Abstract
The transcriptional mediator complex has emerged as an important component of transcriptional regulation, yet it is largely unknown whether its subunits have differential functions in development. We demonstrate that the zebrafish mutation m885 disrupts a subunit of the mediator complex, Crsp34/Med27. To explore the role of the mediator in the control of retinal differentiation, we employed two additional mutations disrupting the mediator subunits Trap100/Med24 and Crsp150/Med14. Our analysis shows that loss of Crsp34/Med27 decreases amacrine cell number, but increases the number of rod photoreceptor cells. In contrast, loss of Trap100/Med24 decreases rod photoreceptor cells. Loss of Crsp150/Med14, on the other hand, only slightly reduces dopaminergic amacrine cells, which are absent from both crsp34m885 and trap100lessen mutant embryos. Our data provide evidence for differential requirements for Crsp34/Med27 in developmental processes. In addition, our data point to divergent functions of the mediator subunits Crsp34/Med27, Trap100/Med24, and Crsp150/Med14 and, thus, suggest that subunit composition of the mediator contributes to the control of differentiation in the vertebrate CNS.
STUDIES investigating the transcriptional control of pattern formation and cell differentiation in development have previously focused predominantly on the role of the combinatorial function of transcription factors and coregulators (Mannervik et al. 1999; reviewed in Blais and Dynlacht 2005). In recent years, the transcriptional mediator complex has been recognized as an additional level of transcriptional regulation in eukaryotes, serving as an adapter between enhancer-bound transcription factors and the machinery of RNA polymerase II (reviewed in Taatjes et al. 2004a). The human mediator comprises ∼25 subunits, depending on source and preparation procedure (Fondell et al. 1996; Rachez et al. 1998, 1999; Naar et al. 1999; Ryu et al. 1999; Malik et al. 2000). In yeast, analysis of deletion strains has revealed differential requirements for individual subunits for different portions of the transcriptome (Holstege et al. 1998). During metazoan evolution, individual subunits have diverged considerably, potentially reflecting the increased complexity of transcriptional regulation (discussed in Boube et al. 2002). Recent studies have demonstrated that combinatorial recruitment of mediator subunits and conformational changes of the complex can regulate transcription in vitro (Taatjes and Tjian 2004; Taatjes et al. 2004b). The emergence of this concept has added further complexity to our understanding of transcriptional regulation and the intrinsic control of developmental events. Further, several subunits have been implied as essential components of signal transduction pathways and developmental processes of metazoan organisms. Subunit Trap220 interacts with a number of nuclear receptors and is required for thyroid hormone signaling in the mouse (Yuan et al. 1998; Ito et al. 2000). Trap100 interacts with Trap220 and serves as a cofactor for Trap220 function (Zhang and Fondell 1999). In zebrafish, Trap100 is required for proliferation of enteric neuron progenitors (Pietsch et al. 2006) and, in mouse fibroblasts, for full transcriptional activation from several promoters (Ito et al. 2002). Trap230 and Trap240 are involved in hedgehog signaling in the Drosophila eye and wing discs, controlling cell differentiation and affinity (Boube et al. 2000; Treisman 2001; Janody et al. 2003). Trap80 on the other hand is essential for cell viability, suggesting a more general role in transcriptional initiation (Tudor et al. 1999; Boube et al. 2000). Thus, a number of in vivo studies have substantiated roles of mediator subunits in controlling developmental decisions. Yet, the majority of subunits have not been functionally characterized in metazoan organisms. Therefore, it remains unclear to what extent mediator composition regulates developmental processes.
Investigating the role of the transcriptional mediator complex in development requires a system that is well understood with respect to mechanisms of cellular differentiation. During the development of the vertebrate central nervous system (CNS), a multitude of neuronal and glial cell types differentiate under tight temporal and spatial control. This control is mediated by an interplay between extrinsic and intrinsic factors, as shown for several neuronal structures, such as the vertebrate retina (Alexiades and Cepko 1997; Belliveau and Cepko 1999; Morrow et al. 1999; Kay et al. 2001; Li et al. 2004; Martinez-Morales et al. 2005). In the retina, six classes of neurons and one class of glia develop from a pool of precursor cells, whose competence decreases during development (Wetts and Fraser 1988). Differentiated cells are stereotypically organized in three layers: the ganglion cell layer (GCL) of ganglion and displaced amacrine cells; the inner nuclear layer (INL) of amacrine, bipolar, and horizontal cells as well as Müller glia; and the outer nuclear layer (ONL), harboring rod and cone photoreceptor cells. Both this well-described anatomy and the availability of molecular markers make the retina a good model for the study of neuronal differentiation in the vertebrate CNS.
Here, we address the roles of the mediator subunits Crsp34 (34 kDa; also called Crsp8, Trap37, Med27), Trap100 (100 kDa; also called Crsp4, Crsp100, Med24), and Crsp150 (150 kDa; also called Crsp2, Trap170, Med14) in the development of the zebrafish retina through comparative analysis of their loss-of-function phenotypes. We describe the positional cloning of the allele crsp34m885 and provide in vivo data for roles of Crsp34 in development. In the retina, loss of Crsp34 leads to decreased formation of amacrine cells but increased formation of rod photoreceptor cells. On the contrary, trap100lessen mutant embryos (Pietsch et al. 2006) exhibit a reduction of rod photoreceptor cells, while crsp150hi2143 (Amsterdam et al. 2004) allows normal development of most retinal cell types. Our data thus demonstrate divergent phenotypes in zebrafish embryos mutant for crsp34, trap100, or crsp150, pointing to subunit-specific functions in retinal development.
MATERIALS AND METHODS
Fish breeding, strains, and mutagenesis:
Zebrafish were maintained and bred under standard conditions at 28.5° (Westerfield 1994). Embryonic stages are defined according to Kimmel et al. (1995). To avoid pigmentation, embryos were raised in 0.2 mm 1-phenyl-2-thiourea (Sigma, St. Louis) prior to fixation. The following strains were used: crsp34m885, trap100lessen (Pietsch et al. 2006), crsp150hi2143 (Amsterdam et al. 2004), and AB/TL wild-type fish. The crsp34m885 allele was derived from a chemical mutagenesis screen; carriers were scored for defects in th expression in the haploid F2 generation at 3 days postfertilization (dpf) (Holzschuh et al. 2003).
Mapping and cloning of crsp34m885 and RT–PCR:
For mapping, crsp34m885 in AB was crossed to the WIK strain. Information on simple sequence length polymorphism (SSLP) markers was obtained from zebrafish.mgh.harvard.edu. PAC clones were derived from BUSM1 PAC library (Amemiya and Zon 1999). Sequence data on genomic traces and contigs were derived from the zebrafish genome project at the Sanger Institute (http://www.ensembl.org). The crsp34 ORF (Sanger Institute, Zv2) was sequenced from 5-dpf cDNA of mutant and wild-type embryos (Superscript II; Invitrogen, San Diego), revealing a C to A exchange at base 333, producing the stop codon TAA.
crsp34_long and crsp34_shortA ORFs were predicted by Ensembl Zv2 (Sanger Institute). 5′ and 3′ RACE (BD Bioscience Smart RACE) with specific primers in proximity of the point mutation amplified the same common 5′ end, the 3′ end of crsp34_long, and an additional 3′ end (of variant crsp34_shortB). Full-length isoforms were amplified from mRNA isolated from 27-hr postfertilization (hpf) embryos (Micro-Fast Track, Invitrogen), using the same forward primer in the common 5′-UTR (5′-AGTGTTCGAGCAGAAGCA-3′) and reverse primers complementary to isoform-specific 3′-UTRs: crsp34_long (GenBank accession no. BC057508) (5′-AGCCGTGGAAAGCTGTTATC-3′; 988-bp product), crsp34_shortA (5′-AAAAGAGACAAATCGTCACAA-3′; 664-bp product), and crsp34_shortB (5′-AAATTCAAATGATACCCAATTC-3′; 640-bp product). Each isoform was cloned into pGEM-T-Easy (Promega, Madison, WI). For analysis of temporal profiles of isoform expression, we used these isoform-specific primer pairs and cDNA reverse transcribed (Superscript II, Invitrogen) from 1.2 μg total RNA (RNeasy; QIAGEN, Valencia, CA; AB/TL embryos).
Genotyping:
m885 mutant embryos were genotyped by genomic PCR and sequencing or, alternatively, by PCR using a perfectly matched forward primer (AGCGTCTCAGCACTTTGGTT) and a mismatch reverse primer (AACTTACTTTATTGGACCATTCG), whose 3′ end hybridizes to the site of the point mutation, forming two 3′ mismatches with the mutant, but only one mismatch base with the wild-type allele (Kwok et al. 1995 and references therein). This primer pair specifically amplifies the wild-type locus. The hi2143 allele was identified by PCR with one primer in the viral sequence (CTGTTCCATCTGTTCCTGAC) and one in the genomic DNA adjacent to the insertion site (GCTACAGCGAACCTATCATGAG) [Amsterdam et al. 2004; N. Hopkins, A. Amsterdam and S. Farrington (MIT), personal communication]. Embryos from heterozygous trap100lessen parents were genotyped according to Pietsch et al. (2006).
Micro-injection of mRNA and morpholino:
ORFs of crsp34 isoforms were cloned from pGEM-T-Easy (Promega) into the EcoRI site of pCS2+ (http://sitemaker.umich.edu/dlturner.vectors). Capped mRNA was transcribed by a mmessage mmachine Sp6 polymerase kit (Ambion, Austin, TX). Morpholino MOcrsp34 (5′-TCACTCCAACATTCATAACATCCGC-3′) was designed and synthesized by GeneTools and targeted to bases +4 to +28 of all known crsp34 transcripts. For injection, crsp34 mRNAs and MOcrsp34 were diluted in RNase-free H2O and 0.05% phenol red. The amount of MOcrsp34 was optimized to exclude nonspecific effects such as brain degeneration (Nasevicius and Ekker 2000). Injection of 0.5, 0.7, and 0.9 pmol MOcrsp34 was followed by brain degeneration in 16/31, 8/9, and 19/19 embryos at 24 hpf, respectively. Injection of 0.3 pmol MOcrsp34 led to brain degeneration in only 1/17 injected embryos. For rescue and phenocopy experiments, we injected 300–600 pg of crsp34_long mRNA or 0.3 pmol MOcrsp34 into 1-cell stage embryos. Injected drop volumes were ∼1 nl, as measured in halocarbon oil (series 27, Halocarbon Products) on a micrometer slide.
Whole-mount in situ hybridization, immunohistochemistry, and TUNEL:
Whole-mount in situ hybridization (WISH) with alkaline phosphatase-based color reaction was performed as described (Hauptmann and Gerster 1994), using the following probes: th (Holzschuh et al. 2001), tphD1 (Bellipanni et al. 2002), pax6.2 (Nornes et al. 1998), mcm5 (Ryu et al. 2005), and red opsin (Robinson et al. 1995). A fragment of the published rhodopsin sequence (NM_131084) was cloned into pGEM-T-Easy (Promega) for probe synthesis. The crsp34 antisense probe was transcribed from crsp34_shortA in pGEM-T-Easy and the crsp34 sense probe from crsp34_shortA in pCS2+. Fluorescent WISH was carried out adapting reported protocols (Denkers et al. 2004; Kosman et al. 2004) to the zebrafish, using tyramide signal amplification (TSA) (Alexa Fluor 488; Molecular Probes, Eugene, OR). Immunohistochemistry (IHC) was performed as described (Solnica-Krezel and Driever 1994), using a monoclonal antibody against 5-HT (Chemicon, Temecula, CA), 1:50; goat polyclonal antibody against ChAT (Chemicon), 1:250 (Arenzana et al. 2005); rabbit polyclonal antibody against GABA (Chemicon), 1:500; zpr1/FRet43, 1:20 (Larison and Bremiller 1990); and zn5, 1:500 (Trevarrow et al. 1990). Anti-5HT, anti-GABA, and zn5 were detected by biotinylated secondary antibodies and ABC–HRP (Vectastain); anti-ChAT and zpr1 were detected by rabbit anti-goat-Alexa555 and goat anti-mouse-Alexa488, respectively (Molecular Probes). For terminal transferase dUTP nick end labeling (TUNEL) assay, embryos were fixed, permeabilized, and treated with Proteinase K as for WISH. TUNEL reaction was performed according to the manufacturer's instructions (ApopTag; Intergen, Purchase, NY).
Histological sections:
Embryos were embedded in 0.4% gelatin, 27% BSA, 18% saccharose, and 14% glutaraldehyde and were cut to 10 μm (rhodopsin), 15 μm (pax6.2, anti-GABA, anti-CAII), or 20 μm (red opsin, TUNEL, zn5) sections with a vibratome. For plastic sections, fixed embryos were embedded in agarose, dehydrated in ethanol, and embedded in Technovit 7100 (Heraeus Kulzer), according to the manufacturer's instructions. Sections were cut to 4 μm with a microtome and counterstained with 1% methylene blue in H2O. For cryosections, embryos were equilibrated in 30% sucrose in PBT, embedded in tissue-freezing medium (Jung), and cut to 16-μm sections with a cryostat (Reichert-Jung Frigocut).
Quantification of retinal cell layers:
Thickness of amacrine and ganglion cell layers was measured in cross-sections of 78-hpf embryos in the proximity of the optic nerve, using Zeiss (Göttingen, Germany) Axiovision software. The GCL was measured as the distance between the edge of the lens and the inner plexiform layer. The layer of amacrine cells was measured in the following sections: anti-GABA, each one m885 mutant and one wild type; anti-Hu (Molecular Probes) and elavl3 WISH (Kudoh et al. 2001), both each one m885 mutant and one wild-type embryo, data not shown. Further, amacrine cell layers were distinguished by morphology in four wild-type and three m885 mutant embryos. Three lessen mutant embryos and three wild-type siblings and three hi2143 embryos and four wild-type siblings were analyzed by pax6.2 WISH (Nornes et al. 1998). We took five measurements each from the central half of the left and the right retinas. The mean value of 10 data points (in rare cases of only one suitable cross-section: 5 data points) was recorded as data entry per embryo. For evaluation of rod cells, confocal three-dimensional (3-D) stacks were recorded (LSM510, Zeiss) from embryos stained by fluorescent WISH to rhodopsin and counterstained with TOTO3 (Molecular Probes). Equal image acquisition settings were applied for each embryo. To determine the volume of stained tissue and average staining intensities per embryo, stacks were measured using Volocity 3.1 software (classifier set to intensity range above background, including all object sizes). P-values were analyzed by an unpaired t-test (StatView 5.0.1).
RESULTS
The mutation m885 ablates the formation of dopaminergic amacrine cells:
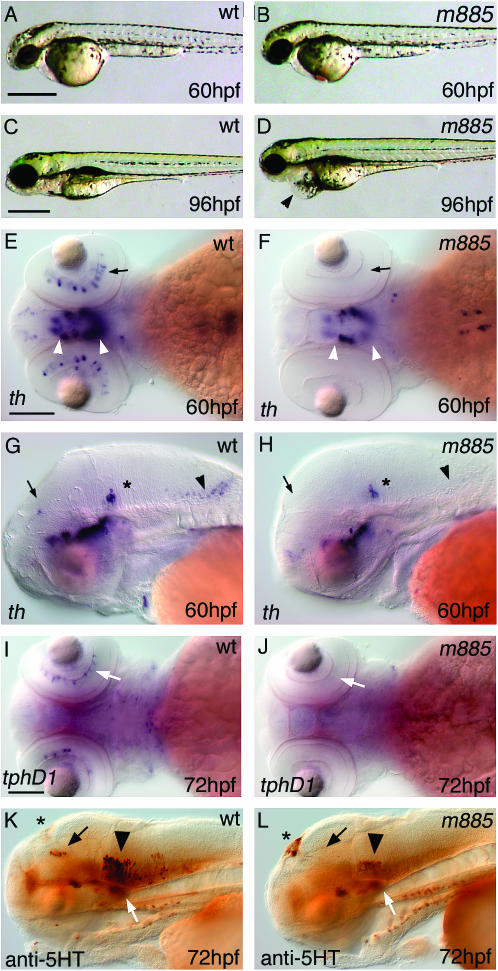
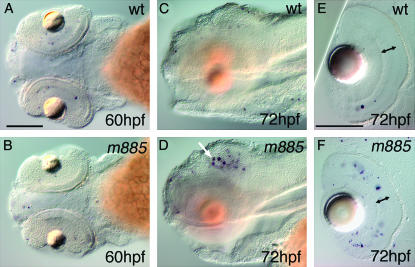
We isolated the mutation m885 from a haploid genetic screen for mutations that affect the development of catecholaminergic neurons in zebrafish. At 60 hpf, live m885 mutant embryos show normal overall morphology (Figure 1, A and B), but entirely lack dopaminergic (DA) amacrine cells in the retina, as revealed by absence of tyrosine hydroxylase (th) expression (Figure 1, E and F). Additionally, m885 affects the development of other catecholaminergic groups in the dorsal and ventral diencephalon and the hindbrain, which are not discussed here (Figure 1, G and H). Further, it appears that all serotonergic groups contain reduced numbers of neurons, as judged from anti-serotonin immunohistochemistry (Figure 1, K and L). In the retina, absence of tphD1 expression reveals that serotonergic amacrine cells are largely missing (Figure 1, I and J). However, cholinoceptive groups of the optic tectum, hindbrain, and spinal cord appear largely normal, as judged by acetylcholine esterase activity assay (not shown). From 3.5 dpf onward, a reduction of head, eye, and jaw size becomes evident, and mutant embryos develop a pericardial edema (Figure 1, C and D). Mutant larvae fail to inflate their swim bladders and die ∼6 dpf.
Figure 1.—
The phenotype of m885 mutant embryos. (A) At 60 hpf, live wild-type embryos resemble their (B) mutant siblings. (C and D) At 96 hpf, mutant larvae exhibit reduced brain and eye size, as well a reduction of the jaw and a pericardial edema (arrowhead in D). (E) WISH to tyrosine hydroxylase (th) labels DA amacrine cells (arrow) of 60 hpf wild-type embryos. (F) In m885 mutant siblings, DA amacrine cells do not form. DA cells of the ventral diencephalon are reduced in mutant embryos when compared to wild-type siblings (arrowheads in E and F). (G) Sixty hpf wild-type embryos develop, among other sites, DA neurons in the pretectum (arrow) and NA neurons in the medulla oblongata (arrowhead). (H) These groups do not form in m885 mutant siblings. NA neurons in the locus coeruleus (asterisk) develop normally. (I) WISH to tryptophane hydroxylase D1 (tphD1) labels serotonergic amacrine cells in 72 hpf wild-type embryos (arrow). (J) These cells are absent from m885 mutant siblings. (K) IHC to serotonin (5-HT) in 72 hpf wild-type embryos labels serotonergic neurons in the dorsal and ventral diencephalon (black and white arrows, respectively) and in the raphe nucleus (arrowhead). (L) These groups are reduced in m885 mutant siblings. Serotonin-producing cells of the epiphysis (asterisk) form normally in the mutant; the weaker signal apparent in this wild-type embryo (K) does not reflect a consistent difference. Bars: A, 500 μm for A–D; E, 100 μm for E–H; I, 100 μm for I–L.
m885 is an allele of crsp34, encoding a subunit of the transcriptional mediator complex:
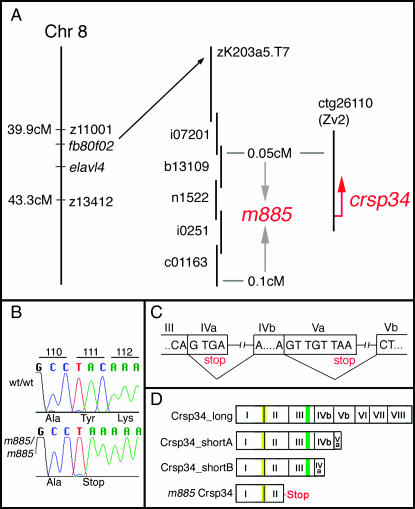
To identify the gene affected by m885, we undertook a positional cloning approach. The mutation was mapped to chromosome 8 by bulked segregant analysis with SSLPs (Michelmore et al. 1991) and localized to a genetic interval of 0.15 cM that was annotated with the crsp34 gene (Sanger Institute Zv2; Figure 2A). Sequencing the ORF from mutant embryos revealed a C to A base exchange, introducing an early stop codon into the second exon of crsp34 (Figure 2, B and D). Using the Ensembl gene prediction (Zv2, Sanger Institute) and data from 5′ and 3′ RACE and RT–PCR, we isolated three zebrafish crsp34 transcripts, which were termed crsp34_long, crsp34_shortA, and crsp34_shortB. These variants are produced by alternative splicing of exons IV and V (Figure 2C). Exons V and VI are not predicted by Ensembl Zv5 (ENSDARG00000009681, Sanger Institute), but share high similarity to human Crsp34 protein. Crsp34 protein contains a coiled coil domain in exons I and II and a bipartite nuclear localization signal in exon III (Figure 2D; domains were predicted by Ensembl Zv5, Sanger Institute). m885 truncates all isoforms of Crsp34 to 110 amino acids, removing the nuclear localization signal.
Figure 2.—
Positional cloning of m885. (A) The mutation was mapped between SSLPs z11001 and z13142 and further fine mapped between EST fb80f02 and the elavl4 gene. fb80f02 aligned to a genomic contig (Sanger Institute, Zv2) that contained BAC end zK203.a5.T7. A genomic walk started from zK203.a5.Sp6 spanned the critical interval by PAC clones b13109, n1522, i0251, and c01163. The interval was defined by one recombinant in 2054 meioses on either side of the interval, representing a genetic distance of 0.15 cM. The closest proximal marker was anchored on contig ctg26110, which contained the crsp34 gene (Sanger Institute, Zv2). (B) Sequencing the ORF of crsp34 revealed a C to A exchange in the m885 allele, introducing a premature stop codon. (C and D) crsp34 exon structure was revealed by alignment of cDNA to genomic sequence (Zv5, Sanger Institute). As exons Vb and VI could not be retrieved from the genome assembly, their boundaries were identified on the basis of the conserved exon structure of the human CRSP34 locus, which we found for exons I–III, IVb, VII, and VIII (human genome assembly, Sanger Institute). (C) Alternative splicing of exons IVa and IVb, and Va and Vb. Inclusion of exon sequences IVa or Va in the mature transcript causes stop codons in the ORF after 4 and 8 bases, respectively. (D) Crsp34 protein is expressed in three isoforms: Crsp34_long (311 aa), encoded by exons I–VIII; Crsp34_shortA (194 aa), encoded by exons I–Va; and Crsp34_shortB (160 aa), encoded by exons I–IVa. m885 truncates all isoforms to 110 aa, retaining a coiled coil domain (depicted in yellow) but removing the nuclear localization signal (depicted in green). Domains were predicted by Ensembl Zv5 (Sanger Institute).
A BLAST search of the zebrafish genome against crsp34 coding sequence retrieved only one significant match (Ensembl Zv5, Sanger Institute). Human, mouse, and zebrafish crsp34 loci are flanked by the rap guanine nucleotide exchange factor 1 and netrin G1 genes (http://www.ncbi.nlm.nih.gov/mapview), revealing synteny conservation. Thus there is no indication of an additional copy of the crsp34 gene in the zebrafish genome. Zebrafish Crsp34_long protein (AAH57508) shares 88% identity with human Crsp34 (NP_004260), which has been isolated as a subunit of the transcriptional mediator complex (Ryu et al. 1999). Crsp34_long homologs are also found in Drosophila melanogaster, Caenorhabditis elegans, and Schizosaccharomyces pombe, yet this subunit is absent from the Saccharomyces cerevisiae mediator (Bourbon et al. 2004; H. M. Bourbon, personal communication).
Rescue and morpholino phenocopy confirm the identity of the mutation:
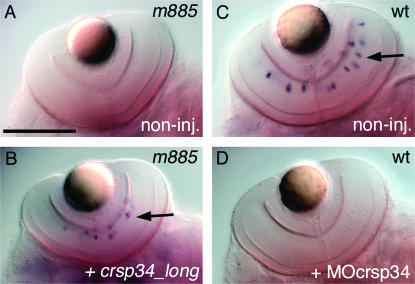
To establish a causal relationship between truncation of Crsp34 and the lack of DA amacrine cells, we tested whether micro-injection of wild-type crsp34 mRNA could rescue the formation of these cells. Homozygous mutant embryos injected with wild-type crsp34_long mRNA consistently formed DA amacrine cells (22/22 mutant embryos), which never occurred in untreated mutant embryos (Figure 3, A and B). In contrast, DA amacrine cells of crsp34 mutant embryos are not rescued by crsp34_shortA or crsp34_shortB mRNA (not shown). At the same time, we did not observe any gain-of-function phenotypes in wild-type siblings (regarding live body shape of larvae and the formation of catecholaminergic groups) injected with any crsp34 isoform (not shown). Thus, Crsp34_long is permissively required for the development of DA amacrine cells in the zebrafish retina. Furthermore, translational inhibition of crsp34 by morpholino MOcrsp34 injection prevented the formation of DA amacrine cells in all injected wild-type embryos (17/17; Figure 3, C and D). Co-injection of MOcrsp34 with MOcrsp34 target sequence fused to GFP entirely ablated green fluorescence in 98/98 injected embryos, while co-injection with control GFP mRNA produced robust fluorescence in 91/91 injected embryos (not shown). This result indicates that MOcrsp34 efficiently and specifically inhibits translation of crsp34 and, together with the loss of the nuclear localization signal in mutant Crsp34 protein, argues that m885 is a null allele. Together, our rescue and phenocopy experiments confirm the causal relationship between the disruption of the crsp34 gene and the absence of DA amacrine cells.
Figure 3.—
Rescue and phenocopy of m885 confirm the identity of the mutation. Embryos were stained by WISH for th. (A) Control m885 mutant embryos at 3 dpf do not form DA amacrine cells. (B) Micro-injection of 600 pg crsp34_long mRNA rescues the formation of some DA amacrine cells in homozygous mutant siblings at 3 dpf (arrow). (C) Untreated wild-type embryos form normal numbers of DA amacrine cells by 80 hpf (arrow), (D) which wild-type siblings injected with 0.3 pmol MOcrsp34 morpholino fail to generate. Bar, 100 μm.
Expression of crsp34 during zebrafish development:
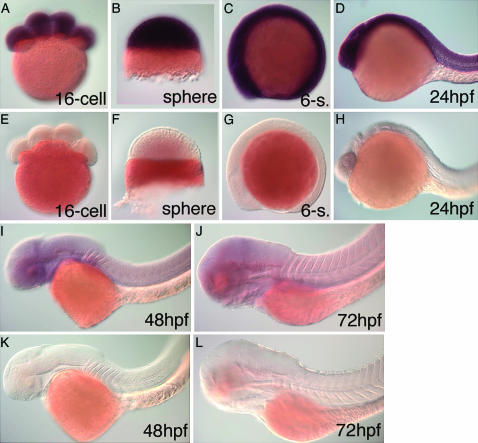
We analyzed the expression of crsp34 during zebrafish development by whole-mount WISH. A riboprobe to all crsp34 isoforms detects maternal contribution of crsp34 mRNA at the 16-cell stage, when zygotic transcription has not yet been initiated (Figure 4A). Expression of crsp34 is ubiquitous at all stages analyzed (Figure 4, B–D, I, and J). At 48 and 72 hpf, the level of expression appears to be lower when compared to that at earlier stages. To allow comparison of staining intensities, embryos of all stages were processed in the same batch. Comparison to a parallel batch of embryos that was hybridized to the sense probe suggests that the weak signal detected at 48 and 72 hpf represents a specific staining (Figure 4, E–H, K, and L).
Figure 4.—
Expression of crsp34 during zebrafish development. WISH was carried out with a common probe for all isoforms. (A) crsp34 mRNA is present at the 16-cell stage, when zygotic transcription has not yet been initiated. crsp34 is ubiquitously expressed at (B) sphere stage, (C) six somites, (D) 24 hpf, (I) 48 hpf, and (J) 72 hpf. (E–H, K, and L) WISH with the sense probe serves as a reference for background staining. Staining time was identical for all embryos and antisense and sense control embryos were processed together in a single tube.
To determine the temporal expression profile of crsp34 isoforms, we performed RT–PCR on mRNA isolated from different developmental stages with isoform-specific primer pairs. crsp34_long is expressed at all embryonic stages analyzed, as well as in the adult fish (not shown). Conversely, expression of crsp34_shortA and crsp34_shortB is detected only at 24 hpf and at 24 and 48 hpf, respectively (not shown). Accordingly, crsp34_long is the predominantly expressed variant, and therefore the ubiquitous signal observed by WISH most likely reflects crsp34_long expression.
Loss of Crsp34 affects the formation of subsets of amacrine cells:
Any in vivo role of mediator subunit Crsp34 had not been previously addressed. Thus, we set out to use crsp34-deficient zebrafish to investigate the role of this subunit in the development of the CNS. As cell types of the zebrafish retina can be easily distinguished both by molecular markers and by the positions of their cell bodies (Malicki 2000), we decided to focus our analysis of Crsp34 on the retina.
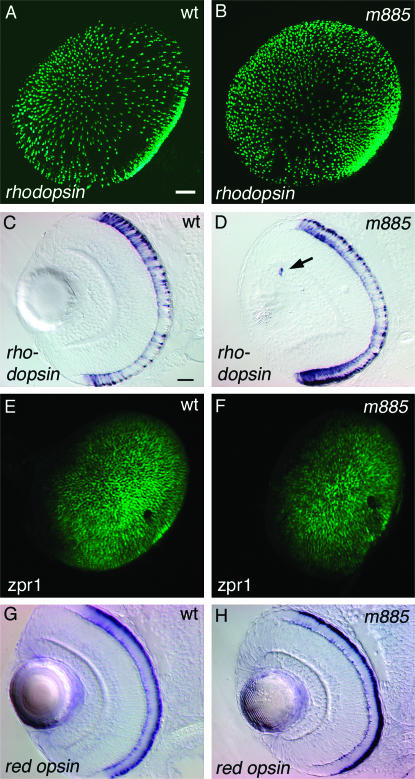
The lack of DA amacrine cells in the crsp34m885 mutant retina could indicate a failure of neurotransmitter production or the absence of this population of cells. To distinguish between these alternatives we assayed for amacrine cells by pan-amacrine features. We identified the amacrine cell layers by the rounded morphology of amacrine cell bodies (Dowling 1987) in retinal cross-sections (Figure 5, A and B, red arrows) and by expression of pax6.2 (Figure 5, C and D, red arrows), which was used as a marker for ganglion and amacrine cell layers (Hirsch and Harris 1997). Quantification of the amacrine cell layer thickness in cross-sections at 78 hpf revealed a 35% reduction in crsp34m885 mutant embryos as compared to wild-type siblings (Table 1). Consistently, we observed a reduction of several amacrine subtypes, including GABAergic cells (Figure 5, E and F), and absence of both cholinergic (Figure 5, G and H) and serotonergic subtypes (Figure 1, I and J).
Figure 5.—
Loss of crsp34 leads to reduction of amacrine and ganglion cell layers. All embryos were fixed at 78 hpf. (A–F) The ganglion cell layer (GCL, white arrow) and amacrine cell layer (ac, red arrow) can be distinguished in sections, counterstained with methylene blue (A and B), stained for pax6.2 expression by WISH (C and D), or by anti-GABA immunohistochemistry (E and F). Quantification of layer thickness revealed a 35% reduction of amacrine cells and a 15% reduction of ganglion cells in mutant vs. wild-type embryos (Table 1). (G) Wild-type embryos develop cholinergic amacrine cells expressing choline acetyl transferase (ChAT). (H) ChAT immunoreactivity is absent from crsp34m885 mutant embryos. (I) Zn5 labels DM-GRASP-expressing ganglion cells and their projections. (J) In crsp34m885 mutant embryos, zn5 staining intensity is reduced. Bar: 20 μm in A for A, B, E, and F and in C for C, D, and G–J.
TABLE 1.
Formation of retinal cells in crsp34m885 mutant embryos and wild-type siblings (78 hpf)
| Assay | wt ± SD | Mutant ± SD | P-value |
|---|---|---|---|
| Thickness of amacrine layer | 24.6 ± 3.2 μm (n = 8) | 16.1 ± 2.7 μm (n = 7) | P < 0.0001 |
| Thickness of ganglion cell layer | 33.4 ± 5.4 μm (n = 13) | 28.3 ± 4.0 μm (n = 14) | P < 0.01 |
| rhodopsin-stained volume | 97,637 ± 28,196 (n = 10) | 165,632 ± 59,793 (n = 8) | P < 0.006 |
| rhodopsin staining intensity (average per pixel) | 187 ± 10 (n = 10) | 190 ± 15 (n = 8) | P = 0.56 |
| Nuclear TOTO3 staining intensity (average per pixel) | 93 ± 8 (n = 10) | 94 ± 6 (n = 8) | P = 0.89 |
| Ectopic rhodopsin-expressing cells | 0.3 ± 0.7 (n = 10) | 6.5 ± 1.8 (n = 8) | P < 0.0001 |
wt, wild type.
Similarly, we quantified the thickness of the ganglion cell layer, comprising ganglion and displaced amacrine cells (Figure 5, A–F, I, and J, white arrows), and found a 15% reduction in crsp34m885 mutant embryos as compared to wild-type siblings (78 hpf; Table 1). It remains unclear whether any loss of displaced amacrine cells contributes to this reduction of the GCL. Although ganglion cells of crsp34m885 mutant embryos show normal expression of pax6.2 and HuC and normal levels of GABA (Figure 5, C–F, and data not shown), they display reduced expression levels of DM-GRASP (Figure 5, I and J), an adhesion molecule involved in fasciculation (Fashena and Westerfield 1999). This indicates that the remaining portion of ganglion cells may harbor additional defects. Taken together, our data suggest that loss of Crsp34 leads to an overall reduction of amacrine cells, including DA cells, and potentially affects some aspects of ganglion cell differentiation.
Crsp34 mutant embryos produce more and ectopic rod photoreceptor cells:
We asked whether loss of Crsp34 similarly affects the formation of other retinal cell types. By 78 hpf, wild-type embryos have formed rod photoreceptor cells across the outer nuclear layer, which can be detected by WISH for rhodopsin (Figure 6A). At the same time, crsp34m885 mutant embryos display a denser arrangement of rods (Figure 6B), which appears indicative of an overproduction of rod cells. To address this possibility, we performed fluorescent WISH for rhodopsin and quantified the stained volume in 3-D stacks of confocal images, taking the total volume of rods as a measure for rod number. This analysis revealed a 70% increase of rod photoreceptor cells in crsp34m885 mutant embryos as compared to their wild-type siblings (Table 1). To exclude artifacts by variations in staining intensities or recording conditions, we analyzed the mean intensities of in situ hybridization signals (Alexa-488) and of nuclear counterstaining by TOTO3. This control for quantification revealed no significant difference between mutant and wild-type embryos (Table 1), suggesting that the detected change indeed represents a difference in stained volume, respectively cell number, but not in staining intensities or recording techniques. Surprisingly, we observed rhodopsin-expressing cells at ectopic locations in crsp34m885 mutant embryos, which occurred only rarely in wild-type siblings (Figure 6, C and D, and Table 1). These ectopic cells reside in the GCL and the innermost region of the INL, which is populated by amacrine cells.
Figure 6.—
Loss of crsp34 differentially affects photoreceptor cells. The analysis was carried out at 78 hpf. (A and B) Rod photoreceptor cells were labeled by fluorescent in situ hybridization to rhodopsin, recorded as 3-D stacks and subsequently processed to 2-D projections. Rhodopsin-expressing cells are denser in the retina of crsp34m885 mutant embryos. Quantification of rhodopsin staining volume revealed a 70% increase in mutant embryos as compared to wild-type siblings (Table 1). (C and D) Mutant embryos produce single rhodopsin-expressing cells at ectopic locations, which rarely occurred in the wild type. (E and F) Double-cone photoreceptors were labeled by zpr1/FRet43 immunohistochemistry. crsp34m885 mutant embryos show slightly reduced zpr1/FRet43 expression. (G and H) Red cones, labeled by red opsin WISH, form normally in crsp34m885 mutant embryos. Bar, 20 μm in A for A, B, E, and F and in C for C, D, G, and H.
Next, we addressed the formation of cone photoreceptor cells. Immunohistochemistry with zpr1/FRet43 (Larison and Bremiller 1990) indicates a slight reduction of double cone photoreceptors in crsp34 mutant embryos (Figure 6, E and F). Conversely, red opsin WISH revealed no difference between wild-type and crsp34m885 mutant embryos, indicating that red cone photoreceptor cells are not affected by loss of Crsp34 (Figure 6, G and H).
Furthermore, we attempted to determine the effect of the mutation on the formation of bipolar cells, detected by WISH for vsx2, and Müller glia, labeled by immunohistochemistry to carbonic anhydrase II. While some cell bodies of Müller glia retained abnormal positions within the INL, we did not observe changes in the population sizes of these cell types (not shown).
In summary, loss of zygotic Crsp34 significantly increases the formation of rod photoreceptor cells but may reduce the formation of double cone photoreceptors, while proportions of bipolar cells, Müller glia, and red cone photoreceptor cells remain unaffected.
The reduction of amacrine cells is not caused by cell death and may reflect a differentiation defect:
Potentially, the reduction of amacrine cells could be due to apoptosis. To investigate this possibility, we labeled apoptotic cells in mutant and wild-type embryos at 24, 48, 60, and 72 hpf by TUNEL assay. Up to 60 hpf, we detected no increase in cell death in crsp34m885 mutant embryos (Figure 7, A and B, and data not shown). Enhanced cell death was first observed at 72 hpf in the optic tectum and was scattered across the INL of the retina (Figure 7, C–F). At 4 dpf, apoptosis is still restricted to optic tectum and retina (not shown). As the lack of DA amacrine cells is overt at 60 hpf, there is no indication for a role of cell death in the establishment of the DA amacrine phenotype in crsp34m885 mutant embryos.
Figure 7.—
m885 mutant embryos have normal apoptosis levels until 60 hpf. (A and B) At 60 hpf, TUNEL staining reveals that apoptosis levels are similar in crsp34m885 mutant and wild-type embryos. (C and D) At 72 hpf, mutant embryos exhibit an increase of cell death in the optic tectum (arrow), as well as in the retina, where apoptotic cells are scattered across the INL (E and F, black arrow). Bar, 100 μm in A for A–D and in E for E and F.
Alternatively, loss of Crsp34 could, directly or indirectly, affect retinal patterning or differentiation. Amacrine cells are reduced concentrically in mutant embryos, which could be indicative of the reduced range of an activator or increased range of an inhibitor, secreted from the ganglion cell layer. A candidate inhibitory factor was sonic hedgehog, which is secreted from ganglion and a subset of amacrine cells (Neumann and Nuesslein-Volhard 2000; Shkumatava et al. 2004) and mediates negative feedback regulation of ganglion cell differentiation (Zhang and Yang 2001). Yet, we found no indication for increased hedgehog signaling by expression analysis of the target genes ptc2 (Stenkamp et al. 2000), gli3 (Tyurina et al. 2005), or hhip1 (Chuang and McMahon 1999) (data not shown). Accordingly, any potential extrinsic or intrinsic factor that mediates the reduction of amacrine cells remains to be identified.
The increased production of rod cells is probably not caused by premature cell cycle exit of progenitors:
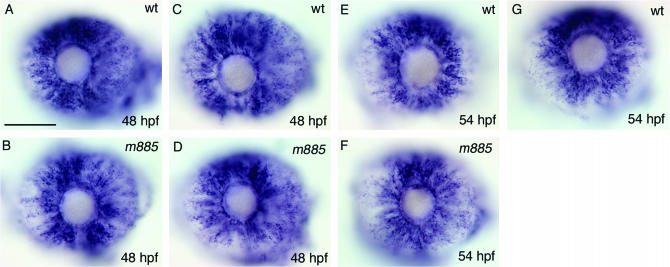
In wild-type embryos, the majority of rod precursor cells become postmitotic between 48 and 54 hpf (Hu and Easter 1999). As differentiation is closely linked to the time point of cell cycle exit (Alexiades and Cepko 1997), we asked whether premature cell cycle exit might account for the overproduction of rod cells in crsp34m885 mutant embryos. To address this issue, we labeled proliferating cells by WISH for mcm5, which is expressed in proliferating cells (Blow and Hodgson 2002). At 48 and 54 hpf, mutant and wild-type embryos display variable expression of mcm5 in the retina. Although embryos with strongest or weakest retinal expression of mcm5 tend to be wild type and homozygous mutant (Figure 8, A and B), respectively, there are mutant and wild-type embryos with indistinguishable patterns and levels of mcm5 expression at 48 hpf (two of three mutants like 3 of 9 wild types; Figure 8, C and D) and 54 hpf (six of six mutants like 7 of 10 wild types; Figure 8, E–G). Accordingly, we do not find a clear correlation, and therefore it seems unlikely that the overproduction of rods in mutant embryos is caused by a proliferation defect.
Figure 8.—
Proliferation is not consistently altered in m885 mutant embryos. (A) At 48 hpf, some wild-type embryos show strong mcm5 expression (6/9 embryos), which is exclusively expressed in proliferating cells. (B) At the same stage, weaker mcm5 expression can occur in crsp34m885 mutant embryos (1/3 embryos). (C and D) Importantly, some wild-type (3/9) and mutant (2/3) embryos exhibit comparable levels of mcm5 expression. (E and F) Similarly, at 54 hpf crsp34m885 mutant embryos (6/6) show similar mcm5 expression as a portion of wild-type siblings (7/10), while some wild-type embryos (3/10) display stronger mcm5 expression levels (G). Genotypes of embryos were determined by genomic PCR and sequencing. Bar, 100 μm.
Loss of mediator subunits Crsp34, Trap100, and Crsp150 differentially affects amacrine cells and rods:
The retinal phenotype of crsp34m885 mutant embryos could reflect a subunit-specific function of Crsp34 or, alternatively, reduced activity of the mediator complex. To examine these alternatives, we investigated whether loss of different mediator subunits has divergent or similar effects on retinal development. To this end, we took advantage of the recent isolation of two zebrafish lines carrying mutations in other subunits of the mediator complex: the hi2143 allele, a retroviral insertion that disrupts the crsp150 gene (Amsterdam et al. 2004), and the lessen allele, carrying an early stop codon in the trap100 gene (Pietsch et al. 2006).
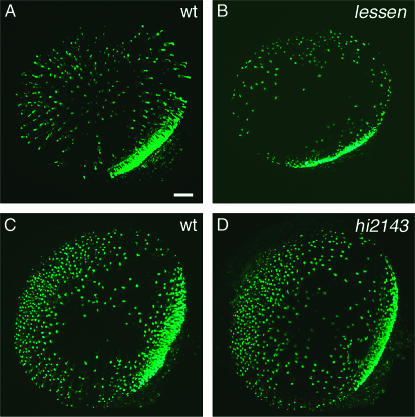
To explore the roles of these two subunits in the formation of amacrine cells, we performed in situ hybridization to pax6.2, labeling ganglion cells as well as amacrine cells (Figure 9, A–C, red arrows). Again, we measured the thickness of amacrine cell layers in cross-sections. At 78 hpf, both trap100lessen and crsp150hi2143 mutant embryos show no significant difference from their wild-type siblings in the extent of pax6.2 expression domains (Tables 2 and 3), in contrast to our results for crsp34m885 mutant embryos. Around the same stage, however, DA amacrine cells are entirely absent from trap100lessen mutant embryos (Figure 9E) and reduced in crsp150hi2143 mutant embryos (Figure 9F).
Figure 9.—
Loss of mediator subunits Trap100 (lessen) and Crsp150 (hi2143) does not affect the thickness of the amacrine cell layer, but impairs the formation of DA amacrine cells. (A–C) The amacrine cell layer (red arrow), as well as the GCL, were labeled by WISH for pax6.2. No difference was observed between wild-type and mutant embryos at 78 hpf. Quantification of layer thickness confirmed the absence of significant changes (Tables 2 and 3). (D) WISH for th shows DA amacrine cells in 3 dpf wild-type embryos (arrow). (E) DA amacrine cells remain absent from trap100lessen mutant embryos at 3 dpf and are reduced in crsp150hi2143 mutant embryos at the same stage (arrow in F). Bars: A, 20 μm for A–C; D, 100 μm for D–F.
TABLE 2.
Formation of retinal cells in trap100lessen mutant embryos and wild-type siblings (78 hpf)
| Assay | wt ± SD | Mutant ± SD | P-value |
|---|---|---|---|
| Thickness of amacrine layer in | 31.2 ± 1.2 μm (n = 3) | 32.7 ± 1.2 μm (n = 3) | P = 0.19 |
| rhodopsin-stained volume | 166,364 ± 38,997 (n = 7) | 55,387 ± 28,192 (n = 7) | P < 0.0001 |
| rhodopsin staining intensity (average per pixel) | 233 ± 4 (n = 7) | 242 ± 10 (n = 7) | P = 0.04 |
| Nuclear TOTO3 staining intensity (average per pixel) | 208 ± 26 (n = 7) | 205 ± 46 (n = 7) | P = 89 |
TABLE 3.
Formation of retinal cells in crsp150hi2143 mutant embryos and wild-type siblings (78 hpf)
| Assay | wt ± SD | Mutant ± SD | P-value |
|---|---|---|---|
| Thickness of amacrine layer | 38.0 ± 2.9 μm (n = 4) | 34.7 ± 2.5 μm (n = 3) | P = 0.18 |
| rhodopsin-stained volume | 209,400 ± 71,627 (n = 8) | 175,621 ± 40,695 (n = 8) | P = 0.27 |
| rhodopsin staining intensity (average per pixel) | 192 ± 11 (n = 8) | 192 ± 14 (n = 8) | P = 0.96 |
| Nuclear TOTO3 staining intensity (average per pixel) | 110 ± 14 (n = 8) | 111 ± 16 (n = 8) | P = 0.89 |
As for crsp34m885, we quantified rod photoreceptor cells in 78-hpf embryos by the stained volume of fluorescent WISH to rhodopsin. In contrast to crsp34m885 mutant embryos, which show a 70% increase (see above), trap100lessen mutant embryos show a 77% decrease of rod photoreceptor cells (Figure 10, A and B, and Table 2). At the same time, rod formation is not significantly affected in crsp150hi2143 mutants (Figure 10, C and D, and Table 3). Further, neither loss of trap100 nor loss of crsp150 leads to the formation of rhodopsin-expressing cells at ectopic locations, as observed in crsp34 mutant embryos.
Figure 10.—
Loss of mediator subunit Trap100 leads to decreased formation of rod photoreceptor cells. Rods were labeled by fluorescent WISH for rhodopsin (78 hpf), recorded as 3-D stacks for quantitative analysis, and presented here as 2-D projections. (A and B) Comparison of rod density in (A) wild-type embryos and (B) trap100lessen mutant siblings reveals a lower density in the central region of the mutant retina. Quantitative analysis of staining volumes indicates a 77% decrease in trap100lessen mutant embryos (Table 2). (C and D) The corresponding analysis of rod formation in crsp150hi2143 mutant embryos does not reveal any apparent change between mutant and wild type. Quantitative analysis confirms the absence of a significant difference (Table 3). Bar, 20 μm.
As before, we tested the possibility of staining or recording artifacts by comparison of signal intensities between mutant and wild-type embryos. Mean intensity values for in situ hybridization signals and nuclear counterstaining are highly similar between sets of lessen or hi2143 mutant embryos and their wild-type siblings, indicating that our data indeed reflect differences in staining volumes (Tables 2 and 3). Furthermore, we measured the thickness of ganglion cell layers in cross-sections and analyzed the formation of bipolar cells and Müller glia in trap100lessen and crsp150hi2143 mutant embryos at 78 hpf. These analyses did not reveal changes in lessen or hi2143 mutant embryos vs. their wild-type siblings (not shown).
In summary, DA amacrine cells are reduced or absent in embryos mutant for crsp34, trap100, and crsp150, while the entire layer of amacrine cells is significantly reduced only in the crsp34 mutant. Surprisingly, the formation of rhodopsin-expressing cells is significantly increased upon loss of Crsp34—both in the ONL and at ectopic positions—but decreased upon loss of Trap100. Rod cell formation is not affected by loss of Crsp150. Thus, we observed both overlapping and divergent mutant phenotypes, indicating both common and distinct functions of Crsp34, Trap100, and Crsp150.
DISCUSSION
As most subunits of the transcriptional mediator complex have not been functionally analyzed in vertebrates, the extent to which subunit composition may contribute to developmental mechanisms has remained unclear. Here, we present the first functional analysis of the subunit Crsp34 in the context of development and cell differentiation of zebrafish. We tested the concept of subunit-specific roles by analyzing the loss-of-function effects of three mediator subunits on the development of the vertebrate CNS: Trap100 (Pietsch et al. 2006), Crsp150 (Amsterdam et al. 2004), and Crsp34 (this study). As an experimental system we chose the developing zebrafish retina, since it represents one of the best-studied units of the CNS, regarding anatomy and control of proliferation, as well as expression of genes involved in patterning and differentiation for zebrafish (reviewed in Malicki 2000). Therefore, analysis of the retina in a mutant embryo allows us to detect developmental defects at high resolution.
We isolated the zebrafish mutation crsp34m885, which disrupts the mediator subunit Crsp34 and reduces the formation of several neuronal groups, including DA neurons of the retina. Micro-injection of wild-type mRNA into crsp34m885 mutant embryos showed that the predominant crsp34 isoform is sufficient to rescue the formation of DA amacrine cells. At the same time, no gain-of-function phenotypes were observed for crsp34 overexpression. Thus, crsp34 does not appear to have instructive functions in development, but may be required to execute instructive functions in conjunction with other developmental signaling, patterning, or differentiation programs.
In the zebrafish retina, loss of Crsp34 leads to a reduction of amacrine cells, including DA, GABAergic, cholinergic, and serotonergic neurons. In the photoreceptor layer of crsp34m885 mutant embryos, the number of rods is significantly increased, while double cones are decreased, and red cones appear unaffected. Our data show that this shift of retinal cell type composition is likely not due to changes in proliferation or apoptosis levels. As ganglion cells are essential for normal development of other retinal cells types (Kay et al. 2001), the subtle defect observed in differentiated ganglion cells of crsp34m885 mutant embryos could potentially play a causative role in the establishment of other retinal defects. Furthermore, as amacrine and rod cells of the rat retina derive from a common precursor population (Alexiades and Cepko 1997), loss of Crsp34 may, directly or indirectly, bias common precursors toward the rod fate.
The ubiquitous expression of crsp34 mRNA may suggest a universal requirement for Crsp34 in transcriptional initiation. Further, maternally contributed wild-type Crsp34 protein may partially rescue zygotic requirements for Crsp34 in mutant embryos during early phases of development. This constellation might mimic a specific function of Crsp34 in the formation of cell types that differentiate late in development, due to lower residual levels of maternal protein (compare to maternal rescue of zygotic functions for the Nodal coreceptor oep and pou5f1/spg genes described in Gritsman et al. 1999 and Lunde et al. 2004). However, in crsp34m885 mutant embryos a reduction of cells is observed in the ganglion and amacrine cell layers, whose progenitors are the first to become postmitotic, as well as in later-born double-cone photoreceptors (Hu and Easter 1999). Several late differentiating cell types—bipolar cells, Müller glia, and red cone photoreceptor cells—appear to form normally, while the number of rod photoreceptor cells is even considerably increased. Accordingly, there is no simple correlation between differentiation defects and the residual level of maternal Crsp34.
The apparent discrepancy between the restricted defects of crsp34m885 mutant embryos and ubiquitous expression of crsp34 mRNA may be explained by dynamic recruitment of Crsp34 protein to the mediator complex. As recruitment of subunits can induce conformational changes and expose differential reactive interfaces (Taatjes et al. 2004a), Crsp34 protein may thus contribute to the regulation of a restricted subset of promoters, despite its ubiquitous mRNA expression. Interestingly, the Crsp34 homolog of S. pombe, Pcm3 (Bourbon et al. 2004; H. M. Bourbon, personal communication), is nonessential for cell viability, supporting a restricted role of this subunit in transcription (Spahr et al. 2001).
In striking contrast to the overproduction of rod cells observed in crsp34m885 mutant embryos, loss of Trap100 leads to a significant reduction of this cell type. At the same time, the layer of amacrine cells is significantly reduced only in crsp34 mutant embryos, but not in embryos mutant for trap100 or crsp150. Yet, DA amacrine cells are reduced or absent in all three mutant lines. These findings allow the following conclusions:
Judged from their loss-of-function phenotypes, Crsp34 and Trap100 appear to play divergent roles in the development of rod photoreceptor cells. Since Trap100 has been linked to thyroid hormone signaling via subunit Trap220 (Zhang and Fondell 1999; Ito et al. 2002), the reduction of rod photoreceptor cells in the trap100lessen mutant retina could be due to a defect in this signaling pathway.
DA amacrine cells are particularly sensitive to the activity of the mediator complex as a whole or to a submodule comprising all three subunits.
The observation that the thickness of amacrine layers is not significantly affected in trap100lessen and crsp150hi2143 may indicate that DA neurons in the zebrafish retina develop as a separate group, as has already been proposed on the basis of their interplexiform projections (Dowling 1987).
Taken together, these data suggest that combinatorial recruitment of mediator subunits specifically modulates developmental decisions and thus represent an additional level of developmental control in the vertebrate CNS.
Acknowledgments
We thank S. Götter for expert zebrafish care, R. Koppa for plastic sections, and the zebrafish community for plasmids and probes. We are grateful to N. Hopkins, A. Amsterdam, and S. Farrington for sharing their crsp150 mutant zebrafish line and for disclosing the genotyping information; and we thank H. M. Bourbon for sharing unpublished results on Crsp34 conservation. We thank Roland Dosch for critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 505-B7 (W.D.), the Landesgraduiertenstipendium (K.D.), the Human Frontiers Long-Term Fellowship (S.R.), the National Institutes of Health (DK067285), and the March of Dimes (5-FY02-270) (I.T.S.).
References
- Alexiades, M. R., and C. L. Cepko, 1997. Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development 124: 1119–1131. [DOI] [PubMed] [Google Scholar]
- Amemiya, C. T., and L. I. Zon, 1999. Generation of a zebrafish P1 artificial chromosome library. Genomics 58: 211–213. [DOI] [PubMed] [Google Scholar]
- Amsterdam, A., R. M. Nissen, Z. Sun, E. C. Swindell, S. Farrington et al., 2004. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 101: 12792–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenzana, F. J., D. Clemente, R. Sanchez-Gonzalez, A. Porteros, J. Aijon et al., 2005. Development of the cholinergic system in the brain and retina of the zebrafish. Brain Res. Bull. 66: 421–425. [DOI] [PubMed] [Google Scholar]
- Bellipanni, G., E. Rink and L. Bally-Cuif, 2002. Cloning of two tryptophan hydroxylase genes expressed in the diencephalon of the developing zebrafish brain. Gene Expr. Patterns 2: 251–256. [DOI] [PubMed] [Google Scholar]
- Belliveau, M. J., and C. L. Cepko, 1999. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development 126: 555–566. [DOI] [PubMed] [Google Scholar]
- Blais, A., and B. D. Dynlacht, 2005. Constructing transcriptional regulatory networks. Genes Dev. 19: 1499–1511. [DOI] [PubMed] [Google Scholar]
- Blow, J. J., and B. Hodgson, 2002. Replication licensing—defining the proliferative state? Trends Cell Biol. 12: 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boube, M., C. Faucher, L. Joulia, D. L. Cribbs and H. M. Bourbon, 2000. Drosophila homologs of transcriptional mediator complex subunits are required for adult cell and segment identity specification. Genes Dev. 14: 2906–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boube, M., L. Joulia, D. L. Cribbs and H. M. Bourbon, 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110: 143–151. [DOI] [PubMed] [Google Scholar]
- Bourbon, H. M., A. Aguilera, A. Z. Ansari, F. J. Asturias, A. J. Berk et al., 2004. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14: 553–557. [DOI] [PubMed] [Google Scholar]
- Chuang, P. T., and A. P. McMahon, 1999. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397: 617–621. [DOI] [PubMed] [Google Scholar]
- Denkers, N., P. Garcia-Villalba, C. K. Rodesch, K. R. Nielson and T. J. Mauch, 2004. FISHing for chick genes: triple-label whole-mount fluorescence in situ hybridization detects simultaneous and overlapping gene expression in avian embryos. Dev. Dyn. 229: 651–657. [DOI] [PubMed] [Google Scholar]
- Dowling, J. E., 1987. The Retina, an Approachable Part of the Brain. The Belknap Press of Harvard University Press, Cambridge, MA.
- Fashena, D., and M. Westerfield, 1999. Secondary motoneuron axons localize DM-GRASP on their fasciculated segments. J. Comp. Neurol. 406: 415–424. [DOI] [PubMed] [Google Scholar]
- Fondell, J. D., H. Ge and R. G. Roeder, 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93: 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsman, K., J. Zhang, S. Cheng, E. Heckscher, W. S. Talbot et al., 1999. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 97: 121–132. [DOI] [PubMed] [Google Scholar]
- Hauptmann, G., and T. Gerster, 1994. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 10: 266. [DOI] [PubMed] [Google Scholar]
- Hirsch, N., and W. A. Harris, 1997. Xenopus Pax-6 and retinal development. J. Neurobiol. 32: 45–61. [PubMed] [Google Scholar]
- Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner et al., 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Holzschuh, J., S. Ryu, F. Aberger and W. Driever, 2001. Dopamine transporter expression distinguishes dopaminergic neurons from other catecholaminergic neurons in the developing zebrafish embryo. Mech. Dev. 101: 237–243. [DOI] [PubMed] [Google Scholar]
- Holzschuh, J., A. Barrallo-Gimeno, A. K. Ettl, K. Durr, E. W. Knapik et al., 2003. Noradrenergic neurons in the zebrafish hindbrain are induced by retinoic acid and require tfap2a for expression of the neurotransmitter phenotype. Development 130: 5741–5754. [DOI] [PubMed] [Google Scholar]
- Hu, M., and S. S. Easter, 1999. Retinal neurogenesis: the formation of the initial central patch of postmitotic cells. Dev. Biol. 207: 309–321. [DOI] [PubMed] [Google Scholar]
- Ito, M., C. X. Yuan, H. J. Okano, R. B. Darnell and R. G. Roeder, 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5: 683–693. [DOI] [PubMed] [Google Scholar]
- Ito, M., H. J. Okano, R. B. Darnell and R. G. Roeder, 2002. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 21: 3464–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janody, F., Z. Martirosyan, A. Benlali and J. E. Treisman, 2003. Two subunits of the Drosophila mediator complex act together to control cell affinity. Development 130: 3691–3701. [DOI] [PubMed] [Google Scholar]
- Kay, J. N., K. C. Finger-Baier, T. Roeser, W. Staub and H. Baier, 2001. Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal homolog. Neuron 30: 725–736. [DOI] [PubMed] [Google Scholar]
- Kimmel, C. B., W. W. Ballard, S. R. Kimmel, B. Ullmann and T. F. Schilling, 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203: 253–310. [DOI] [PubMed] [Google Scholar]
- Kosman, D., C. M. Mizutani, D. Lemons, W. G. Cox, W. McGinnis et al., 2004. Multiplex detection of RNA expression in Drosophila embryos. Science 305: 846. [DOI] [PubMed] [Google Scholar]
- Kudoh, T., M. Tsang, N. A. Hukriede, X. Chen, M. Dedekian et al., 2001. A gene expression screen in zebrafish embryogenesis. Genome Res. 11: 1979–1987. [DOI] [PubMed] [Google Scholar]
- Kwok, S., S. Y. Chang, J. Sninsky and A. Wang, 1995. Design and use of mismatched and degenerate primers, pp. 144–146 in PCR Primer, a Laboratory Manual, edited by W. C. Dieffenbach and G. S. Dveksler. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Larison, K. D., and R. Bremiller, 1990. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development 109: 567–576. [DOI] [PubMed] [Google Scholar]
- Li, S., Z. Mo, X. Yang, S. M. Price, M. M. Shen et al., 2004. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron 43: 795–807. [DOI] [PubMed] [Google Scholar]
- Lunde, K., H. G. Belting and W. Driever, 2004. Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr. Biol. 14: 48–55. [DOI] [PubMed] [Google Scholar]
- Malicki, J., 2000. Harnessing the power of forward genetics—analysis of neuronal diversity and patterning in the zebrafish retina. Trends Neurosci. 23: 531–541. [DOI] [PubMed] [Google Scholar]
- Malik, S., W. Gu, W. Wu, J. Qin and R. G. Roeder, 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5: 753–760. [DOI] [PubMed] [Google Scholar]
- Mannervik, M., Y. Nibu, H. Zhang and M. Levine, 1999. Transcriptional coregulators in development. Science 284: 606–609. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales, J. R., F. Del Bene, G. Nica, M. Hammerschmidt, P. Bovolenta et al., 2005. Differentiation of the vertebrate retina is coordinated by an FGF signaling center. Dev. Cell 8: 565–574. [DOI] [PubMed] [Google Scholar]
- Michelmore, R. W., I. Paran and R. V. Kesseli, 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, E. M., T. Furukawa, J. E. Lee and C. L. Cepko, 1999. NeuroD regulates multiple functions in the developing neural retina in rodent. Development 126: 23–36. [DOI] [PubMed] [Google Scholar]
- Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon et al., 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398: 828–832. [DOI] [PubMed] [Google Scholar]
- Nasevicius, A., and S. C. Ekker, 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26: 216–220. [DOI] [PubMed] [Google Scholar]
- Neumann, C. J., and C. Nuesslein-Volhard, 2000. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science 289: 2137–2139. [DOI] [PubMed] [Google Scholar]
- Nornes, S., M. Clarkson, I. Mikkola, M. Pederson, A. Bardsley et al., 1998. Zebrafish contains two Pax6 genes involved in eye development. Mech. Dev. 77: 185–196. [DOI] [PubMed] [Google Scholar]
- Pietsch, J., J. M. Delalande, B. Jakaitis, J. D. Stensby, S. Dohle et al., 2006. lessen encodes a zebrafish Trap100 required for enteric nervous system development. Development 133: 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez, C., Z. Suldan, J. Ward, C. P. Chang, D. Burakov et al., 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12: 1787–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble et al., 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398: 824–828. [DOI] [PubMed] [Google Scholar]
- Robinson, J., E. A. Schmitt and J. E. Dowling, 1995. Temporal and spatial patterns of opsin gene expression in zebrafish (Danio rerio). Vis. Neurosci. 12: 895–906. [DOI] [PubMed] [Google Scholar]
- Ryu, S., S. Zhou, A. G. Ladurner and R. Tjian, 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397: 446–450. [DOI] [PubMed] [Google Scholar]
- Ryu, S., J. Holzschuh, S. Erhardt, A. K. Ettl and W. Driever, 2005. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc. Natl. Acad. Sci. USA 102: 18467–18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkumatava, A., S. Fischer, F. Muller, U. Strahle and C. J. Neumann, 2004. Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development 131: 3849–3858. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel, L., and W. Driever, 1994. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development 120: 2443–2455. [DOI] [PubMed] [Google Scholar]
- Spahr, H., C. O. Samuelsen, V. Baraznenok, I. Ernest, D. Huylebroeck et al., 2001. Analysis of Schizosaccharomyces pombe mediator reveals a set of essential subunits conserved between yeast and metazoan cells. Proc. Natl. Acad. Sci. USA 98: 11985–11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp, D. L., R. A. Frey, S. N. Prabhudesai and P. A. Raymond, 2000. Function for Hedgehog genes in zebrafish retinal development. Dev. Biol. 220: 238–252. [DOI] [PubMed] [Google Scholar]
- Taatjes, D. J., and R. Tjian, 2004. Structure and function of CRSP/Med2; a promoter-selective transcriptional coactivator complex. Mol. Cell 14: 675–683. [DOI] [PubMed] [Google Scholar]
- Taatjes, D. J., M. T. Marr and R. Tjian, 2004. a Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell. Biol. 5: 403–410. [DOI] [PubMed] [Google Scholar]
- Taatjes, D. J., T. Schneider-Poetsch and R. Tjian, 2004. b Distinct conformational states of nuclear receptor-bound CRSP-Med complexes. Nat. Struct. Mol. Biol. 11: 664–671. [DOI] [PubMed] [Google Scholar]
- Treisman, J., 2001. Drosophila homologues of the transcriptional coactivation complex subunits TRAP240 and TRAP230 are required for identical processes in eye-antennal disc development. Development 128: 603–615. [DOI] [PubMed] [Google Scholar]
- Trevarrow, B., D. L. Marks and C. B. Kimmel, 1990. Organization of hindbrain segments in the zebrafish embryo. Neuron 4: 669–679. [DOI] [PubMed] [Google Scholar]
- Tudor, M., P. J. Murray, C. Onufryk, R. Jaenisch and R. A. Young, 1999. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 13: 2365–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurina, O. V., B. Guner, E. Popova, J. Feng, A. F. Schier et al., 2005. Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling. Dev. Biol. 277: 537–556. [DOI] [PubMed] [Google Scholar]
- Westerfield, M., 1994. The Zebrafish Book. University of Oregon Press, Eugene, OR.
- Wetts, R., and S. E. Fraser, 1988. Multipotent precursors can give rise to all major cell types of the frog retina. Science 239: 1142–1145. [DOI] [PubMed] [Google Scholar]
- Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu and R. G. Roeder, 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95: 7939–7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and J. D. Fondell, 1999. Identification of mouse TRAP100: a transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol. Endocrinol. 13: 1130–1140. [DOI] [PubMed] [Google Scholar]
- Zhang, X. M., and X. J. Yang, 2001. Regulation of retinal ganglion cell production by Sonic hedgehog. Development 128: 943–957. [DOI] [PMC free article] [PubMed] [Google Scholar]