Abstract
Exposure of Salmonella enterica to sodium cholate, sodium deoxycholate, sodium chenodeoxycholate, sodium glychocholate, sodium taurocholate, or sodium glycochenodeoxycholate induces the SOS response, indicating that the DNA-damaging activity of bile resides in bile salts. Bile increases the frequency of GC → AT transitions and induces the expression of genes belonging to the OxyR and SoxRS regulons, suggesting that bile salts may cause oxidative DNA damage. S. enterica mutants lacking both exonuclease III (XthA) and endonuclease IV (Nfo) are bile sensitive, indicating that S. enterica requires base excision repair (BER) to overcome DNA damage caused by bile salts. Bile resistance also requires DinB polymerase, suggesting the need of SOS-associated translesion DNA synthesis. Certain recombination functions are also required for bile resistance, and a key factor is the RecBCD enzyme. The extreme bile sensitivity of RecB−, RecC−, and RecA− RecD− mutants provides evidence that bile-induced damage may impair DNA replication.
AS a defense against bacterial pathogens, higher eukaryotes produce DNA-damaging agents such as nitric oxide (Vazquez-Torres and Fang 2000) and reactive oxygen species (O'Rourke et al. 2003). As a consequence, the pathogen relies on protective functions to prevent injuries caused by host-synthesized compounds and on DNA repair functions to repair them. Known examples of DNA repair functions required for bacterial virulence include base excision repair (BER) in Helicobacter pylori (O'Rourke et al. 2003) and Salmonella enterica (Suvarnapunya et al. 2003), mismatch repair in Listeria monocytogenes (Merino et al. 2002), nucleotide excision repair (NER) in Mycobacterium tuberculosis (Darwin and Nathan 2005), and homologous recombination in S. enterica (Buchmeier et al. 1993; Cano et al. 2002; Schapiro et al. 2003).
Bile is a digestive secretion produced in the mammalian liver and stored in the gall bladder. The composition of bile is complex and includes bile salts, cholesterol, bilirubin, phospholipids, and a variety of proteins (Hofmann 1998). Bile salts act as detergents and have strong antibacterial activity (Gunn 2000). A decrease in bile production, which can be caused either by malnourishment or by biliary pathological conditions, increases the susceptibility of the individual to a variety of bacterial pathogens (Gunn 2000). However, enteric bacteria are resistant to bile. Envelope structures act as barriers that reduce intake of bile salts (Picken and Beacham 1977; Prouty et al. 2002; Pucciarelli et al. 2002; Ramos-Morales et al. 2003); in addition, efflux pumps transport bile salts outside the cell. One such pump, encoded by the acrAB operon of Escherichia coli (Rosenberg et al. 2003), is also involved in resistance to multiple antibiotics and other antibacterial compounds (Ma et al. 1994). Another E. coli efflux system involved in expulsion of both bile and antibiotics is EmrAB (Lomovskaya and Lewis 1992).
Bile is not merely an antibacterial secretion; certain enteric pathogens use bile as a signal for the control of virulence genes (Pope et al. 1995; Prouty and Gunn 2000; Prouty et al. 2004; Hung and Mekalanos 2005). An especially interesting case of bile/pathogen interaction is found in S. enterica, which encounters bile in two different situations: (i) in the lumen of the mammalian intestine, where concentrations of bile salts range from 0.2 to 2% (Gunn 2000), and (ii) in the gall bladder, where much higher concentrations of bile are found (Hofmann 1998). The presence of S. enterica serovar Typhi in the gall bladder is characteristic of the asymptomatic “carrier state” of typhoid. Furthermore, hepatobiliar infections caused by S. enterica serovar Typhi and other Salmonella serovars cause acute cholecystitis (Lalitha and John 1994).
An investigation of the causes of bile sensitivity in DNA adenine methylase (Dam−) mutants of S. enterica serovar Typhimurium indicated that bile causes damage to Salmonella DNA and increases the rates of gene rearrangements and point mutations (Prieto et al. 2004). Bile may have similar effects on E. coli DNA, as indicated by the observation that exposure to bile salts induces genes of the SOS network (H. Bernstein et al. 1999). Evidence that bile salts likewise can be mutagenic in eukaryotic cells has existed for decades (Cook et al. 1940). A recent, comprehensive review on bile-induced mutagenesis in humans suggests that high levels of bile salts increase the frequencies of several types of cancer (Bernstein et al. 2005). In turn, epidemiologic studies have shown a relationship between Salmonella-induced gallstone formation and the development of hepatobiliary carcinomas (Dutta et al. 2000). Below we describe investigations aimed at identifying the DNA-damaging compounds contained in bile, the origin of nucleotide substitutions caused by bile exposure, and the DNA repair mechanisms that contribute to bile resistance in S. enterica.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacteriophages, and strain construction:
All the S. enterica strains listed in Table 1 belong to serovar Typhimurium. Unless otherwise indicated, all strains derive from the mouse virulent strain ATCC 14028. Strain DA7974 was obtained from Dan I. Andersson (Swedish Institute for Infectious Disease Control, Solna, Sweden). The alleles recA1 and recF522∷ Tn5 were provided by J. R. Roth (Section of Microbiology, University of California, Davis, CA). Other recombination alleles used in this study are recB497∷MudJ and recC498∷MudJ (Mahan and Roth 1989), recD543∷Tn10dTc (Miesel and Roth 1994), and recJ504∷MudCm (Mahan et al. 1992). Transfer of transposon-tagged mutations to ATCC 14028 was performed by transductional crosses with phage P22 HT 105/1 int201 (Schmieger 1972), followed by lysogen disposal on green plates (Chan et al. 1972). The sbcB21 allele, originally in the background of strain LT2, was introduced into ATCC 14028 by P22-mediated cotransduction with the zeb-6314∷Tn10dTc insertion. The recombination-deficient phenotypes of newly constructed strains were verified using tests described elsewhere (Mahan and Roth 1989; Benson and Roth 1994; Miesel and Roth 1994; Garzón et al. 1996).
TABLE 1.
Strains of S. enterica serovar Typhimurium
| Strain | Genotype |
|---|---|
| SV3019a | proA692∷MudQ/F′ lacZ101 |
| SV3020a | proA692∷MudQ/F′ lacZ102 |
| SV3021a | proA692∷MudQ/F′ lacZ103 |
| SV3022a | proA692∷MudQ/F′ lacZ104 |
| SV3023a | proA692∷MudQ/F′ lacZ105 |
| SV3024a | proA692∷MudQ/F′ lacZ106 |
| SV4579 | recD541∷Tn10dCm recJ504∷MudJ |
| SV4844 | recB497∷MudJ |
| SV4851 | 14028/pGE108 |
| SV4869 | srl203∷Tn10dCm recA1 |
| SV4870 | srl203∷Tn10dCm recA1/pGE108 |
| SV4992 | Δnei∷Cm |
| SV4994 | Δnth∷Cm |
| SV4996 | Δnfo∷Cm |
| SV4998 | ΔxthA∷Cm |
| SV5029 | ΔxthA∷Cm Δnfo∷Km |
| SV5033 | Δnth∷Cm Δnei∷Km |
| SV5038 | ΔmutM |
| SV5039 | ΔmutM Δnei∷Cm Δnth∷Km |
| SV5043 | lexA39(Ind−)/pGE108 |
| SV5074 | recJ504∷MudCm recF522∷Tn5 |
| SV5075 | recD543∷Tn10dTc recF525∷Tn5 |
| SV5076 | recJ504∷MudCm |
| SV5078 | recA1 srl-211∷Tn5 recJ504∷MudCm |
| SV5079 | sbcB22 zeb-6314∷Tn10dTc recB497∷MudJ |
| SV5080 | recF522∷Tn5 |
| SV5081 | recA1 srl-203∷Tn10dCm recB497∷MudJ |
| SV5082 | recC498∷MudJ |
| SV5107 | Δada |
| SV5108 | Δtag |
| SV5109 | Δogt |
| SV5110 | ΔalkA |
| SV5111 | Δada Δtag |
| SV5112 | ΔalkA Δtag |
| SV5141 | Δada∷Cm Δogt |
| SV5142 | ΔuvrB∷Cm |
| SV5144 | ΔumuDC∷Km |
| SV5147 | ΔmutT∷Km |
| SV5149 | ΔkatG |
| SV5150 | Δdps |
| SV5153 | ΔfumC |
| SV5154 | φ(fumC-′lacZY) |
| SV5157 | φ(katG′-lacZY) |
| SV5158 | φ(dps′-lacZY) |
| SV5159 | φ(nfo-′lacZY) |
| SV5166 | recD543∷Tn10dTc |
| SV5167 | mutY131∷Tn5 |
| SV5168 | recA1 srl∷Tn10dCm recD∷Tn10dTc |
| SV5209 | φ(nei′-lacZY) |
| SV5210 | φ(nth′-lacZY) |
| SV5213 | ΔmutM mutY131∷Tn5 |
| DA7974 | dinB1013∷Cm |
LT2 background.
Media and chemicals:
NCE, a modification of E medium (Vogel and Bonner 1956) lacking citric acid, was used as the standard minimal medium for S. enterica. Carbon sources for NCE were either 0.2% glucose or 1% lactose. The rich medium was Luria–Bertani broth (LB). Solid media contained agar at 1.5% final concentration. Sodium choleate (ox bile extract) and sodium salts of deoxycholic acid, cholic acid, chenodeoxycholic acid, glycocholic acid, taurocholic acid, and glycochenodeoxycholic acid were purchased from Sigma Chemical (St. Louis). Antibiotics were used at the final concentrations described elsewhere (Garzón et al. 1996). Green plates were prepared using the original recipe (Chan et al. 1972), except that methyl blue (Sigma Chemical) substituted for aniline blue.
Construction of S.enterica mutants by gene targeting:
Disruption of selected genes in the S. enterica chromosome was achieved by adapting to S. enterica a gene-targeting method previously described in E. coli (Datsenko and Wanner 2000). Primers designed to eliminate specific DNA stretches, based on the LT2 nucleotide sequence (McClelland et al. 2001), are shown in supplemental Table 1 at http://www.genetics.org/supplemental/. When necessary, the kanamycin resistance cassette introduced by the gene-targeting procedure was eliminated by recombination with plasmid pCP20 (Datsenko and Wanner 2000). Pairs of additional external PCR primers were used to verify the predicted gene deletions (supplemental Table 1 at http://www.genetics.org/supplemental/).
Construction of transcriptional lac fusions in the Salmonella chromosome:
FRT sites generated by excision of Kmr cassetes (Datsenko and Wanner 2000) were used to integrate plasmid pCE37 (Ellermeier et al. 2002) to generate transcriptional fusions in the dps, katG, nfo, and fumC genes of S. enterica.
Minimal inhibitory concentrations of sodium deoxycholate and ox bile extract:
Exponential cultures in LB broth were prepared. Samples containing ∼3 × 102 colony-forming-units were transferred to polypropylene microtiter plates (Soria Genlab, Valdemoro, Spain) containing known amounts of sodium deoxycholate or ox bile extract. After 12 hr incubation at 37°, growth was monitored visually. Assays were carried out in triplicate. Student's t-test was used to analyze every minimal inhibitory concentration (MIC). The null hypothesis was that MICs were not significantly different from the MIC for the wild type. P-values of ≤0.01 were considered significant.
Estimation of mutation rates using lac alleles:
Aliquots containing ∼106 cells were added to tubes of LB containing 15% bile. The cultures were incubated at 37° until saturation (∼109 cells/ml), washed twice with phosphate-buffered saline (PBS), and concentrated 25-fold. Aliquots were then spread on NCE–lactose plates. Lac+ revertants were scored after 48 hr incubation at 37°. Viable cell counts were carried out on NCE–glucose plates. Mutation rates were calculated using the median method (Lea and Coulson 1949).
β-Galactosidase assays:
Levels of β-galactosidase activity were assayed using the CHCl3–sodium dodecyl sulfate permeabilization procedure (Miller 1972). Plate tests for monitoring β-galactosidase activity were carried out as described elsewhere (Prieto et al. 2004), using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (“X-gal,” Sigma Chemical) as indicator. To monitor the β-galactosidase activities of dps∷lac, katG∷lac, nfo∷lac, and fumC∷lac fusions in response to sodium deoxycholate, exponential cultures in LB were washed with PBS buffer and resuspended in PBS containing 5% sodium deoxycholate. The bacterial suspensions were incubated with shaking at 37° for 3 hr and washed twice with PBS before measuring β-galactosidase activities.
RESULTS
DNA-damaging activity of bile salts:
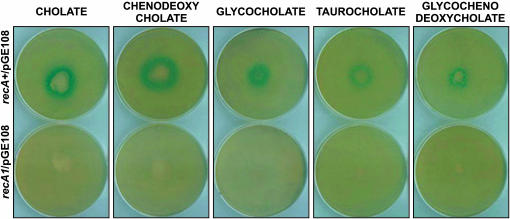
A previous study showed that the DNA-damaging ability of bile could be reproduced with an individual bile component, sodium deoxycholate (Prieto et al. 2004). To ascertain whether other bile salts were able to cause DNA damage, we tested their DNA-damaging capacity. The occurrence of DNA damage was examined by monitoring the activity of an SOS-inducible fusion (cea∷lacZ) carried on plasmid pGE108, as previously described (Prieto et al. 2004). Exposure to any of five individual bile salts (sodium cholate, sodium chenodeoxycholate, sodium gycocholate, sodium taurocholate, and sodium glycochenodeoxycholate) turned on the expression of the cea∷lacZ fusion in a RecA+ background (Figure 1). Absence of cea∷lacZ expression in a RecA− background confirmed the occurrence of canonical, RecA-dependent SOS induction (Figure 1). Hence, the DNA-damaging activity of bile appears to reside in bile salts. Evidence that bile salts cause DNA damage has also been obtained in eukaryotic cells (Bernstein et al. 2005).
Figure 1.—
Plate tests of SOS induction by bile salts. Each plate was seeded with >107 cells (which form a lawn after growth). A small amount of bile salt powder was placed in the center of the plate. Plates were incubated overnight at 37°. The plates at the top correspond to strain SV4851 (recA+/pGE108); the bottom plates correspond to SV4870 (recA1/pGE108).
Specificity of substitution mutations induced by bile:
Exposure of S. enterica to bile causes both nucleotide substitutions and −1 frameshifts; the latter, however, are largely reduced in a LexA(Ind−) mutant, suggesting that frameshift mutations are a consequence of SOS induction (Prieto et al. 2004). It is well known that translesion DNA synthesis by the SOS-associated DinB polymerase causes −1 frameshifts in E. coli (Kim et al. 2001). To determine the specificity of nucleotide substitutions caused by treatment with bile, we used a collection of lacZ alleles that allow detection of the six base substitutions (Cupples and Miller 1989). Results from 20 independent experiments can be summarized as follows:
One of the six alleles (lacZ102) showed a >20-fold increase in reversion to Lac+ in the presence of bile. The estimated mutation rates of lacZ102 were 2.32 × 10−10 in LB and 5.61 × 10−9 in LB containing 15% bile.
Reversion of the lacZ101, lacZ103, lacZ104, lacZ105, and lacZ106 alleles was not affected by bile (data not shown).
These experiments indicated that exposure to bile induces GC → AT transitions, a pattern previously described for DNA-alkylating agents such as ethyl methanesulfonate and N-methyl-N′-nitro-N-nitrosoguanidine (Cupples and Miller 1989) and also for DNA-oxidizing agents (Kreutzer and Essigmann 1998). Although the detailed spectrum of a given mutagen ideally requires the use of multiple sites to monitor each specific mutagenic event, the occurrence of GC → AT transitions provided preliminary evidence that bile salts could be either alkylating agents or oxidizing agents.
Genetic evidence that bile salts cause DNA oxidative damage:
If bile caused oxidation of DNA, we reasoned, S. enterica mutants lacking functions involved in repair of DNA oxidation could be expected to be bile sensitive. The same rationale was applied to repair of DNA alkylation. On these grounds, appropriate mutants were constructed and tested for growth in the presence of sodium deoxycholate and ox bile. These surveys provided evidence that bile salts may cause DNA oxidation rather than DNA alkylation:
A mutant of S. enterica lacking exonuclease III (XthA) and endonuclease IV (Nfo) was extremely sensitive to bile salts (Table 2). In E. coli, the AP endonucleases XthA and Nfo are required for repair of oxidative damage, and their absence renders E. coli sensitive to DNA-oxidizing agents (Demple et al. 1986). However, XthA and Nfo are also required to repair DNA damage caused by alkylating agents (Cunningham et al. 1986).
A S. enterica strain lacking the 3-methyladenine DNA glycosylases I and II (TagA and AlkA, respectively) was not bile sensitive (Table 2). In E. coli, TagA− AlkA− mutants are extremely sensitive to alkylation exposure (Clarke et al. 1984). This observation provided evidence that bile salts do not cause DNA alkylation.
In E. coli, Ada and Ogt are required for the reversal of DNA lesions caused by N-methyl-N′-nitro-N-nitrosoguanidine, N-methyl-N-nitrosourea, and other alkylating agents (Samson 1992). As a consequence, Ada− Ogt− mutants of E. coli are sensitive to alkylating agents (Samson 1992). In contrast, a S. enterica strain lacking Ada and Ogt methyltransferases proved to be bile resistant (Table 2), providing further evidence against bile-induced DNA alkylation.
TABLE 2.
Minimal inhibitory concentrations (g/100 ml) of sodium deoxycholate and ox bile extract in S. enterica mutants lacking DNA repair functions
| Strain | Genotype | Sodium deoxycholate | Ox bile extract |
|---|---|---|---|
| 14028 | Wild type | 7 | 14 |
| SV4992 | Δnei∷Cm | 6 | 13 |
| SV4994 | Δnth∷Cm | 7 | 14 |
| SV4996 | Δnfo∷Cm | 6 | 13 |
| SV4998 | ΔxthA∷Cm | 7 | 14 |
| SV5008 | Δnei∷Km Δnth∷Cm | 6 | 13 |
| SV5029 | Δnfo∷Km ΔxthA∷Cm | 0.4a | 6a |
| SV5039 | Δnei∷Km Δnth∷Cm ΔmutM | 6 | 13 |
| SV5110 | ΔalkA | 7 | 14 |
| SV5111 | Δada Δtag | 6 | 12 |
| SV5112 | ΔalkA Δtag | 6 | 12 |
| SV5141 | Δada∷Cm Δogt | 6 | 13 |
| SV5142 | ΔuvrB | 7 | 14 |
| SV5147 | ΔmutT∷Km | 7 | 14 |
| SV5149 | ΔkatG | 6 | 13 |
| SV5150 | Δdps | 6 | 13 |
| SV5153 | ΔfumC | 7 | 14 |
| SV5167 | mutY131∷Tn5 | 6 | 12 |
| SV5213 | mutY131∷Tn5 ΔmutM | 6 | 12 |
| SV5043 | lexA39(Ind−) | 2a | 8a |
| SV5144 | ΔumuDC∷Km | 6 | 13 |
| DA7974 | dinB1013∷Cm | 0.4a | 6a |
Data are based on an average of more than three independent experiments.
MIC showing significant difference from the MIC for the wild type (P < 0.01, Student's t-test).
A frequent outcome of oxidative damage is the production of 7,8-dihydro-8-oxoguanine (8-oxoG), a mutagenic base analog (Shibutani et al. 1991). Protection against 8-oxoG relies on the GO system, composed of three genes: mutM (also known as fpg), encoding formamidopyrimidine DNA glycosylase (Boiteux et al. 1987); mutT, encoding nucleoside triphosphate pyrophosphohydrolase (Bhatnagar et al. 1991); and mutY, encoding MutY DNA adenine glycosylase (Michaels and Miller 1992). A priori, it seemed unlikely that bile salts might increase the pool of 8-oxoG, since the latter causes mainly AT → CG transversions (Fowler et al. 2003) while exposure to bile increases GC → AT transitions. Despite this evidence against the occurrence of 8-oxoG as a bile-induced lesion, we tested whether mutations in the GO system caused bile sensitivity. For this purpose, we estimated the MICs of sodium deoxycholate and ox bile extract for MutT−, MutY−, and MutM− MutY− mutants. None of them was bile sensitive (Table 2), providing further evidence that exposure to bile salts does not result in 8-oxoG formation. An alternative possibility is that bile salts may cause cytosine oxidation, which does increase GC → AT transitions (Kreutzer and Essigmann 1998). Whatever the primary lesion(s), the observation that an Nth− Nei− mutant of S. enterica is resistant to bile salts (Table 2) may suggest that such lesions are not substrates for endonucleases III (Nth) and VIII (Nei), two BER enzymes with overlapping substrate specificities for oxidative damage products (Purmal et al. 1998; Dizdaroglu et al. 2001).
Induction of the S. enterica OxyR and SoxS regulons in the presence of bile:
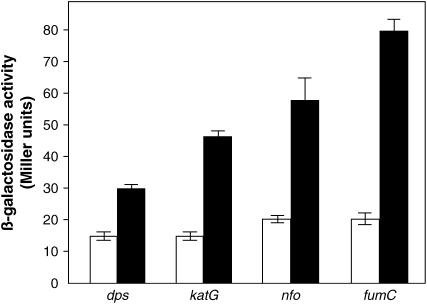
In both E. coli and S. enterica, oxidative stress induces a variety of defense functions (Demple 1999). Key factors in inducible oxidative stress responses are the OxyR and SoxR transcription factors that control expression of the OxyR and SoxRS regulons, respectively (Storz and Imlay 1999; Volkert and Landini 2001). If bile salts cause oxidative damage as suggested by the mutant analyses described in Genetic evidence that bile salts cause DNA oxidative damage, exposure of S. enterica to bile should induce the OxyR and SoxRS regulons. On these grounds, we monitored the expression of selected OxyR- and SoxRS-regulated genes in response to sodium deoxycholate. Transcriptional lac fusions were constructed in dps and katG, two genes of the OxyR regulon that undergo strong induction upon oxidative damage (Zheng et al. 2001), and in nfo and fumC, two genes of the SoxRS regulon (Liochev and Fridovich 1992; Li and Demple 1994). All the fusions increased their expression in the presence of sodium deoxycholate (Figure 2), indicating that exposure to bile does induce the OxyR and SoxRS regulons. These observations provide additional evidence that bile salts may be DNA-oxidizing agents. Deoxycholate-mediated activation of promoters that respond to oxidative stress has been also described in E. coli (C. Bernstein et al. 1999) and in human cell lines (H. Bernstein et al. 1999).
Figure 2.—
Induction of genes in the oxyR and soxRS regulons by sodium deoxycholate. The strains shown, from left to right, are SV5158 (dps∷lacZ), SV5157 (katG∷lacZ), SV5159 (nfo∷lacZ), and SV5154 (fumC∷lacZ). Open bars show β-galactosidase activities of the fusions in LB. Solid bars show β-galactosidase activities of the fusions after exposure to 5% sodium deoxycholate. The data shown are means and standard deviations of four independent experiments.
Role of base excision repair in bile resistance:
In most organisms, the primary means of restoring the correct DNA base sequence is by the DNA BER pathway (Mol et al. 2000). In S. enterica, BER is required for infection in the murine typhoid fever model and plays a role in the repair of DNA damage within macrophages (Suvarnapunya et al. 2003). The possibility that BER might be likewise involved in bile resistance was therefore examined. For this purpose, we estimated the MICs of bile and sodium deoxycholate for mutants carrying mutations in one or more of the following genes: xthA, encoding exonuclease III (White et al. 1976); nei, encoding endonuclease VIII (Saito et al. 1997); nth, encoding endonuclease III (Saito et al. 1997); and nfo, encoding endonuclease IV (Cunningham et al. 1986). The results can be summarized as follows:
Individual XthA−, Nei−, Nth−, and Nfo− mutants showed MICs similar to that of the wild type: ∼140 mg/ml of ox bile and ∼70 mg/ml sodium deoxycholate (Table 2). Hence, none of the individual exonucleases involved (exonuclease III, endonuclease VIII, endonuclease III, and endonuclease IV) is essential for bile resistance.
Combinations of two or even three DNA glycosylase mutations did not cause bile sensitivity, suggesting that S. enterica does not harbor a DNA glycosylase that specifically recognizes bile-induced damage. It is well known that many forms of DNA damage, particularly those caused by the interaction of DNA with environmental agents, are not recognized by specific DNA glycosylases (Friedberg et al. 1995).
Lack of both exonuclease III (XthA) and endonuclease IV (Nfo) caused extreme bile sensitivity (Table 2). This observation suggests that bile-induced DNA lesions cannot be repaired by base excision repair in the absence of both exonuclease III and endonuclease IV, probably because the free DNA ends required for repair are not produced.
Nucleotide excision repair is dispensable for bile resistance:
To investigate whether NER plays a role in bile resistance, we tested the effect of uvrB gene disruption. UvrB is a subunit of excision nuclease and is part of the UvrABC excision complex (Braun and Grossman 1974; Seeberg 1978). The MICs of bile and sodium deoxycholate were similar in a S. enterica UvrB− mutant and in the wild type (Table 2), indicating that nucleotide excision repair is dispensable for bile resistance. NER is usually involved in repair of DNA distortions involving bulky base adducts (Friedberg et al. 1995). Hence, the observation that NER is dispensable for bile resistance suggests that the DNA lesions caused by bile salts are nondistortive. Oxidized bases, as proposed above, would certainly fit in this category.
Role of SOS induction in bile resistance:
Exposure of S. enterica to bile triggers SOS induction in a dose-dependent manner (Prieto et al. 2004). To examine whether SOS induction was required for bile resistance, we compared the minimal inhibitory concentration of bile in the wild type and in an isogenic LexA(Ind−) mutant. The latter was found to be bile sensitive (Table 2), suggesting that SOS induction is indeed required to cope with bile-induced DNA damage. We also examined whether the SOS-induced DNA polymerases DinB (Pol IV) and UmuDC (Pol V) were involved in bile resistance. A DinB− mutant was found to be bile sensitive while an UmuD− mutant was bile resistant (Table 2). These observations suggest that DNA replication in the presence of bile salts requires Pol IV-mediated translesion synthesis, while Pol V is not involved. It is well known that UmuDC and DinB have distinct abilities to replicate over DNA templates, depending on the nature of the lesions produced (Sutton et al. 2000; Kokubo et al. 2005). Hence, our data suggest that lesions caused by bile may impair DNA replication and that translesion synthesis of bile-damaged DNA may occur via Pol IV. This view is consistent with the observation that exposure to bile increases −1 frameshifts in a LexA-dependent manner (Prieto et al. 2004).
The involvement of the SOS response in bile resistance might also reflect the need of homologous recombination for survival after bile exposure (see below). In E. coli, certain recombination functions (e.g., RecA and RecN) are known to be part of the SOS regulon (Khil and Camerini-Otero 2002).
Role of homologous recombination in bile resistance:
To identify recombination functions of S. enterica required for bile resistance, strains carrying recA, recB, recC, recD, recF, and recJ mutations (individually or combined) were tested for sensitivity to ox bile extract and to sodium deoxycholate. The MICs obtained are shown in Table 3. Relevant observations are as follows:
Among single mutants, the most sensitive to bile were RecB− and RecC−. RecA− mutants were also bile sensitive, but to a lesser extent than RecB− and RecC−. These bile sensitivity patterns indicate that the RecBCD enzyme is a crucial function for bile resistance. In E. coli, SOS induction by certain DNA-damaging agents requires RecBCD (McPartland et al. 1980). However, a recB mutation does not abolish SOS induction by bile in S. enterica (data not shown). Hence, we tentatively interpret that RecBCD may be required to repair DNA damage during DNA replication, either by recombination or by degradation of double-stranded ends (Uzest et al. 1995; Michel et al. 1997).
The involvement of RecA in bile resistance may indicate the need of double-strand break repair via the RecBCD pathway, the need of SOS induction for survival in the presence of bile, or both. The additive effect of recA and recB mutations on bile sensitivity (Table 3) argues in favor of a RecB-independent RecA activity such as SOS induction.
A RecA− RecD− double mutant was sensitive to bile, providing further evidence that elimination of both the RecBCD pathway and RecB-mediated degradative repair may prevent repair of bile-induced lesions that impair DNA replication.
A recD mutation did not cause bile sensitivity, indicating that the exonuclease activity of the RecBCD enzyme is dispensable for bile resistance. An interpretation may be that RecBCD-mediated recombination is sufficient under such conditions and that the degradative pathway is not needed.
RecF−, RecJ−, and RecF− RecJ− mutants were bile resistant. Hence, aside from recA, mutations that disrupt the RecF pathway of homologous recombination do not cause bile sensitivity.
sbcB mutations, which suppress the recombination defect of RecBC− strains by activating the RecF pathway (Benson and Roth 1994), were unable to suppress bile sensitivity in a RecB− background (Table 3). These experiments provided further evidence that the RecF pathway is not required for bile resistance.
The observation that a RecD− RecJ− mutant was bile sensitive (Table 3) provided additional evidence for the need of double-strand break repair by the RecBCD pathway, since ExoV and ExoIX can provide alternative exonuclease activities in E. coli (Viswanathan and Lovett 1998) and Salmonella (Garzón et al. 1996). Functional redundancy may explain why single RecD− and RecJ− mutants of S. enterica are bile resistant.
TABLE 3.
Minimal inhibitory concentrations (g/100 ml) of sodium deoxycholate and ox bile extract in recombination mutants of S. enterica
| Strain | Genotype | Sodium deoxycholate | Ox bile extract |
|---|---|---|---|
| 14028 | Wild type | 7 | 14 |
| SV4869 | recA1 | 3a | 9a |
| SV4844 | recB497∷MudJ | 1a | 7a |
| SV5082 | recC498∷MudJ | 1a | 7a |
| SV5166 | recD543∷Tn10dTc | 7 | 14 |
| SV5080 | recF522∷Tn5 | 6 | 13 |
| SV5076 | recJ504∷MudCm | 6 | 13 |
| SV5081 | recA1 recB497∷MudJ | 0.4a | 4a |
| SV5168 | recA1 recD543∷Tn10dTc | 2a | 5a |
| SV5078 | recA1 recJ504∷MudCm | 3a | 9a |
| SV5079 | recB497∷MudJ sbcB21 | 1a | 7a |
| SV5075 | recD543∷Tn10dTc recF522∷Tn5 | 6 | 12 |
| SV4579 | recD543∷Tn10dTc recJ504∷MudCm | 0.4a | 6a |
| SV5074 | recF522∷Tn5 recJ504∷MudCm | 6 | 13 |
Data are based on an average of more than three independent experiments.
MIC showing significant difference from the MIC for the wild type (P < 0.01, Student's t-test).
DISCUSSION
In addition to its widely known role as a detergent that disrupts the bacterial envelope (Gunn 2000), bile causes DNA-damaging activity on S. enterica and induces DNA rearrangements and point mutations (Prieto et al. 2004). This study provides evidence that, like its detergent activity, the DNA-damaging capacity of bile resides in bile salts. The specific nature of the damage caused by bile salts to S. enterica DNA remains speculative at this stage. However, several lines of evidence indicate that bile salts may cause oxidative damage: (i) exposure to bile induces GC → AT transitions; (ii) sodium deoxycholate activates transcription of genes belonging to the oxidative-damage-responsive OxyR and SoxSR regulons; and (iii) S. enterica mutants lacking exonuclease III (XthA) and endonuclease IV (Nfo) are extremely sensitive to bile (Table 2). Evidence that bile salts cause base oxidative damage also has been obtained in eukaryotic cells (Bernstein et al. 2005).
A common outcome of oxidative damage is the formation of 8-oxoG. However, the absence of transversions, especially GC → TA (Fowler et al. 2003), among bile-induced mutations suggests that 8-oxoG is not the major primary lesion caused by bile. This hypothesis is further supported by the observation that S. enterica mutants lacking functions involved in removal of oxidized forms of guanine (Michaels and Miller 1992; Fowler et al. 2003) are not sensitive to bile. Hence, we tentatively propose that the GC → AT transitions induced by bile salts may result from the formation of oxidized forms of cytosine, as previously described for other oxidizing agents (Kreutzer and Essigmann 1998).
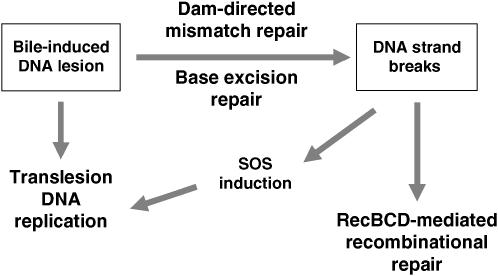
Whatever the primary lesion caused by bile salts, S. enterica appears to require an ample repertoire of DNA repair functions to cope with the resulting DNA damage. The potential roles of such functions are accommodated in the tentative model in Figure 3. Primary lesions may trigger base excision repair, and the activity of either endonuclease III or exonuclease IV can be expected to produce DNA strand breaks as an intermediate step in the DNA repair process. These DNA strand breaks may impair progression of replication forks, inducing the SOS response; the latter may allow DinB-mediated translesion synthesis. It is also conceivable that bile-induced lesions could directly block DNA replication, thus inducing the SOS response in a direct fashion. In such a scenario, the need of homologous recombination mediated by the RecBCD enzyme might reflect the occurrence of stalled DNA replication forks (Uzest et al. 1995; Michel et al. 1997; Seigneur et al. 1998). An alternative explanation for the need of RecA, RecBCD, and DinB polymerase is that exposure to bile might trigger stress-induced mutagenesis, a physiological condition that relies on the same repertoire of DNA repair functions (Ponder et al. 2005).
Figure 3.—
Model for repair of bile-induced damage in S. enterica. Primary lesions (perhaps oxidized cytosines) are repaired by Dam-directed mismatch repair and by base excision repair. Either process generates single-stranded DNA that can induce the SOS response. Direct SOS induction can also occur if primary lesions impair DNA replication. Furthermore, single-strand breaks generated by DNA repair can give rise to double-strand breaks during DNA replication. Upon SOS induction, translesion DNA replication by the DinB polymerase may help to overcome DNA replication blockage. In turn, RecBCD may rescue arrested replication forks by either recombinational repair or degradation of double-strand ends.
Bile resistance also requires Dam-directed mismatch repair (Pucciarelli et al. 2002; Prieto et al. 2004), which in E. coli is known to repair oxidative DNA lesions (Wyrzykowski and Volkert 2003). The observation that Dam− mutants of S. enterica are bile sensitive (Pucciarelli et al. 2002) while MutHLS− mutants are bile resistant (Prieto et al. 2004) can be explained as follows: in MutHLS− mutants, the combined activities of DNA repair systems other than Dam/MutHLS (e.g., base excision repair, SOS translesion synthesis, and RecBC-mediated recombinational repair) may be sufficient to resist bile. In Dam− mutants, however, absence of DNA strand discrimination results in the formation of MutHLS-mediated double-strand breaks that render the cell bile sensitive (Prieto et al. 2004). This view is supported by other studies showing that dam mutations sensitize E. coli to a variety of DNA-damaging agents (Glickman and Radman 1980; Fram et al. 1985; Nowosielska and Marinus 2005).
During animal infection, S. enterica encounters bile salts in the gut, where bile concentrations are low and changing (Hofmann 1998). Because bile is not a strong mutagen, it seems, a priori, unlikely that bile-induced DNA damage can be a significant source of genetic polymorphism during intestinal infection (Prieto et al. 2004). However, Salmonella serovars that cause systemic and chronic infections colonize the gall bladder, where bile can reach a steady concentration of 15% or higher (Gunn 2000). Furthermore, the formation of biofilms on gallstones exposes Salmonella to even higher concentrations of bile salts (Prouty et al. 2003). Thus, it seems conceivable that bile-induced DNA damage might play a role in the evolution of Salmonella populations in the gall bladder. For instance, the genome rearrangements commonly found in S. typhi strains (Echeita and Usera 1998; Ng et al. 1999; Kothapalli et al. 2005) might be favored by exposure to bile salts, whose ability to induce DNA rearrangements has been previously shown (Prieto et al. 2004). Bile-induced mutagenesis might also increase the polymorphism of Salmonella populations in the harsh environment of the mammalian gall bladder, perhaps providing an example of stress-induced genetic variability (Rosenberg and Hastings 2003; Saint-Ruf and Matic 2006).
The mutagenic effect of bile salts in vivo may be relieved by the presence of bilirubin, which represents ∼0.3% of the bile composition of healthy humans (Hofmann 1998) and has been shown to possess antioxidant activity (Stocker et al. 1987). However, the protective effect of bilirubin may be only partial, as indicated by the observation that ox bile extract has DNA-damaging capacity (Prieto et al. 2004). Furthermore, epidemiologic studies have unambiguously shown that increased levels of bile, associated either with cholecystitis or with other hepatobiliar disorders, increase the incidence of several types of cancer (Bernstein et al. 2005). Salmonella can play a direct role in some such conditions, either by causing acute cholecystitis or by persisting in the gall bladder of chronic typhoid carriers. In fact, Salmonella-induced gallstone formation has been shown to favor the development of hepatobiliary carcinomas (Dutta et al. 2000).
Acknowledgments
We are grateful to Susan Lovett, Martin Marinus, Pablo Radicella, Bénédicte Michel, Andrés Aguilera, and Shoshy Altuvia for helpful discussions. This study was supported by grant BIO2004-3455-CO2-02 from the Spanish Ministry of Education and Science and the European Regional Fund. A. I. Prieto was the recipient of a predoctoral fellowship from the Fundación Ramón Areces.
References
- Benson, N. R., and J. Roth, 1994. Suppressors of recB mutations in Salmonella typhimurium. Genetics 138: 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, C., H. Bernstein, C. M. Payne, S. E. Beard and J. Schneider, 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 39: 68–72. [DOI] [PubMed] [Google Scholar]
- Bernstein, H., C. M. Payne, C. Bernstein, J. Schneider, S. E. Beard et al., 1999. Activation of the promoters of genes associated with DNA damage, oxidative stress, ER stress and protein malfolding by the bile salt, deoxycholate. Toxicol. Lett. 108: 37–46. [DOI] [PubMed] [Google Scholar]
- Bernstein, H., C. Bernstein, C. M. Payne, K. Dvorakova and H. Garewal, 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 589: 47–65. [DOI] [PubMed] [Google Scholar]
- Bhatnagar, S. K., L. C. Bullions and M. J. Bessman, 1991. Characterization of the mutT nucleoside triphosphatase of Escherichia coli. J. Biol. Chem. 266: 9050–9054. [PubMed] [Google Scholar]
- Boiteux, S., T. R. O'Connor and J. Laval, 1987. Formamidopyrimidine-DNA glycosylase of Escherichia coli: cloning and sequencing of the fpg structural gene and overproduction of the protein. EMBO J. 6: 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, A., and L. Grossman, 1974. An endonuclease from Escherichia coli that acts preferentially on UV-irradiated DNA and is absent from the uvrA and uvrB mutants. Proc. Natl. Acad. Sci. USA 71: 1838–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier, N. A., C. J. Lipps, M. Y. So and F. Heffron, 1993. Recombination-deficient mutants of Salmonella typhimurium are avirulent and sensitive to the oxidative burst of macrophages. Mol. Microbiol. 7: 933–936. [DOI] [PubMed] [Google Scholar]
- Cano, D. A., M. G. Pucciarelli, F. Garcia-del Portillo and J. Casadesús, 2002. Role of the RecBCD recombination pathway in Salmonella virulence. J. Bacteriol. 184: 592–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, R. K., D. Botstein, T. Watanabe and Y. Ogata, 1972. Specialized transduction of tetracycline by phage P22 in Salmonella typhimurium. II. Properties of a high frequency transducing lysate. Virology 50: 883–898. [DOI] [PubMed] [Google Scholar]
- Clarke, N. D., M. Kvaal and E. Seeberg, 1984. Cloning of Escherichia coli genes encoding 3-methyladenine DNA glycosylases I and II. Mol. Gen. Genet. 197: 368–372. [DOI] [PubMed] [Google Scholar]
- Cook, J. W., E. L. Kennaway and N. M. Kennaway, 1940. Production of tumours in mice by deoxycholic acid. Nature 145: 627. [Google Scholar]
- Cunningham, R. P., S. M. Saporito, S. G. Spitzer and B. Weiss, 1986. Endonuclease IV (nfo) mutant of Escherichia coli. J. Bacteriol. 168: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples, C. G., and J. H. Miller, 1989. A set of lacZ mutations that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86: 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin, K. H., and C. F. Nathan, 2005. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect. Immun. 73: 4581–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko, K. A., and B. L. Wanner, 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 90: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple, B., 1999. Radical ideas: genetic responses to oxidative stress. Clin. Exp. Pharmacol. Physiol. 26: 64–68. [DOI] [PubMed] [Google Scholar]
- Demple, B., A. Johnson and D. Fung, 1986. Exonuclease III and endonuclease IV remove 3′ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc. Natl. Acad. Sci. USA 83: 7731–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu, M., S. M. Burgess, P. Jaruga, T. K. Hazra, H. Rodriguez et al., 2001. Substrate specificity and excision kinetics of Escherichia coli endonuclease VIII (Nei) for modified bases in DNA damaged by free radicals. Biochemistry 40: 12150–12156. [DOI] [PubMed] [Google Scholar]
- Dutta, U., P. K. Garg, R. Kumar and R. K. Tandon, 2000. Typhoid carriers among patients with gallstones are at increased risk for carcinoma of the gall bladder. Am. J. Gastroenterol. 95: 784–787. [DOI] [PubMed] [Google Scholar]
- Echeita, M. A., and M. A. Usera, 1998. Chromosomal rearrangements in Salmonella enterica serovar Typhi affecting molecular typing in outbreak investigations. J. Clin. Microbiol. 36: 2123–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier, C. D., A. Janakiram and J. M. Slauch, 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290: 153–161. [DOI] [PubMed] [Google Scholar]
- Fowler, R. G., S. J. White, C. Koyama, S. C. Moore, R. L. Dunn et al., 2003. Interactions among the Escherichia coli mutT, mut M, and mutY damage prevention pathways. DNA Repair 2: 159–173. [DOI] [PubMed] [Google Scholar]
- Fram, R. J., P. S. Cusick, J. M. Wilson and M. G. Marinus, 1985. Mismatch repair of cis-diamminedichloroplatinum(II)-induced DNA damage. Mol. Pharmacol. 28: 51–55. [PubMed] [Google Scholar]
- Friedberg, E. C., G. C. Walker and W. Siede, 1995. DNA Repair and Mutagenesis. American Society of Microbiology, Washington, DC.
- Garzón, A., C. R. Beuzón, M. J. Mahan and J. Casadesús, 1996. recB recJ mutants of Salmonella typhimurium are deficient in transductional recombination, DNA repair and plasmid maintenance. Mol. Gen. Genet. 270: 570–580. [DOI] [PubMed] [Google Scholar]
- Glickman, B. W., and M. Radman, 1980. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl. Acad. Sci. USA 77: 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn, J. S., 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2: 907–913. [DOI] [PubMed] [Google Scholar]
- Hofmann, A. F., 1998. Bile secretion and enterohepatic circulation of bile acids, pp. 937–948 in Sleisenger and Fordtran's Gastrointestinal Disease, Ed. 6, edited by M. Feldman, B. F. Scharschmidt and W. B. Sleisenger. W. B. Saunders, Philadelphia.
- Hung, D. T., and J. J. Mekalanos, 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. USA 102: 3028–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil, P. P., and D. Camerini-Otero, 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44: 89–115. [DOI] [PubMed] [Google Scholar]
- Kim, S. R., K. Matsui, M. Yamada, P. Gruz and T. Nohmi, 2001. Roles of chromosomal and episomal dinB genes encoding DNA Pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266: 207–215. [DOI] [PubMed] [Google Scholar]
- Kokubo, K., M. Yamada, Y. Kanke and T. Nohmi, 2005. Roles of replicative and specialized DNA polymerases in frameshift mutagenesis: mutability of Salmonella typhimurium strains lacking one or all SOS-inducible polymerases to 26 chemicals. DNA Repair 4: 1160–1171. [DOI] [PubMed] [Google Scholar]
- Kothapalli, S., S. Nair, S. Alokam, T. Pang, R. Khakhria et al., 2005. Diversity of genome structure in Salmonella enterica serovar Typhi populations. J. Bacteriol. 187: 2638–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutzer, D. A., and J. M. Essigmann, 1998. Oxidized, deaminated cytosines are a source of C-T transitions in vivo. Proc. Natl. Acad. Sci. USA 95: 3578–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalitha, M. K., and R. John, 1994. Unusual manifestations of salmonellosis—a surgical problem. Q. J. Med. 87: 301–309. [PubMed] [Google Scholar]
- Lea, D., and C. A. Coulson, 1949. The distribution of numbers of mutants in bacterial populations. J. Genet. 49: 264–285. [DOI] [PubMed] [Google Scholar]
- Li, Z., and B. Demple, 1994. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J. Biol. Chem. 269: 18371–18377. [PubMed] [Google Scholar]
- Liochev, S. I., and I. Fridovich, 1992. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc. Natl. Acad. Sci. USA 89: 5892–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya, O., and K. Lewis, 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89: 8938–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D., D. N. Cook and H. Nikaido, 1994. Efflux pumps and drug resistance in gram-negative bacteria. Trends Microbiol. 2: 489–493. [DOI] [PubMed] [Google Scholar]
- Mahan, M. J., and J. R. Roth, 1989. The recB and recJ genes of Salmonella typhimurium. J. Bacteriol. 171: 612–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan, M. J., J. Casadesús and J. R. Roth, 1992. The Salmonella typhimurium recJ function permits growth of P22 abc phage on recBCD+ hosts. Mol. Gen. Genet. 232: 470–478. [DOI] [PubMed] [Google Scholar]
- McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille et al., 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413: 852–856. [DOI] [PubMed] [Google Scholar]
- McPartland, A., L. Green and H. Echols, 1980. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell 20: 731–737. [DOI] [PubMed] [Google Scholar]
- Merino, D., H. Reglier-Poupet, P. Berche and A. Charbit, 2002. A hypermutator phenotype attenuates the virulence of Listeria monocytogenes. Mol. Microbiol. 44: 877–887. [DOI] [PubMed] [Google Scholar]
- Michaels, M. L., and J. H. Miller, 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174: 6321–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, B., S. D. Ehrlich and M. Uzest, 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 17: 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesel, L., and J. R. Roth, 1994. Salmonella typhimurium recD mutations increase recombination in a short-sequence transduction assay. J. Bacteriol. 176: 4092–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. H., 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Mol, C. D., D. J. Hosfield and J. A. Tainer, 2000. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in base excision repair: the 3′ ends justify the means. Mutat. Res. 460: 211–229. [DOI] [PubMed] [Google Scholar]
- Ng, I., L. Shu-Lin and K. E. Sanderson, 1999. Role of genomic rearrangements in producing new ribotypes of Salmonella typhi. J. Bacteriol. 181: 3536–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowosielska, A., and M. G. Marinus, 2005. Cisplatin induces double-strand break formation in Escherichia coli dam mutants. DNA Repair 4: 773–781. [DOI] [PubMed] [Google Scholar]
- O'Rourke, E. J., C. Chevalier, A. V. Pinto, J. M. Thiberge, L. Ielpi et al., 2003. Pathogen DNA as target for host-generated oxidative stress: role for repair of DNA damage in Helicobacter pylori colonization. Proc. Natl. Acad. Sci. USA 100: 2789–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picken, R. N., and I. R. Beacham, 1977. Bacteriophage-resistant mutants of Escherichia coli K-12. Location of receptors within the lypopolysaccharide. J. Gen. Microbiol. 102: 305–318. [DOI] [PubMed] [Google Scholar]
- Ponder, R. G., N. C. Fonville and S. M. Rosenberg, 2005. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19: 791–804. [DOI] [PubMed] [Google Scholar]
- Pope, L. M., K. E. Reed and S. M. Payne, 1995. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect. Immun. 63: 3642–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto, A. I., F. Ramos-Morales and J. Casadesús, 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 168: 1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty, A. M., and J. S. Gunn, 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68: 6763–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty, A. M., J. C. van Velkinburgh and J. S. Gunn, 2002. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184: 1270–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty, A. M., W. H. Schwesinger and J. S. Gunn, 2003. Biofilm formation and interaction with the surface of gallstones by Salmonella spp. Infect. Immun. 70: 2640–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty, A. M., I. E. Brodsky, J. Manos, R. Belas, S. Falkow et al., 2004. Transcriptional regulation of Salmonella enterica serovar Typhimurium genes by bile. FEMS Immunol. Med. Microbiol. 41: 177–185. [DOI] [PubMed] [Google Scholar]
- Pucciarelli, M. G., A. I. Prieto, J. Casadesús and F. García-del Portillo, 2002. Envelope instability in DNA adenine methylase mutants of Salmonella enterica. Microbiology 148: 1171–1182. [DOI] [PubMed] [Google Scholar]
- Purmal, A. A., G. W. Lampman, J. P. Bond, Z. Hatahet and S. S. Wallace, 1998. Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J. Biol. Chem. 273: 10026–10035. [DOI] [PubMed] [Google Scholar]
- Ramos-Morales, F., A. I. Prieto, C. Beuzón, D. W. Holden and J. Casadesús, 2003. Role for Salmonella enterobacterial common antigen in bile resistance and virulence. J. Bacteriol. 185: 5328–5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, S. M., and P. J. Hastings, 2003. Modulating mutation rates in the wild. Science 300: 1382–1383. [DOI] [PubMed] [Google Scholar]
- Rosenberg, E. Y., D. Bertenthal, M. W. Nilles, K. P. Bertrand and H. Nikaido, 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48: 1609–1619. [DOI] [PubMed] [Google Scholar]
- Saint-Ruf, C., and I. Matic, 2006. Environmental tuning of mutation rates. Environ. Microbiol. 8: 193–199. [DOI] [PubMed] [Google Scholar]
- Saito, Y., F. Uraki, S. Nakajima, A. Asaeda, K. Ono et al., 1997. Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J. Bacteriol. 179: 3783–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, L., 1992. The suicidal DNA repair methyltransferases of microbes. Mol. Microbiol. 7: 825–831. [DOI] [PubMed] [Google Scholar]
- Schapiro, J. M., S. J. Libby and F. C. Fang, 2003. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc. Natl. Acad. Sci. USA 100: 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger, H., 1972. Phage P22 mutants with increased or decreased transducing abilities. Mol. Gen. Genet. 119: 75–88. [DOI] [PubMed] [Google Scholar]
- Seeberg, E., 1978. Reconstitution of an Escherichia coli repair endonuclease activiy from the separated uvrA and uvrB/uvrC gene products. Proc. Natl. Acad. Sci. USA 75: 2569–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur, M., V. Bidnenko, S. D. Ehrlich and B. Michel, 1998. RuvA acts at arrested replication forks. Cell 95: 419–430. [DOI] [PubMed] [Google Scholar]
- Shibutani, S., M. Takeshita and A. P. Grollman, 1991. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349: 431–434. [DOI] [PubMed] [Google Scholar]
- Stocker, R., Y. Yamamoto, A. F. McDonagh, A. N. Glazer and B. N. Ames, 1987. Bilirubin is an antioxidant of possible physiological importance. Science 235: 1043–1046. [DOI] [PubMed] [Google Scholar]
- Storz, G., and A. Imlay, 1999. Oxidative stress. Curr. Opin. Microbiol. 2: 188–194. [DOI] [PubMed] [Google Scholar]
- Sutton, M. D., B. T. Smith, V. G. Godoy and G. C. Walker, 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34: 479–497. [DOI] [PubMed] [Google Scholar]
- Suvarnapunya, A. E., H. A. D. Lagassé and M. A. Stein, 2003. The role of DNA base excision repair in the pathogenesis of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 48: 549–559. [DOI] [PubMed] [Google Scholar]
- Uzest, M., S. D. Ehrlich and B. Michel, 1995. Lethality of rep recB and rep recC double mutants in Escherichia coli. Mol. Microbiol. 17: 1177–1188. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres, A., and F. C. Fang, 2000. Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol. 9: 29–33. [DOI] [PubMed] [Google Scholar]
- Viswanathan, M., and S. T. Lovett, 1998. Single-strand DNA-specific exonucleases in E. coli: roles in repair and mutation avoidance. Genetics 149: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, H., and D. Bonner, 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218: 97–106. [PubMed] [Google Scholar]
- Volkert, M. R., and P. Landini, 2001. Transcriptional responses to DNA damage. Curr. Opin. Microbiol. 4: 178–185. [DOI] [PubMed] [Google Scholar]
- White, B. J., S. J. Hochhauser, N. M. Cintron and B. Weiss, 1976. Genetic mapping of xthA, the structural gene for exonuclease III in Escherichia coli K-12. J. Bacteriol. 126: 1082–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrzykowski, J., and M. R. Volkert, 2003. The Escherichia coli methyl-directed mismatch repair system repairs base pairs containing oxidative lesions. J. Bacteriol. 185: 1701–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa et al., 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183: 4562–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]