total cholesterol (mmol) | Sex | 2018 | 97.59 | 15.23 | 5.81 | 14.08 |
| Biochemistry | Total cholesterol (mmol) | Weight | 2018 | 5.43 | 0.66 | — | — |
| Biochemistry | Total protein (g/liter) | Cage density | 1882 | — | — | 5.86 | 10.40 |
| Biochemistry | Total protein (g/liter) | Month | 1565 | 16.74 | 5.29 | 12.86 | 40.36 |
| Biochemistry | Total protein (g/liter) | Season | 1565 | 12.91 | 3.23 | 8.38 | 40.76 |
| Biochemistry | Total protein (g/liter) | Sex | 1882 | 12.49 | 2.34 | 5.71 | 20.71 |
| Biochemistry | Total protein (g/liter) | Study day | 1565 | — | — | 5.86 | 0.02 |
| Biochemistry | Total protein (g/liter) | Weight | 1882 | 8.64 | 1.57 | — | — |
| Biochemistry | Triglycerides (mmol) | Cage density | 1738 | — | — | 7.30 | 10.48 |
| Biochemistry | Triglycerides (mmol) | Month | 1448 | — | — | 6.31 | 20.79 |
| Biochemistry | Triglycerides (mmol) | Sex | 1738 | 85.67 | 16.34 | — | — |
| Biochemistry | Urea (mmol) | Cage density | 1992 | — | — | 8.79 | 13.88 |
| Biochemistry | Urea (mmol) | Litter | 1982 | — | — | 10.54 | 11.86 |
| Biochemistry | Urea (mmol) | Month | 1673 | 6.80 | 2.44 | 8.58 | 24.17 |
| Biochemistry | Urea (mmol) | Season | 1673 | 5.68 | 1.35 | — | — |
| Biochemistry | Urea (mmol) | Sex | 1992 | — | — | 10.62 | 27.54 |
| Context freezing | Time freezing to context (sec) | Apparatus | 1671 | 18.88 | 4.13 | — | — |
| Context freezing | Time freezing to context (sec) | Experimenter | 1671 | 5.22 | 1.70 | — | — |
| Context freezing | Time freezing to context (sec) | Sex | 1671 | 24.20 | 5.14 | — | — |
| Context freezing | Time freezing to context (sec) | Weight | 1671 | 6.04 | 1.14 | — | — |
| Cue conditioning | Fecal boli after cue | Sex | 1768 | 4.80 | 0.98 | — | — |
| Cue conditioning | Time freezing after cue (sec) | Age | 1791 | 5.08 | 1.05 | — | — |
| Cue conditioning | Time freezing after cue (sec) | Apparatus | 1665 | 43.68 | 10.98 | — | — |
| Cue conditioning | Time freezing during cue (sec) | Apparatus | 1665 | 46.03 | 10.54 | — | — |
| Elevated plus maze | Closed-arm entries | Experimenter | 2229 | 4.57 | 1.44 | — | — |
| Elevated plus maze | Closed-arm entries | Weight | 2229 | 5.68 | 0.89 | — | — |
| Elevated plus maze | Closed-arm time | Month | 2221 | — | — | 4.98 | 15.61 |
| Elevated plus maze | Open-arm distance | Experimenter | 2261 | 7.14 | 1.71 | — | — |
| Elevated plus maze | Open-arm distance | Month | 2260 | — | — | 7.52 | 17.06 |
| Elevated plus maze | Open-arm distance | Weight | 2261 | 4.82 | 0.63 | — | — |
| Elevated plus maze | Open-arm entries | Weight | 2261 | 5.90 | 0.80 | — | — |
| Elevated plus maze | Open-arm time | Experimenter | 2261 | 6.78 | 1.69 | — | — |
| Elevated plus maze | Open-arm time | Month | 2260 | — | — | 7.18 | 17.03 |
| Elevated plus maze | Open-arm time | Weight | 2261 | 5.30 | 0.72 | — | — |
| Fear potentiated startle | Startle response | Age | 2005 | 5.40 | 0.82 | — | — |
| Fear potentiated startle | Startle response | Apparatus | 2005 | 31.59 | 5.54 | — | — |
| Fear potentiated startle | Startle response | Sex | 2005 | 15.69 | 2.65 | — | — |
| Fear potentiated startle | Startle response | Study day | 2005 | 4.86 | 0.73 | — | — |
| Fear potentiated startle | Startle response | Weight | 2005 | 7.53 | 1.19 | — | — |
| Fear potentiated startle | Startle response | Year | 2005 | 5.60 | 0.85 | — | — |
| Glucose tolerance test | AUC-G (mg/dl) | Age | 2130 | — | — | 5.36 | 0.94 |
| Glucose tolerance test | AUC-G (mg/dl) | Cage density | 2130 | — | — | 8.35 | 12.51 |
| Glucose tolerance test | AUC-G (mg/dl) | Experimenter | 2130 | 9.45 | 2.11 | 8.77 | 26.37 |
| Glucose tolerance test | AUC-G (mg/dl) | Hour | 2130 | — | — | 5.04 | 16.56 |
| Glucose tolerance test | AUC-G (mg/dl) | Litter | 2117 | — | — | 12.48 | 11.96 |
| Glucose tolerance test | AUC-G (mg/dl) | Month | 2130 | 4.74 | 1.59 | 18.71 | 36.65 |
| Glucose tolerance test | AUC-G (mg/dl) | Season | 2130 | 5.10 | 1.01 | 7.80 | 25.18 |
| Glucose tolerance test | AUC-G (mg/dl) | Study day | 2130 | — | — | 5.60 | 0.01 |
| Glucose tolerance test | AUC-G (mg/dl) | Weight | 2130 | — | — | 6.26 | 0.53 |
| Glucose tolerance test | AUC-IRI (ng/ml) | Cage density | 2105 | — | — | 5.10 | 7.54 |
| Glucose tolerance test | AUC-IRI (ng/ml) | Month | 2105 | — | — | 10.38 | 24.65 |
| Glucose tolerance test | AUC-IRI (ng/ml) | Season | 2105 | — | — | 4.63 | 16.35 |
| Glucose tolerance test | AUC-IRI (ng/ml) | Sex | 2105 | — | — | 7.95 | 21.33 |
| Glucose tolerance test | AUC-IRI (ng/ml) | Weight | 2105 | 6.66 | 1.07 | — | — |
| Glucose tolerance test | AUC-IRI/AUC-G | Cage density | 1982 | — | — | 6.51 | 10.92 |
| Glucose tolerance test | AUC-IRI/AUC-G | Litter | 1970 | — | — | 5.85 | 8.22 |
| Glucose tolerance test | AUC-IRI/AUC-G | Month | 1982 | — | — | 10.08 | 26.85 |
| Glucose tolerance test | AUC-IRI/AUC-G | Season | 1982 | — | — | 5.08 | 19.65 |
| Glucose tolerance test | AUC-IRI/AUC-G | Sex | 1982 | — | — | 6.70 | 19.96 |
| Glucose tolerance test | DG (mg/dl) | Age | 2131 | — | — | 5.88 | 1.01 |
| Glucose tolerance test | DG (mg/dl) | Cage density | 2131 | — | — | 8.53 | 12.82 |
| Glucose tolerance test | DG (mg/dl) | Experimenter | 2131 | 11.18 | 2.43 | 9.80 | 28.15 |
| Glucose tolerance test | DG (mg/dl) | Hour | 2131 | — | — | 5.36 | 17.29 |
| Glucose tolerance test | DG (mg/dl) | Litter | 2118 | — | — | 12.75 | 12.24 |
| Glucose tolerance test | DG (mg/dl) | Month | 2131 | 5.15 | 1.68 | 18.64 | 36.72 |
| Glucose tolerance test | DG (mg/dl) | Season | 2131 | 5.79 | 1.13 | 7.61 | 25.02 |
| Glucose tolerance test | DG (mg/dl) | Study day | 2131 | — | — | 5.87 | 0.01 |
| Glucose tolerance test | DG (mg/dl) | Weight | 2131 | — | — | 6.62 | 0.56 |
| Glucose tolerance test | DIRI (ng/ml) | Cage density | 2107 | — | — | 5.14 | 7.64 |
| Glucose tolerance test | DIRI (ng/ml) | Litter | 2095 | — | — | 5.02 | 6.49 |
| Glucose tolerance test | DIRI (ng/ml) | Month | 2107 | — | — | 9.93 | 24.02 |
| Glucose tolerance test | DIRI (ng/ml) | Season | 2107 | — | — | 4.85 | 17.02 |
| Glucose tolerance test | DIRI (ng/ml) | Sex | 2107 | — | — | 7.48 | 20.05 |
| Glucose tolerance test | DIRI (ng/ml) | Weight | 2107 | 6.66 | 1.06 | — | — |
| Glucose tolerance test | DIRI/DG | Cage density | 1984 | — | — | 6.53 | 10.85 |
| Glucose tolerance test | DIRI/DG | Litter | 1972 | — | — | 6.95 | 9.39 |
| Glucose tolerance test | DIRI/DG | Month | 1984 | 4.86 | 1.71 | 11.17 | 28.02 |
| Glucose tolerance test | DIRI/DG | Season | 1984 | — | — | 6.05 | 21.73 |
| Glucose tolerance test | DIRI/DG | Sex | 1984 | — | — | 6.74 | 19.65 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Age | 2225 | 4.66 | 0.43 | 22.81 | 1.50 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Cage density | 2225 | 4.71 | 0.43 | 18.00 | 12.92 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Experimenter | 2225 | 23.96 | 2.95 | 24.04 | 34.21 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Hour | 2225 | — | — | 12.95 | 22.62 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Litter | 2212 | 13.63 | 1.38 | 19.37 | 19.71 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Month | 2225 | 8.09 | 1.42 | 46.92 | 41.41 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Season | 2225 | 5.89 | 0.72 | 26.62 | 39.04 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Sex | 2225 | 158.57 | 20.42 | 16.37 | 24.82 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Study day | 2225 | 14.64 | 1.51 | 24.49 | 0.01 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Weight | 2225 | — | — | 15.79 | 0.72 |
| Glucose tolerance test | Glucose 0 (mg/dl) | Year | 2225 | 28.29 | 3.09 | 10.67 | 30.40 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Age | 2204 | — | — | 11.08 | 1.30 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Cage density | 2204 | — | — | 15.31 | 18.04 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Experimenter | 2204 | 53.70 | 8.43 | 16.69 | 30.82 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Hour | 2204 | 5.29 | 1.16 | 6.02 | 19.58 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Litter | 2192 | 8.02 | 1.12 | 9.18 | 15.06 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Month | 2204 | 4.72 | 1.40 | 31.77 | 44.55 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Season | 2204 | 5.54 | 0.96 | 19.27 | 40.90 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Sex | 2204 | 15.01 | 2.21 | 7.35 | 21.40 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Study day | 2204 | 4.79 | 0.63 | 18.57 | 0.01 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Weight | 2204 | — | — | 5.56 | 0.55 |
| Glucose tolerance test | Glucose 15 (mg/dl) | Year | 2204 | 12.49 | 1.82 | 5.74 | 36.94 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Age | 2187 | — | — | 6.94 | 0.94 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Cage density | 2187 | — | — | 11.78 | 16.14 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Experimenter | 2187 | 21.23 | 4.04 | 11.68 | 29.41 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Hour | 2187 | — | — | 5.59 | 20.08 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Litter | 2174 | 4.68 | 0.67 | 8.05 | 11.02 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Month | 2187 | 5.03 | 1.59 | 19.07 | 34.35 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Season | 2187 | 5.72 | 1.08 | 11.18 | 31.27 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Sex | 2187 | 9.87 | 1.53 | 4.84 | 18.00 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Study day | 2187 | — | — | 10.33 | 0.01 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Weight | 2187 | — | — | 5.30 | 0.63 |
| Glucose tolerance test | Glucose 30 (mg/dl) | Year | 2187 | — | — | 4.79 | 35.33 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Age | 2153 | — | — | 7.87 | 1.02 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Cage density | 2153 | — | — | 4.99 | 7.52 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Experimenter | 2153 | 10.63 | 2.14 | 17.74 | 37.77 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Hour | 2153 | — | — | 7.83 | 20.67 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Litter | 2140 | — | — | 10.28 | 10.05 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Month | 2153 | 5.90 | 1.70 | 18.07 | 32.97 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Season | 2153 | 7.51 | 1.33 | 7.83 | 23.33 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Sex | 2153 | 34.87 | 5.67 | 5.24 | 15.93 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Study day | 2153 | — | — | 6.82 | 0.01 |
| Glucose tolerance test | Glucose 75 (mg/dl) | Weight | 2153 | — | — | 9.78 | 0.69 |
| Glucose tolerance test | Insulin 0 (ng/ml) | Cage density | 2206 | — | — | 11.14 | 14.52 |
| Glucose tolerance test | Insulin 0 (ng/ml) | Experimenter | 2206 | — | — | 12.19 | 28.49 |
| Glucose tolerance test | Insulin 0 (ng/ml) | Hour | 2206 | — | — | 11.02 | 32.11 |
| Glucose tolerance test | Insulin 0 (ng/ml) | Litter | 2193 | — | — | 9.26 | 15.06 |
| Glucose tolerance test | Insulin 0 (ng/ml) | Month | 2206 | 11.67 | 2.91 | 26.56 | 42.42 |
| Glucose tolerance test | Insulin 0 (ng/ml) | Season | 2206 | 7.28 | 1.37 | 15.28 | 34.80 |
| Glucose tolerance test | Insulin 0 (ng/ml) | Sex | 2206 | 12.78 | 2.05 | 9.06 | 24.42 |
| Glucose tolerance test | Insulin 0 (ng/ml) | Weight | 2206 | 5.08 | 0.74 | — | — |
| Glucose tolerance test | Insulin 15 (ng/ml) | Cage density | 2197 | — | — | 6.44 | 10.52 |
| Glucose tolerance test | Insulin 15 (ng/ml) | Experimenter | 2197 | — | — | 7.73 | 20.84 |
| Glucose tolerance test | Insulin 15 (ng/ml) | Litter | 2185 | — | — | 9.16 | 10.53 |
| Glucose tolerance test | Insulin 15 (ng/ml) | Month | 2197 | 8.96 | 2.37 | 13.45 | 27.78 |
| Glucose tolerance test | Insulin 15 (ng/ml) | Season | 2197 | 5.95 | 1.12 | 6.03 | 20.13 |
| Glucose tolerance test | Insulin 15 (ng/ml) | Sex | 2197 | — | — | 7.91 | 20.73 |
| Glucose tolerance test | Insulin 15 (ng/ml) | Weight | 2197 | 4.96 | 0.72 | 5.71 | 0.57 |
| Glucose tolerance test | Insulin 30 (ng/ml) | Cage density | 2178 | — | — | 6.23 | 9.10 |
| Glucose tolerance test | Insulin 30 (ng/ml) | Experimenter | 2178 | — | — | 5.51 | 16.19 |
| Glucose tolerance test | Insulin 30 (ng/ml) | Litter | 2166 | — | — | 7.56 | 9.10 |
| Glucose tolerance test | Insulin 30 (ng/ml) | Month | 2178 | 7.05 | 2.03 | 14.18 | 29.40 |
| Glucose tolerance test | Insulin 30 (ng/ml) | Season | 2178 | — | — | 8.50 | 24.71 |
| Glucose tolerance test | Insulin 30 (ng/ml) | Sex | 2178 | — | — | 7.23 | 18.84 |
| Glucose tolerance test | Insulin 30 (ng/ml) | Weight | 2178 | 4.62 | 0.67 | — | — |
| Glucose tolerance test | Insulin 75 (ng/ml) | Cage density | 2112 | 4.88 | 0.70 | 7.15 | 9.48 |
| Glucose tolerance test | Insulin 75 (ng/ml) | Experimenter | 2112 | — | — | 5.65 | 17.04 |
| Glucose tolerance test | Insulin 75 (ng/ml) | Hour | 2112 | — | — | 8.55 | 22.87 |
| Glucose tolerance test | Insulin 75 (ng/ml) | Litter | 2100 | — | — | 6.11 | 12.60 |
| Glucose tolerance test | Insulin 75 (ng/ml) | Month | 2112 | 6.86 | 1.96 | 16.98 | 30.66 |
| Glucose tolerance test | Insulin 75 (ng/ml) | Season | 2112 | — | — | 6.91 | 21.29 |
| Glucose tolerance test | Insulin 75 (ng/ml) | Sex | 2112 | 30.76 | 5.18 | 7.85 | 18.15 |
| Glucose tolerance test | Insulin 75 (ng/ml) | Weight | 2112 | 11.85 | 1.87 | — | — |
| Glucose tolerance test | Insulin slope | Sex | 1122 | 5.58 | 1.83 | — | — |
| Glucose tolerance test | K (glucose slope) | Sex | 1953 | 11.19 | 2.22 | — | — |
| Glucose tolerance test | K (glucose slope) | Year | 1953 | 4.83 | 0.87 | — | — |
| Growth | Growth slope | Cage density | 2418 | — | — | 20.00 | 15.53 |
| Growth | Growth slope | Litter | 2462 | — | — | 26.05 | 20.10 |
| Growth | Growth slope | Sex | 2474 | 135.72 | 17.45 | 19.14 | 26.19 |
| Hematology | Hemoglobin (g/dl) | Month | 1870 | — | — | 8.59 | 23.95 |
| Hematology | Hemoglobin (g/dl) | Season | 1870 | — | — | 5.06 | 18.96 |
| Hematology | Hemoglobin (g/dl) | Sex | 1870 | 9.07 | 1.73 | — | — |
| Hematology | Hemoglobin (g/dl) | Weight | 1870 | 6.22 | 1.14 | 4.86 | 0.40 |
| Hematology | Lymphocytes | Age | 1833 | 4.58 | 0.70 | — | — |
| Hematology | Lymphocytes | Litter | 1822 | 7.18 | 1.15 | 7.79 | 8.82 |
| Hematology | Lymphocytes | Month | 1833 | — | — | 10.72 | 25.40 |
| Hematology | Lymphocytes | Season | 1833 | — | — | 10.24 | 25.19 |
| Hematology | Lymphocytes | Sex | 1833 | — | — | 5.90 | 18.71 |
| Hematology | Lymphocytes | Study day | 1833 | 14.16 | 2.40 | — | — |
| Hematology | Lymphocytes | Year | 1833 | 6.04 | 0.96 | — | — |
| Hematology | Mean cellular Hb concentration (%) | Age | 1863 | 9.85 | 1.40 | 26.04 | 1.83 |
| Hematology | Mean cellular Hb concentration (%) | Cage density | 1862 | — | — | 33.52 | 29.32 |
| Hematology | Mean cellular Hb concentration (%) | Litter | 1852 | — | — | 39.31 | 35.19 |
| Hematology | Mean cellular Hb concentration (%) | Month | 1863 | 68.92 | 11.39 | 58.19 | 54.51 |
| Hematology | Mean cellular Hb concentration (%) | Season | 1863 | 28.14 | 4.54 | 40.00 | 58.58 |
| Hematology | Mean cellular Hb concentration (%) | Sex | 1863 | 6.02 | 0.84 | 19.55 | 39.16 |
| Hematology | Mean cellular Hb concentration (%) | Study day | 1863 | 5.96 | 0.82 | 19.79 | 0.07 |
| Hematology | Mean cellular Hb concentration (%) | Weight | 1863 | — | — | 14.68 | 0.92 |
| Hematology | Mean cellular Hb concentration (%) | Year | 1863 | — | — | 31.10 | 82.94 |
| Hematology | Mean cellular volume (fl) | Cage density | 1875 | — | — | 7.63 | 12.47 |
| Hematology | Mean cellular volume (fl) | Litter | 1865 | — | — | 4.68 | 5.79 |
| Hematology | Mean cellular volume (fl) | Month | 1876 | 13.39 | 3.18 | 15.63 | 30.61 |
| Hematology | Mean cellular volume (fl) | Season | 1876 | — | — | 7.50 | 21.56 |
| Hematology | Mean cellular volume (fl) | Sex | 1876 | — | — | 8.89 | 20.90 |
| Hematology | Mean cellular volume (fl) | Study day | 1876 | 6.15 | 0.90 | — | — |
| Hematology | Mean cellular volume (fl) | Weight | 1876 | — | — | 5.16 | 0.45 |
| Hematology | Mean cellular volume (fl) | Year | 1876 | 6.67 | 0.99 | — | — |
| Hematology | Mean corpuscular hemoglobin (pg) | Age | 1871 | — | — | 15.80 | 1.25 |
| Hematology | Mean corpuscular hemoglobin (pg) | Cage density | 1870 | — | — | 13.30 | 16.26 |
| Hematology | Mean corpuscular hemoglobin (pg) | Litter | 1860 | — | — | 16.42 | 16.51 |
| Hematology | Mean corpuscular hemoglobin (pg) | Month | 1871 | 32.68 | 6.77 | 10.21 | 22.91 |
| Hematology | Mean corpuscular hemoglobin (pg) | Season | 1871 | 19.94 | 3.62 | 8.85 | 25.86 |
| Hematology | Mean corpuscular hemoglobin (pg) | Sex | 1871 | — | — | 11.35 | 27.44 |
| Hematology | Mean corpuscular hemoglobin (pg) | Study day | 1871 | — | — | 9.47 | 0.04 |
| Hematology | Mean corpuscular hemoglobin (pg) | Weight | 1871 | — | — | 5.59 | 0.55 |
| Hematology | Mean corpuscular hemoglobin (pg) | Year | 1871 | — | — | 8.52 | 44.24 |
| Hematology | Plateletcrit (%) | Age | 1839 | — | — | 5.10 | 0.73 |
| Hematology | Plateletcrit (%) | Month | 1839 | 14.39 | 4.06 | 6.26 | 19.33 |
| Hematology | Plateletcrit (%) | Season | 1839 | 4.85 | 1.11 | 4.59 | 19.65 |
| Hematology | Plateletcrit (%) | Sex | 1839 | 36.50 | 7.50 | 8.92 | 27.53 |
| Hematology | Platelets (n/μl) | Month | 1863 | 16.43 | 4.18 | 10.19 | 24.81 |
| Hematology | Platelets (n/μl) | Season | 1863 | 6.62 | 1.36 | 7.75 | 26.64 |
| Hematology | Platelets (n/μl) | Sex | 1863 | 30.92 | 5.76 | 10.78 | 27.67 |
| Hematology | Platelets (n/μl) | Study day | 1863 | 9.44 | 1.60 | — | — |
| Hematology | Platelets (n/μl) | Weight | 1863 | 5.77 | 0.93 | 5.54 | 0.50 |
| Hematology | Platelets (n/μl) | Year | 1863 | 5.56 | 0.89 | — | — |
| Hematology | Red blood cell count (n/μl) | Month | 1870 | — | — | 11.88 | 29.98 |
| Hematology | Red blood cell count (n/μl) | Season | 1870 | — | — | 5.84 | 21.52 |
| Hematology | Red blood cell count (n/μl) | Sex | 1870 | 9.32 | 1.77 | 4.95 | 19.31 |
| Hematology | Red blood cell count (n/μl) | Weight | 1870 | 5.90 | 1.07 | 4.94 | 0.48 |
| Hematology | Red cell distribution width | Litter | 1850 | — | — | 4.64 | 8.40 |
| Hematology | Red cell distribution width | Month | 1861 | 8.27 | 2.18 | 10.68 | 23.56 |
| Hematology | Red cell distribution width | Season | 1861 | — | — | 5.74 | 17.99 |
| Hematology | Red cell distribution width | Sex | 1861 | 12.88 | 1.99 | — | — |
| Hematology | Red cell distribution width | Weight | 1861 | — | — | 4.86 | 0.42 |
| Hematology | White blood cell count (n/μl) | Cage density | 1875 | — | — | 5.44 | 12.88 |
| Hematology | White blood cell count (n/μl) | Litter | 1865 | 6.24 | 1.00 | 11.57 | 12.54 |
| Hematology | White blood cell count (n/μl) | Month | 1876 | — | — | 15.14 | 33.46 |
| Hematology | White blood cell count (n/μl) | Season | 1876 | — | — | 15.64 | 36.77 |
| Hematology | White blood cell count (n/μl) | Sex | 1876 | — | — | 6.89 | 23.62 |
| Hematology | White blood cell count (n/μl) | Study day | 1876 | 10.18 | 1.71 | 6.86 | 0.03 |
| Hematology | White blood cell count (n/μl) | Weight | 1876 | — | — | 5.48 | 0.58 |
| Hematology | White blood cell count (n/μl) | Year | 1876 | — | — | 6.33 | 42.86 |
| Hematology | Hematocrit (%) | Month | 1873 | — | — | 10.13 | 27.26 |
| Hematology | Hematocrit (%) | Season | 1873 | — | — | 4.92 | 19.31 |
| Hematology | Hematocrit (%) | Sex | 1873 | 12.18 | 2.39 | 5.01 | 18.41 |
| Hematology | Hematocrit (%) | Weight | 1873 | 5.59 | 1.02 | 6.60 | 0.48 |
| Immunology | %B220+ | Age | 1723 | — | — | 9.73 | 2.70 |
| Immunology | %B220+ | Cage density | 1677 | — | — | 7.72 | 10.49 |
| Immunology | %B220+ | Litter | 1713 | — | — | 9.38 | 14.26 |
| Immunology | %B220+ | Month | 1723 | 11.68 | 2.84 | 28.30 | 41.28 |
| Immunology | %B220+ | Season | 1723 | — | — | 23.99 | 46.06 |
| Immunology | %B220+ | Sex | 1723 | 13.02 | 2.07 | 10.71 | 28.81 |
| Immunology | %CD3+ | Litter | 1723 | 5.00 | 0.78 | 6.68 | 13.19 |
| Immunology | %CD3+ | Month | 1733 | 16.65 | 4.07 | 19.89 | 37.17 |
| Immunology | %CD3+ | Season | 1733 | 5.47 | 1.12 | 14.69 | 35.98 |
| Immunology | %CD3+ | Sex | 1733 | — | — | 11.57 | 30.57 |
| Immunology | %CD4+ | Cage density | 1673 | — | — | 4.92 | 8.94 |
| Immunology | %CD4+ | Litter | 1721 | — | — | 11.42 | 18.10 |
| Immunology | %CD4+ | Month | 1731 | 14.99 | 3.94 | 23.00 | 44.64 |
| Immunology | %CD4+ | Season | 1731 | — | — | 18.16 | 44.24 |
| Immunology | %CD4+ | Sex | 1731 | — | — | 9.74 | 28.88 |
| Immunology | %CD4+/CD3+ | Age | 1732 | — | — | 6.12 | 1.24 |
| Immunology | %CD4+/CD3+ | Cage density | 1674 | — | — | 5.40 | 6.88 |
| Immunology | %CD4+/CD3+ | Litter | 1722 | — | — | 15.81 | 10.27 |
| Immunology | %CD4+/CD3+ | Month | 1732 | 25.71 | 4.65 | 13.23 | 21.82 |
| Immunology | %CD4+/CD3+ | Season | 1732 | 8.50 | 1.35 | 8.04 | 17.89 |
| Immunology | %CD8+ | Age | 1733 | — | — | 5.32 | 1.08 |
| Immunology | %CD8+ | Month | 1733 | 14.17 | 2.57 | 7.57 | 14.65 |
| Immunology | %CD8+ | Season | 1733 | 10.53 | 1.46 | — | — |
| Immunology | %CD8+ | Sex | 1733 | — | — | 5.30 | 12.61 |
| Immunology | %NK cells | Cage density | 1667 | — | — | 5.29 | 10.41 |
| Immunology | %NK cells | Litter | 1714 | — | — | 11.43 | 16.79 |
| Immunology | %NK cells | Month | 1724 | 35.27 | 8.68 | 16.15 | 32.91 |
| Immunology | %NK cells | Season | 1724 | 9.04 | 2.06 | 16.04 | 37.88 |
| Immunology | %NK cells | Sex | 1724 | 5.22 | 0.95 | — | — |
| Immunology | CD4+/CD8+ | Age | 1729 | — | — | 6.02 | 1.07 |
| Immunology | CD4+/CD8+ | Cage density | 1671 | — | — | 4.85 | 5.83 |
| Immunology | CD4+/CD8+ | Litter | 1719 | — | — | 10.62 | 7.45 |
| Immunology | CD4+/CD8+ | Month | 1729 | 17.55 | 3.26 | 9.38 | 17.39 |
| Immunology | CD4+/CD8+ | Season | 1729 | 8.52 | 1.29 | 4.78 | 12.21 |
| Length | Body length (cm) | Age | 1942 | — | — | 7.56 | 0.81 |
| Length | Body length (cm) | Litter | 1932 | — | — | 10.25 | 8.30 |
| Length | Body length (cm) | Month | 1942 | 16.46 | 3.22 | 11.29 | 21.08 |
| Length | Body length (cm) | Season | 1942 | 5.61 | 0.91 | 13.42 | 24.69 |
| Length | Body length (cm) | Sex | 1942 | 35.12 | 5.17 | — | — |
| Length | Body length (cm) | Study day | 1942 | — | — | 4.91 | 0.02 |
| Length | Body length (cm) | Weight | 1942 | 87.23 | 13.94 | — | — |
| Length | Body length (cm) | Year | 1942 | 5.94 | 0.76 | — | — |
| New home-cage activity | Fine movement | Age | 2294 | 5.14 | 0.72 | — | — |
| New home-cage activity | Fine movement | Sex | 2294 | 7.62 | 1.13 | — | — |
| New home-cage activity | Total beam breaks (30 min) | Experimenter | 2290 | — | — | 6.49 | 17.79 |
| New home-cage activity | Total beam breaks (30 min) | Month | 2290 | — | — | 7.05 | 18.22 |
| New home-cage activity | Total beam breaks (30 min) | Season | 2290 | — | — | 4.87 | 16.40 |
| New home-cage activity | Total beam breaks (first 5 min) | Experimenter | 2289 | 11.94 | 2.90 | 5.06 | 15.50 |
| New home-cage activity | Total beam breaks (first 5 min) | Month | 2289 | — | — | 11.44 | 24.53 |
| New home-cage activity | Total beam breaks (first 5 min) | Season | 2289 | — | — | 9.49 | 23.45 |
| New home-cage activity | Total beam breaks (last 5 min) | Hour | 2275 | 4.99 | 1.41 | — | — |
| Open field | Fecal boli | Month | 2304 | 5.34 | 1.75 | — | — |
| Open field | Fecal boli | Sex | 2304 | — | — | 5.40 | 16.38 |
| Open field | Total activity | Experimenter | 2302 | 4.99 | 1.34 | — | — |
| Open field | Total activity | Season | 2302 | — | — | 6.11 | 16.96 |
| Open field | Total activity | Sex | 2302 | 7.60 | 1.07 | — | — |
| Plethysmography | Enhanced pause (baseline) | Age | 2169 | — | — | 15.55 | 2.18 |
| Plethysmography | Enhanced pause (baseline) | Cage density | 2169 | — | — | 7.42 | 8.81 |
| Plethysmography | Enhanced pause (baseline) | Hour | 2169 | — | — | 18.39 | 31.96 |
| Plethysmography | Enhanced pause (baseline) | Litter | 2157 | 6.97 | 0.91 | 24.63 | 25.45 |
| Plethysmography | Enhanced pause (baseline) | Month | 2169 | 77.44 | 11.87 | 24.29 | 31.43 |
| Plethysmography | Enhanced pause (baseline) | Season | 2169 | 48.57 | 7.17 | 13.71 | 27.43 |
| Plethysmography | Enhanced pause (baseline) | Sex | 2169 | 15.80 | 2.24 | 11.77 | 25.38 |
| Plethysmography | Enhanced pause (baseline) | Study day | 2169 | — | — | 20.59 | 0.09 |
| Plethysmography | Enhanced pause (baseline) | Year | 2169 | 5.51 | 0.72 | 12.08 | 48.49 |
| Plethysmography | Enhanced pause (metacholine) | Age | 1943 | — | — | 7.64 | 1.41 |
| Plethysmography | Enhanced pause (metacholine) | Hour | 1943 | — | — | 20.37 | 36.68 |
| Plethysmography | Enhanced pause (metacholine) | Litter | 1931 | — | — | 10.94 | 20.86 |
| Plethysmography | Enhanced pause (metacholine) | Month | 1943 | 16.72 | 3.76 | 19.83 | 37.13 |
| Plethysmography | Enhanced pause (metacholine) | Season | 1943 | 11.87 | 2.12 | 15.67 | 36.31 |
| Plethysmography | Enhanced pause (metacholine) | Sex | 1943 | 21.63 | 3.56 | 9.84 | 26.78 |
| Plethysmography | Enhanced pause (metacholine) | Weight | 1943 | 6.51 | 0.97 | — | — |
| Plethysmography | Enhanced pause (metacholine) | Year | 1943 | — | — | 8.18 | 50.39 |
| Plethysmography | Expiratory time (baseline) | Hour | 2165 | — | — | 17.65 | 34.38 |
| Plethysmography | Expiratory time (baseline) | Litter | 2153 | — | — | 9.09 | 10.52 |
| Plethysmography | Expiratory time (baseline) | Month | 2165 | 17.72 | 3.94 | 11.84 | 26.41 |
| Plethysmography | Expiratory time (baseline) | Season | 2165 | 13.74 | 2.42 | 10.14 | 26.77 |
| Plethysmography | Expiratory time (baseline) | Sex | 2165 | — | — | 9.43 | 25.60 |
| Plethysmography | Expiratory time (baseline) | Study day | 2165 | — | — | 5.30 | 0.03 |
| Plethysmography | Expiratory time (baseline) | Weight | 2165 | 4.77 | 0.68 | 6.66 | 0.57 |
| Plethysmography | Expiratory time (baseline) | Year | 2165 | 9.98 | 1.53 | — | — |
| Plethysmography | Expiratory time (metacholine) | Hour | 1935 | — | — | 5.56 | 15.75 |
| Plethysmography | Expiratory time (metacholine) | Month | 1935 | 5.31 | 1.78 | — | — |
| Plethysmography | Expiratory time (metacholine) | Sex | 1935 | — | — | 6.15 | 17.12 |
| Plethysmography | Inspiratory time (baseline) | Cage density | 2174 | 5.18 | 0.74 | — | — |
| Plethysmography | Inspiratory time (baseline) | Hour | 2174 | — | — | 17.02 | 32.09 |
| Plethysmography | Inspiratory time (baseline) | Litter | 2162 | — | — | 12.07 | 12.87 |
| Plethysmography | Inspiratory time (baseline) | Month | 2174 | 15.47 | 3.57 | 9.87 | 24.28 |
| Plethysmography | Inspiratory time (baseline) | Season | 2174 | 10.83 | 1.95 | 10.09 | 27.17 |
| Plethysmography | Inspiratory time (baseline) | Sex | 2174 | 12.32 | 1.95 | 12.50 | 29.61 |
| Plethysmography | Inspiratory time (baseline) | Study day | 2174 | — | — | 4.99 | 0.03 |
| Plethysmography | Inspiratory time (baseline) | Weight | 2174 | — | — | 9.22 | 0.71 |
| Plethysmography | Inspiratory time (metacholine) | Hour | 1946 | — | — | 7.07 | 18.98 |
| Plethysmography | Inspiratory time (metacholine) | Month | 1946 | 5.41 | 1.64 | — | — |
| Plethysmography | Inspiratory time (metacholine) | Season | 1946 | 4.58 | 0.86 | — | — |
| Plethysmography | Inspiratory time (metacholine) | Sex | 1946 | 20.64 | 3.33 | — | — |
| Plethysmography | Inspiratory time (metacholine) | Weight | 1946 | 6.35 | 0.93 | — | — |
| Plethysmography | PenH difference | Age | 1934 | — | — | 7.34 | 1.63 |
| Plethysmography | PenH difference | Cage density | 1934 | — | — | 5.67 | 9.31 |
| Plethysmography | PenH difference | Hour | 1934 | — | — | 19.08 | 35.57 |
| Plethysmography | PenH difference | Litter | 1922 | — | — | 9.53 | 18.03 |
| Plethysmography | PenH difference | Month | 1934 | 8.20 | 2.32 | 21.79 | 42.75 |
| Plethysmography | PenH difference | Season | 1934 | 5.00 | 1.00 | 17.35 | 42.74 |
| Plethysmography | PenH difference | Sex | 1934 | 14.92 | 2.52 | 12.02 | 29.38 |
| Plethysmography | PenH difference | Weight | 1934 | — | — | 6.27 | 0.60 |
| Plethysmography | PenH difference | Year | 1934 | — | — | 10.72 | 58.13 |
| Plethysmography | Respiratory rate (baseline) | Cage density | 2163 | 4.82 | 0.69 | — | — |
| Plethysmography | Respiratory rate (baseline) | Hour | 2163 | — | — | 18.19 | 33.50 |
| Plethysmography | Respiratory rate (baseline) | Litter | 2151 | — | — | 11.93 | 12.53 |
| Plethysmography | Respiratory rate (baseline) | Month | 2163 | 21.39 | 4.60 | 10.40 | 24.62 |
| Plethysmography | Respiratory rate (baseline) | Season | 2163 | 16.14 | 2.84 | 9.65 | 26.40 |
| Plethysmography | Respiratory rate (baseline) | Sex | 2163 | 6.22 | 0.92 | 11.01 | 27.80 |
| Plethysmography | Respiratory rate (baseline) | Study day | 2163 | — | — | 6.20 | 0.04 |
| Plethysmography | Respiratory rate (baseline) | Weight | 2163 | — | — | 6.07 | 0.57 |
| Plethysmography | Respiratory rate (baseline) | Year | 2163 | 6.34 | 0.94 | — | — |
| Plethysmography | Respiratory rate (metacholine) | Hour | 1928 | — | — | 5.87 | 17.38 |
| Plethysmography | Respiratory rate (metacholine) | Season | 1928 | 5.21 | 1.08 | — | — |
| Plethysmography | Tidal minute volume (baseline) | Age | 2158 | — | — | 5.04 | 0.68 |
| Plethysmography | Tidal minute volume (baseline) | Cage density | 2158 | 7.51 | 0.89 | — | — |
| Plethysmography | Tidal minute volume (baseline) | Hour | 2158 | — | — | 11.54 | 19.48 |
| Plethysmography | Tidal minute volume (baseline) | Litter | 2146 | — | — | 6.21 | 7.69 |
| Plethysmography | Tidal minute volume (baseline) | Month | 2158 | 13.49 | 2.54 | 12.97 | 20.28 |
| Plethysmography | Tidal minute volume (baseline) | Season | 2158 | — | — | 8.70 | 19.41 |
| Plethysmography | Tidal minute volume (baseline) | Sex | 2158 | 64.43 | 9.12 | — | — |
| Plethysmography | Tidal minute volume (baseline) | Weight | 2158 | 68.90 | 9.81 | — | — |
| Plethysmography | Tidal minute volume (metacholine) | Hour | 1930 | — | — | 6.17 | 11.81 |
| Plethysmography | Tidal minute volume (metacholine) | Litter | 1918 | — | — | 5.61 | 5.33 |
| Plethysmography | Tidal minute volume (metacholine) | Month | 1930 | 18.51 | 3.03 | 4.64 | 10.60 |
| Plethysmography | Tidal minute volume (metacholine) | Season | 1930 | 10.81 | 1.45 | — | — |
| Plethysmography | Tidal minute volume (metacholine) | Sex | 1930 | 105.05 | 14.76 | 5.95 | 12.00 |
| Plethysmography | Tidal minute volume (metacholine) | Weight | 1930 | 71.25 | 9.55 | 5.82 | 0.35 |
| Plethysmography | Tidal volume (baseline) | Age | 2149 | — | — | 16.84 | 1.43 |
| Plethysmography | Tidal volume (baseline) | Cage density | 2149 | — | — | 6.01 | 4.08 |
| Plethysmography | Tidal volume (baseline) | Hour | 2149 | — | — | 20.24 | 22.32 |
| Plethysmography | Tidal volume (baseline) | Litter | 2137 | — | — | 16.96 | 16.06 |
| Plethysmography | Tidal volume (baseline) | Month | 2149 | 39.79 | 5.07 | 20.32 | 22.38 |
| Plethysmography | Tidal volume (baseline) | Season | 2149 | 20.90 | 2.41 | 10.96 | 20.87 |
| Plethysmography | Tidal volume (baseline) | Sex | 2149 | 131.89 | 16.98 | 7.75 | 13.06 |
| Plethysmography | Tidal volume (baseline) | Study day | 2149 | — | — | 7.27 | 0.03 |
| Plethysmography | Tidal volume (baseline) | Weight | 2149 | 87.94 | 10.73 | 6.12 | 0.32 |
| Plethysmography | Tidal volume (metacholine) | Age | 1932 | — | — | 9.74 | 0.80 |
| Plethysmography | Tidal volume (metacholine) | Hour | 1932 | — | — | 10.72 | 15.83 |
| Plethysmography | Tidal volume (metacholine) | Litter | 1920 | — | — | 8.86 | 7.08 |
| Plethysmography | Tidal volume (metacholine) | Month | 1932 | 26.81 | 3.64 | 5.22 | 10.32 |
| Plethysmography | Tidal volume (metacholine) | Season | 1932 | 18.02 | 2.07 | — | — |
| Plethysmography | Tidal volume (metacholine) | Sex | 1932 | 141.45 | 18.43 | 6.03 | 11.35 |
| Plethysmography | Tidal volume (metacholine) | Weight | 1932 | 85.28 | 10.30 | — | — |
| Plethysmography | Tidal volume (metacholine) | Year | 1932 | 6.15 | 0.59 | — | — |
| Weight | Body mass index | Age | 1925 | 5.87 | 0.79 | — | — |
| Weight | Body mass index | Month | 1925 | 8.83 | 2.16 | 7.41 | 16.75 |
| Weight | Body mass index | Season | 1925 | 6.21 | 1.07 | 9.93 | 21.20 |
| Weight | Body mass index | Sex | 1925 | 113.93 | 20.32 | — | — |
| Weight | Body mass index | Weight | 1925 | 19.91 | 3.03 | — | — |
| Weight | Weight, 10 wk (g) | Cage density | 2319 | — | — | 5.37 | 3.29 |
| Weight | Weight, 10 wk (g) | Litter | 2307 | — | — | 10.27 | 6.10 |
| Weight | Weight, 10 wk (g) | Sex | 2320 | Inf | 41.37 | — | — |
| Weight | Weight, 6 wk (g) | Cage density | 2432 | 5.57 | 0.32 | 20.89 | 9.30 |
| Weight | Weight, 6 wk (g) | Litter | 2498 | — | — | 39.27 | 16.69 |
| Weight | Weight, 6 wk (g) | Sex | 2511 | Inf | 30.63 | 12.63 | 12.87 |
| Weight | Weight, 7 wk (g) | Cage density | 2405 | 5.04 | 0.26 | 8.22 | 4.40 |
| Weight | Weight, 7 wk (g) | Litter | 2457 | 6.16 | 0.32 | 21.11 | 7.79 |
| Weight | Weight, 7 wk (g) | Sex | 2470 | Inf | 35.40 | 7.13 | 8.27 |
| Weight | Weight, 8 wk (g) | Litter | 2290 | — | — | 10.46 | 2.53 |
| Weight | Weight, 8 wk (g) | Sex | 2302 | Inf | 44.52 | — | — |
| Wound healing | Ear hole area (mm2) | Cage density | 2185 | 10.45 | 1.37 | 9.10 | 9.56 |
| Wound healing | Ear hole area (mm2) | Litter | 2172 | 6.00 | 0.76 | 9.72 | 10.59 |
| Wound healing | Ear hole area (mm2) | Month | 2185 | 24.17 | 4.34 | 9.94 | 18.33 |
| Wound healing | Ear hole area (mm2) | Season | 2185 | 17.81 | 2.67 | 6.08 | 14.33 |
| Wound healing | Ear hole area (mm2) | Sex | 2185 | 13.93 | 1.91 | 4.87 | 12.85 |
| Wound healing | Ear hole area (mm2) | Study day | 2185 | — | — | 9.67 | 0.04 |
| Wound healing | Ear hole area (mm2) | Year | 2185 | — | — | 6.42 | 32.64 |
Note that not all combinations of phenotype and covariate were available in the study. Log P denotes the logarithm to the base 10 of the P-value. Inf denotes a P-value that was computationally indistinguishable from zero. For brevity, results are omitted for effects with log P's of <4.55 (i.e., the corrected 5% significance level).
We estimated the significances and effects of the remaining covariates by adding each to a model that already included family, sex, and weight. Significant main effects of covariates were more common in physiological than behavioral phenotypes (33% of the time vs. 13%; see Table 6). Overall, 21 of the 258 significant effects explained >10% of the variance; the five cases of when a covariate explained >25% of the variance involved sex. Table 6 provides a summary for each covariate, splitting results by category of phenotype. Figure 1 plots log P-values and the percentage of phenotypic variance explained by significant covariates. Figure 2 summarizes the variance explained by significant covariates for the 16 subcategories of phenotype.
TABLE 6.
Summary of main effects
| Physiological phenotypes
|
Behavioral phenotypes
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Covariate | Median log P | Mean % variance | SD | No. observed (significant/all) | Median log P | Mean % variance | SD | No. observed (significant/all) |
| Age | 0.82 | 0.98 | 0.40 | 6/65 | 0.73 | 0.86 | 0.17 | 3/18 |
| Apparatus | — | — | — | — | 31.59 | 7.80 | 3.47 | 4/5 |
| Cage density | 1.01 | 0.68 | 0.34 | 9/70 | 0.84 | — | — | 0/18 |
| Experimenter | 2.04 | 3.30 | 2.44 | 7/25 | 2.50 | 1.80 | 0.56 | 6/20 |
| Hour | 1.55 | 1.16 | — | 1/29 | 1.51 | 1.41 | — | 1/20 |
| Litter | 0.97 | 0.90 | 0.31 | 9/70 | 0.88 | — | — | 0/18 |
| Month | 8.96 | 3.56 | 2.24 | 51/65 | 2.14 | 1.75 | — | 1/18 |
| Season | 5.47 | 1.90 | 1.25 | 38/65 | 1.57 | — | — | 0/18 |
| Sex | 12.41 | 9.47 | 11.62 | 48/70 | 2.06 | 2.19 | 1.79 | 5/18 |
| Study day | 1.00 | 2.03 | 1.60 | 15/65 | 1.27 | 0.73 | — | 1/18 |
| Test order | 0.37 | — | — | 0/25 | 0.72 | — | — | 0/16 |
| Weight | 2.92 | 3.06 | 3.89 | 27/65 | 2.06 | 0.90 | 0.23 | 6/18 |
| Year | 1.87 | 1.30 | 0.71 | 19/65 | 1.61 | 0.85 | — | 1/18 |
Variances (means and standard deviations) refer only to effects that were significant at log P > 4.55.
Figure 2.—
Main and interaction effects of covariates on 88 phenotypes from 16 experimental tests. The y-axis gives the percentage variance explained by significant covariates; the x-axis lists the test performed with the number of phenotypes measured from that test in parentheses. Physiological tests are listed first and behavioral tests second. Boxes show the median (central line) and interquartile range (IQR; box perimeter), whiskers indicate the furthest data point <1.58 IQRs from the median, and circles show outliers.
We then extended our model to test for gene-by-covariate interactions, taking the main-effects models reported above and then assessing how much adding interaction terms improved the fit. We found 389 significant interaction effects. Figure 3 illustrates the interaction between sex and family on the percentage of B-cells (%B220+) among white blood cells. It shows that the effect of sex is often marked within families but its direction can vary between families. Similarly, Figure 4 illustrates the interaction between family and season on mean adrenal weight measured at 10 weeks. It shows seasonal means (spring in green, summer in red, autumn in brown, and winter in blue) for 28 families. In 11 families, adrenal glands are heaviest when harvested in winter, whereas in 9 families they are heaviest in summer. The seasonal effects are strong within but inconsistent across families, reflecting the greater importance of interaction over main effects.
Figure 3.—
Interaction between sex and family for the immunological phenotype percentage of B-cells among lymphocytes in 2056 mice. For each of 69 families (x-axis) we plot means (solid circles) and standard errors (bars) of the phenotype value for males (blue) and females (pink). The y-axis gives the phenotype as the square root of the percentage of white blood cells presenting B220. The graph shows that sex can have a strong effect within families but that the direction of the effect varies between families (interaction log P = 10.7). For example, in families plotted on the left, males are enriched in the B-cell compared with females, whereas for families on the right this sex effect is reversed. The graph also illustrates the marginal effects on the trait of family (differing overall heights; heritability = 59.9%) and sex (females higher overall; main effect log P = 13.0).
Figure 4.—
Interaction between season and family for the physiological phenotype mean adrenal weight in 696 mice. For each of 28 families (x-axis) we plot the seasonal means (solid circles) and standard errors (bars) of the phenotype for animals phenotyped in winter (blue), spring (green), summer (red), and autumn (brown). The y-axis gives the phenotype as the logarithm to the base 10 of the mean weight in grams of adrenal glands at 10 weeks old. The graph shows that the effect of season is consistent within family but can vary between families. For example, for the rightmost family adrenal glands are lightest in animals tested in summer and heaviest in autumn. Yet the rank order of seasons varies considerably through the graph.
The distribution of the 389 significant interaction effects differed from that of the main effects (Figure 1 and appendix). Remarkably, half of the effects could explain >20% of the variance. In nine cases the interaction could explain >50% of the variance. The largest numbers of interactions were with month (65 significant effects), season (55), sex (53), litter (51), and cage density (40). There were only 13 significant interactions with experimenter.
Physiological phenotypes showed the largest number of interactions with covariates (56% of interactions tested were significant; Table 7). Largest effects were found on mean cellular hemoglobin concentration, serum sodium and serum chloride concentrations, and plethysmography measures. There were fewer interactions with behavioral phenotypes (5% of interactions tested were significant, amounting to 11 in total), although the effect sizes were much the same on average (mean of 18.1% for behavior compared with a mean of 18.6% for physiology; see Figure 2).
TABLE 7.
Summary of interaction effects between covariates and family
| Physiological phenotypes
|
Behavioral phenotypes
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Covariate | Median log P | Mean % variance | SD | No. observed (significant/all) | Median log P | Mean % variance | SD | No. observed (significant/all) |
| Age | 2.39 | 1.22 | 0.59 | 26/65 | 0.25 | — | — | 0/18 |
| Apparatus | — | — | — | — | 0.00 | — | — | 0/5 |
| Cage density | 5.05 | 10.80 | 4.45 | 40/70 | 0.37 | — | — | 0/18 |
| Experimenter | 4.28 | 26.41 | 6.89 | 11/25 | 2.35 | 16.65 | 1.62 | 2/20 |
| Hour | 6.17 | 23.69 | 7.78 | 21/29 | 1.96 | — | — | 0/20 |
| Litter | 9.17 | 13.58 | 7.21 | 51/70 | 0.53 | — | — | 0/18 |
| Month | 11.84 | 29.94 | 11.63 | 60/65 | 3.87 | 18.49 | 3.50 | 5/18 |
| Season | 8.04 | 29.24 | 11.66 | 52/65 | 2.25 | 18.94 | 3.92 | 3/18 |
| Sex | 6.82 | 22.33 | 6.66 | 52/70 | 2.21 | 16.38 | — | 1/18 |
| Study day | 2.86 | 0.03 | 0.02 | 22/65 | 0.39 | — | — | 0/18 |
| Test order | 0.21 | — | — | 0/25 | 0.00 | — | — | 0/16 |
| Weight | 3.88 | 0.59 | 0.14 | 28/65 | 1.39 | — | — | 0/18 |
| Year | 2.09 | 39.77 | 15.72 | 15/65 | 0.69 | — | — | 0/18 |
Variances (means and standard deviations) refer only to effects that were significant at log P > 4.55.
DISCUSSION
We have carried out the first systematic analysis of a range of covariates across multiple phenotypes (see appendix). We have estimated the heritability of 88 phenotypes, assessed the impact of a number of environmental factors, and measured the size of gene-by-environment interactions. Our large data set provides the most robust assessments to date of these measures in both behavioral and physiological domains.
We found large interactions between gene and environment and report that the effects are not restricted to behavioral phenotypes (see appendix). We do not believe this is an artifact of our analysis. Our calculations of percentage variance for random interaction effects and for fixed main effects are only roughly comparable with each other (see methods) and the interaction effects are subject to a slight upward bias. However, that is not sufficient to account for the substantially higher effect of significant interactions (18.6%) compared with significant main effects (3.7%). Second, inhomogeneity of phenotype variance across families is also unlikely to account for our findings since in many cases the rank order of covariate effects differs between families (Ungerer et al. 2003) as illustrated in Figures 3 and 4.
We report the effects of covariates as the percentage of phenotypic variance they explain and in doing so provide one assessment of how environmental covariates influence a phenotype. But the true nature of this interaction is more complex. For example, the concentration of alanine transaminase is subject to gene-by-environment interactions of month, accounting for 48.49% of the phenotypic variance, of season, accounting for 45.51%, and of litter, accounting for 18.17%. Yet these effects combine, with further covariates, to produce 100%. How is this possible?
The correlational structure of our data complicates an assessment of the relative importance of different covariates and interactions. The observed phenotypic variance is the sum of the variances of the covariates minus twice the covariances between the covariates. This means that two covariates could have individual effects of 50% but a summed effect of 60% if they are positively correlated (or one of <50% if they are negatively correlated). An observed covariate effect, just like an observed QTL effect, therefore includes a portion of the effect of any element that correlates with it; an actual month effect will partly manifest as observed litter and season effects and vice versa. A more comprehensive analysis would build a complete picture of each phenotype in the context of a path diagram or structural equation model that enumerated all relationships, both raw effects and correlations, between actors (e.g., Lynch and Walsh 1998).
The importance of gene-by-environment interactions has been emphasized in the analysis of mouse behavior and largely ignored in studies of mouse physiology. In the light of this, we designed our phenotyping protocol to minimize the effects of covariates on behavioral measures. All such tests were automated, so that the experimenter's intervention was limited to placing animals in the apparatus. This may explain why some covariates, previously suspected to influence behavioral phenotypes, were found to make a small contribution to the variance: time of day (hour) was a nonsignificant (or hardly significant with negligible effect) contributor to all measures including those that utilize exploration as a measure of anxiety (elevated plus maze, which had observations from 9 different hours of the day, and open field, which had observations from 10), despite the fact that exploratory activity has been reported to vary throughout the day (Aschoff 1981). The order in which animals are tested is also considered to have an important effect on behavior (Harro 1997), but we found no evidence for this: its effect was nonsignificant on all phenotypes measured.
Physiological phenotypes were not so controlled. There are no automated ways of administering an intraperitoneal glucose tolerance test, for example, and we observed large experimenter effects on these tests. This raises the question as to whether some phenotypes are more susceptible to interaction effects than others. Differences in the assessment protocols cannot be the only factor that accounts for the smaller number of interactions in behavioral tests. There are a number of covariates common to all phenotypes whose effects we could not ameliorate: month, season, year, sex, and weight. All of these covariates impinge more on physiological than on behavioral phenotypes (Tables 6 and 7).
Importantly, we observed many significant and large gene-by-environment interactions in our analysis of physiological phenotypes. Biochemical measures showed strong (>10% effect) gene-by-environment interactions with month (in 14 of 16 biochemical phenotypes), sex (12), season (9), and litter (8). We saw a similar pattern of strong seasonal and sex effects for hematology, immunology, plethysmography (which also had a strong hour interaction), and the glucose tolerance test (which also had a strong experimenter interaction). This has profound implications for QTL studies.
QTL detection experiments suffer when covariates are not adequately accommodated in the experimental design and subsequent analysis. First, a QTL may owe some, or indeed all, of its significance to an environmental effect confounded with the allelic variant. When a phenotype is strongly affected by who performed the experiment, any nonfunctional variant that correlates with the experimenter will manifest as a significant, but spurious, effect. The random nature of recombination means that in any experimental cross a fully balanced design is impossible and so confounds of this type are ineluctable. While the impact of covariates can be minimized by regressing out their effects prior to mapping (e.g., Valdar et al. 2006), this is highly conservative, since in the converse scenario, where experimenter acts as a surrogate variable for an actual QTL effect, the QTL will be missed.
Second, an interaction between a QTL and an environmental covariate may conceal the effect of both, even when covariate and QTL are in the model. For instance, if mice with allele a fear experimenter John more than experimenter Alice, but mice with allele A fear Alice more than John and all four conditions occur in about equal proportion, then neither experimenter nor QTL will have an observed effect. To recover the genetic effect in this case it is necessary to model the interaction in the mapping procedure (e.g., Wang et al. 2006).
Our analyses are limited by the relatively small number of covariates that we collected. We have no information on temperature fluctuation and humidity levels [shown to be important for behavioral tests of nociception (Chesler et al. 2002a,b)], which might explain month and seasonal effects. We have no information on noise levels that are significantly increased during working hours (Milligan et al. 1993). The predominance of significant temporal covariates reflects the importance of many other unknown environmental factors whose effect is moderated through the animals' genotypes. Thus the dissection of complex phenotypes in the mouse will require far more sophisticated observation and analysis of these interactions than has hitherto been attempted.
Acknowledgments
W.V. gratefully acknowledges receipt of an Access to Research Infrastructures fellowship under Örjan Carlborg, Uppsala University, Sweden, and additionally thanks Mike Neale, Tom Price, and Peter Visscher for helpful discussions. This work was funded by grants from the Wellcome Trust and the European Union Framework 6 Programme, contract no. LHSG-CT-2003-503265.
References
- Aschoff, J., 1981. Handbook of Behavioral Neurobiology: Vol. 4. Biological Rhythms. Plenum Press, New York/London.
- Biddinger, S. B., K. Almind, M. Miyazaki, E. Kokkotou, J. M. Ntambi et al., 2005. Effects of diet and genetic background on sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase 1, and the development of the metabolic syndrome. Diabetes 54: 1314–1323. [DOI] [PubMed] [Google Scholar]
- Blom, G., 1958. Statistical Elements and Transformed Beta Variables. Wiley, New York.
- Brown, S. D., P. Chambon and M. H. de Angelis, 2005. EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat. Genet. 37: 1155. [DOI] [PubMed] [Google Scholar]
- Cabib, S., C. Orsini, M. Le Moal and P. V. Piazza, 2000. Abolition and reversal of strain differences in behavioral responses to drugs of abuse after a brief experience. Science 289: 463–465. [DOI] [PubMed] [Google Scholar]
- Champy, M. F., M. Selloum, L. Piard, V. Zeitler, C. Caradec et al., 2004. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm. Genome 15: 768–783. [DOI] [PubMed] [Google Scholar]
- Chesler, E. J., S. G. Wilson, W. R. Lariviere, S. L. Rodriguez-Zas and J. S. Mogil, 2002. a Identification and ranking of genetic and laboratory environment factors influencing a behavioral trait, thermal nociception, via computational analysis of a large data archive. Neurosci. Biobehav. Rev. 26: 907–923. [DOI] [PubMed] [Google Scholar]
- Chesler, E. J., S. G. Wilson, W. R. Lariviere, S. L. Rodriguez-Zas and J. S. Mogil, 2002. b Influences of laboratory environment on behavior. Nat. Neurosci. 5: 1101–1102. [DOI] [PubMed] [Google Scholar]
- Clifford, D., and P. McCullagh, 2005. regress: Gaussian linear models with linear covariance structure. R package version 0.4 (http://galton.uchicago.edu/∼clifford).
- Cooper, R. M., and J. P. Zubek, 1958. Effects of enriched and restricted early environments on the learning ability of bright and dull rats. Can. J. Psychol. 12: 159–164. [DOI] [PubMed] [Google Scholar]
- Crabbe, J. C., D. Wahlsten and B. C. Dudek, 1999. Genetics of mouse behavior: interactions with laboratory environment. Science 284: 1670–1672. [DOI] [PubMed] [Google Scholar]
- Demarest, K., J. Koyner, J. McCaughran, Jr., L. Cipp and R. Hitzemann, 2001. Further characterization and high-resolution mapping of quantitative trait loci for ethanol-induced locomotor activity. Behav. Genet. 31: 79–91. [DOI] [PubMed] [Google Scholar]
- Ehrich, T. H., T. Hrbek, J. P. Kenney-Hunt, L. S. Pletscher, B. Wang et al., 2005. Fine-mapping gene-by-diet interactions on chromosome 13 in a LG/J × SM/J murine model of obesity. Diabetes 54: 1863–1872. [DOI] [PubMed] [Google Scholar]
- Harro, J., 1997. Measurements of exploratory behavior in rodents, pp. 359–377 in Methods in Neuroscience, edited by P. M. Conn. Academic Press, New York.
- Henderson, N. D., 1970. Genetic influences on the behavior of mice can be obscured by laboratory rearing. J. Comp. Physiol. Psychol. 72: 505–511. [DOI] [PubMed] [Google Scholar]
- Kafkafi, N., Y. Benjamini, A. Sakov, G. I. Elmer and I. Golani, 2005. Genotype-environment interactions in mouse behavior: a way out of the problem. Proc. Natl. Acad. Sci. USA 102: 4619–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Milligan, S. R., G. D. Sales and K. Khirnykh, 1993. Sound levels in rooms housing laboratory animals: an uncontrolled daily variable. Physiol. Behav. 53: 1067–1076. [DOI] [PubMed] [Google Scholar]
- Nyberg, J., K. Sandnabba, L. Schalkwyk and F. Sluyter, 2004. Genetic and environmental (inter)actions in male mouse lines selected for aggressive and nonaggressive behavior. Genes Brain Behav. 3: 101–109. [DOI] [PubMed] [Google Scholar]
- Olejnik, S., and J. Algina, 2003. Generalized eta and omega squared statistics: measures of effect size for some common research designs. Psychol. Methods 8: 434–447. [DOI] [PubMed] [Google Scholar]
- Pinheiro, J. C., and D. M. Bates, 2000. Mixed Effects Models in S and S-PLUS. Springer-Verlag, New York.
- R Development Core Team, 2004. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna.
- Reifsnyder, P. C., G. Churchill and E. H. Leiter, 2000. Maternal environment and genotype interact to establish diabesity in mice. Genome Res. 10: 1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, H., and M. Ageel, 2000. Analysis of Variance: Fixed, Random and Mixed Models. Birkhauser, Boston.
- Self, S. G., and K. Y. Liang, 1987. Asymptotic properties of maximum likelihood estimators and likelihood ratio tests under nonstandard conditions. J. Am. Stat. Assoc. 82: 605–610. [Google Scholar]
- Solberg, L. C., W. Valdar, D. Gauguier, G. Nunez, A. Taylor et al., 2006. A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm. Genome 17: 129–146. [DOI] [PubMed] [Google Scholar]
- Ungerer, M. C., S. S. Halldorsdottir, M. D. Purugganan and T. F. Mackay, 2003. Genotype-environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics 165: 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar, W., L. C. Solberg, D. Gauguier, S. Burnett, P. Klenerman et al., 2006. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat. Genet. 38: 879–887. [DOI] [PubMed] [Google Scholar]
- Venables, W. N., and B. D. Ripley, 2002. Modern Applied Statistics. Springer, New York.
- Wahlsten, D., P. Metten, T. J. Phillips, S. L. Boehm, II, S. Burkhart-Kasch et al., 2003. Different data from different labs: lessons from studies of gene-environment interaction. J. Neurobiol. 54: 283–311. [DOI] [PubMed] [Google Scholar]
- Wang, S., N. Yehya, E. E. Schadt, H. Wang, T. A. Drake et al., 2006. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, B., A. A. Truett, M. P. Monteiro, S. J. Barry, C. H. Warden et al., 1999. Gene-environment interaction: a significant diet-dependent obesity locus demonstrated in a congenic segment on mouse chromosome 7. Mamm. Genome 10: 457–462. [DOI] [PubMed] [Google Scholar]







