Abstract
Intercrosses between inbred lines provide a traditional approach to analysis of polygenic inheritance in model organisms. Chromosome substitution strains (CSSs) have been developed as an alternative to accelerate the pace of gene identification in quantitative trait mapping. We compared a classical intercross and three CSS intercrosses to examine the genetic architecture underlying plasma high-density lipoprotein cholesterol (HDL) levels in the C57BL/6J (B) and A/J (A) mouse strains. The B × A intercross revealed significant quantitative trait loci (QTL) for HDL on chromosomes 1, 4, 8, 15, 17, 18, and 19. A CSS survey revealed that many have significantly different HDL levels compared to the background strain B, including chromosomes with no significant QTL in the intercross and, in some cases (CSS-1, CSS-17), effects that are opposite to those observed in the B × A intercross population. Intercrosses between B and three CSSs (CSS-3, CSS-11, and CSS-8) revealed significant QTL but with some unexpected differences from the B × A intercross. Our inability to predict the results of CSS intercrosses suggests that additional complexity will be revealed by further crosses and that the CSS mapping strategy should be viewed as a complement to, rather than a replacement for, classical intercross mapping.
THE genomewide distribution of allelic variation in an intercross population, as in human populations, provides great potential for epistatic interactions that can mask or enhance the impact of allelic variation at multiple loci. This complexity may hinder our ability to detect the genes and alleles that are most important. One approach to circumvent this possibility is to reduce the number of potential interactions by reducing the amount of segregating variation in a cross. Chromosome substitution strain (CSS, also known as consomic) panels achieve this goal by restricting variation to a single chromosome (Nadeau et al. 2000; Cowley et al. 2004; Fernandes et al. 2004; Bevova et al. 2006). In these panels each strain is derived from a “background” inbred strain except a single chromosome that is derived from a distinct “donor” inbred strain. Consequently any genetic variation seen among CSSs is presumably due to allelic variation on the donor chromosome. It is possible that some differences between the CSSs may be due to recent mutations (Cook et al. 2006). However, it is estimated that only 3% of polymorphisms among strains have arisen within the past 100 years. Thus, most of the differences between inbred strains, and specifically for this report between C57BL/6J (B) and A/J (A), are ancestral and differences among consomic strains due to recent mutations are expected to be quite rare (Frazer et al. 2004).
Ischemic cardiovascular disease (ICD) is the leading cause of morbidity and mortality in developed nations. Major risk factors for ICD are high levels of low-density lipoprotein cholesterol (LDL) and low levels of high-density lipoprotein cholesterol (HDL) in plasma (Fruchart and Duriez 2002). High levels of plasma HDL provide protection against heart disease as shown in both human (Gordon et al. 1989; Wilson et al. 1994; Nissen et al. 2003) and animal studies (Badimon et al. 1990; Rubin et al. 1991; Plump et al. 1994; Sugano et al. 1998; Okamoto et al. 2000; Rittershaus et al. 2000). A successful route toward identifying genes that affect quantitative phenotypes such as HDL levels is through the use of inbred mouse strains and quantitative trait locus (QTL) analysis. A combination of genetic tools and techniques is helping to identify these QTL genes (Korstanje and Paigen 2002; Dipetrillo et al. 2005; Hillebrandt et al. 2005; Wang et al. 2005). A total of 22 mapping crosses have identified >130 loci for HDL and these localize to 37 unique QTL (Wang and Paigen 2005). Only a small number of gene-by-gene interactions have been reported for HDL (Ishimori et al. 2004), in comparison to other traits such as obesity (Cheverud et al. 2001; Stylianou et al. 2006). Thus either HDL interactions are rare or they have been too complex to detect with current methods.
In this study we carried out a standard F2 intercross between B and A and detected QTL on seven chromosomes (Chr) including both main-effect and interacting QTL. We then surveyed the chromosome substitution strains and carried out intercrosses between B and three CSSs. The results showed that CSSs do not simply partition the genetic variation as expected. The unique genetic backgrounds present in the CSSs and parental line intercrosses alter the genetic architecture of HDL regulation. Some QTL are clearly shared among the different mapping populations but many are not. We demonstrate that epistatic interactions among multiple loci are buffering the potential for phenotypic variation in the parental genomes.
MATERIALS AND METHODS
Mice:
Mouse strains were obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained on a 14 hr light/10 hr dark cycle. Mice were housed in individually pressurized cages (Thoren Caging Systems), containing pine shaving bedding and topped with a polyester filter, and allowed ad libitum access to acidified water and food (18% protein rodent diet, 6% fat, product 2018; Harlan Teklad, Madison, WI). Animal protocols were reviewed and approved by the Animal Care and Use Committee at The Jackson Laboratory.
Crosses:
F2 progeny were obtained by intercrossing (B6 × A/J) F1 mice to produce 343 F2 males and 271 females. These two crosses were carried out at different times for different purposes; however, the conditions under our control were consistent for both crosses. In particular, both crosses were carried out using a standard chow (4% fat) diet. HDL was measured in both crosses and the data were combined for this report.
The C57BL/6J with A/J chromosome substitution strains whose nomenclature is B-Chr#A are simplified to CSS-# throughout this article. The B × CSS intercrosses were generated using F1 mice from B6 females mated to CSS-3, CSS-8, or CSS-11 males. These F1 mice were intercrossed to generate F2 animals. The CSS-3 intercross population consisted of 146 mice (65 females and 65 males), the CSS-8 intercross consisted of 150 F2 mice (77 females and 71 males), and the CSS-11 intercross consisted of 179 F2 mice (84 females and 70 males).
Genotyping:
Genotyping was performed for each CSS intercross using evenly spaced MIT microsatellite markers as previously described (Stylianou et al. 2004). SNP genotyping was performed for the larger B × A intercross. For SNP genotyping genomic DNA was isolated from the tail of each mouse and genotyped with markers initially evenly spaced throughout the genome with additional markers added in regions of interest. A total of 97 markers for males and 174 for females were used. SNPs were chosen from a panel designed to facilitate genotyping in inbred mouse strains (Petkov et al. 2004) and it was performed by the Allele-Typing Service at The Jackson Laboratory in conjunction with KBiosciences (Hoddesdon, UK).
Phenotyping:
Blood samples from mice fasted for 4 hr in the morning were collected in tubes containing EDTA and centrifuged at 9000 rpm for 5 min. Plasma was placed into a fresh tube and frozen at −20° until analyzed. Plasma samples were thawed, vortexed, and analyzed within a week of being collected. Plasma HDL concentrations from each blood sample were measured directly, using an enzymatic reagent kit (no. 650207, Beckman Coulter) according to manufacturer's recommendations on the Synchron CX Delta System (Beckman Coulter).
CSS strain survey:
For the strain survey, 8–10 mice of each sex from each strain were collected and phenotyped over a 6-month period. Control animals were tested concurrently with the CSSs. Each strain and sex were represented by animals from at least three independent litters. Statistical significance of HDL between strain B and each CSS was determined using student's t-test. Two-way analysis of variance was used to test for sex-specific QTL. Multiple testing was accounted for using a Bonferroni correction within sex. Homogeneity of variances was assessed using Bartlett's and Levene's tests.
QTL and statistical analyses:
QTL analysis was performed using Pseudomarker 2.03 (http://www.jax.org/staff/churchill/labsite/software/pseudomarker). The two parental intercrosses were combined and marker locations were exact as base pair positions of the markers were known. Approximate centimorgan coordinates for QTL analysis were obtained by dividing megabase pair positions by a factor of 2 (Mouse Genome build 33). The validity of this approximation was confirmed by comparison to estimated map positions in Pseudomarker. For consistency positions of microsatellite markers used to genotype the B × CSS intercrosses were obtained as the base pair position of the amplified fragments by doing a BLAST with the primer sequences. Again megabase pair positions were converted into centimorgans by dividing by 2. Genetic maps, recombination frequencies, and segregation analysis were performed within Pseudomarker for quality control. Approximate confidence intervals were computed using the maximum posterior density method (Sen and Churchill 2001). QTL names were assigned by the Mouse Genome Nomenclature Committee and are listed in Table 1.
TABLE 1.
Multilocus models of HDL QTL
| Multiple-QTL analysis
|
Details of main-effect QTL
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Intercross | Effect/QTL | d.f.a | Type III SSa | % vara | P(f)b | QTL name | Chr | Peak (cM)c | C.I. (cM)c | LODc | Position of marker (Mb)cd | High allele | Mode of inheritance |
| Sex | 1 | 20.05 | 48.1 | 0 | |||||||||
| B × A | Chr 1 at 2 cM | 6 | 0.70 | 1.7 | 0.00034 | Hdlq40 | 1 | 2 | — | 1.5 | 4 | Heterozygous | Interactive |
| Chr 1 at 48 cM | 6 | 0.74 | 1.8 | 0.00019 | Hdlq41 | 1 | 48 | — | 5.0 | 96 | A/J | Interactive | |
| Chr 1 at 76 cM | 6 | 0.53 | 1.3 | 0.0041 | Hdlq42 | 1 | 76 | 54–92 | 6.4 | 152 | A/J | Dominant | |
| Chr 4 at 52 cM | 2 | 0.66 | 1.6 | 8.95 × 10−6 | Hdlq43 | 4 | 52 | 34–66 | 4.4 | 104 | B6 | Recessive | |
| Chr 8 at 30 cM | 6 | 2.11 | 5.1 | 1.60 × 10−13 | Hdlq44 | 8 | 30 | 20–40 | 9.4 | 60 | B6 | Dominant | |
| Chr 15 at 22 cM | 2 | 0.33 | 0.8 | 0.0026 | Hdlq45 | 15 | 22 | 2–30 | 2.6 | 44 | B6 | Additive | |
| Chr 17 at 26 cM | 2 | 0.30 | 0.7 | 0.0043 | Hdlq46 | 17 | 26 | 12–42 | 3.4 | 52 | B6 | Recessive | |
| Chr 18 at 26 cM | 2 | 0.35 | 0.8 | 0.0018 | Hdlq47 | 18 | 26 | 16–34 | 3.0 | 52 | B6 | Recessive | |
| Chr 19 at 4 cM | 2 | 0.28 | 0.7 | 0.0063 | Hdlq48 | 19 | 4 | 0–14 | 2.1 | 8 | B6 | Overdominant | |
| Chr 1 at 2 cM × Chr 1 at 48 cM | 4 | 0.52 | 1.2 | 0.00096 | |||||||||
| Chr 1 at 76 cM × Chr 8 at 30 cM | 4 | 0.44 | 1.0 | 0.0035 | |||||||||
| Error | 583 | 16.06 | |||||||||||
| B × CSS-3 | Sex | 1 | 3.61 | 68.17 | |||||||||
| Chr 3 at 16 cM | 2 | 0.22 | 4.15 | 6.9 × 10−5 | Hdlq49 | 3 | 16 | 10–28 | 4.2 | 32 | B6 | Additive | |
| Error | 126 | 5.29 | |||||||||||
| B × CSS-8 | Sex | 1 | 3.09 | 52.14 | |||||||||
| Chr 8 at 50 cM | 2 | 0.86 | 14.5 | 1.78 × 10−11 | Hdlq50 | 8 | 50 | 44–54 | 10.8 | 100 | B6 | Dominant | |
| Error | 144 | 2.17 | |||||||||||
| B × CSS-11 | Sex | 1 | 2.85 | 64.85 | |||||||||
| Chr 11 at 22 cM | 2 | 0.10 | 2.34 | 0.0056 | Hdlq51 | 11 | 22 | 6–32 | 2.3 | 44 | B6 | Dominant | |
| Error | 150 | 1.45 | |||||||||||
—, confidence intervals cannot be accurately estimated due to multiple linked QTL.
Includes contributions of the interactions of main effects.
Based on regression F-test for this term.
Obtained from the single-QTL genome scans.
Megabase positions based on mapped position of peak marker to NCBI build 33.
HDL values were natural log transformed. The log transform reduced the right skew in the HDL distribution but, of greater importance, it also resulted in constant variance among the crosses. When cross populations are combined for analysis purposes, if one population has a greater phenotypic variance, it will tend to dominate the results. All crosses were analyzed with sex as an additive covariate, in addition to performing separate analyses that included a sex-by-QTL interaction to examine sex-specific effects. Genomewide significance was determined using 1000 permutations with stratification by sex (Churchill and Doerge 1994). Other statistical analyses were performed using JMP version 6.0 (SAS Institute, Cary, NC).
RESULTS
B × A F2 intercross:
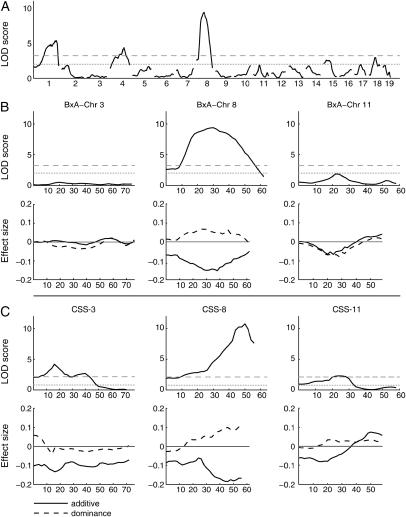
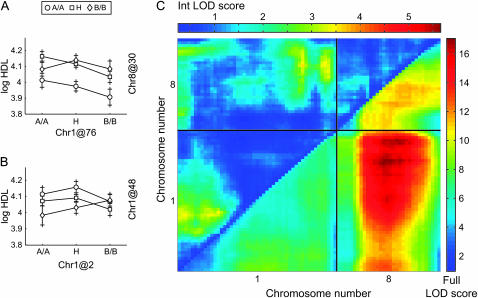
A total of 578 B × A F2 were measured for HDL cholesterol. A genomewide scan with an adjustment for sex detected significant QTL on Chrs 1, 4, and 8 and several suggestive QTL (Figure 1A). A pairwise genome scan indicated several potential QTL interactions (Figure 2). Joint modeling of all main effects and interactions was performed by merging all significant and suggestive results and then removing one QTL or interaction at a time until all terms remaining in the multiple QTL model were significant (P < 0.01). The final model (Table 1) includes QTL on Chr 1, 4, 8, 15, 17, 18, and 19 and two interaction terms, Chr 1 at 76 cM by Chr 8 at 30 cM (Figure 2, A and C) and Chr 1 at 2 cM by Chr 1 at 48 cM (Figure 2, B and C). Sex explained 48% and the Chr 8 QTL explained 5% of the total variance. In a cross of this size we can detect QTL that explain as little as 1% of the total variation after accounting for sex. Note that it is not possible to distinguish sex effects from cross effects in this study but it is important to account for this source of variation by using an additive covariate in the genome scans. In the supplemental data (supplemental Figure 1 at http://www.genetics.org/supplemental/) we show that fitting “sex-by-QTL” interaction reveals that loci on Chr 11 and 17 are further influenced by sex.
Figure 1.—
Interval mapping. (A) Genomewide scan of the B × A intercross for HDL with sex as an additive covariate. (B) Chromosome-specific plots of Chr 3, Chr 8, and Chr 11 from the B × A intercross with genomewide thresholds [top horizontal lines indicate significant (P < 0.01) and the bottom lines indicate suggestive thresholds (P < 0.63)]. (C) Mainscan plots for each of the F2 consomic crosses, CSS-3, CSS-8, and CSS-11, along with chromosomewide thresholds [top horizontal lines indicate significant (P < 0.01) and bottom lines indicate suggestive thresholds (P < 0.63)] (Lander and Kruglyak 1995). Effect size estimates are shown at each putative QTL location. Additive effect is half of the difference between the AA and BB genotype classes. Positive values indicate that A is the high allele. Dominance effect is the difference between the mean of the AB genotype class and the midpoint of the AA and BB genotype class means. Positive values indicate dominance of the B allele. All analyses are performed on natural log-transformed HDL. Positions are given in centimorgans.
Figure 2.—
Interaction analysis. Effect sizes of the two interactions between (A) Chr 1 at 2 cM-by-Chr 1 at 48 cM and (B) Chr 1 at 76 cM-by-Chr 8 at 30 cM. (C) Heat map from part of the genomewide pairwise scan focusing on Chrs 1 and 8. Int LOD score is the interaction LOD score and applies to the area above the diagonal. Full LOD score is the LOD score for the full model and applies to the area below the diagonal.
CSS strain survey:
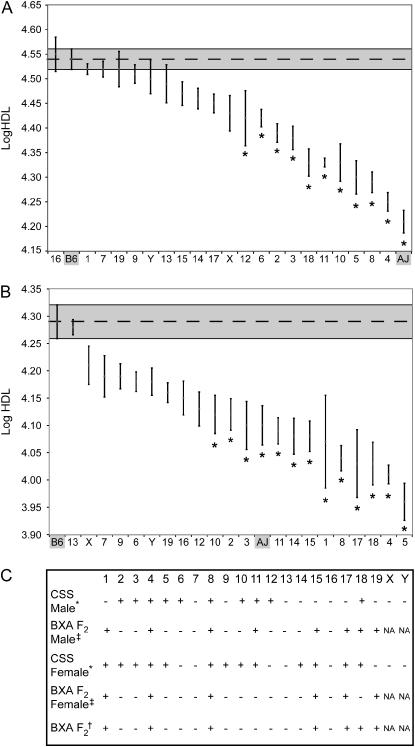
Males and females for each of the 21 strains of the CSS panel and both parental strains A and B were analyzed for HDL levels (Figure 3). When compared to parental strain B, the CSSs show evidence for a QTL in at least one sex for all chromosomes except Chr 7 and 13 (P < 0.05), and, after accounting for multiple testing using a Bonferroni correction [P < 0.002 (P < 0.05/N = 21)], approximately half the strains within each sex are still significantly different from the B strain. With sample sizes of 10 per group, it should be possible to detect differences between CSS-# and B with 80% power of 1.5 standard deviation units at the α = 0.01 significance level and 1.25 standard deviation units at α = 0.05 significance. Despite some differences in QTL detection between male and female CSSs, tests for sex-specific QTL did not exceed the multiple-test adjusted α = 0.05 significance threshold; however, there is suggestive evidence for sex-specific QTL on CSS-1, -14, -15, -16, and -17. The variance heterogeneity apparent in Figure 3 is statistically significant (P < 0.001) for both males and females, suggesting that genetic background may influence individual variability in HDL levels.
Figure 3.—
Chromosome substitution strain survey. HDL natural log transformed for (A) male and (B) female CSSs and parental B and A mice. The mean of strain B is given by the horizontal broken line and the shaded area represents the ±1 SEM boundaries; an * indicates significance between the CSS-# and B6 after correction for multiple testing. N = 8–10 for each group. (C) Summary of the presence (+) or absence (−) of QTL as detected in the CSS survey compared with the B × A intercross. For B × A intercross animals, presence (+) indicates at least a suggestive QTL as discussed in the text, and for the CSS animals presence (+) indicates a significant difference from parental strain B after a Bonferroni correction for multiple testing [P < 0.002 (P < 0.05/N = 21)].
B × CSS F2 intercrosses:
Intercrosses between B and three CSSs were performed. We chose CSS-11 because it showed a suggestive QTL in the B × A intercross that would presumably become significant in the B × CSS-11 intercross. We chose CSS-8 because it had the strongest QTL in the parental intercross, and we chose CSS-3 because Chr 3 had no QTL in the parental F2 intercross but the CSS-3 was significantly different from B (Figure 3).
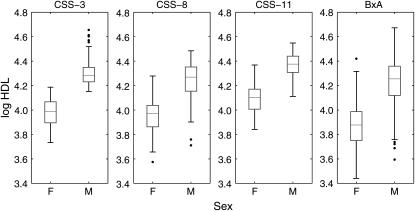
The HDL values were natural log transformed to obtain stable variances (Figure 4). Prior to transformation, the standard deviation in HDL of male mice is roughly twice that of females. Following the natural log transformation, male and female standard deviations are approximately equal. The trait variances in each of the intercross populations reflect an environmental component plus contributions due to segregating genetic variance (Table 2). As expected, the variance in each of the intercross populations is greater than the within-strain variance in the survey population. The trait variance in the B × A intercross is largest, reflecting the genetic complexity of this population. The B × CSS-8 cross is intermediate in variability due to segregation of a QTL with large effect. The B × CSS-3 and B × CSS-11 intercrosses are segregating small QTL and have correspondingly smaller standard deviations.
Figure 4.—
Box plot HDL variation in intercross populations. Natural log-transformed HDL for each of the four intercrosses B × CSS-3, B × CSS-8, B × CSS-11, and B × A is shown.
TABLE 2.
Summary statistics and significant thresholds
| Log HDL
|
Threshold
|
|||||
|---|---|---|---|---|---|---|
| Cross | Sex | N | Mean | SD | Suggestive (P < 0.63) | Significance (P < 0.05) |
| B × CSS-3 | F | 65 | 3.98 | 0.11 | 0.8 | 2.2 |
| M | 65 | 4.31 | 0.11 | |||
| B × CSS-8 | F | 77 | 3.96 | 0.13 | 0.8 | 2.1 |
| M | 71 | 4.24 | 0.16 | |||
| B × CSS-11 | F | 84 | 4.1 | 0.11 | 0.8 | 2.1 |
| M | 70 | 4.37 | 0.09 | |||
| B × A | F | 270 | 3.87 | 0.19 | 2.0 | 3.2 |
| M | 341 | 4.24 | 0.19 | 1.9 | 3.2 | |
N, the number of mice in each cross.
All crosses were analyzed to detect sex-specific effects by fitting sex-by-QTL interaction in the genome scans. We also fit models with one, two, and three linked QTL on each chromosome. Power to detect linked QTL when linkage is tight (with 10–20 cM) can be limited and it is always possible that an apparently singular QTL is the result of two or more linked polymorphisms. There was no evidence to support either sex-specific effects or multiple linked QTL in any of the B × CSS intercrosses.
Thresholds for significant (0.05) and suggestive (0.63) QTL were calculated using 1000 permutations as described in materials and methods. The smaller target genome size of the B × CSS intercrosses substantially reduced the significance thresholds from 3.2 in the B × A cross to ∼2.2 in the B × CSS intercrosses as summarized in Table 2. We note that the chromosome-specific thresholds for the B × A cross are the same as those for the B × CSS thresholds (0.8 and 2.1 for Chr 8 and Chr 11, 0.8 and 2.2 for Chr 3), indicating that the reduced target genome size is the most important factor for determining significance thresholds.
B × CSS-3:
The main-effect genome scan (Figure 1C) shows one significant QTL with a peak at 16 cM (LOD 4.2) and there is a second peak at 40 cM; however, a two-QTL analysis does not find support for a second QTL. The B allele is associated with higher HDL both in the CSS-3 strain (Figure 3) and in the B × CSS-3 intercross. The B × A intercross data did not indicate any QTL on Chr 3 (Figure 1, A and B). This surprising difference could be explained by a complex epistatic interaction requiring B alleles at multiple locations. This genotype would occur in only a few animals in a B × A intercross population.
B × CSS-8:
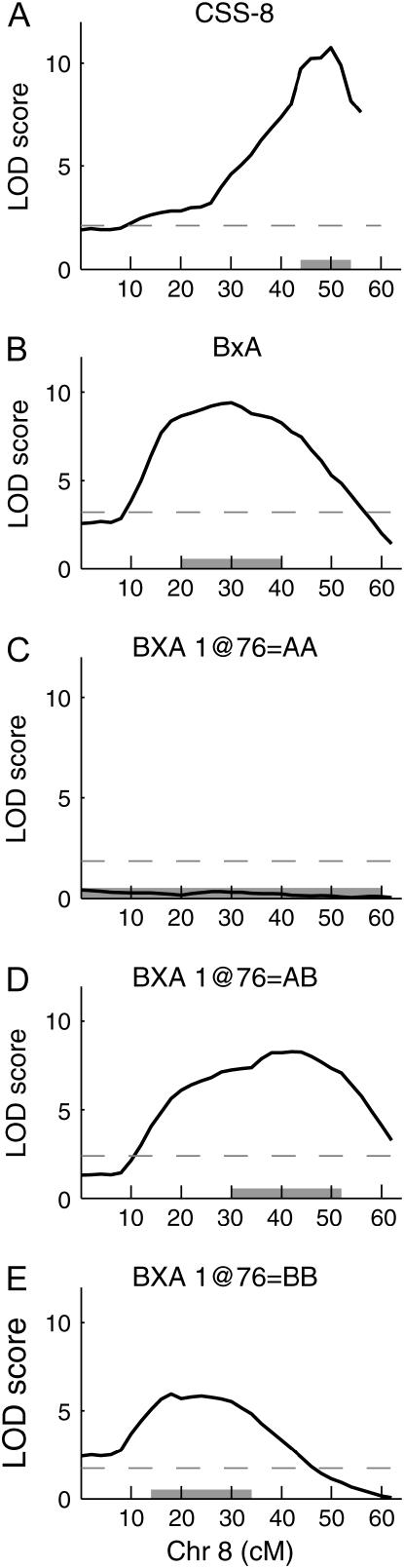
This cross confirms both the data from the CSS survey, where the CSS-8 strain is significantly different from B6 for both sexes, and the B × A intercross where the Chr 8 QTL was highly significant. The Chr 8 QTL from the B × A intercross has a peak at 30 cM (95% C.I., 20–40 cM); however, the LOD plot is rather broad, suggesting the influence of multiple linked QTL. The QTL peak from the CSS-8 intercross population is at 50 cM and its 95% C.I. is outside that of the B × A intercross Chr 8 QTL. As mentioned above, within the B × A intercross there is a significant interaction between the HDL QTL on Chr 1 at 76 cM with Chr 8 at 30 cM (Figure 2, A and C). When homozygous A/J alleles are fixed within the B × A population at Chr 1 at 76 cM, the Chr 8 at 30 cM QTL is absent, seemingly being fully dependent on at least one B allele being present at the Chr 1 location (Figure 5). This demonstrates that two QTL exist on Chr 8: one seen in the B × A intercross and one seen in the B × CSS-8 intercross. Both QTL are large, highly significant, and dependent on the background genotype. The B × A Chr 8 at 30 cM QTL is dependent on B alleles on Chr 1 at 76 cM, and the CSS-8 Chr 8 at 50 cM QTL is dependent on B6 alleles at locations that were not elucidated from these crosses.
Figure 5.—
Chromosome 8 QTL analysis. Mainscan plots of Chr 8 intercrosses for (A) B × CSS-8; (B) B × A; (C) the B × A, where alleles at the interacting locus on Chr 1 at 76 cM are homozygous for A/J, (D) heterozygous, and (E) homozygous for B alleles. Shaded horizontal bars indicate the 95% confidence intervals and dashed lines indicate the significance threshold.
B × CSS-11:
The main-effect genome scan (Figure 1C) looks almost identical to that of the B × A intercross (Figure 1B) with some minor differences in the effect plots. Whereas the Chr 11 QTL in the B × A intercross is only suggestive, the QTL in the B × CSS-11 intercross is significant. The LOD threshold for significance is 2.1 in the CSS intercross compared to the genomewide threshold of 3.2 in the B × A intercross.
DISCUSSION
Chromosome substitution strains have been developed as tools for genetic analysis. They complement recombinant inbred, recombinant congenic, and other types of strain resources. A benefit of using CSS is that fine-mapping crosses can be accelerated due to the fixed genetic background. Typically 5–10 backcross generations are required to isolate a QTL on a fixed background and generate a congenic model, breeding that is already completed for CSS. The CSS panel with C57BL/6J background and A/J donor chromosomes is available to the research community through The Jackson Laboratory, a panel of C57BL/6J with PWD donor chromosomes will soon become available, and a third panel of DBA/2 with donor chromosomes from an obese strain Du6i is being generated by Bevova et al. (2006). These three panels of mouse CSSs will enhance the potential for mapping QTL since each panel will harbor different allelic variation.
In this report we demonstrate that using the CSS lines to dissect genetic architecture may not be as straightforward as was originally proposed (Belknap 2003; Singer et al. 2005). Combining a QTL analysis of a large intercross between the donor strains with a CSS survey and intercrosses among three of the B × CSSs indicates that a variety of outcomes are possible. The most straightforward outcome was the Chr 11 QTL in the B × CSS-11 intercross. This QTL illustrated that, consistent with predictions made by Belknap (2003), Singer et al. (2005), and others, CSS intercrosses can provide greater power to detect QTL of small effect. It is worth noting that the Chr 11 QTL has a similar LOD profile in both crosses and that it exceeds the chromosomewide significance criteria in the B × A intercross. Thus, the difference in background genetic variation in the B × CSS-11 intercross had little impact on our ability to detect this particular QTL, and the difference between the B × CSS-11 intercross and the B × A intercross is largely due to the reduced target genome, which provides a lower significance threshold. Confidence intervals for the QTL location are similar in the two crosses.
The assumption underlying theoretical analyses of the CSS strategy is that QTL with the same effect size will be present in both the parental and the CSS intercross population and that the only difference will be a reduction of the background genetic variation in the latter (Belknap 2003; Singer et al. 2005). This was observed for the B × CSS-11 intercross but is not consistent with our findings on Chr 3, where a QTL appears in the B × CSS-3 intercross that was not present in the B × A intercross. It is not the case that we simply fail to detect it in the B × A intercross. The estimated effect size of the QTL in the B × CSS-3 intercross (−0.12 log HDL additive effect) is sufficiently large to detect if it were present in the B × A intercross. It is likely that the Chr 3 QTL is masked by epistatic loci segregating in the mixed background of the B × A intercross.
On Chr 8, we observe QTL in both the B × A and CSS intercrosses, but the position is changed and the pattern of epistatic interactions is at odds with our expectations. The QTL in the B × A intercross is broad, which suggests that multiple QTL may be present. Examination of the effect plots (Figure 1, B and C) suggests that the QTL with a peak at 30 cM identified in the B × A intercross may be present in the B × CSS-8 intercross. However, the peak of the QTL in the B × CSS-8 intercross at 50 cM indicates a new QTL with a larger dominance and additive effect than the QTL at 30 cM. Epistatic effects exist between Chr 1 and Chr 8 in the B × A intercross; this interaction requires at least one B allele on Chr 1 for the QTL on Chr 8 at 30 cM to yield increased HDL. In the B × CSS-8 intercross homozygous B alleles are fixed on Chr 1 and thus we would expect to see a large peak on Chr 8 at 30 cM. This indicates that alleles on other chromosomes that we did not detect in the B × A intercross may be interacting with one or more loci on Chr 8.
The Chr 1 × Chr 8 interaction seems critical to the Chr 8 QTL since the presence of homozygous A alleles at Chr 1 at 76 cM neutralizes the Chr 8 QTL (Figure 5). Ishimori et al. (2004) used a C57BL/6J × 129S1/SvImJ F2 cross and observed an interaction between Chr 1 at 104 cM by Chr 2 at 90 cM. It appears that Chr 1 may be a key player in the epistatic genetics of HDL QTL.
The genetic architecture of HDL in CSS lines is altered in comparison to the parental intercross presumably due to multiple epistatic interactions. We can pinpoint one such interaction (Chr 1 × Chr 8) in the intercross and we can detect how these changes in genetic architecture affect other QTL. For example, in the B × A intercross the QTL located at Chr 1 at 48 cM, Chr 1 at 76 cM, and Chr 17 at 26 cM are caused by high A/J alleles. The CSS survey indicates, however, that there must also be alleles for reducing HDL on chromosomes 1 and 17 or at least the absence of interactions, since CSS-1 and CSS-17 females actually have significantly lower HDL than B6 (P < 0.0001) (Figure 3), not higher, as would have been expected from the B × A intercross results.
Is the CSS strategy effective?
It has been proposed that the CSS strains can be used to map QTL using fewer mice. Sample size is an important parameter in determining statistical power and confidence interval size in any statistical application. The sample size required to achieve a desired power or precision scales in proportion to the variance of the sample data. Phenotypic variances are smaller in the CSS intercrosses than in the B × A intercross population (Table 2) and we can obtain an empirical estimate of the relative efficiencies of the cross designs by computing the ratio of variances (square of SDs). Comparing CSS-3, -8, and -11 to the B × A cross, the estimated relative efficiencies are 0.35, 0.58, and 0.28, respectively. This means that 35 mice in the CSS-3 population will provide the same power and precision as 100 mice in the B × A intercross. This efficiency of the CSS cross is a consequence of reduced genetic background variation (Belknap 2003). Multiple-locus mapping procedures (Zeng 1994) could improve the relative efficiency of the B × A cross but would not close the gap completely. Another factor to consider is the genomic coverage provided by the different types of crosses. A CSS cross provides information only for a single chromosome, and the number of CSS crosses required to cover the whole genome will vary depending on the outcome of the strain survey. In the extreme case where crosses are made for each of 20 chromosomes, as many as 700 animals would be required to obtain the same precision and coverage as 100 B × A intercross animals.
Using ∼600 mice in the B × A intercross we detect nine QTL on seven chromosomes and two interactions (Table 1). The CSS strategy employs two stages. First, the strain survey, with a total of 371 mice, indicated that almost all chromosomes carried HDL QTL. We confirmed three of these QTL with B × CSS intercrosses using 130–154 mice for each cross. The total number of mice in the CSS study was ∼900. Given the different objectives and outcomes of the two cross designs, it is difficult to formulate a fair basis for the numbers of mice needed to obtain the same information. However, when we compare the results from both cross types we find significant complementary information about the genetics of the HDL trait. Genetic background can have unpredictable effects and this comparison provides specific insights into the nature of genetic architecture. We conclude that the combination of different cross designs provides novel information that is not available from either approach considered alone.
Acknowledgments
We are grateful to Sarah Langley for assistance in phenotyping and genotyping and to Jesse Hammer for assistance with graphics. This work was supported by The American Heart Association grant 0525816T and the National Institutes of Health grants HL77796 (to B.J.P) and GM070683 (to G.C).
References
- Badimon, J. J., L. Badimon and V. Fuster, 1990. Regression of atherosclerotic lesions by high density lipoprotein plasma fraction in the cholesterol-fed rabbit. J. Clin. Invest. 85: 1234–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap, J. K., 2003. Chromosome substitution strains: some quantitative considerations for genome scans and fine mapping. Mamm. Genome 14: 723–732. [DOI] [PubMed] [Google Scholar]
- Bevova, M. R., Y. S. Aulchenko, S. Aksu, U. Renne and G. A. Brockmann, 2006. Chromosome-wise dissection of the genome of the extremely big mouse line DU6i. Genetics 172: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud, J. M., T. T. Vaughn, L. S. Pletscher, A. C. Peripato, E. S. Adams et al., 2001. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm. Genome 12: 3–12. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, D. N., G. S. Whitehead, L. H. Burch, K. G. Berman, Z. Kapadia et al., 2006. Spontaneous mutations in recombinant inbred mice: mutant Toll-like receptor 4 (Tlr4) in BXD29 mice. Genetics 172: 1751–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley, Jr., A. W., R. J. Roman and H. J. Jacob, 2004. Application of chromosomal substitution techniques in gene-function discovery. J. Physiol. 554: 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPetrillo, K., X. Wang, I. M. Stylianou and B. Paigen, 2005. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 21: 683–692. [DOI] [PubMed] [Google Scholar]
- Fernandes, C., L. Liu, J. L. Paya-Cano, S. Gregorova, J. Forejt et al., 2004. Behavioral characterization of wild derived male mice (Mus musculus musculus) of the PWD/Ph inbred strain: high exploration compared to C57BL/6J. Behav. Genet. 34: 621–630. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., C. M. Wade, D. A. Hinds, N. Patil, D. R. Cox et al., 2004. Segmental phylogenetic relationships of inbred mouse strains revealed by fine-scale analysis of sequence variation across 4.6 mb of mouse genome. Genome Res. 14: 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruchart, J. C., and P. Duriez, 2002. HDL and triglyceride as therapeutic targets. Curr. Opin. Lipidol. 13: 605–616. [DOI] [PubMed] [Google Scholar]
- Gordon, D. J., J. L. Probstfield, R. J. Garrison, J. D. Neaton, W. P. Castelli et al., 1989. High-density lipoprotein cholesterol and cardiovascular disease. Four prospective American studies. Circulation 79: 8–15. [DOI] [PubMed] [Google Scholar]
- Hillebrandt, S., H. E. Wasmuth, R. Weiskirchen, C. Hellerbrand, H. Keppeler et al., 2005. Complement factor 5 is a quantitative trait gene that modifies liver fibrogenesis in mice and humans. Nat. Genet. 37: 835–843. [DOI] [PubMed] [Google Scholar]
- Ishimori, N., R. Li, P. M. Kelmenson, R. Korstanje, K. A. Walsh et al., 2004. Quantitative trait loci analysis for plasma HDL-cholesterol concentrations and atherosclerosis susceptibility between inbred mouse strains C57BL/6J and 129S1/SvImJ. Arterioscler. Thromb. Vasc. Biol. 24: 161–166. [DOI] [PubMed] [Google Scholar]
- Korstanje, R., and B. Paigen, 2002. From QTL to gene: the harvest begins. Nat. Genet. 31: 235–236. [DOI] [PubMed] [Google Scholar]
- Lander, E., and L. Kruglyak, 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11: 241–247. [DOI] [PubMed] [Google Scholar]
- Nadeau, J. H., J. B. Singer, A. Matin and E. S. Lander, 2000. Analysing complex genetic traits with chromosome substitution strains. Nat. Genet. 24: 221–225. [DOI] [PubMed] [Google Scholar]
- Nissen, S. E., T. Tsunoda, E. M. Tuzcu, P. Schoenhagen, C. J. Cooper et al., 2003. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial. J. Am. Med. Assoc. 290: 2292–2300. [DOI] [PubMed] [Google Scholar]
- Okamoto, H., F. Yonemori, K. Wakitani, T. Minowa, K. Maeda et al., 2000. A cholesteryl ester transfer protein inhibitor attenuates atherosclerosis in rabbits. Nature 406: 203–207. [DOI] [PubMed] [Google Scholar]
- Petkov, P. M., Y. Ding, M. A. Cassell, W. Zhang, G. Wagner et al., 2004. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 14: 1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plump, A. S., C. J. Scott and J. L. Breslow, 1994. Human apolipoprotein A-I gene expression increases high density lipoprotein and suppresses atherosclerosis in the apolipoprotein E-deficient mouse. Proc. Natl. Acad. Sci. USA 91: 9607–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittershaus, C. W., D. P. Miller, L. J. Thomas, M. D. Picard, C. M. Honan et al., 2000. Vaccine-induced antibodies inhibit CETP activity in vivo and reduce aortic lesions in a rabbit model of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 20: 2106–2112. [DOI] [PubMed] [Google Scholar]
- Rubin, E. M., R. M. Krauss, E. A. Spangler, J. G. Verstuyft and S. M. Clift, 1991. Inhibition of early atherogenesis in transgenic mice by human apolipoprotein AI. Nature 353: 265–267. [DOI] [PubMed] [Google Scholar]
- Sen, S., and G. A. Churchill, 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, J. B., A. E. Hill, J. H. Nadeau and E. S. Lander, 2005. Mapping quantitative trait loci for anxiety in chromosome substitution strains of mice. Genetics 169: 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou, I. M., J. K. Christians, P. D. Keightley, L. Bunger, M. Clinton et al., 2004. Genetic complexity of an obesity QTL (Fob3) revealed by detailed genetic mapping. Mamm. Genome 15: 472–481. [DOI] [PubMed] [Google Scholar]
- Stylianou, I. M., R. Korstanje, R. Li, S. Sheehan, B. Paigen et al., 2006. Quantitative trait locus analysis for obesity reveals multiple networks of interacting loci. Mamm. Genome 17: 22–36. [DOI] [PubMed] [Google Scholar]
- Sugano, M., N. Makino, S. Sawada, S. Otsuka, M. Watanabe et al., 1998. Effect of antisense oligonucleotides against cholesteryl ester transfer protein on the development of atherosclerosis in cholesterol-fed rabbits. J. Biol. Chem. 273: 5033–5036. [DOI] [PubMed] [Google Scholar]
- Wang, X., and B. Paigen, 2005. Genetics of variation in HDL cholesterol in humans and mice. Circ. Res. 96: 27–42. [DOI] [PubMed] [Google Scholar]
- Wang, X., M. Ria, P. M. Kelmenson, P. Eriksson, D. C. Higgins et al., 2005. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat. Genet. 37: 365–372. [DOI] [PubMed] [Google Scholar]
- Wilson, P. W., K. M. Anderson, T. Harris, W. B. Kannel and W. P. Castelli, 1994. Determinants of change in total cholesterol and HDL-C with age: the Framingham Study. J. Gerontol. 49: M252–M257. [DOI] [PubMed] [Google Scholar]
- Zeng, Z. B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]