Abstract
Centromeres are difficult to map even in species where genetic resolution is excellent. Here we show that junctions between repeats provide reliable single-copy markers for recombinant inbred mapping within centromeres and pericentromeric heterochromatin. Repeat junction mapping was combined with anti-CENH3-mediated ChIP to provide a definitive map position for maize centromere 8.
THE centromeres in most plants and animals contain hundreds of kilobases of simple repeats and interspersed retroelements. Such repetitive domains evolve at remarkable rates—continually expanding, contracting, and generating new arrays (Schueler et al. 2001; Henikoff 2002; Nagaki et al. 2004; Lee et al. 2005; Ma and Bennetzen 2006). This process tends to homogenize centromeres and drive out single- and low-copy sequences that are necessary for sequencing and molecular-marker-based mapping (Henikoff 2002; Dawe 2005). Two relatively simple rice centromeres have been bridged and characterized (Nagaki et al. 2004; Wu et al. 2004; Zhang et al. 2004), but as a rule centromeres fall into large and poorly resolved genetic gaps. In maize, the centromere positions are estimates based on rough interval or trisomic mapping and visual comparisons to the cytogenetic map (Weber and Helentjaris 1989; Schneerman et al. 1998; Lin et al. 2001).
Another complexity of centromere mapping is that sequence alone cannot be used to accurately predict where centromeres begin and end. Maize centromeres contain long arrays of the tandem repeat CentC as well as clusters of specialized transposons known as centromeric retroelements (generally known as CR elements and called CRM in maize; Jiang et al. 2003). However, arrays and clusters of CentC and CRM extend well outside of the domains identified by centromere/kinetochore protein centromeric histone H3 (CENH3; Zhong et al. 2002; Jin et al. 2004). These and other data suggest that much of the pericentromeric DNA in higher eukaryotes is derived from old and discarded centromere repeats, presumably displaced as new repeats are generated in the CENH3-binding core (Schueler et al. 2001; Henikoff 2002). New mapping procedures especially adapted to the unique genetics of centromeres will be required to complete the physical maps of large-genome species like maize.
In a prior effort by Nagaki et al. (2003), several BACs containing CentC were selected at random from a library prepared from the inbred Mo17. One of these contains a large quantity of CentC (BAC16H10) and a multitude of CRM elements (Zhong et al. 2002; Nagaki et al. 2003). Another BAC (BAC06E22, not described by Nagaki et al. 2003) contains only six monomers of CentC and no CRM elements. These two BACs are likely to represent the types of sequence in centromeric chromatin (BAC16H10) and pericentromeric heterochromatin (BAC06E22), respectively.
As is typical of centromere and pericentromere regions, there are no single-copy areas that could be used for generating markers in either BAC (aside from a gene at one end of BAC06E22). We wondered if there are any other forms of usable markers in such repetitive regions. A unique feature of cereal centromeres is the presence of CR elements and other retroelements, both nested within each other and among satellite arrays (Nagaki et al. 2003, 2004). Maize centromeres differ substantially in size among inbreds, suggesting that such insertions may be polymorphic (Kato et al. 2004). Further, prior evidence had shown that retroelements generally do not target specific sequences or nucleotides. Assuming random insertion, there are 13,282 ways that a CRM retroelement can insert into another identical copy of itself (assuming a 6641-bp target and two possible orientations at each nucleotide). The potential variation is exponentially higher when we consider that thousands of different maize retroelement families have been inserting into each other for over ∼6 million years (SanMiguel et al. 1998). We reasoned that a marker system that exploited this variation might be particularly useful for centromere mapping.
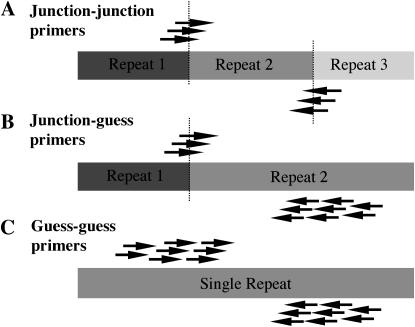
Among the sequence of the BACs 16H10 and 06E22, we identified seven areas where repeats had inserted close to the end (120–400 bp) of another repeat such that two junctions were created in close proximity (“junction–junction” markers). Several primers were designed for each junction (Figure 1). DNA from 20 recombinant inbred (RI) lines from the Intermated B73 and Mo17 population (referred to as IBM; Lee et al. 2002) were used as test templates, anticipating that dominant single-copy markers would segregate in an ∼1:1 ratio (Figure 2). By these criteria, usable markers were identified for six of the seven junction–junction sites chosen. The likelihood of empirical success for junction–junction primer pairs was ∼11.3% (11 of 97 primer pairs gave clear plus/minus results). The results show that within the BACs chosen, junction–junction sites are generally single copy and polymorphic.
Figure 1.—
Types of markers tested. (A) Junction–junction primers that flank repeat junctions were most effective, yielding usable markers with 10.3% of the primer pairs tried. (B) Junction–guess primers and (C) guess–guess primers were much less effective, yielding a 0.4% success rate. At least three primers were designed for each end of all amplicons.
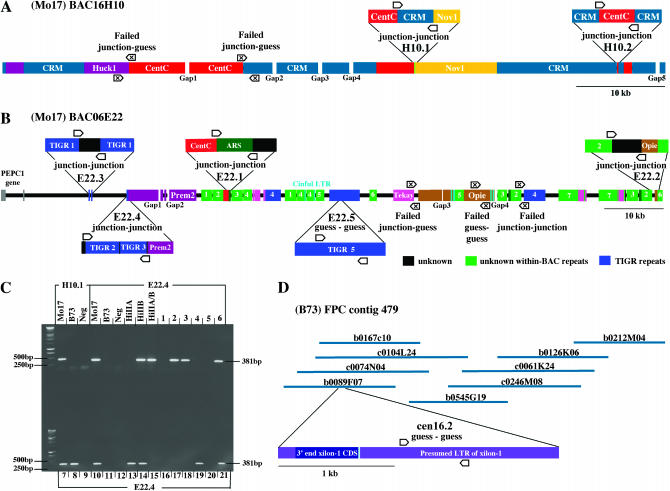
Figure 2.—
BACs analyzed in this study and locations of markers. (A) The annotation of BAC16H10 was adapted from Nagaki et al. (2003). (B) BAC06E22 was sequenced at 10× coverage (see Nagaki et al. 2003 for sequencing details), ordered and finished by PCR where possible, and annotated as shown (see also GenBank AC114395). The approximate size of gaps 1, 3, and 4 are 350, 250, and 500 bp, respectively. Gap 2 could not be confirmed but we can infer its position by process of elimination. The seven junction–junction markers and five markers involving guess primers are labeled accordingly. The Institute for Genome Research (http://www.tigr.org) repeats are as follows: (1) fam_29305_C1; (2) fam_21448_C2; (3) fam_33660_C1; (4) Prem1_276N13-2; (5) diguus_123C01-1. (C) Mapping gel for marker Mo17-E22.4. Mo17-E22.4 is not detectable in B73 but is observed in Mo17 and about half of the (numbered) RI lines derived from B73 and Mo17. HiIIA and HiIIB are complex hybrids of the inbreds B73 and A188. (D) Marker B73-cen16.2 lies in a CRM-rich FPC contig. Contig 479 (version 8.2 of the Arizona Genomics Institute maize assembly; http://www.genome.arizona.edu/fpc/WebAGCoL/maize/WebFPC/) contains >200 clones, but only the nearest CRM-containing BACs are shown here. Primers used, amplified fragments, and PCR conditions can be found in the following GenBank STS entries: Mo17-H10.1: BV686428; Mo17-H10.2: BV686429; Mo17-E22.1: BV686430; Mo17-E22.2: BV686431; Mo17-E22.3: BV686432; Mo17-E22.4: BV686434; Mo17-E22.5: BV686433; and B73-cen16.2: DQ863276.
We also attempted to identify polymorphisms by designing “guess” primers over poorly conserved regions. Five sites were targeted by this approach: three with a repeat junction on one side and a poorly conserved region on the other (“junction—guess”) and two over regions with no junctions but unusually low homology (<70%) to other known repeats. Only guess–guess site Mo17-E22.5 provided a useful single-copy polymorphism, and only 1 of 104 primer pairs at this site provided a clear dominant marker. The overall frequency of success for primers involving a guess site was ∼0.4% (1 of 246 primer pairs gave clear plus/minus results). These data suggest that sites involving a guess primer are much less useful for identifying polymorphisms than junction–junction sites.
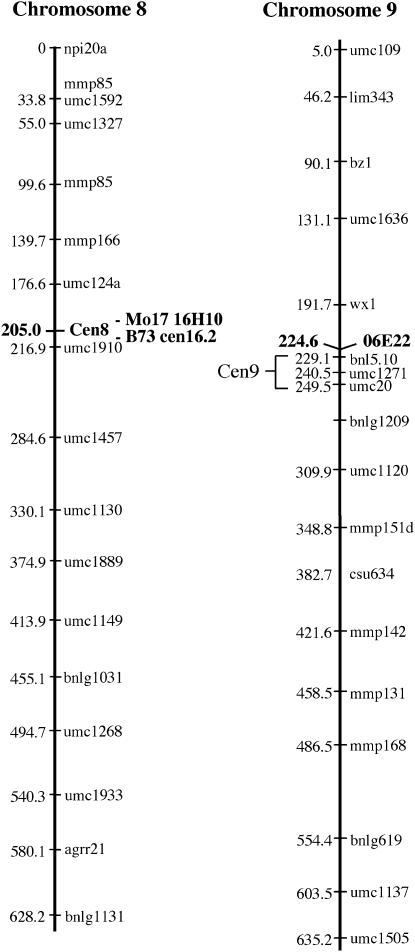
Three primer pairs, Mo17-H10.1, Mo17-H10.2, and Mo17-E22.1, were used to score a complete 94-sample IBM population (Lee et al. 2002; Sanchez-Villeda et al. 2003). The segregation patterns for Mo17-H10.1 and Mo17-H10.2 were identical for all 94 DNA samples, indicating that different markers from a single BAC provide consistent and reproducible data. Marker Mo17-H10.1 maps to locus 205 on chromosome 8 in a region that is broadly consistent with the location of the centromere (although information on centromere 8 is limited). Marker Mo17-E22.1 maps to chromosome 9 in a region that is close to, but not within, the centromere (Lin et al. 2001; Figure 3). Although there are several maps based on the intermated B73 × Mo17 inbreds, the IBM2 map is considered the current standard (http://www.maizegdb.org). To simplify display and discussion, the Community IBM Mapping Service (CIMDE) map data were interpolated into the IBM2 framework (see Figure 3 legend).
Figure 3.—
Map positions of BAC16H10, B73-cen16.2, and BAC06E22. Reference IBM2 2004 maps for chromosomes 8 and 9 are shown along with the positions of newly mapped loci (boldface type). Map positions were calculated and provided by CIMDE (Community IBM Mapping Service; http://www.maizemap.org/CIMDE/cIBMmap.htm). We have interpolated the position for centromere 8 reported to us by CIMDE (169.0) into the IBM2 framework using linked markers that are common to both the CIMDE and the IBM2 map. These numerical values may change as linkage maps improve. Markers Mo17-H10.1 and Mo17-H10.2 (BAC16H10) map to chromosome 8 with a LOD score of 25.9, marker B73-cen16.2 maps to chromosome 8 with a LOD score of 27.3, and marker Mo17-E22.1 (BAC06E22) maps to chromosome 9S with a LOD score of 14.9. Centromere 9 has been placed by translocations to a position several IBM2 units proximal to BAC06E22 (Lin et al. 2001).
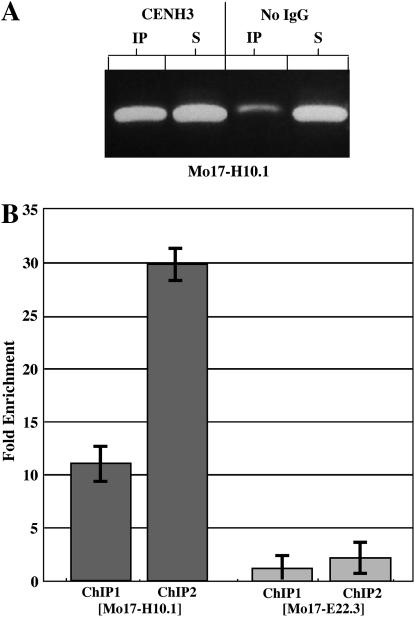
Placing centromere repeats on the genetic map does not necessarily map the centromere, since arrays of centromere repeats often extend outside of the kinetochore-binding region (Jin et al. 2004). Chromatin immunoprecipitation (ChIP) using antisera to histone H3 variant CENH3 is an accepted method for showing that a locus associates with the kinetochore (Zhong et al. 2002; Nagaki et al. 2004). We combined antisera to maize CENH3 (Zhong et al. 2002), native ChIP (Topp et al. 2004), and real-time PCR to determine whether BACs 16H10 and 06E22 lie within functional centromeres. Markers Mo17-H10.1 and Mo17-E22.3 were assayed in two independent ChIP experiments. Mock treatments were used to control for background signal. We were unable to detect an interaction between CENH3 and Mo17-E22.3, supporting the view that BAC06E22 lies in pericentromeric heterochromatin. In contrast, we detected a significant association between CENH3 and marker Mo17-H10.1 (Figure 4). These results indicate that BAC16H10 lies within the functional centromere and confirm that centromere 8 maps to locus 205 on the current IBM2 map. Our data revise the location of centromere 8 by >100 IBM2 MU (the prior estimate was 99.7; see IBM2 2004 neighbors 8 at http://www.maizegdb.org).
Figure 4.—
ChIP–PCR analyses. Nuclei from Mo17 root tips were subjected to native ChIP as described previously (Topp et al. 2004). Markers Mo17-H10.1 and Mo17-E22.3 were assayed in two independent ChIP experiments using an MJ Research (Watertown, MA) Chromo 4 real-time system. (A) Agarose gels showing anti-CENH3-mediated ChIP–PCR results after 35 cycles. (B) Bar graphs depicting the enrichment of markers relative to no immunoglobulin G controls. Error bars show the variation associated with real-time PCR (standard deviation, n = 2). A ΔΔCt calculation (Livak and Schmittgen 2001) using Mo17-E22.3 as a negative control for Mo17-H10.1 gave a relative fold enrichment of 9.4 ± 1.9.
In a final set of experiments, we tested whether the position of centromere 8 in Mo17 accurately represents the position of centromere 8 in the B73 inbred, which is currently being sequenced. Coauthor Presting and colleagues are in the process of low-pass (2×) sequencing a number of centromeric BACs from B73. Among the chosen BACs is b0089F07, which contains CRM sequence (≥7%) but no CentC, a composition that is typical of the transition areas between centromeres and flanking heterochromatin (Jin et al. 2004). BAC b0089F07 is embedded within a fingerprint (FPC) contig that contains numerous other BACs with homology to CRM (Figure 3D).
A single-copy marker (B73-cen16.2, of the guess–guess type) from BAC b0089F07 yielded an amplification pattern complementary to that of Mo17-H10.1; i.e., the B73-cen16.2 band was absent in recombinant inbred lines that yielded an Mo17-H10.1 band, and vice versa. The only exception to this pattern among the 94 recombinant inbreds was M0281, which contained both bands and may represent a rare heterozygote or single recombination event separating the two markers. Thus, B73-cen16.2 maps to the same position as Mo17-H10.1 and Mo17-H10.2. These data support the assertion that centromere 8 maps genetically to a single small region in both Mo17 and B73.
Acknowledgments
This work was supported by a grant from the National Science Foundation to R.K.D. (0421671). We thank Michael Gerau and Mary Polacco of the CIMDE resource for their help and advice with recombinant inbred mapping.
References
- Dawe, R. K., 2005. Centromere renewal and replacement in the plant kingdom. Proc. Natl. Acad. Sci. USA 102: 11573–11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., 2002. Near the edge of a chromosome's ‘black hole’. Trends Genet. 18: 165–167. [DOI] [PubMed] [Google Scholar]
- Jiang, J., J. A. Birchler, W. A. Parrott and R. K. Dawe, 2003. A molecular view of plant centromeres. Trends Plant Sci. 8: 570–575. [DOI] [PubMed] [Google Scholar]
- Jin, W., J. R. Melo, K. Nagaki, P. B. Talbert, S. Henikoff et al., 2004. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, A., J. C. Lamb and J. A. Birchler, 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. R., W. Zhang, T. Langdon, W. Jin, H. Yan et al., 2005. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102: 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., N. Sharopova, W. D. Beavis, D. Grant, M. Katt et al., 2002. Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol. Biol 48: 453–461. [DOI] [PubMed] [Google Scholar]
- Lin, B. Y., S. J. Chang and H. M. Lin, 2001. RFLP mapping of the centromere of chromosomes 1, 6 and 9 by B-A translocations in maize. Bot.. Bull. Acad. Sinica 42: 273–279. [Google Scholar]
- Livak, K. J., and T. D. Schmittgen, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the Delta Delta C(T) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2006. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. USA 103: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., J. Song, R. Stupar, A. S. Parokonny, Q. Yuan et al., 2003. Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163: 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki, K., Z. Cheng, S. Ouyang, P. B. Talbert, M. Kim et al., 2004. Sequencing of a rice centromere reveals active genes. Nat. Genet. 36: 138–145. [DOI] [PubMed] [Google Scholar]
- Sanchez-Villeda, H., S. Schroeder, M. Polacco, M. McMullen, S. Havermann et al., 2003. Development of an integrated laboratory information management system for the maize mapping project. Bioinformatics 19: 2022–2030. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., B. Gaut, A. Tikhonov, Y. Nakajima and J. Bennetzen, 1998. The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43–45. [DOI] [PubMed] [Google Scholar]
- Schneerman, M. C., W. S. Lee, G. Doyle and D.F. Weber, 1998. RFLP mapping of the centromere of chromosome 4 in maize using isochromosomes for 4S. Theor. Appl. Genet. 96: 361–368. [DOI] [PubMed] [Google Scholar]
- Schueler, M. G., A. W. Higgins, M. K. Rudd, K. Gustashaw and H. F. Willard, 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109–114. [DOI] [PubMed] [Google Scholar]
- Topp, C. N., C. X. Zhong and R. K. Dawe, 2004. Centromere-encoded RNAs are integral components of the maize kinetochore. Proc. Natl. Acad. Sci. USA 101: 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, D., and T. Helentjaris, 1989. Mapping RFLP loci in maize using B-A translocations. Genetics 121: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., H. Yamagata, M. Hayashi-Tsugane, S. Hijishita, M. Fujisawa et al., 2004. Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16: 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Y. Huang, L. Zhang, Y. Li, T. Lu et al., 2004. Structural features of the rice chromosome 4 centromere. Nucleic Acids Res. 32: 2023–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C. X., J. B. Marshall, C. Topp, R. Mroczek, A. Kato et al., 2002. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14: 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]