Abstract
Somatic loss of tumor suppressor gene function comprising the second hit of Knudson's two-hit hypothesis is important in human cancer. A genetic screen was performed in zebrafish (Danio rerio) to find mutations that cause genomic instability (gin), as scored by Streisinger's mosaic-eye assay that models this second hit. The assay, based on a visible test for loss of wild-type gene function at a single locus, golden, is representative of genomewide events. Twelve ENU-induced genomic instability (gin) mutations were isolated. Most mutations showed weak dominance in heterozygotes and all showed a stronger phenotype in homozygotes. Trans-heterozygosity for 7 of these mutations showed greatly enhanced instability. A variety of spontaneous tumors were found in heterozygous adults from all gin lines, consistent with the expectation that genomic instability (mutator) mutations can accelerate carcinogenesis. The incidence of spontaneous cancer at 30–34 months was increased 9.6-fold in heterozygotes for the mutant with the strongest phenotype, gin-10. Tumors were seen in skin, colon, kidney, liver, pancreas, ovary, testis, and neuronal tissues, with multiple tumors in some fish. The study of these mutants will add to our understanding of the mechanisms of somatic loss of gene function and how those mechanisms contribute to cancer susceptibility.
CANCER is the result of an accumulation of somatic events, including mutations, which affect gene function. Somatic mutations occur spontaneously at a low rate due to the inherent inaccuracy of genomic processing or exposure to endogenous and exogenous mutagens (for a review, see Bertram 2000). According to the mutator hypothesis (Nowell 1976; Loeb et al. 2003), an elevated rate of mutation (mutator phenotype) is required to account for the multiple mutations characteristic of most cancers. The genomic instability associated with a mutator phenotype may result from defects in genome maintenance functions such as DNA repair, DNA replication, recombination, chromosome segregation, and checkpoint control (Cheng and Loeb 1993; Lengauer et al. 1998; Loeb et al. 2003) or in epigenetic regulation (Feinberg et al. 2002).
Genetic screens have advanced our understanding of mutation and its relationship to cancer. For example, work with mutator mutants of bacteria (Cox 1976) and yeast (Kolodner et al. 2002) led to the identification of components of the mutHLS mismatch correction system and genes involved in the human cancer syndrome hereditary nonpolyposis colon cancer (Marra and Boland 1995). Genetic defects in DNA repair and recombination cause autosomal recessive human cancer syndromes such as xeroderma pigmentosum, ataxia telangiectasia, Bloom's syndrome, Fanconi's anemia, and Werner's syndrome (Hoeijmakers 2001; Charames and Bapat 2003). To increase understanding of genomic instability in human cancers and the genes involved and how they relate to cancer progression and susceptibility, it is desirable to pursue mutator screens in diploid organisms that develop cancers.
Knudson's two-hit hypothesis for loss of tumor suppressor gene function in retinoblastoma (Knudson 1971) motivated our screen. In this model, inheritance of one tumor suppressor gene mutation (the first hit) must be followed by loss of function of the remaining wild-type allele (the second hit) to yield tumorigenic clones of cells (Figure 1A). These two hits apply to any cancer involving tumor suppressor genes and can occur by genetic (Cavenee et al. 1983) or epigenetic mechanisms (Feinberg et al. 2002). In spontaneous retinoblastoma, both hits occur after birth. In familial cases, individuals carrying retinoblastoma mutations from birth show enhanced susceptibility to cancer (dominant susceptibility), since only one additional somatic loss of gene function event is needed. Our N-ethyl-N-nitrosourea (ENU) screen for zebrafish genomic instability (gin) mutants models the somatic “second hit” events.
Figure 1.—
(A) Mechanisms of somatic mutation and loss of gene function. When a tumor suppressor locus is heterozygous for a mutation (Knudson's “first hit,” indicated by an “×” on the gray chromosome), loss of gene function (the “second hit”) can occur by multiple mechanisms, including chromosome loss (with or without reduplication), recombination (including simple crossover and gene conversion), independent mutation (including point mutation, deletion, or insertion), or epigenetic mechanisms (including chromatin or DNA-modification-based effects on gene expression). LOH, while often erroneously implied to include all of these mechanisms, occurs only by the first two mechanisms. Adapted from Cavenee et al. (1983). (B) Mutagenesis scheme and screening protocol used for isolating gin mutations. Wild-type zebrafish males were treated with ENU and outcrossed to homozygous golden females. Resulting gin?/+; gol/+ G1 females were screened for gin mutations by scoring for mosaic eyes among half-tetrad parthenogenetic progeny. Once identified, gin/+; gol/+ G1 were outcrossed to homozygous golden males to confirm Mendelian inheritance and to create gin families for further study. (C) Mosaic-eye pigmentation in zebrafish embryos. The eyes of three 72-hr postfertilization (hpf) half-tetrad embryos from a gin-1/+; gol/+ female are shown. The gol/gol embryo has the light golden-brown color characteristic of this pigment mutation. A black wild-type eye is visible in the +/+ or gol/+ sibling. The top embryo with a gin/gin; gol/+ genotype has mosaic-eye pigmentation due to loss of function of the wild-type gol allele in the light cells. (D) EP in female carriers of gin and golden creates half-tetrads with the gin/gin; gol/+ genotype required for the mosaic-eye assay. At the top are two sister-chromatid pairs of a primordial egg cell, after DNA replication, from a gol/+ G1 mother carrying a gin mutation (B). The bottom four egg genotypes result from the indicated combinations of meiosis I crossovers. These egg genotypes are preserved using EP, in which UV inactivation of sperm is used to prevent paternal contribution, and hydraulic pressure prevents the reduction to haploidy normally caused by meiosis II. The black chromosomes are from the mutagenized male founder (B). The gray chromosomes are from the nonmutagenized golden founder female. The chromosomes to the left (illustrated as short only for clarity) represent the linkage group containing the gin gene originally mutated in the founder male. The chromosomes to the right represent the linkage group containing the golden gene (the mutant allele deriving from the founder female). In half-tetrads, genetic markers close to the centromere tend to remain homozygous, due to a low probability of crossovers, while markers close to the telomere, such as golden, tend to become heterozygous from a crossover. The progeny illustrated represent the 89% half-tetrads that are gol/+ (Streisinger et al. 1986); recombination involving gin is not illustrated.
A readily scorable assay was sought to measure genomic instability in somatic tissue. We adopted the mosaic-eye assay (Streisinger 1984), which uses the golden locus to measure somatic loss of gene function. At 2 days postfertilization, wild-type embryos and embryos heterozygous for the null goldenb1 mutation (henceforth golden, or gol) show cell-autonomous black pigmentation in the retinal pigment epithelium (RPE) and somatic melanophores (Streisinger et al. 1989). In contrast, golden homozygotes have a much lighter golden-brown pigmentation. Using the mosaic-eye assay, Streisinger (1984) showed that somatic loss of function of the wild-type allele in golden heterozygotes can be induced by physical or chemical mutagens. Wild-type and golden pigmentation can be readily distinguished in adjacent zebrafish RPE cells. To detect a somatic mutator phenotype, we screened the RPE of golden heterozygotes for lightly pigmented cells, indicating loss of function of the wild-type golden allele in these cells (Figure 1). A half-tetrad screen was used to provide the necessary genetic configuration (Cheng and Moore 1997). The screen yielded 12 gin mutations. At least one causes a striking dominant susceptibility to spontaneous cancer. These results represent, to the best of our knowledge, the first direct genetic screen in a vertebrate for somatic mutators based on a locus-specific assay.
MATERIALS AND METHODS
Stocks:
Outbred wild-type zebrafish were obtained from Ekk Will Waterlife Resources (Gibsonton, FL), Michael Rust (University of Washington), and Lyles Tropical Fish (Ruskin, FL). Wild-type (AB and WIK) and homozygous golden stocks were obtained from Zebrafish International Resource Center (Eugene, OR). Additional wild type (WIK) stocks were a gift from Michael Granato (University of Pennsylvania).
ENU mutagenesis:
Our forward genetic screen for somatic mutators using the mosaic-eye assay required a specific genetic configuration: simultaneous homozygosity for newly induced mutator (gin) mutations and heterozygosity at the target locus (golden). Early pressure parthenogenesis (EP) in zebrafish was used to create this configuration in an efficient manner (Streisinger et al. 1981). EP produces zygotes that are half-tetrads containing the sister-chromatid pairs obtained after meiosis I (Figure 1D). In EP progeny of heterozygous mothers, loci close to centromeres tend to be homozygous and loci far from centromeres tend to be heterozygous (Streisinger et al. 1986). In our screen, we generated females heterozygous for both the golden mutation and a newly induced potential gin mutation and scored their gol/+ EP offspring for somatic loss of the wild-type golden allele. The vast majority (89%) of the EP offspring of these heterozygous golden mothers are gol/+, consistent with the finding that golden lies close to a telomere (Streisinger et al. 1986; Gestl et al. 1996; Lamason et al. 2005). Somatic loss of a single wild-type golden allele cannot be scored in the remaining 11% of embryos because half are homozygous golden and half homozygous are wild type. Assuming that mutator alleles may be recessive and that all gol/+ embryos homozygous for a mutator allele (*) will produce mosaic eyes, we can calculate the probability that some EP progeny will be gol/+; */*, and thus scorable in our assay. A Poisson calculation indicates that screening 75 or 40 EP embryos would yield a 98 or 87% chance, respectively, of detecting a newly induced telomeric gin mutation that would be homozygous among 5% of the EP progeny (Cheng and Moore 1997). Mutator alleles at more centromere-proximal locations could be readily detected with screening of even fewer EP progeny.
Thirty 6- to 9-month-old wild-type zebrafish males (from Ekk Will and Rust) were chosen for mutagenesis by their ability to fertilize clutches of 50 or more eggs (Moore et al. 2004). ENU (Sigma, St. Louis) was dissolved in 0.03% Instant Ocean solution (Aquarium Systems, Mentor, OH) in 10 mm sodium phosphate, pH 6.5, as described in Solnica-Krezel et al. (1994), except that ENU was dissolved by crushing and stirring and used within 1 hr. Fish were mutagenized at 25° for 1 hr every 3 days, for a total of three treatments (Mullins et al. 1994).
Six fish were buffer-only controls, 12 were treated with 2.5 mm ENU, and 12 with 3.0 mm ENU. Mutagenized males were bred weekly for 2 weeks to eliminate mature sperm exposed to ENU that would otherwise cause mosaic embryos representing strand-specific mutations in the golden gene rather than genomic instability (Grunwald and Streisinger 1992). Males mutagenized with 3.0 mm ENU produced more abnormally developing embryos than did the 2.5 mm ENU-treated males. Mutagenized males were bred every 1 to 2 weeks with golden homozygous females to yield G1 potential carriers (Figure 1B).
To measure the effectiveness of ENU mutagenesis, all 48-hr-old embryos fathered by the mutagenized males were examined and scored as either golden (gol/gol*; due to a new ENU-induced golden allele, gol*) or wild type (gol/+). No golden embryos were seen in the 1460 offspring of the 0.0 mm ENU control group of zebrafish males, indicating a frequency of spontaneous mutation at this locus of <0.07%. Two golden embryos of 1818 produced by two separate 2.5 mm males and five golden embryos of 1905 from four separate 3.0 mm males represent 0.1 and 0.3% specific-locus mutation frequencies, respectively. No mosaic-eye embryos were observed, consistent with the expectation that the ENU-induced golden mutations detected were fixed on both DNA strands of the sperm.
Screening for mutations:
Early-pressure parthenogenesis (Streisinger et al. 1981; Westerfield 1995; Gestl et al. 1996) was performed on eggs from gin?/+; gol/+ G1 females (Figure 1B). Embryos were maintained at ∼28° and scored at 48 and 72 hr for mosaic-eye pigmentation (Moore et al. 2004). At later stages, RPE mosaicism becomes obscured by the opacity of scleral iridophores and choroidal melanocytes. When scoring gin embryos for mosaicism, golden pigmentation can be scored with confidence in as few as two cells in one eye, although both eyes can be affected (Figure 2). Mosaic embryos were categorized as having only a single patch or multiple patches of lightly pigmented RPE cells. Pigmentation of the melanophores on the trunk and tail of the embryo was not used to score mosaicism, due to normal variation in number and position. For storage, mosaic embryos were manually dechorionated, euthanized, fixed in 4% paraformaldehyde (PFA), and stored in 100% methanol, following a protocol for in situ hybridization (Westerfield 1995).
Figure 2.—
A range of mosaic-eye phenotypes is associated with gin mutations. The strength of the mosaic-eye phenotype can vary from unilateral mosaics, here with three cells that have lost the wild-type golden allele (A), to eyes that are almost all golden with only a few wild-type cells present (B). Mosaicism can also occur bilaterally, where each patch of golden cells may indicate an independent somatic mutation event (C). Mosaic eyes were also found in gin-2/gin-2; gol/+; albino/+ (D) and gin-5/gin-5; gol/+; pig-5/+ (E) embryos. Loss-of-function mosaic patches were determined to be in albino and pig-5, respectively, because the light patches did not darken to golden brown on day 4. The shadows in the top right of A and the bottom left of B are caused by the contralateral eye.
G1 females were identified as gin/+ on the basis of their ability to produce parthenogenetic offspring with mosaic eyes and outcrossed to golden homozygotes (Figure 1B). G2 and later gin carriers were identified by either EP (Table 1) or crosses with known carriers. Carriers were outcrossed as follows: golden homozygotes were outcrossed to either wild type (AB), wild type (Lyle), or wild type (WIK); golden heterozygotes were usually outcrossed to homozygous golden stocks.
TABLE 1.
Mosaicism among half-tetrad progeny of gin/+; gol/+ females
| Mutation (no. of females screened) | No. of gol/+ embryos scoreda | % single-patch mosaics | % multiple-patch mosaics | Total % mosaics |
|---|---|---|---|---|
| gin-1 (8) | 1,067 | 0.4 | 1.6 | 2.0 |
| gin-2 (17) | 2,290 | 0.8 | 1.4 | 2.2 |
| gin-3 (6) | 768 | 0.5 | 1.4 | 1.9 |
| gin-4 (13) | 1,712 | 0.6 | 1.2 | 1.8 |
| gin-5 (6) | 1,090b | 0.4 | 1.7 | 2.1 |
| gin-6 (4) | 448 | 0 | 1.3 | 1.3 |
| gin-7 (12) | 1,442 | 0.7 | 1.2 | 1.8 |
| gin-8 (10) | 947 | 0.7 | 1.6 | 2.3 |
| gin-9 (9) | 740 | 0.8 | 1.9 | 2.7 |
| gin-10 (10) | 868 | 2.3 | 2.0 | 4.3 |
| gin-11 (2) | 357 | 0 | 0.6 | 0.6 |
| gin-12 (9) | 946 | 1.6 | 2.1 | 3.7 |
| Total | 12,762 | 0.8 | 1.5 | 2.3 |
The first column includes the founder female and female progeny from subsequent generations identified as carriers. Females were scored as being a carrier if they had any EP progeny with mosaic eyes.
Calculated number of gol/+ embryos after adjustment for the frequency of gol heterozygosity after EP. EP treatment of eggs from a gol/+ mother produces 89% gol/+, 5.5% +/+, and 5.5% gol/gol embryos (Streisinger et al. 1986). We multiplied the number of dark embryos by 89/(89 + 5.5) = 0.942 to calculate the number of gol/+ embryos.
This number does not include the 12 mosaic/256 dark embryos (4.7%) scored from a gin/+; gol/+female also heterozygous for an ENU-induced pigmentation mutation (pig-5). The number of eggs scored could not be adjusted for the frequency of pigmentation heterozygosity after EP because the map position of the mutation is unknown.
We screened 49 clutches of at least 40 half-tetrad embryos each, yielding 9 of the 12 gin mutations. Decreased fertility was observed in daughters of males treated at the higher ENU dose, resulting in fewer of their offspring being screened. Of the 49 clutches, 36 were descendants of males treated with 2.5 mm ENU and 13 were descended from 3.0 mm ENU-treated males. The remaining three gin mutations were found in smaller clutches derived from 2.5 mm ENU-treated males. The mutations were derived from eight different ENU-mutagenized males, all but one from the 2.5 mm dose. The gin-4 and gin-5 mutations were recovered from one ENU-treated male; gin-8, gin-9, and gin-11 were derived from another.
Genetic interactions:
Trans-heterozygotes were generated to detect potential interactions between different gin mutations by crossing pairs of carriers (Table 2). Most of these gin carrier crosses were between golden heterozygotes and golden homozygotes, ensuring that all darkly pigmented embryos would be gol/+, and thus scorable in the mosaic-eye assay. For some crosses, only golden heterozygotes were available. Since only two-thirds of the darkly pigmented embryos from these crosses are expected to be golden heterozygotes, all embryos with wild-type pigmentation were scored, and only two-thirds of that number was used as the number scored. In cases where both gin carriers were heterozygous at golden, the incidence of mosaics was not significantly different from that seen in crosses between gol heterozygotes and gol homozygotes (0.5 > P > 0.1, by χ2 and G-test).
TABLE 2.
Frequency of mosaic-eye embryos from outcrosses and intercrosses
| +/+ (35503) | gin-1/+ (5749) | gin-2/+ (7305) | gin-3/+ (6448) | gin-4/+ (6779) | gin-5/+ (9681) | gin-6/+ (3854) | gin-7/+ (7310) | gin-8/+ (3847) | gin-9/+ (7043) | gin-10/+ (14166) | gin-11/+ (4718) | gin-12/+ (8137) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gin-1/+ | 0.2 + 0 = 0.2% (2564) | 0.7 + 0.3 =1% (591) | |||||||||||
| gin-2/+ | 0.2 + 0 = 0.2% (2622) | 0 + 0 = <1% (83) | 1 + 1 = 2% (495) | ||||||||||
| gin-3/+ | 0 + 0 = <0.03% (3171) | 0.5 + 0 = 0.5% (419) | 0 + 0 = <0.4% (231) | 0.4 + 0.3 =0.7% (1209) | |||||||||
| gin-4/+ | 0.2 + 0 = 0.2% (4224) | 0 + 0 = <0.7% (146) | 0 + 0 = <1% (95) | 0 + 0 = <0.7% (144) | 0.3 + 0.4 = 0.7% (966) | ||||||||
| gin-5/+ | 0.7 + 0.1 =0.8% (3039) | 0.4 + 0 = 0.4% (278) | 1.7 + 1.3 =3.0% (824) | 0.6 + 0 = 0.6% (341) | 0.8 + 0 = 0.8% (247) | 1.3 + 0.5 =1.8% (742) | |||||||
| gin-6/+ | 0 + 0 = <0.06% (1584) | 0 + 0 = <0.46% (218) | 0 + 0 = <1% (70) | 0.3 + 0 = 0.3% (329) | ND | 0 + 0 = <0.6% (157) | 0.7 + 0.7 =1.4% (855) | ||||||
| gin-7/+ | 0.3 + 0 = 0.3% (2675) | 0.9 + 0 = 0.9% (216) | 0.7 + 0.7 =1.4% (809) | ND | 9.5 + 1.5 =11% (137) | 2.8 + 1 =3.8% (572) | 0.6 + 0 = 0.6% (156) | 1 + <0.7 =1% (786) | |||||
| gin-8/+ | 0.1 + 0 = 0.1% (1617) | 0 + 0 = <0.3% (297) | 0 + 0 = <0.5% (182) | 0 + 0 = <0.8% (123) | ND | 0 + 0 = <0.9% (106) | 0 + 0 = <6% (17) | 3 + 0 = 3% (31) | 0.9 + <0.2 =0.9% (634) | ||||
| gin-9/+ | 0.2 + 0 = 0.2% (2734) | 0 + 0 = <0.6% (179) | 1 + 0.5 =1.5% (215) | 0.4 + 0.4 =0.8% (233) | 0 + 0 = <2% (63) | 0.2 + 0 = 0.2% (547) | 0 + 0 = <0.5% (201) | 0.4 + 0 = 0.4% (228) | 0 + 0 = <1% (103) | 0.9 + 0.5 =1.4% (812) | |||
| gin-10/+ | 1.4 + 0.03 =1.4% (5736) | 1 + 0 = 1% (246) | 5.2 + 14.6 = 19.8% (1073) | 2 + 0 = 2% (248) | 2.5 + 2.8 =5.3% (679) | 3.1 + 6.9 =10% (1488) | 2 + 0.4 =2.4% (255) | 2.5 + 12.7 =15.2% (1146) | 4 + 0 = 4% (80) | 3.8 + 5.5 =9.3% (1117) | 2.0 + 0.7 =2.7% (894) | ||
| gin-11/+ | 0.04 + 0 = 0.04% (2580) | 0.4 + 0 = 0.4% (280) | ND | ND | ND | ND | ND | 1.54 + 0 = 1.54% (65) | 0 + 0 = <0.2% (411) | 0.3 + 0 = 0.3% (292) | 1 + 0 = 1% (102) | 0.7 + 0.3 =1.0% (988) | |
| gin-12/+ | 0.37 + 0 = 0.37% (2957) | 0.4 + 0 = 0.4% (232) | 4.8 + 1.3 =6.1% (606) | ND | 6 + 1 =7% (78) | 4.9 + 2.2 =7.1% (1340) | 0 + 0 = <8% (12) | 2.7 + 1.2 =3.9% (489) | 0.4 + 0 = 0.4% (246) | 5 + 3 =8% (319) | 3.8 + 17.2 =21.0% (1102) | ND | 0.4 + 0.5 =0.9% (756) |
The outcross (+/+) column includes data from both male and female carrier outcrosses. There was no significant difference in mosaic frequencies on the basis of the sex of the carrier for gin-10 outcrosses, which was the mutation with the strongest effect on the frequency of mosaic progeny. The numbers in parentheses in column headings indicate the total number of embryos scored in that gin line. Low fertility in some gin lines resulted in the data gaps. Numbers shown include the percentages of single-patch (nonboldface), multiple-patch (boldface), and total (underlined) mosaic-eye embryos, followed by the number (n) of gol/+ embryos scored. Single patch (nonboldface) plus multiple patch (boldface) equals total mosaics (underlined); number of embryos scored is in parentheses. In each clutch, golden embryos were not counted, and if a mixture of gol/+ and +/+ embryos was present, the numbers of gol/+ were calculated from expected Mendelian ratios. The golden genotype (heterozygous or homozygous) in the parents had no significant effect on mosaic frequencies. In crosses between gin heterozygotes, 25% are homozygous or trans-heterozygous gin. ND, not determined.
Half-tetrad mapping:
Centromere-linkage mapping was done as described (Mohideen et al. 2000). Simple sequence repeats (SSRs) from the centromeric region of each linkage group were tested for heterozygosity in the germ lines of individual gin/+ females by analyzing DNA from pools of nonmosaic sibling EP progeny. Informative (heterozygous) markers were tested on panels of genomic DNA from 7 to 12 individual half-tetrad mosaic embryos. Mosaic embryos with multiple patches were used for preliminary mapping of gin mutations, as these mosaics were nearly always gin homozygotes (Table 3, column J). Single-patch mosaics were also analyzed due to their large number and because many of these are also homozygous gin (Table 3, column H).
TABLE 3.
Effect of gin mutation copy number on mosaicism
|
gin/+ × +/+ outcrosses
|
gin/+ × gin/+ homoallelic crosses
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Single-patch mosaics
|
Multiple-patch mosaics
|
|||||||||
| A | B | C | D | E | F | G | H | I | J | |
| Mutation | % single-patch mosaics | % multiple-patch mosaics | % single-patch mosaics | % multiple-patch mosaics | % gin/gin single-patch mosaics
|
% gin/gin multiple- patch mosaics
|
Prs(gin/+)
|
Prs(gin/gin)
|
Prm(gin/+)
|
Prm(gin/gin)
|
| C − A | D − B | 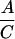 |
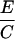 |
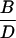 |
 |
|||||
| gin-1 | 0.2 | 0 (0/2564) | 0.7 | 0.3 | 0.5 | 0.3 | 0.29 | 0.71 | 0 | 1 |
| gin-2 | 0.2 | 0 (0/2622) | 1 | 1 | 0.8 | 1 | 0.20 | 0.80 | 0 | 1 |
| gin-3 | 0 (0/3171) | 0 (0/3171) | 0.4 | 0.3 | 0.4 | 0.3 | 0.00 | 1.00 | 0 | 1 |
| gin-4 | 0.2 | 0 (0/4224) | 0.3 | 0.4 | 0.1 | 0.4 | 0.67 | 0.33 | 0 | 1 |
| gin-5 | 0.7 | 0.1 (3/3039) | 1.3 | 0.5 | 0.6 | 0.4 | 0.54 | 0.46 | 0.2 | 0.8 |
| gin-6 | 0 (0/1584) | 0 (0/1584) | 0.7 | 0.7 | 0.7 | 0.7 | 0.00 | 1.00 | 0.0009 | 1 |
| gin-7 | 0.3 | 0 (0/2675) | 1 | 0 (0/786) | 0.7 | 0 | 0.30 | 0.70 | NA | NA |
| gin-8 | 0.1 | 0 (0/1617) | 0.9 | 0 (0/634) | 0.8 | 0 | 0.11 | 0.89 | NA | NA |
| gin-9 | 0.2 | 0 (0/2734) | 0.9 | 0.5 | 0.7 | 0.5 | 0.22 | 0.78 | 0 | 1 |
| gin-10 | 1.4 | 0.03 (2/5736) | 2 | 0.7 | 0.6 | 0.7 | 0.70 | 0.30 | 0.04 | 1 |
| gin-11 | 0.04 | 0 (0/2580) | 0.7 | 0.3 | 0.66 | 0.3 | 0.06 | 0.94 | 0 | 1 |
| gin-12 | 0.55 | 0 (0/2957) | 1.1 | 0.98 | 0.55 | 0.98 | 0.50 | 0.50 | 0 | 1 |
Heterozygote outcrosses were performed between gin/+ and +/+ parents, yielding 50% gin/+ and 50% +/+ progeny. Mosaics from these outcrosses (A and B) are presumed to have arisen among the gin/+ progeny. Homoallelic crosses between gin/+ parents yield 50% gin/+, 25% gin/gin, and 25% +/+ progeny. Mosaics from these crosses (C and D) are presumed to be due to both the gin/+ and the gin/gin progeny. The mosaics contributed by the gin/gin progeny (E and F) were calculated by subtracting the contribution of gin/+ embryos as determined from the outcrosses (A and B). The probability that single-patch mosaics from crosses between heterozygotes are due to gin heterozygosity, Prs(gin/+) (G), is the percentage of mosaic embryos arising among gin/+ progeny (A) divided by the percentage of mosaics among both gin/+ and gin/gin progeny (C). Derivation of probabilities of single-patch and multiple-patch mosaics Prm, with each genotype parallels this reasoning. The actual numbers of mosaic embryos and embryos scored are shown in parentheses when the percentage of mosaics approached 0. The data for columns A–D are from Table 2.
A gin mutation was preliminarily mapped to a linkage group if (1) a specific centromeric SSR allele was homozygous in the majority of mosaic half-tetrad embryos (some single-patch mosaics are expected be gin/+) and (2) at least one informative marker linked to the SSR on one chromosome arm was also usually homozygous in mosaics.
Reactions were run on 6% polyacrylamide gels (Sequel 6, American Bioanalytical; Natick, MA) and visualized with X-ray film with an intensifying screen.
Histology and loss of heterozygosity analysis:
Zebrafish adults were prepared for paraffin sectioning using either 4% PFA or 10% neutral buffered formalin as fixatives, followed by decalcification in 0.5 m EDTA (Moore et al. 2002, 2004). Genomic DNA for analysis was extracted from tissue scraped from unstained, deparaffinized tissue sections (Talbot and Schier 1999; Moore et al. 2002). Genomic DNA was prepared from normal and tumor cells and SSRs were scored as described above. Zebrafish SSR primers used for this analysis were linkage group 1 (LG1): z1351, z6592, z7353, z9294, z9395, z21260; LG4: z6977; LG12: z4188; LG14: z9017; LG15: z21452; LG18: z5479, z7256, z6098, z9194, z10312, z11270, z13426, z13692, z15417, z20015, z20602, z20605, z20981, z25661, z27145, z33808. Primer sequences and map positions were obtained from the database at http://zebrafish.mgh.harvard.edu/zebrafish/index.htm.
RESULTS
Zebrafish genomic instability (gin) mutations were induced by ENU in wild-type males and recovered from G1 generation gin?/+; gol/+ females using a screen for mosaic eyes in their parthenogenetic half-tetrad embryos (Figure 1B). In this screen, golden served as a locus-specific indicator of genomewide events caused by gin mutations. We also asked whether the gin mutations increased susceptibility to spontaneous cancer, due to their effect on cancer genes.
Screening for gin mutations:
All G1 gol/+ embryos, produced by matings between gol/gol females and ENU-mutagenized G0 males, were scored for mosaic-eye pigmentation to find potentially dominant genomic instability mutations. However, no mosaics were found among 1818 or 1905 G1 embryos derived from 2.5 or 3.0 mm ENU-mutagenized males, respectively. Therefore, the frequency of dominant gin mutations with 100% penetrance was <0.03%.
Adult G1 females were screened for mutations that cause genomic instability by examining their half-tetrad progeny for the presence of mosaic eyes (Figure 1B). Individual clutches were scored as positive for a gin mutation upon finding at least one patch of light cells in one eye of an otherwise normal parthenogenetic embryo. We expected recessive mutations to be detected when made homozygous by EP, and weak dominant mutations to be detected as either heterozygotes or homozygotes. A total of 12 gin mutations, designated gin-1–gin-12, were identified using the mosaic-eye assay.
The fraction of half-tetrad embryos with mosaic eyes was expected to depend upon the distance of the mutation from the centromere and the penetrance of the mutation in the mosaic-eye assay. Average frequencies of mosaic embryos in half-tetrad clutches ranged from 0.6 to 4.3% of gol/+ embryos, averaging 2.3% (Table 1). Histological examination (Tsao-Wu et al. 1998) of a set of mosaic-eye embryos for each mutant family showed that the mosaicism was not due to an underlying defect in eye development.
The mosaic-eye assay was expected to reflect genomewide events. In support of this idea, gin-mutation-induced mosaicism was also observed in embryos harboring either of two incidental ENU-induced pigment mutations. One was a new allele of albino (identified by noncomplementation with albino) in the gin-2 line and the second was a new pigmentation mutation in the gin-5 line that complemented both golden and albino (Figure 2, D and E).
Phenotype of gin heterozygotes:
To determine the frequency of mosaicism in gin/+ heterozygotes, embryos from outcrosses between gin carriers and noncarriers were scored for mosaic eyes (Table 2, +/+ column). No golden mosaicism was observed among gin-3/+ or gin-6/+ offspring. In contrast, a small but measurable fraction, up to 1.4%, of embryos heterozygous for any of the other gin mutations had mosaic eyes, indicating a very weak dominant mutator effect.
Penetrance of the mosaic-eye phenotype in gin homozygotes:
The expected frequency of homozygous gin embryos from crosses between gin carriers is 25%. However, the average observed frequency of mosaic embryos was only 1.3% (Table 2), indicating a penetrance of ∼5.2%. As loss of the wild-type golden allele was scored only in the ∼1000 cells of the RPE in each embryo, the simplest explanation is that the somatic mutation rates at the golden locus are lower than 1/1000 cell divisions. Consider, for example, a gin mutation associated with a mutation rate of 1/10,000 cell divisions. Since ∼1000 cells are scored per embryo, mosaics would be present, on average, in only 1 of 10 gin homozygotes. Thus, only 2.5%, rather than 25%, of mosaic-eye embryos would be derived from a cross between such heterozygotes, corresponding to a penetrance of 10%. The observed penetrance of mosaicism associated with the gin mutations is consistent with somatic mutation rates of ∼1 in 20,000 cell divisions in gin homozygotes.
Crosses between carriers of the same gin mutation were used to calculate the frequency of mosaicism in gin homozygotes (Table 3, columns C and D). The progeny of these crosses were expected to consist of 25% +/+ wild type, 50% gin/+ heterozygotes, and 25% gin/gin homozygotes. The wild-type progeny do not contribute mosaics, since no mosaics were seen in the 5183 +/+; gol/+ F1 scored (see materials and methods). The contribution of gin homozygotes to the total incidence of mosaicism was derived by subtracting the mosaicism contributed by the gin/+ heterozygotes, extrapolated from the outcross data, from the total mosaicism observed in these crosses (Table 3, columns E and F). The results indicate that gin homozygotes tend to have a higher frequency of mosaicism than gin heterozygotes.
We classified the mosaic eyes by the number of patches as a measure of phenotypic strength (expressivity). Mosaics containing only one patch of golden cells were called single-patch mosaics, while mosaics with multiple patches, regardless of whether one or both eyes were affected, were called multiple-patch mosaics. We calculated the probability that a single- or multiple-patch mosaic embryo is either heterozygous or homozygous for gin (Table 3). In general, single-patch mosaics can be either homozygous or heterozygous gin. For all of the gin mutations except gin-5, a multiple-patch mosaic has nearly a 100% probability of being gin homozygous. The specificity of this phenotype is critical for both identifying carriers and mapping the mutations.
Phenotype of gin trans-heterozygotes:
To detect potential interactions between the 12 gin mutations in trans-heterozygotes, crosses were done between carriers of each mutation (Table 2). A range of interactions is evident among the trans-heterozygous crosses. The highest fractions of mosaics among trans-heterozygotes involved gin-10 and a subset of the other mutations, designated the “gin-10-interacting group” (Table 4). Distinct patterns of maternal and zygotic gene expression were revealed upon separation of these crosses by sex of the carrier. Crosses between gin-10 heterozygous females and males heterozygous for gin-2, -5, -7, or -12 yielded frequencies of mosaic embryos of 16–27%, much higher than found in any homoallelic cross (0.7–2%). The reciprocal crosses (between gin-10 males and gin-2, -5, -7, or -12 females) showed a smaller degree of interaction. While the gin-4 and gin-9 mutations also show an increased frequency of mosaic embryos when bred to a gin-10 heterozygous female, these interactions are weaker (5.4 and 4% mosaics, respectively) than those of the other members of the group. On the basis of mosaic frequencies, the gin-1, -3, -6, -8, and -11 mutations do not appear to be members of the gin-10-interacting group.
TABLE 4.
Maternal and zygotic contributions to mosaicism in the gin-10-interacting group
| Female parent | Male parent
|
|||||
|---|---|---|---|---|---|---|
| gin-10/+ | gin-2/+ | gin-5/+ | gin-7/+ | gin-9/+ | gin12/+ | |
| gin-10/+ | 2.1 + 0.6 = 2.7% (894) | 7.0 + 20.8 = 27.8% (629) | 4.8 + 12 = 16.8% (725) | 2.4 + 24.1 = 26.5% (597) | 2.3 + 2.5 = 4.8% (471) | 5.0 + 20.4 = 25.4% (760) |
| gin-2/+ | 2.7 + 5.9 = 8.6% (444) | 1 + 1 = 2% (495) | 1 + 0.5 =1.5% (408) | 1 + 1 = 2% (440) | 0 + 0 = <1% (90) | 5.1 + 2 = 7.1%.(312) |
| gin-5/+ | 1.6 + 1.8 = 3.4% (763) | 2.4 + 2 = 4.4% (416) | 1.3 + 0.5 = 1.8% (742) | 2.7 + 1 = 3.7% (445) | 0.3 + 0 = 0.3% (289) | 5.9 + 4.0 = 9.9% (607) |
| gin-7/+ | 2.7 + 0.4 = 3.1% (549) | 0.3 + 0 = 0.3% (369) | 3 + 0 = 3% (127) | 1 + 0 = 1% (786) | 0 + 0 = <5% (20) | 0.3 + 0.3 = 0.6% (338) |
| gin-9/+ | 4.8 + 7.6 = 12.4% (646) | 2 + 0.8 = 2.8% (125) | 0 + 0 = <0.4% (258) | 0 + 0.5 = 0.5% (208) | 0.9 + 0.5 = 1.4% (812) | 12 + 8 = 20% (112) |
| gin-12/+ | 2 + 15 = 17% (342) | 4.4 + 0.7 = 5.1% (294) | 4.0 + 0.8 = 4.8% (733) | 8 + 3 = 11% (151) | 0 + 1 = 1% (207) | 0.4 + 0.5 = 0.9% (756) |
Numbers represent percentages of single-patch (nonboldface), multiple-patch (boldface), and total (underlined) mosaic-eye embryos, followed by the number (n) of gol/ + embryos scored. Data are included from two types of crosses between gin carriers. In crosses between gol/gol and gol/+ carriers of gin, all pigmented embryos are gol/+, allowing scoring of all pigmented embryos for mosaic eyes. In crosses between two gol/+ carriers of gin, two-thirds of pigmented embryos are gol/+ and 1/3 +/+. While all pigmented embryos from these crosses were scored, we use two-thirds of the number of pigmented embryos as the number of gol/+ embryos (n) scored.
Both single-patch and multiple-patch mosaics were found among progeny of crosses between carriers of different gin mutations (Table 5). Analogous to results of crosses among homoallelic carriers, single-patch mosaics may be either heterozygous or trans-heterozygous for gin (Table 5, columns I and J). In contrast, multiple-patch mosaic offspring of adults heterozygous for different gin mutations of the gin-10-interacting group have a 99–100% chance of being trans-heterozygotes (Table 5, columns G and H).
TABLE 5.
Mosaicism in gintrans-heterozygotes from the gin-10-interacting group
|
gin/+ × +/+ outcrosses
|
Female gin-10/+ × male gin-x/+ crosses
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Single-patch mosaics
|
Multiple-patch mosaics
|
|||||||||
| A | B | C | D | E | F | G | H | I | J | |
| Male mutation (x) | % single- patch mosaics | % multiple- patch mosaics | % single- patch mosaics | % multiple- patch mosaics | % gin-10/gin-x single-patch mosaics
|
% gin-10/gin-x multiple-patch mosaics
|
Prs(gin-10 or -x/+)
|
Prs(gin-10/gin-x)
|
Prm(gin-10 or -x/+)
|
Prm(gin-10/gin-x)
|
| Cx − 0.5(A10 + Ax) | Dx − 0.5(B10 + Bx) | 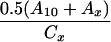 |
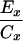 |
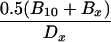 |
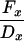 |
|||||
| gin-2 | 0.2 | 0 (0/2622) | 7.0 | 20.8 | 6.2 | 20.79 | 0.11 | 0.89 | 0.0007 | 0.9993 |
| gin-5 | 0.7 | 0.1 (3/3039) | 4.8 | 12 | 3.75 | 11.94 | 0.22 | 0.78 | 0.01 | 0.99 |
| gin-7 | 0.3 | 0 (0/2675) | 2.4 | 24.1 | 1.55 | 24.09 | 0.35 | 0.65 | 0.0006 | 0.9994 |
| gin-9 | 0.2 | 0 (0/2374) | 2.3 | 2.5 | 1.5 | 2.49 | 0.35 | 0.65 | 0.006 | 0.994 |
| gin-10 | 1.4 | 0.03 (2/5736) | 2.1 | 0.6 | 0.7 | 0.57 | 0.67 | 0.33 | 0.05 | 0.95 |
| gin-12 | 0.55 | 0 (0/2957) | 5.0 | 20.4 | 4.03 | 20.39 | 0.20 | 0.81 | 0.0007 | 0.9993 |
Crosses were performed between gin-2/+, gin-5/+, gin-7/+, gin-9/+, gin-10/+, or gin-12/+ males and either +/+ or gin-10/+ females and the incidence of single-patch and multiple-patch mosaics among gol/+ heterozygous progeny was scored. Crosses between two gin mutation heterozygotes yield 25% gin-10/gin-x, 25% gin-10/+, 25% gin-x/+, and 25% wild-type progeny. Mosaic progeny from these crosses can result from the first three of these genotypes. In the following equations, the subscript “x” designates the male gin line, while the subscript “10” designates the female gin-10 line. For example, in column E, for the gin-2 line, Cx = 7, A10 = 1.4, and Ax = 0.2, which gives 7 − 0.5(1.4 + 0.2) = 6.2. Mosaicism in trans-heterozygotes (gin-10/gin-x) can therefore be calculated (E and F) by correcting for the contributions to mosaicism due to the 25% heterozygotes for gin-10 (from the female) and the 25% heterozygotes for gin-x (from the male), alone. Since mosaicism due to 25% heterozygotes is half of the mosaicism due to 50% heterozygotes from outcrosses, we use 0.5 of the outcross values (A or B) for the corrections to yield mosaicism caused only by trans-heterozygosity (gin-10/gin-x). As in Table 2, these frequencies can be used to calculate the corresponding probabilities of specific genotypes in single-patch or multiple-patch mosaics (G–J). The data for columns A and B are from Table 2. The data for columns C and D are from Table 4.
Viability with genomic instability:
In an attempt to establish gin homozygous lines, mosaic embryos were raised to adulthood from crosses between heterozygotes. Of 38 mosaic embryos produced from multiple gin lines in the first few generations after the mutagenesis, only 8 survived to adulthood (“mosaic adults”), suggesting a decrease in viability. Crosses between mosaic-eye adults carrying the gin-3, -6, or -10 mutation and known heterozygotes of the same mutation yielded significantly higher frequencies of mosaic-eye progeny than crosses between known heterozygotes (data not shown, P < 0.05), consistent with the interpretation that homozygotes of these gin mutations were viable. No other homozygous gin adults have been unambiguously identified thus far. In subsequent recent generations, gin-10 and gin-12 multiple-patch mosaics have survived to the juvenile stage. Their homozygous status remains to be confirmed by crosses.
Mapping gin mutations:
Preliminary map positions for the mutations in the gin-10-interacting group were obtained. Half-tetrad mosaic embryos from gin-2, -4, -5, -7, -9, -10, and -12 were used to map these mutations (Johnson et al. 1995; Mohideen et al. 2000). Preliminary linkage group assignments were made for gin-2 (linkage group 25), gin-4 (linkage group 13), gin-7 (linkage group 10), and gin-12 (linkage group 4) and were confirmed using adjacent markers. Interestingly, the preliminary assignments for gin-5, -9, and -10 were to linkage group 18, potentially on the same chromosome arm as golden. Low penetrance prevented an assessment of distance of the gin mutations from their respective centromeres using half-tetrads. Fine mapping remains to be done.
The gin-10A “high-instability” line:
The fourth generation of one of three lines derived from the original founder of gin-10, the gin-10A line, included progeny that generated significantly higher frequencies of mosaic embryos among EP and intercross progeny. We call this the “high-instability” line. The frequency of mosaic gol/+ embryos in crosses between gin-10 carriers is typically 1.3%. Intercrosses among seventh-generation members of this “high-instability” line produced a remarkable 83–96% mosaics. Mosaic frequencies of 25% or greater were even observed in outcrosses of gin-10A to wild-type fish. The high mosaic frequency observed among progeny of crosses between members of the A line cannot be accounted for solely by assuming that all members of the A line are gin-10 homozygotes and therefore all progeny scored for mosaicism were homozygotes. First, some sets of parents were derived from outcrosses and were therefore heterozygotes. Second, many mosaics, including multiple-patch mosaics, were produced from outcrosses and therefore could not be homozygous. In sum, members of the gin-10A line produce frequencies of mosaics that are much higher than “ordinary” gin-10 crosses. We suspect the introduction of a new modifier into the gin-10A line that increases the penetrance of the gin-10 mutation.
Spontaneous tumor susceptibility in gin heterozygotes:
The incidence of a familial cancer is typically elevated by the inheritance of tumor suppressor gene mutations or mutator gene mutations in humans (Marsh and Zori 2002). We therefore asked whether gin carriers show increased susceptibility to spontaneous cancers. Families of fish derived from outcrosses of the gin mutations, not subjected to any type of carcinogen or tumor-promoting agents, were examined for the appearance of tumors at 2.5–3 years. Specific tumor types were not expected, since there was no reason, a priori, to expect gin genes to be expressed in any one tissue type.
Spontaneous tumors were found in adult carriers of all 12 gin mutations. Since the gin-10 mutation showed a strong phenotype and was central to the gin-10-interacting group, the gin-10 mutant line was surveyed to determine the frequency and variety of spontaneous cancers in these families. A set of 50 gin-10 family and 67 wild-type fish were sectioned for histology at 30–34 months of age. The control fish were descended from G1 gin?/+; gol/+ females that did not yield mosaic EP progeny. A greater frequency and variety of cancers were seen in gin-10 as compared with non-gin control families (Table 6). Both epithelial and stromal tumors were found; the most common were adenocarcinomas. Representative gross and microscopic photos of the encountered tumors are shown in Figure 3. Microscopic images of six other tumors are shown with their diagnoses in Figure 4. The raw frequency of cancers was increased ∼6-fold from ∼5 to 28%. By adjusting for the expected fraction of carriers and the background frequency of tumors in the control families, we estimate that heterozygosity for gin-10 causes a 9.6-fold increase in tumor incidence in 30- to 34-month-old fish.
TABLE 6.
Tumor incidence in gin-10 and wild-type families
| % gin fisha (n) | % wild typeb (n) | |
|---|---|---|
| Intestinal adenocarcinoma | 10 (5) | 0 |
| Pancreatic adenocarcinoma | 2 (1) | 3 (2) |
| Cholangiocarcinoma | 8 (4) | 0 |
| Testicular neoplasm (invasive) | 6 (2)c | 0 |
| Malignant peripheral nerve sheath tumor d | 2 (1) | 0 |
| Primitive neuroectodermal tumor d | 2 (1) | 0 |
| Poorly differentiated renal cell tumor | 0 | 1.5 (1) |
| Total tumors | 14/50 = 28% | 3/67 = 5% |
| Adjustment for 5.0% background | (14 − 2) = 12 | |
| Adjustment for gin carrier frequency | 12 × 2 = 24 | |
| Corrected tumor frequencye | 24/50 = 48% | |
| = 9.6-fold increase in tumor incidence, P < 0.001 | ||
A total of 50 gin-10 family fish were included in this experiment, 32 male and 18 female, between 30 and 34 months of age. These fish were a mixture of gin-10/+ and +/+ genotypes with a predicted distribution of 50% gin carriers.
A total of 67 wild-type fish were included in this experiment, 29 male and 38 female, between 30 and 34 months of age. These fish were also descended from ENU males from which the gin mutants were derived, but had not exhibited mosaic eyes. They were outcrossed to lab stocks for several generations.
This frequency is calculated from the number of males in this group (2/32).
These tumors occurred in gin fish that were known to be carriers.
This is likely an underestimate, since loss of fish at an earlier age is expected to include more tumor-bearing animals (presumed gin carriers) that were undetected than would be seen in wild-type animals.
Figure 3.—
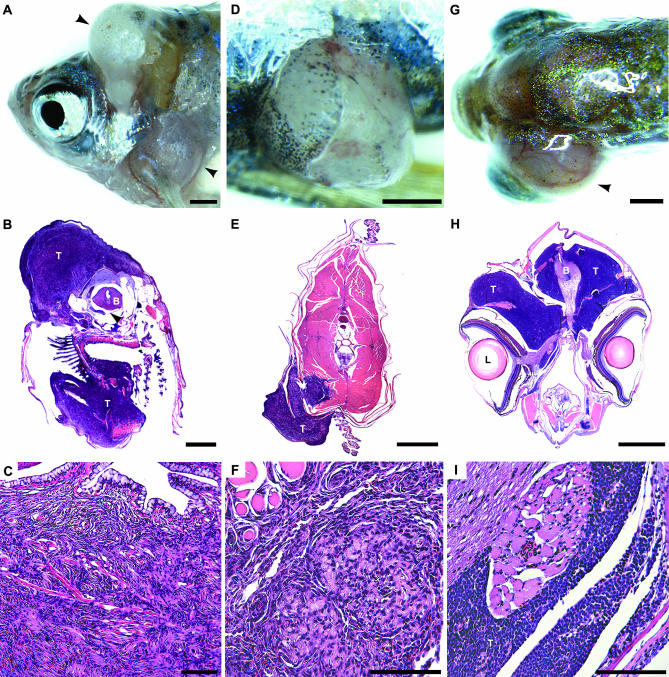
Examples of spontaneous tumors found among adult progeny from gin carrier outcrosses. (A–C) Malignant peripheral nerve sheath tumor first seen in a 6-month-old gin-1 outcross family female (photographed at 9 months). (A) Protruding tumor on the head and down the side of the fish as indicated by arrowheads. Histological analysis was performed at 13 months of age. (B) A transverse section stained with hematoxylin and eosin shows that the tumor (“T”) had spread from the head, down the side of the body and into the heart; “B” indicates brain. The top left tumor mass corresponds to the top left arrowhead in A. (C) Higher magnification reveals one of the common patterns found in this tumor type, similar to the Antoni A pattern found in human schwannomas (Cotran et al. 1999). (D–F) Images of a carcinoma of the skin found in a 17-month-old gin-10 outcross family male. (D) The tumor was located adjacent to the ventral fin. (E) The tumor (“T”) arose on the surface of the skin and then invaded into the underlying muscle (pink). (F) At higher magnification, invasion of muscle (top left) is clearly visible. (G–I) Tumors of a 23-month-old gin-5 outcross family female. (G) Primitive neuroectodermal tumor of the brain, causing severe and moderate exopthalmos of the left (black arrowhead) and right eyes, respectively. (H) A low-power view shows invasion of the tumor (“T”) around the brain (“B”); “L” indicates left lens. An even greater volume of tumor pushes out the eye on the left; invasion is also evident above the right eye. (I) Note the invasion of tumor surrounding the brain (H) and optic nerve (I). Bars, 1 mm in A, B, D, E, G, H; 100 μm in C, F, and I.
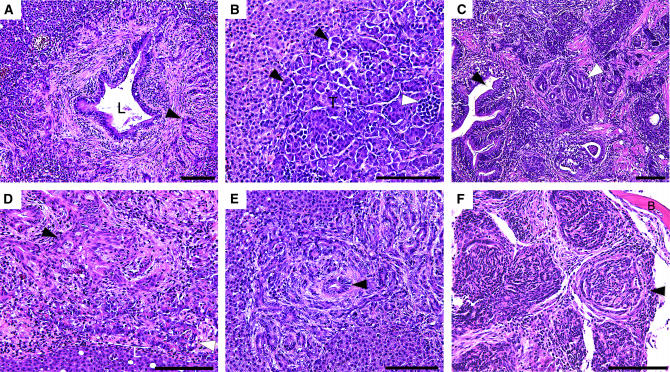
Figure 4.—
Additional examples of tumors found in adult progeny of gin heterozygotes. A–E are from outcross progeny of gin-10 carriers; F is from a gin-6 outcross. (A) One of two tumors from a 31-month-old gin-10 family male, an adenocarcinoma of the bile duct, showing a lumen (L) lined by abnormal cells and invasive ductal cells indicated by the arrowhead. (B) Invasive seminoma from the same gin-10 male as in A, showing an advancing front of malignant germ cells (black arrowheads) invading from the bottom right toward the liver (top left). Scattered clusters of spermatocytes (white arrowhead) are present in the body of the tumor. (C) Adenocarcinoma of intestine from a 25-month-old gin-10 heterozygote with atypical hyperplasia/carcinoma in situ in the intestine (black arrowhead) and clusters of invasive glands (white arrowhead) showing gland-in-gland formation. (D) Mixed adenocarcinoma in a 30-month-old gin-10 family female containing both ductal (black arrowhead) and acinar (white arrowhead) components. (E) Cholangiocarcinoma showing malignant ducts in a 31-month-old gin-10 family male. A cross-sectional cut of a malignant duct is indicated by the black arrowhead. (F) Renal cell adenocarcinoma, showing balls of malignant glands (black arrowhead) invading to vertebral bone (B) in a 28-month-old gin-6 family male. Bars, 100 μm.
The majority of externally visible tumors, seen in almost all the gin lines, were malignant peripheral nerve sheath tumors. These fast-growing tumors often break through the skin of the fish, appearing as firm white growths (Figure 3A). Histological examination of these tumors shows extensive infiltration into adjacent organs (Figure 3, B and C). Other externally visible tumors include primitive neuroectodermal tumors (Figure 3, G–I) and a carcinoma of the skin (Figure 3, D–F). Tumors that were not externally visible include adenocarcinomas of the gastrointestinal tract, including the liver, pancreas (Figure 4, A and C–E), and kidney (Figure 4F).
An unexpected finding was the high proportion of testicular masses in both wild-type and gin-10 families. These growths are smooth, white, noninvasive, and distinct from other internal organs and can be quite large (>1 cm in maximum dimension). They ultimately appear to compromise the viability of the fish by compression of other vital organs. Several histologically distinguishable types of noninvasive testicular neoplasms (tumors) were found. Some contain the full range of cell types expected in spermatogenesis and therefore may be regarded as hyperplasias (overgrowth of normal tissue). The two accepted criteria of malignancy are invasion and metastasis (Cotran et al. 1999). A lack of these features, despite their large size, suggests that these lesions may be considered benign (noninvasive) neoplasms. Testicular neoplasms containing predominantly one cell type, consistent with clonal growth of that cell type deriving from an early stage in spermatogenesis, were seen in 53% (17/32) of the gin-10 males, compared with 17% (5/29) of the control males. An appropriate name for these growths is benign seminomas. Two testicular tumors showed invasion, one into the liver (Figure 4B) and the other into skin (not shown); both were from a gin-10 family. It is unclear why there is a lower frequency of testicular hyperplasias in the gin males (25%, 8/32) compared with wild-type males (48%, 14/29) (P < 0.01).
The early appearance of tumors is a marker of cancer susceptibility. In contrast to the rare tumors from our control fish, many tumors in gin carriers were obvious at a young age. Malignant peripheral nerve sheath tumors were seen in a 6-month-old gin-1 female (Figure 3, A–C) and a 12-month-old gin-9 female. Intestinal adenocarcinomas were found in carriers of several gin lines: one 8-month-old and one 9-month-old gin-8, two 12-month-old gin-10's, and one 12-month-old gin-12. A rare carcinoma of the skin was apparent in a 17-month-old gin-10 male (Figure 3, E–G).
Loss of heterozygosity in gin tumors:
Some tumors found in gin heterozygotes may be associated with loss of function of the remaining wild-type gin allele, which, in turn, would be expected to greatly elevate the rate of mutation in the affected somatic cells (mutator phenotype). Since the mutations have not yet been positionally cloned, point mutation and epigenetic mechanisms of loss cannot be detected at this time. However, simple sequence repeat markers are useful in loss of heterozygosity (LOH) analysis for the detection of recombination or deletion in the study of human tumors (Mendelsohn 2001). In this type of analysis, markers that are heterozygous in normal tissues can be genotyped in tumors to detect the loss of one parental allele. Preliminary mapping data and evidence of linkage to golden placed the gin-5 and gin-10 mutations on linkage group 18 (data not shown). Using this information, PCR-based LOH analysis was done for seven tumors: two malignant peripheral nerve sheath tumors (gin-5 and gin-10), two primitive neuroectodermal tumors (gin-5 and gin-10), a mixed adenocarcinoma (gin-5), a pancreatic adenocarcinoma (gin-10), and a carcinoma of the skin (gin-10) (Figure 3, D–F). On the basis of preliminary map positions of the gin mutations, 26 SSR markers on linkage groups 1, 4, 12, 14, 15, and 18 were amplified from tumor and nontumor DNA samples from each fish. We detected loss of one parental allele for marker Z20981 (preliminarily mapped near gin-10) in a carcinoma of the skin (Figure 5) from a gin-10 carrier, while adjacent normal cells maintained both parental alleles for five proximal and two distal markers on LG18. This result is consistent with recombination or regional deletion.
Figure 5.—
Loss of heterozygosity in the gin-10 region. SSRs were used to analyze the genotype of the carcinoma of the skin shown in Figure 3, D–F. Marker z20981, whose tentative map position lies within the gin-10 region, amplifies both parental alleles in adjacent normal tissue (N), but only one allele in tumor DNA (T). The bracket indicates the top allele, which was lost in the tumor. Each allele is represented by a ladder of bands, as is typical for dinucleotide repeat markers (Odelberg and White 1993). The nonbolded markers on the map showed preservation of both parental alleles in the tumor.
DISCUSSION
Cancer is a disease in which normal somatic cells mutate into a form that can kill their host. The mutator hypothesis, described conceptually by Nowell (1976) and elaborated by Loeb (1991), postulates that the number of mutations necessary for the development of many human cancers cannot be explained by normal rates of mutation, but can be accounted for by the acquisition of a mutator phenotype. The mutator phenotype is caused by mutations in genes involved in genome maintenance. The affected processes include chromosome segregation, recombination, checkpoint control, and the fidelity of DNA replication and repair (Cheng and Loeb 1993; Gollin 2005). We reasoned that a genetic screen that can detect any of these mechanisms might provide insight into the possible genes and mechanisms involved in genomic instability. Such a screen should also independently test the association of mutator mutations with cancer susceptibility and eventually provide an indication of the most vulnerable mechanisms in a vertebrate model. Here, we have described the generation of somatic mutator mutants from a zebrafish ENU screen, estimated the number of potential genes that can be detected, and provided evidence that such mutations increase susceptibility to spontaneous cancer.
A somatic mutator screen in zebrafish:
Our direct genetic screen for genomic instability mutants in zebrafish yielded 12 gin mutants with increased frequencies of somatic mutation as detected using the mosaic-eye assay (Streisinger 1984). We estimate from crosses between gin lines and preliminary genetic mapping data that these mutations reside in 5–12 genes.
Since our 12 gin mutants were identified from screening 49 potential carrier females, a larger number of mutants can be expected by screening more potential carriers. The observed frequency of 0.1% mutations at golden using 2.5 mm ENU suggests that onefold coverage of the zebrafish genome requires screening of ∼1000 females. On the basis of finding 8 gin mutants from clutches of at least 40 eggs from 36 females derived from the 2.5 mm ENU dose, we estimate that screening 1000 females would yield ∼200 gin mutations, potentially corresponding to nearly as many genes among the estimated 21,500 genes of the zebrafish genome (http://www.ensembl.org/Danio_rerio/index.html). The low penetrance of our mutations and the centromere bias of half-tetrad screens suggest that this may be an underestimate. The large number is not surprising, given the multiple known mechanisms of genomic maintenance, each associated with a family of genes (Figure 1A).
Penetrance affects the probability of detecting a mutator mutant. A very strong mutator (high penetrance) would always give multiple-patch mosaics when homozygous or heterozygous, depending on its dominance. However, a full range of phenotypes was observed for each of the 12 mutators, ranging from multiple- and single-patch mosaics to no phenotype, indicating incomplete penetrance. More complete penetrance may be expected using assays that score more than the 1000 cells used in the mosaic-eye assay.
The identification of gin mutations relied upon a screen for mosaic-eye pigmentation caused by loss of wild-type gene function at the golden locus in golden heterozygotes. Pigmentation associated with golden is cell autonomous and readily distinguishable from the wild-type color at 48 hr. The eventual development of color in golden mutant cells of mosaic-eye embryos indicates that the lack of pigment was due to lack of golden function, rather than an absence of RPE cells (Malicki et al. 1996). The recent cloning of the golden gene (Lamason et al. 2005) will facilitate study of somatic mutation and/or gene inactivation in the gin mutants.
Factors influencing mosaicism:
The interpretation of single- and multiple-patch mosaic eyes requires an understanding of the timing and frequency of loss-of-function events impacting the golden gene. A single small patch of pale cells most likely represents a clone derived from one loss-of-function event at the golden locus that occurred late during development of the RPE. There are two likely explanations for multiple small patches. It may be that a single early LOH event produces multiple daughter cells that eventually establish distinct golden patches in eyes, as observed following irradiation of gol/+ heterozygous embryos at the two-cell stage (Streisinger et al. 1989). The multiple patches that we observed, however, were most common in crosses yielding a high frequency of mosaic eyes, a finding more consistent with multiple, late, and independent loss-of-function events.
Frequencies of mosaicism in gin heterozygotes, homozygotes, and trans-heterozygotes:
Mosaics arising in crosses between heterozygotes may be due to either gin/gin or gin/+ genotypes. To determine the frequency of gin/gin among those mosaics, it was therefore necessary to know the contribution of gin/+ to mosaicism. The heterozygous mosaic frequency was derived from outcrosses of gin carriers. These crosses produced low frequencies of mosaics, primarily with single small patches (Table 2, +/+ column). The results indicate a weak dominant effect in most gin heterozygous embryos.
When compared with gin heterozygous embryos, gin homozygous embryos had a higher frequency of mosaics and multiple patches (Table 3). The calculations show that multiple-patch mosaics are largely the result of gin homozygosity (I and J), but that single-patch mosaics may be either homozygous or heterozygous for gin (G and H). This knowledge is essential for positional cloning.
A striking enhancement of mosaicism was associated with trans-heterozygosity in the gin-10-interacting group. The basis of these interactions is not clear, but likely involves a common mechanism such as chromosome loss or recombination. The gin products may interact via a multimeric complex or common metabolic pathway. Another possibility is that these gin mutations affect duplicate pathways. Knocking down the function of one pathway would not cause strong genomic instability if another can compensate. This would account for the relatively low rates of mosaic eyes in gin homozygotes. Positional cloning will be required to distinguish between these and other possibilities.
Spontaneous tumor susceptibility in gin heterozygotes:
Our tumor susceptibility studies focused on gin-10 since it was the center of the gin-10-interacting group. The frequency of spontaneous cancers was increased nearly 10-fold in 30- to 34-month-old gin-10/+ adults. Other tumors arose in unusually young fish. These findings, together with the presence of multiple tumors in some fish, comprise the classic characteristics of a germ-line tumor susceptibility mutation (Marsh and Zori 2002) and suggest an acceleration of tumor progression by the gin-10 mutation. Since most cancers are due to loss of function in more than one gene (Lengauer et al. 1998), our results are consistent with the mutator hypothesis in which a state of genomic instability predates the acquisition of the full complement of mutations required for cancer (Loeb et al. 2003).
Two levels of instability may be expected during the life span of a gin heterozygote. First, the mosaicism that we observed in gin heterozygotes suggests that a mild mutator phenotype may apply in all cells of a heterozygote throughout its life span, which in other settings is referred to as haplo-insufficiency (Largaespada 2001; Amsterdam et al. 2004). Second, a stronger mutator phenotype may arise in some somatic cells whenever the function of the wild-type gin allele of heterozygotes is lost (Knudson's “second hit”). The resultant increase in global mutation rate would also be expected to affect cancer genes, leading to an elevated frequency of cancer. Different levels of mutation associated with homo- and heterozygosity may also apply to some human genomic instability syndromes.
Two previously reported genetic screens done in vertebrates were indirectly found to show evidence of genomic instability, although neither assayed mutation rates at a specific marker locus. Shepard et al. (2005) screened zebrafish for cell cycle mutants on the basis of changes in the pattern of phosphohistone H3 staining and found evidence of chromosome instability in the mutant embryos. There was no detectable increase in susceptibility to spontaneous cancer at 1 year, but susceptibility to carcinogen-induced cancer was increased about twofold. It remains possible that an increase in spontaneous cancer would be detectable at a later time. Shima et al. (2003) found mouse mutants with an increase in micronucleus formation in lymphocytes, but susceptibility to spontaneous cancer has not yet been reported.
The high frequency of gin mutants found in an embryonic screen indicates that zebrafish is a good vertebrate model to directly screen for somatic mutator genes. The dominant susceptibility to spontaneous cancer in adults conferred by gin-10 confirms the relevance of such mutations to cancer. Mechanistic studies and the identification and study of the affected genes may provide insight into the relationships among somatic loss of gene function, tumor progression, and cancer susceptibility in all vertebrates, including humans.
Acknowledgments
The authors thank Lynn Budgeon for her histology work, Peggy Hubley for fish husbandry, Victor Canfield for insightful discussion, Larry Loeb and John Kreider for their support, past members of the Cheng lab for their help, and many colleagues for their comments on the manuscript. We are especially grateful for editor David Grunwald's extensive and valuable discussions and suggestions. This work was supported by grants from the American Cancer Society (JFRA-581), the National Science Foundation (MCB-93198174), the National Institutes of Health (RO1-CA73935 and RO1-HD40179), the Four Diamonds Fund, and the Pennsylvania Tobacco Settlement Funds to K.C.C., the National Research Service Award F32-GM119794 to J.L.M., and the Jake Gittlen Memorial Golf Tournament.
We dedicate this work to the memory of DeLill Nasser, whose interest in new genetic approaches was essential to this project's success.
References
- Amsterdam, A., K. C. Sadler, K. Lai, S. Farrington, R. T. Bronson et al., 2004. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2: E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, J. S., 2000. The molecular biology of cancer. Mol. Aspects Med. 21: 167–223. [DOI] [PubMed] [Google Scholar]
- Cavenee, W. K., T. P. Dryja, R. A. Phillips, W. F. Benedict, R. Godbout et al., 1983. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 305: 779–784. [DOI] [PubMed] [Google Scholar]
- Charames, G. S., and B. Bapat, 2003. Genomic instability and cancer. Curr. Mol. Med. 3: 589–596. [DOI] [PubMed] [Google Scholar]
- Cheng, K. C., and L. A. Loeb, 1993. Genomic instability and tumor progression: mechanistic considerations. Adv. Cancer Res. 60: 121–147. [DOI] [PubMed] [Google Scholar]
- Cheng, K. C., and J. L. Moore, 1997. Genetic dissection of vertebrate processes in the zebrafish: a comparison of uniparental and two-generation screens. Biochem. Cell Biol. 75: 525–533. [PubMed] [Google Scholar]
- Cotran, R. S., V. Kumar, T. Collins and S. L. Robbins, 1999. Robbins Pathologic Basis of Disease. Saunders, Philadelphia.
- Cox, E. C., 1976. Bacterial mutator genes and the control of spontaneous mutation. Annu. Rev. Genet. 10: 135–156. [DOI] [PubMed] [Google Scholar]
- Feinberg, A. P., H. Cui and R. Ohlsson, 2002. DNA methylation and genomic imprinting: insights from cancer into epigenetic mechanisms. Semin. Cancer Biol. 12: 389–398. [DOI] [PubMed] [Google Scholar]
- Gestl, E. E., E. J. Kauffman, J. L. Moore and K. C. Cheng, 1996. New conditions for generation of gynogenetic half-tetrad embryos in zebrafish. J. Hered. 88: 76–79. [Google Scholar]
- Gollin, S. M., 2005. Mechanisms leading to chromosomal instability. Semin. Cancer Biol. 15: 33–42. [DOI] [PubMed] [Google Scholar]
- Grunwald, D. J., and G. Streisinger, 1992. Induction of recessive lethal and specific locus mutations in the zebrafish with ethyl nitrosourea. Genet. Res. 59: 103–116. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers, J. H., 2001. Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. [DOI] [PubMed] [Google Scholar]
- Johnson, S. L., D. Africa, S. Horne and J. H. Postlethwait, 1995. Half-tetrad analysis in zebrafish: mapping the ros mutation and the centromere of linkage group I. Genetics 139: 1727–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson, A. G., Jr., 1971. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA 68: 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner, R. D., C. D. Putnam and K. Myung, 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557. [DOI] [PubMed] [Google Scholar]
- Lamason, R. L., M. A. Mohideen, J. R. Mest, A. C. Wong, H. L. Norton et al., 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Largaespada, D. A., 2001. Haploinsufficiency for tumor suppression: the hazards of being single and living a long time. J. Exp. Med. 193: F15–F18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer, C., K. W. Kinzler and B. Vogelstein, 1998. Genetic instabilities in human cancers. Nature 396: 643–649. [DOI] [PubMed] [Google Scholar]
- Loeb, L. A., 1991. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 51: 3075–3079. [PubMed] [Google Scholar]
- Loeb, L. A., K. R. Loeb and J. P. Anderson, 2003. Multiple mutations and cancer. Proc. Natl. Acad. Sci. USA 100: 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki, J., S. C. Neuhauss, A. F. Schier, L. Solnica-Krezel, D. L. Stemple et al., 1996. Mutations affecting development of the zebrafish retina. Development 123: 263–273. [DOI] [PubMed] [Google Scholar]
- Marra, G., and C. R. Boland, 1995. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J. Natl. Cancer Inst. 87: 1114–1125. [DOI] [PubMed] [Google Scholar]
- Marsh, D., and R. Zori, 2002. Genetic insights into familial cancers—update and recent discoveries. Cancer Lett. 181: 125–164. [DOI] [PubMed] [Google Scholar]
- Mendelsohn, J., 2001. The Molecular Basis of Cancer. Saunders, Philadelphia.
- Mohideen, M. A., J. L. Moore and K. C. Cheng, 2000. Centromere-linked microsatellite markers for linkage groups 3, 4, 6, 7, 13, and 20 of zebrafish (Danio rerio). Genomics 67: 102–106. [DOI] [PubMed] [Google Scholar]
- Moore, J. L., M. Aros, K. G. Steudel and K. C. Cheng, 2002. Fixation and decalcification of adult zebrafish for histological, immunocytochemical, and genotypic analysis. Biotechniques 32: 296–298. [DOI] [PubMed] [Google Scholar]
- Moore, J. L., E. E. Gestl and K. C. Cheng, 2004. Mosaic eyes, genomic instability mutants, and cancer susceptibility. Methods Cell Biol. 76: 555–568. [DOI] [PubMed] [Google Scholar]
- Mullins, M. C., M. Hammerschmidt, P. Haffter and C. Nusslein-Volhard, 1994. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr. Biol. 4: 189–202. [DOI] [PubMed] [Google Scholar]
- Nowell, P. C., 1976. The clonal evolution of tumor cell populations. Science 194: 23–28. [DOI] [PubMed] [Google Scholar]
- Odelberg, S. J., and R. White, 1993. A method for accurate amplification of polymorphic CA-repeat sequences. PCR Methods Appl. 3: 7–12. [DOI] [PubMed] [Google Scholar]
- Shepard, J. L., J. F. Amatruda, H. M. Stern, A. Subramanian, D. Finkelstein et al., 2005. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc. Natl. Acad. Sci. USA 102: 13194–13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, N., S. A. Hartford, T. Duffy, L. A. Wilson, K. J. Schimenti et al., 2003. Phenotype-based identification of mouse chromosome instability mutants. Genetics 163: 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnica-Krezel, L., A. F. Schier and W. Driever, 1994. Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics 136: 1401–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger, G., 1984. Attainment of miminal biological variability and measurements of genotoxicity: production of homozygous diploid zebrafish. Natl. Cancer Inst. Monogr. 65: 53–58. [PubMed] [Google Scholar]
- Streisinger, G., C. Walker, N. Dower, D. Knauber and F. Singer, 1981. Production of clones of homozygous diploid zebrafish (Brachydanio rerio). Nature 291: 293–296. [DOI] [PubMed] [Google Scholar]
- Streisinger, G., F. Singer, C. Walker, D. Knauber and N. Dower, 1986. Segregation analyses and gene-centromere distances in zebrafish. Genetics 112: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger, G., F. Coale, C. Taggart, C. Walker and D. J. Grunwald, 1989. Clonal origins of cells in the pigmented retina of the zebrafish eye. Dev. Biol. 131: 60–69. [DOI] [PubMed] [Google Scholar]
- Talbot, W. S., and A. F. Schier, 1999. Positional cloning of mutated zebrafish genes. Methods Cell Biol. 60: 259–286. [DOI] [PubMed] [Google Scholar]
- Tsao-Wu, G. S., C. H. Weber, L. R. Budgeon and K. C. Cheng, 1998. Agarose-embedded tissue arrays for histologic and genetic analysis. Biotechniques 25: 614–618. [DOI] [PubMed] [Google Scholar]
- Westerfield, M., 1995. The Zebrafish Book. University of Oregon Press, Eugene, OR.