Abstract
Eukaryote nuclear ribosomal DNA (rDNA) typically exhibits strong concerted evolution: a pattern in which several hundred rDNA sequences within any one species show little or no genetic diversity, whereas the sequences of different species diverge. We report a markedly different pattern in the genome of the grasshopper Podisma pedestris. Single individuals contain several highly divergent ribosomal DNA groups. Analysis of the magnitude of divergence indicates that these groups have coexisted in the Podisma lineage for at least 11 million years. There are two putatively functional groups, each estimated to be at least 4 million years old, and several pseudogene groups, many of which are transcribed. Southern hybridization and real-time PCR experiments show that only one of the putatively functional types occurs at high copy number. However, this group is scarcely amplified under standard PCR conditions, which means that phylogenetic inference on the basis of standard PCR would be severely distorted. The analysis suggests that concerted evolution has been remarkably ineffective in P. pedestris. We propose that this outcome may be related to the species' exceptionally large genome and the associated low rate of deletion per base pair, which may allow pseudogenes to persist.
THE molecular evolution of ribosomal DNA (rDNA) differs fundamentally from that of single-copy genes. Individual rDNA copies do not evolve independently from each other but are homogenized horizontally, so that little differentiation is typically observed between rDNA sequences within a tandem array of any one species. Conversely, differences do accumulate between species. This pattern is referred to as concerted evolution and may involve a number of molecular processes such as unequal crossing over and gene conversion (Elder and Turner 1995; Graur and Li 2000). For phylogenetic applications, in particular, it has often been assumed that concerted evolution has essentially eliminated within-individual rDNA variation. However, an increasing number of studies indicate that, in many taxa, the rate of homogenization may be too low to prevent significant levels of intraspecific rDNA polymorphisms. Often this variation is restricted to the noncoding rDNA regions such as the internal transcribed spacers (ITS1 and ITS2) or the intergenic spacer (IGS), which separates the transcribed units (Crease 1995; Copenhaver and Pikaard 1996; O'Donnell and Cigelnik 1997; Fenton et al. 1998; Gernandt and Liston 1999; Hugall et al. 1999; Denduangboripant and Cronk 2000; Harris and Crandall 2000; Whang et al. 2002; Parkin and Butlin 2004). However, there are also numerous reports of rDNA pseudogenes where the coding regions are freed from functional constraints (Buckler et al. 1997; Hartmann et al. 2001; Mayol and Rossello 2001; Muir et al. 2001; Márquez et al. 2003; Razafimandimbison et al. 2004; Ruggiero and Procaccini 2004).
Substantial differentiation within individuals, corresponding to several million years of divergence, has been found in plants. These large differences, however, may be attributed to divergence that has accumulated in different species and has then been brought together by hybridization (Lim et al. 2000; Hartmann et al. 2001; Muir et al. 2001). The rDNA of hybrids can diverge rapidly from either parent within two or three generations (Kovarik et al. 2004). For this reason, and because of the divergence between different rDNA arrays and pseudogenes, some authorities consider rDNA to be a poor choice for phylogenetic analysis (Álvarez and Wendel 2003). Nevertheless, rDNA sequences are still extensively used in a wide range of taxa: a citation search detects >3000 papers since 2000.
More important than these concerns are the dynamics of sequence homogenization and translocation, which are of evolutionary interest in themselves. There is some evidence that the chromosomal location of rDNA arrays is particularly labile in the Orthoptera, with loci differing in number and genomic location in different populations of the same species (Flanagan et al. 1999; Cabrero et al. 2003). In the grasshopper Podisma pedestris, there is additional interest in the rDNA because evidence from restriction digests shows strong linkage disequilibrium between a particular rDNA variant and a chromosomal fusion (Dallas et al. 1988), which cannot be explained by close physical proximity to the main rDNA locus (Bella et al. 1991). This disequilibrium hints at the possibility of selection maintaining the association. Further, P. pedestris has an exceptionally large genome of 18,000 Mb (Westerman et al. 1987), which may be a direct consequence of the low relative rate of DNA loss observed in this species. Bensasson et al. (2001) have shown that, in P. pedestris, pseudogenes are likely to be slowly eroded by substitutions, whereas in species with high rates of DNA loss (e.g., Drosophila) they will not persist long enough for much genetic divergence to accumulate.
In this study, we surveyed cloned DNA to characterize variation in the ITS1 of the rDNA genic region within and between individuals of P. pedestris. To characterize the diversity within the genome, we have compared the sequences obtained from genomic DNA by standard PCR and high-stringency PCR and from ribosomal RNA by reverse transcription PCR (RT–PCR). Pseudogenes have been distinguished from putatively functional sequences by DNA sequence analysis and timings estimated for the date of separation of different lineages. We assess different possible explanations for, and implications of, the extraordinarily deep divergence that we detect among the different lineages.
MATERIALS AND METHODS
Study species and sample collection:
The grasshopper species P. pedestris (Orthoptera, Acrididae, Melanoplinae) has a boreal and subalpine distribution. The species includes two races, which are called “fused” (or neo-XY) and “unfused” (or XO), respectively, due to differences in their X chromosomes (John and Hewitt 1970). The two races meet and interbreed along a narrow zone that runs from east to west following the ridges of the Alpes Maritimes in southern France (Barton and Hewitt 1981). This hybrid zone is likely to have existed at its present location for >8000 years (Nichols and Hewitt 1986).
Samples of P. pedestris were collected in 2003 and 2004 in the Alpes Maritimes from six pure populations on each side of the hybrid zone (Table 1). The insects were killed and the hind legs stored in 100% ethanol. For RNA analysis, testicular tissue was stored in RNAlater buffer (Ambion, Austin, TX) at 4°. As outgroups, we included sequences from eight other species of the subfamily Melanoplinae that had been obtained by I. C. Chintauan-Marquier as part of her Ph.D. thesis (Table 1).
TABLE 1.
18S partial sequences and ITS1 sequences obtained in this study
| Sequences
|
||||||
|---|---|---|---|---|---|---|
| Species | Authority | Origin | Individual | 18S | ITS1 | Accession nos. |
| Melanoplinae | Scudder, 1897 | |||||
| Podismini | Yakobson and Bianchi, 1905 | |||||
| P. pedestris | Linnaeus, 1758 | France | ||||
| Unfused | Belledonne | PPD1 | 1 | 1 | AM295160 | |
| Crête des Mélèzes | CM1 | 27 | 26 | AM183598/600/603/604/ 612/621–623/636/ 648–665 | ||
| Crête des Mélèzes | CM2 | 3 | 3 | AM183609/614/615 | ||
| Crête des Mélèzes | CM3 | 1 | 1 | AM183611 | ||
| Crête des Mélèzes | CM4 | 1 | 1 | AM183630 | ||
| Crête des Mélèzes | CM5 | 3 | 3 | AM183606/625/626 | ||
| Col Bas | CB1 | 1 | 3 | AM183620/AM238436/437 | ||
| Lac Noir | LN1 | 2 | 3 | AM183607/632/AM238438 | ||
| Fused | Blayeul | BL1 | 29 | 26 | AM183588/593/594/597/ 601/602/608/616–619/ 624/628/629/631/634/ 635/637–647/666 | |
| Blayeul | BL2 | 3 | 3 | AM183589/590/595 | ||
| Blayeul | BL3 | 1 | 1 | AM183596 | ||
| Blayeul | BL4 | 1 | 1 | AM183599 | ||
| Blayeul | BL5 | 2 | 2 | AM183627/633 | ||
| Chabanon | CH1 | 2 | 2 | AM183587/591 | ||
| La Tartonne | TA1 | 3 | 3 | AM183592/605/610 | ||
| Cophopodisma pyrenaea | Fischer, 1853 | Aude | CPD1 | 1 | 1 | AM295161 |
| Italopodisma acuminata | La Greca, 1969 | Italy: Abruzzo | IPAA1 | 1 | 1 | AM295162 |
| Sinopodisma punctata | Mistshenko, 1954 | Japan: Tatsugou | SPP1 | 1 | 1 | AM295163 |
| Melanoplini | Scudder, 1897 | |||||
| Phaulotettix compressus | Scudder, 1897 | Mexico: Hidalgo | PLC1 | 1 | 1 | AM295164 |
| Melanoplus frigidus | Boheman, 1846 | France: Lautaret | MLF1 | 1 | 1 | AM295165 |
| Dactylotini | Rehn and Randell, 1963 | |||||
| Dactylotum bicolor | Charpentier, 1843 | Mexico | DTBB1 | 1 | 1 | AM295166 |
| Jivarini | Ronderos, 1983 | |||||
| Jivarus sp. | Giglio-Tos, 1898 | South America | JSP1 | 1 | 1 | AM295167 |
| Dichroplini | Liebermann, 1949 | |||||
| Dichroplus sp. | Stal, 1973 | Ecuador: Tungurahua | DCHSP1 | 1 | 1 | AM295168 |
Our study was divided into two main parts. First, the general diversity of rDNA was characterized by cloning and sequencing of PCR products. And second, on the basis of these results, we developed assays to investigate the frequency of specific rDNA variants.
The rDNA diversity in P. pedestris
DNA extraction and PCR:
DNA was extracted from muscle tissue of one or both hind legs using either a high salt extraction method (Aljanabi and Martinez 1997) or a QIAGEN (Chatsworth, CA) DNeasy tissue kit according to the manufacturer's instructions. ITS1 was PCR amplified using the primers 18S(f) (5′-CCTTTGTACACACCGCCCGT) and ITS6(r) (5′-GTTCATGTGTCCTGCAGTTCAC), which target the coding regions flanking ITS1 (Sharpe et al. 2000). The resulting PCR product was ∼650 bp in length and included 119 bp from the 3′-end of the 18S coding region and all of ITS1. PCR amplifications were carried out in volumes of 25 μl using a PTC-220 DNA Engine Dyad Peltier thermal cycler (MJ Research, Watertown, MA). Each reaction contained ∼5–10 ng of genomic DNA, 3 mm MgCl2, 0.12 mm of each dNTP, 12.5 pmol of each primer, 0.25 units of BIOTAQ DNA polymerase (Bioline) and the associated NH4 buffer at 1× concentration. An initial denaturation step of 5 min at 95° was followed by 35 cycles of amplification (30 sec at 94°, 30 sec at 62°, 1 min at 72°) and a final elongation step of 10 min at 72°.
The diversity of ITS1 was studied in particular detail in two individuals, one from the fused (individual BL1) and one from the unfused race (individual CM1). Several steps were taken to avoid problems that may arise in PCR surveys of multigene families such as rDNA. PCR can give misleading results due to the preferential amplification of certain target sequences and/or the formation of PCR artifacts. The impact of stochastic events can be reduced by stopping the PCR at an early stage and pooling several independent amplifications before further analysis (Wagner et al. 1994; Kanagawa 2003). For the cloning and sequencing of rDNA in individuals BL1 and CM1, we combined the PCR products of four independent reactions, each of which had included only 29 cycles of amplification. There may also be a systematic bias in the efficiency with which different target sequences are amplified. For example, it has been shown that nonfunctional rDNA copies (i.e., pseudogenes) may be favored under standard PCR conditions (Buckler et al. 1997). For this reason, PCR was also performed under highly denaturing reaction conditions by adding 5% dimethyl sulfoxide (DMSO). This approach has been found to enhance the amplification of functional rDNA copies, possibly because they have high secondary structure stability (Buckler et al. 1997).
Similarly, the primers and stringent conditions used to obtain the outgroup sequences had been specifically developed to amplify only putatively functional sequences in Orthoptera. The strategy was successful in Podisma since it recovered group 1 sequence (see below; Figure 2). It is notable that, as in Podisma, the genic region of the outgroup sequences is identical to that of mouse (with the exception of Melanoplus sp., which was therefore treated separately in the analysis). This sequence conservation suggests strong selective constraints consistent with functionality.
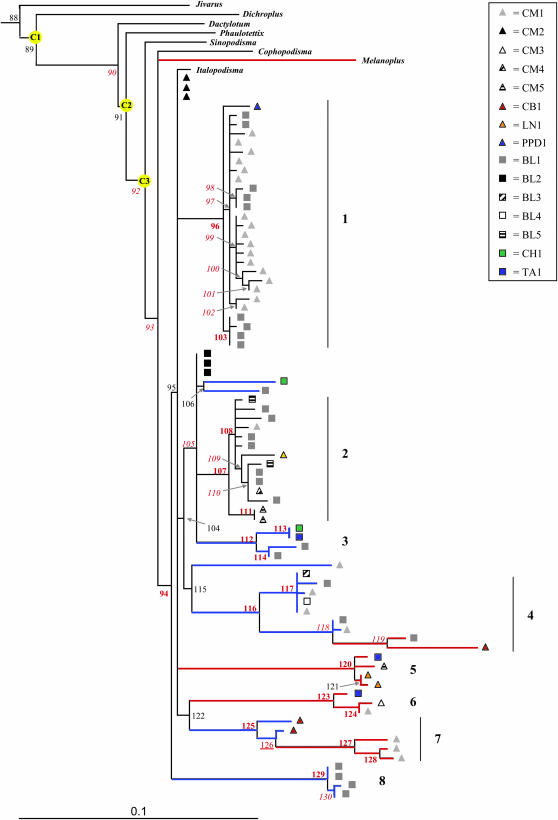
Figure 2.—
Maximum-likelihood tree of ITS1 sequences from P. pedestris and eight related species. Numbers in red indicate nodes that are well supported in both Bayesian and weighted maximum-parsimony analysis (boldface type), in Bayesian analysis only (italic), or in weighted maximum-parsimony analysis only (underlined). Sequences from individuals of the unfused race are represented by triangles; sequences from individuals of the fused race by squares. C1–3 indicate the nodes used as calibrations (see Table 3). Different symbols are used for sequences from different individuals. To estimate divergence times, the branches in blue were assumed to evolve under rate R1 and the branches in red under rate R2.
RNA extraction and RT–PCR:
For the individuals BL1 and CM1 we also studied ITS1 variation in RNA to investigate which copies are actually transcribed. The testicular tissue stored in RNAlater (Ambion) was homogenized by applying it to a QIAshredder column (QIAGEN) in a solution of 100 μl lysis buffer and 0.7 μl β-mercaptoethanol. RNA was then extracted using an Absolutely RNA RT–PCR Miniprep kit (Stratagene, La Jolla, CA) following the manufacturer's instructions with the suggested modifications for extremely small samples. The extraction protocol included an on-column DNase treatment.
cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen, San Diego) in a volume of 20 μl. Each reaction contained 400 units Superscript II, the associated first-strand buffer at 1× concentration, 10 mm DTT, 10 mm of each dNTP (dNTP set, Amersham Biosciences), 40 units RNasin Plus RNase inhibitor (Promega, Madison, WI), 50 pmol each of primers 18S(f) and ITS6(r) and 1.75 μl RNA template. The reaction was incubated at 37° for 1 hr. The cDNA was then used as template in a standard PCR as described above.
Cloning and sequencing:
Before cloning, 20 μl of PCR product was run on a 1.5% agarose gel, and the band was excised and purified using a QIAquick gel extraction kit (QIAGEN). The PCR fragments were then ligated into a pGEM-T Easy vector (Promega) overnight. Two microliters of the ligation reaction were used to transform JM109 High Efficiency Competent Cells (Promega) according to the manufacturer's recommendations. Individual positive clones were then used as a template for PCR amplification of ITS1 with the primers and conditions described above. The PCR products were cleaned and sequenced on an ABI 3700 sequencer. Table 1 lists the number of sequences obtained per individual and population. Note that differences between the number of 18S and ITS1 sequences arise in cases where either the beginning or the end of a sequence was of poor quality and omitted from further analyses. The EMBL accession numbers of the sequences are listed in Table 1.
Phylogenetic analyses:
All sequences were split into two parts representing the 3′-end of the 18S and the ITS1, respectively. Partial 18S sequences were aligned in BioEdit 7.0.1 (Hall 1999) with published sequences from the grasshopper Chorthippus parallelus (EMBL accession no. AY585651), the fruit fly Drosophila melanogaster (M21017), and the mouse Mus musculus (J00623).
The ITS1 sequences were edited manually in Sequence Navigator 1.01 (PE Applied Biosystems) and multiple alignments were generated using ClustalX 1.81 (Thompson et al. 1997). Different gap opening/extension costs (10/0.2, 10/2, 10/5, 15/0.2, 15/5, 18/0.2, and 18/5) were used to identify the hypervariable and “more conserved” regions in different alignments of the same data. The final, unambiguous alignment contained 87 sequences of ∼437 bp of the ITS1 from different melanoplines (supplemental Table 1 at http://www.genetics.org/supplemental/).
The phylogenetic analyses were conducted for the ingroup alone as well as for ingroup and outgroups combined. As outgroups, we selected eight species representing six of the seven tribes of the Melanoplinae subfamily (Table 1) that had been suggested as close relatives of P. pedestris by previous molecular studies (Litzenberger and Chapco 2001; Amedegnato et al. 2003; I. C. Chintauan-Marquier, unpublished data).
Characteristics of the molecular data:
Prior to phylogenetic reconstruction, the homogeneity of base frequencies across taxa was evaluated with a chi-square goodness-of-fit test using PAUP*4.0b10 (Swofford 2002) because compositional bias among sequences can interfere with tree topology (Pereira et al. 2002). We included all sites and variable sites only to assess the potentially confounding effects of invariant sites (Waddell et al. 1999). The parameters allowed to vary in model fitting (e.g., base composition, substitution rates, and rate heterogeneity across sites) were estimated under the most likely model of evolution suggested by the Akaike information criterion (Akaike 1974) in Modeltest 3.0 (Posada and Crandall 1998).
Maximum parsimony:
Heuristic searches were implemented in PAUP*4.0b10 (Swofford 2002) under the maximum-parsimony (MP) criteria with the accelerated transformation (ACCTRAN) option to optimize the state of unordered (Fitch) characters, 100 random sequence addition replicates, the tree bisection–reconnection (TBR) branch swapping, and gaps treated as a fifth character. Unweighted MP methods do not always take full advantage of the information contained in DNA sequences due to the presence of homoplasious characters. To deal with this problem, a weighted parsimony analysis was also conducted using the rescaled consistency index on an initial tree in successive approximations of character weighting (Farris 1969).
Maximum likelihood:
The maximum-likelihood (ML) search was conducted using PAUP*4.0b10 as follows: We first evaluated the model of DNA substitution that best fit our data partition, using the Akaike information criterion in Modeltest 3.0 (Posada and Crandall 1998). Then we calculated the ML model parameters using the data partition of interest and a neighbor-joining topology. These estimated values were fixed, and a full heuristic ML search was conducted with 10 random addition sequence replicates, retaining all minimal trees, and TBR branch swapping. After this first ML search was completed, we reestimated model parameters on the ML tree and used these new values to search again. We repeated this process until the new search found the same topology as the previous search and all parameter estimates were identical.
Bayesian analyses:
Bayesian Markov chain Monte Carlo (MCMC) analyses were performed with MrBayes 2.01 (Huelsenbeck and Ronquist 2001) with the GTR + I + Γ model. To assess the coverage of tree space, we performed three independent Metropolis-coupled MCMC runs started from different random points in parameter space (Huelsenbeck and Bollback 2001; Huelsenbeck et al. 2002). We applied different temperatures (0.2°, 0.5°, 1°) to influence the rate of switching between chains. Each run had 10 million iterations, with one cold and three incrementally heated chains per run and with trees sampled every 500 generations. The number of trees to be discarded as burn-in (Huelsenbeck and Ronquist 2001) was determined graphically. The average log-likelihood values (±SD) at stationarity for the independent runs were calculated with Microsoft Excel and compared for convergence (Huelsenbeck and Bollback 2001). The set of model substitution parameters were analyzed to determine the mean, variance, and 95% credible interval. To estimate posterior probabilities (PPs) of recovered branches (Larget and Simon 1999; Huelsenbeck and Ronquist 2001), 50% majority rule consensus trees were calculated from the remaining trees in MrBayes 2.01.
Node support:
Support for internal branches was assessed in PAUP*4.0b10 (Swofford 2002) by nonparametric bootstrapping (nBT) (Felsenstein 1985) with 2000 pseudoreplicates for MP, using full heuristic searches with 30 random addition sequence replicates, TBR branch swapping, and one tree held at each step during stepwise addition (DeBry and Olmstead 2000). PPs for individual nodes were estimated in a Bayesian MCMC framework with the runs executed as above. Nodes with nBT values >65% and PPs >75% were shown as resolved (Hillis and Bull 1993; Jordan et al. 2003).
Molecular dating:
A likelihood-ratio test (LRT; Felsenstein 1981; Goldman 1993a,b; Yang and Roberts 1995) showed that the data did not conform to the model of a constant molecular clock (LRT = 283, P = 0, 85 d.f.). We therefore implemented Yoder and Yang's local clock method (Yoder and Yang 2000). We made use of the general time reversible plus Γ-distributed rate variation (GTR + Γ) option with local clocks implemented in PAML v3.13 (Yang 1997). Each branch was assigned to a rate category after inspection of the branch lengths in Figure 2 and separate rates (R1–R3) were estimated for each. The branches in each of the three categories are identified in Figure 2: R1 applies to the branches in blue, R2 to those in red, and R3 to the remainder.
We used three alternative calibrations of the molecular clock to cross-check the timings estimated from the genetic divergence. Each calibration was based on a date that had previously been proposed for a particular node on the phylogeny: (1) the separation of the South American melanoplines is thought to correspond to an incursion into North America 34.43–74.05 MYA (Chapco et al. 2001; Amedegnato et al. 2003; I. C. Chintauan-Marquier, unpublished data); (2) more recently, the North American melanoplines are thought to have split from the Eurasian in the early Tertiary/late Cretaceous 20.53–66 MYA (Chapco et al. 2001; Amedegnato et al. 2003; I. C. Chintauan-Marquier, unpublished data); and (3) the segregation of the European from the Asian melanopline fauna has been estimated at 16.13–39.81 MYA (Litzenberger and Chapco 2001; I. C. Chintauan-Marquier, unpublished data). In each case, the date required by PAML's maximum-likelihood dating procedure was taken at the midpoint of the above ranges. The ages of the geological periods and chronostratigraphic references have been taken from the 1999 Geological Time Scale of the Geological Society of America (http://www.geosociety.org/science/timescale/timescl.pdf).
Detection of potential recombinants:
We used two different statistical approaches to detect recombinant sequences. The RDP method (Martin et al. 2005) and the MaxChi method (Maynard Smith 1992; Posada and Crandall 2001) were applied to the 77 complete sequences.
Frequency of specific variants
Restriction digests:
On the basis of the results of the sequencing it was possible to identify restriction enzymes that were expected to cut only one particular group of sequences. AvaII (G^GWCC) is expected to cut group 1 sequences (see below) into two fragments of ∼150 and 500 bp. AvaII restriction sites are also present in three other sequences. However, digestion of these sequences is expected to produce fragments of a different length. The restriction enzyme AlwNI (CAGNNN^CTG) is expected to cut only sequences belonging to group 2 and to produce two fragments of ∼400 and 250 bp. There are no AlwNI recognition sites in any non-group 2 sequences.
To investigate the within-individual frequency of the different sequence variants in more detail, DNA from all P. pedestris individuals used for sequencing (except PPD1; Table 1) was analyzed by restriction digestion. PCR was carried out as described above for the individuals BL1 and CM1 using both standard and highly denaturing reaction conditions. A total of 25 μl of PCR reaction was incubated with 10 units of AvaII or AlwNI (New England Biolabs, Beverly, MA) at 37° for 18 hr. The restriction fragments were then separated on a 1.5% agarose gel.
Southern hybridization:
Southern blot analyses were performed to estimate the relative frequency of group 1 sequences in the genome of P. pedestris. A total of 3–5 μg of genomic DNA from seven different individuals was digested to completion with AvaII. The digested DNA was ethanol precipitated and resuspended in 25 μl TE buffer before electrophoresis through a 1.2% agarose minigel for ∼8 hr. The DNA fragments were then blotted onto a nylon membrane (Hybond-N+, Amersham Biosciences) following the protocol of Sambrook et al. (1989). As a probe, ITS1 was amplified as described above and ∼100 ng of purified PCR product was labeled with [32P]CTP using Ready-To-Go DNA labeling beads (Amersham Biosciences) according to the manufacturer's instructions. Prehybridization and hybridization of the membranes were carried out at 42° in solutions containing 50% formamide, 6× SSC, 5× Denhardt's reagent (prehybridization solution only), 0.5% SDS, and 100 μg/ml denatured, fragmented salmon sperm DNA. After overnight hybridization, membranes were washed in large volumes of 2× SSC and 0.5% SDS twice for 5 min, in 2× SSC and 0.1% SDS for 15 min, and in 0.1× SSC and 0.5% SDS for 30 min at room temperature and again for 30 min at 65°. They were then exposed to a storage phosphor screen for 1–2 hr, and the results were analyzed on a Typhoon 9200 scanner (Amersham Biosciences).
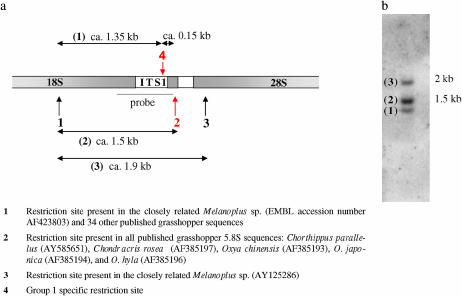
On the basis of published grasshopper sequences it was possible to predict the lengths of AvaII restriction fragments (Figure 1a). Unmethylated group 1 sequences are expected to produce two bands at ∼1.35 and 0.15 kb, while all other rDNA variants should give one band at ∼1.5 kb. In methylated DNA, restriction sites 2 and 4 could be blocked, resulting in just one fragment of ∼1.9 kb (Figure 1a).
Figure 1.—
(a) Location of AvaII restriction sites in the rDNA genic region predicted on the basis of our own and published grasshopper sequences as referenced in the legend. The restriction sites indicated in red (2 and 4) can potentially be blocked by methylation. Site 4 in the ITS1 is characteristic of group 1 sequences. (b) AvaII digest of genomic DNA from P. pedestris.
Quantitative PCR with allele-specific primers:
Quantitative PCR (qPCR) was used to estimate the relative frequency of the main groups of rDNA sequence. Primers specific for group 1 and group 2, respectively, were designed on the basis of sequencing results. We selected positions that were diagnostic of one particular group, i.e., where all sequences in that group shared a nucleotide that was not present in any of the other sequences. Both the forward and reverse primers were designed to end on such a diagnostic base and show a perfect match only to the target sequence and a 3′ mismatch to all others. The group 1-specific forward and reverse primers were 5′-GCTCCTCTCAAAACTAACGCAC and 5′-CAGAATAYATACAGGCGGGACC, and the group 2-specific forward and reverse primers were 5′-CGTGTCGCGCATTTGACAG and 5′-CGAAGCTCGCAGAATACATCT. To monitor the specificity of the primers, clones with known inserts (either group 1 or group 2) were used as PCR templates. Further, we sequenced PCR products to confirm allele-specific amplification from genomic DNA.
Since total rDNA content is thought to vary considerably among individuals (as indicated by fluorescent in situ hybridization on cytogenetic preparations; not shown), we standardized the qPCR results for the main groups by comparison with those for a 180-bp fragment from the 3′-end of the 18S coding region. This fragment was amplified using the primer 18S(f) described above and the new primer 18S(r) (5′-GATCCTTCCGCAGGTTCAC). These primers are expected to show a perfect match to all functional sequences and a good match to relatively young pseudogenes, which have not yet accumulated mutations in the highly conserved region.
The qPCR analysis was performed in a PTC-200 Peltier thermal cycler (MJ Research) with a Chromo4 fluorescence detector (GRI). Reactions were carried out in a 20-μl volume using a DyNAmo HS SYBR Green qPCR kit (New England Biolabs) according to the manufacturer's recommendations. Four replicates of each sample were analyzed with each set of primers. The cycling protocol consisted of an initial denaturation step of 15 min at 95°, 40 cycles of 10 sec at 94°, 30 sec at a primer-specific annealing temperature (TA), 30 sec at 72° followed by a data acquisition step, and a final elongation of 5 min at 72°. The annealing temperatures for the different primer pairs were as follows: 18S, TA = 57°; group 1 specific, TA = 60°; group 2 specific, TA = 56°.
For each sample, the qPCR software determined the number of cycles, C(t), after which the fluorescence intensity crossed a threshold that was set at five standard deviations above the baseline fluorescence. From these C(t) values, the initial template concentration was estimated by comparing the samples to a serial dilution of a standard included in each run. As this standard, we used 16 different dilutions (in a twofold dilution series) of PCR products obtained from clones containing either a group 1 or a group 2 insert. These 16 dilutions spanned the template concentrations typically observed for genomic samples. Both series were included in runs with the 18S primers. On the basis of data from 12 individuals from both races, we estimated the average copy number of group 1 and group 2 sequences relative to the number of conserved 18S sequences.
Direct sequencing:
To further investigate the genomic frequency of the different rDNA variants, DNA extractions from two individuals were sequenced directly without a preceding PCR step. 18S(f) was used as a sequencing primer. The electrophoretogram was analyzed for evidence of multiple peaks at the sites of SNPs that distinguish the variants.
RESULTS
High levels of intraspecific variability:
The ITS1 sequences obtained from P. pedestris revealed a remarkable level of intraspecific variation with an average pairwise difference of 7.7% (SD = 3.8%). Only two cDNA sequences from individual BL1 were identified as potential recombinants by MaxChi, while RDP did not find any evidence of recombination (data not shown). Figure 2 shows a maximum-likelihood tree based on ITS1, including sequences from eight related species.
The P. pedestris sequences form eight principal clusters that are well supported in all trees (Figure 2, Table 3). Clusters 1 and 2 have no more than one difference in the last 58 bp of 18S genic DNA from the conserved sequence found in Drosophila, Chorthippus, and Mus. They were therefore identified as putatively functional. The other six groups (groups 3–8) are identified as pseudogenes because they contain multiple differences in this highly conserved region. Most of these substitutions are shared by several sequences within the group. The three different methods for obtaining sequences recovered different proportions from each category (Table 2). Standard PCR and cDNA analysis recovered a high frequency of pseudogenes as well as members of the putatively functional groups 1 and 2. The frequencies obtained by these two methods were not significantly different (χ2-test, P = 0.14). In contrast, PCR with DMSO recovered almost exclusively putatively functional group 1 sequences in a significantly higher proportion than the other methods (χ2-test, P < 0.001).
TABLE 3.
Maximum-likelihood estimates of divergence times (in million years) under a local clock using three calibrations (C1, C2, and C3) and support of the nodes based on Bayesian (PP) and weighted maximum-parsimony analysis (BT)
| Divergence times
|
95% C.I.
|
Support of nodes
|
||||||
|---|---|---|---|---|---|---|---|---|
| Node | C1 | C2 | C3 | Mean | Lower limit | Upper limit | PP | BT |
| 88 | 60.593 | 107.581 | 95.119 | 87.764 | 60.219 | 115.309 | NS | NS |
| 89 | 54 | 95.876 | 84.77 | 78.215 | 53.667 | 102.764 | NS | NS |
| 90 | 26.563 | 47.162 | 41.699 | 38.475 | 26.399 | 50.550 | 100 | NS |
| 91 | 24.219 | 43 | 38.019 | 35.079 | 24.070 | 46.089 | 50 | NS |
| 92 | 17.836 | 31.668 | 28 | 25.835 | 17.726 | 33.943 | 81 | NS |
| 93 | 13.483 | 23.939 | 21.166 | 19.529 | 13.400 | 25.659 | 75 | NS |
| 94 | 11.25 | 19.974 | 17.66 | 16.295 | 11.181 | 21.409 | 87 | 94 |
| 95 | 9.914 | 17.603 | 15.564 | 14.360 | 9.853 | 18.868 | NS | NS |
| 96 | 3.739 | 6.638 | 5.869 | 5.415 | 3.716 | 7.115 | 100 | 87 |
| 97 | 2.5 | 4.438 | 3.924 | 3.621 | 2.485 | 4.757 | 86 | 54 |
| 98 | 0.63 | 1.118 | 0.989 | 0.912 | 0.626 | 1.198 | 100 | 60 |
| 99 | 1.912 | 3.395 | 3.002 | 2.770 | 1.900 | 3.639 | 87 | NS |
| 100 | 1.509 | 2.679 | 2.368 | 2.185 | 1.500 | 2.871 | 95 | 64 |
| 101 | 0.91 | 1.617 | 1.429 | 1.319 | 0.904 | 1.733 | 91 | 60 |
| 102 | 1.399 | 2.484 | 2.196 | 2.026 | 1.390 | 2.662 | 94 | 63 |
| 103 | 0.53 | 0.942 | 0.833 | 0.768 | 0.527 | 1.010 | 90 | 86 |
| 104 | 9.425 | 16.734 | 14.796 | 13.652 | 9.367 | 17.936 | 54 | NS |
| 105 | 6.645 | 11.798 | 10.431 | 9.625 | 6.604 | 12.645 | 92 | NS |
| 106 | 4.814 | 8.547 | 7.557 | 6.973 | 4.784 | 9.161 | NS | NS |
| 107 | 4.335 | 7.698 | 6.806 | 6.280 | 4.308 | 8.251 | 100 | 94 |
| 108 | 3.77 | 6.693 | 5.917 | 5.460 | 3.747 | 7.173 | 86 | 66 |
| 109 | 3.201 | 5.684 | 5.025 | 4.637 | 3.181 | 6.092 | 76 | 63 |
| 110 | 1.767 | 3.138 | 2.774 | 2.560 | 1.756 | 3.363 | 97 | 63 |
| 111 | 0 | 0 | 0 | 0.000 | 0.000 | 0.000 | 100 | 98 |
| 112 | 2.096 | 3.721 | 3.29 | 3.036 | 2.083 | 3.988 | 100 | 99 |
| 113 | 0 | 0 | 0 | 0.000 | 0.000 | 0.000 | 100 | 100 |
| 114 | 1.253 | 2.224 | 1.966 | 1.814 | 1.245 | 2.383 | 95 | 71 |
| 115 | 9.12 | 16.192 | 14.316 | 13.209 | 9.064 | 17.355 | NS | NS |
| 116 | 5.338 | 9.478 | 8 | 7.605 | 5.231 | 9.979 | 100 | 93 |
| 117 | 0.435 | 0.772 | 0.683 | 0.630 | 0.432 | 0.828 | 100 | 100 |
| 118 | 2.275 | 4.039 | 3.571 | 3.295 | 2.261 | 4.329 | 100 | NS |
| 119 | 1.326 | 2.354 | 2.081 | 1.920 | 1.318 | 2.523 | 100 | NS |
| 120 | 0.618 | 1.098 | 0.971 | 0.896 | 0.614 | 1.177 | 100 | 100 |
| 121 | 0.171 | 0.303 | 0.268 | 0.247 | 0.170 | 0.325 | 60 | 53 |
| 122 | 7.866 | 13.965 | 12.348 | 11.393 | 7.818 | 14.968 | NS | NS |
| 123 | 1.023 | 1.817 | 1.606 | 1.482 | 1.017 | 1.947 | 100 | 100 |
| 124 | 0.238 | 0.423 | 0.374 | 0.345 | 0.237 | 0.453 | 100 | 98 |
| 125 | 3.93 | 6.977 | 6.169 | 5.692 | 3.906 | 7.478 | 100 | 96 |
| 126 | 3.049 | 5.414 | 4.787 | 4.417 | 3.030 | 5.803 | 55 | 78 |
| 127 | 1.118 | 1.986 | 1.756 | 1.620 | 1.111 | 2.129 | 100 | 100 |
| 128 | 0.341 | 0.605 | 0.535 | 0.494 | 0.339 | 0.648 | 100 | 98 |
| 129 | 0.289 | 0.513 | 0.453 | 0.418 | 0.287 | 0.550 | 100 | 100 |
| 130 | 0.124 | 0.22 | 0.194 | 0.179 | 0.123 | 0.236 | 96 | 64 |
Node, number of a particular node as indicated in Figure 2; 95% C.I., 95% confidence interval; PP, posterior probability; BT, bootstrap support; NS, node not supported.
TABLE 2.
Putatively functional sequences and pseudogenes obtained by cloning the PCR product from three different protocols: standard PCR, high-stringency PCR (with DMSO), and (RT–PCR)
| Putatively functional
|
Pseudogenes: | ||
|---|---|---|---|
| Group 1 | Group 2 | Groups 3–8 | |
| Standard PCR | 3 | 9 | 23 |
| PCR with DMSO | 18 | 1 | 0 |
| RT–PCR | 5 | 4 | 7 |
The eight groups represent well-supported clades in a maximum-likelihood tree, of which two are putatively functional (groups 1 and 2) and the rest are pseudogenes (Figure 2).
Different rDNA variants have coexisted in the P. pedestris genome for >10 million years:
The age of the different nodes was estimated using the three distinct calibration points, all of which gave comparable results (Figure 2, Table 3). We found that the most recent common ancestor between some of the most divergent rDNA groups is likely to have existed between 10 and 20 million years ago in the Miocene (nodes 94 and 95, Table 3). This includes the split between the two putatively functional groups 1 and 2 (see previous paragraph). All eight well-supported groups (Figure 2) are likely to have been evolving independently since at least the late Miocene, with the most recent split 6.6–12.6 million years ago between groups 2 and 3 (node 105). Lineages within both groups 1 and 2 have been diverging for at least 3.7 million years (nodes 96 and 107).
One putatively functional group occurs in much higher copy number than the other:
The results of the Southern blots indicate that unmethylated group 1 sequences are found at a relatively high copy number but may not be the most common rDNA type. Digests with AvaII always resulted in three bands of ∼1.3, 1.5, and 1.9 kb (Figure 1b). The middle band at 1.5 kb was always the brightest, followed by the band at 1.9 kb and finally the band at 1.3 kb. The intensity of the two weaker bands was generally very similar and amounted to 34–57% of the intensity of the brightest band. In two of the seven individuals, there were three additional, very weak bands at ∼1, 0.75, and 0.4 kb attributable to other restriction sites. The size of the three bright bands corresponds to that predicted from published sequences (Figure 1a). Band 3 at 1.9 kb would be produced from methylated DNA and band 2 at 1.5 kb from sequences lacking the ITS1 site, while band 1 at 1.3 kb would result from digests of group 1 sequences (Figure 1b). The expected 0.17-kb fragment from group 1 sequences was not detected, most probably because of its short overlap with the probe. Our interpretation is, therefore, that nonmethylated group 1 sequences (band 1) are common, but make up less than half the total rDNA. This provides a lower boundary for the genomic frequency of group 1. The true frequency might be underestimated considerably if bands 2 and 3 contain a high proportion of methylated group 1 sequences.
The qPCR experiments show that group 1 does make up the majority (88%; SE 7%) of rDNA units with relatively well-conserved 18S (i.e., functional rDNA and young pseudogenes), whereas group 2 sequences are very rare, accounting for <1%. For all the standard calibration series, we observed an almost perfect linear relationship between C(t) values and the log of the initial copy number (R2 ≥ 0.99 in all cases). This result indicates that the initial copy numbers of the three different targets could indeed be estimated reliably for the genomic samples by qPCR.
The results of the direct sequencing of genomic DNA further supported the high frequency of group 1 sequences among the sequences with a conserved 18S. Without the initial PCR step, which has been shown to selectively amplify some variants (see below), the electrophoretogram traces were interpretable for only short runs (<200 bp). These were, however, sufficient to show that all SNP loci were unambiguously of the group 1 type, with no detectable evidence of the alternative nucleotides found in the other rDNA groups.
PCR conditions strongly influence the amplification efficiency of different rDNA variants:
The investigation of the sequences amplified under different PCR conditions confirmed that group 1 sequences are the most common product under the denaturing PCR conditions produced by DMSO. In each of the 14 individuals, AvaII digestion produced a very strong double band corresponding to the expected group 1 products, with only a very faint undigested band indicating other groups. There was no visible digestion by the group 2-specific enzyme, AlwNI—in agreement with the qPCR results suggesting that group 2 is relatively rare.
PCR under standard conditions resulted in a significantly different and very variable composition, corresponding to the diversity of sequences that had previously been cloned. Group 2-specific bands were detectable in all 14 individuals, whereas the proportion of group 1 sequences was generally lower and varied widely between individuals. In fact, group 1 sequences were completely undetectable in individuals from population BL, even though they were clearly present in the sequences amplified by PCR with DMSO.
DISCUSSION
Intragenomic rDNA diversity:
We have detected eight distinct groups of ITS1 sequences in P. pedestris individuals. Phylogenetic analysis indicates that they have been evolving independently for >11 million years. Mutations at 18S sites that are otherwise perfectly conserved between disparate species provide convincing evidence that six of the Podisma groups consist of pseudogenes, while two might be functional (groups 1 and 2). Southern blots indicate that the genomic frequency of pseudogenes is possibly higher than that of functional units. One of the putatively functional variants (group 1) also occurs at high copy number and is clearly the most frequent among sequences with a relatively well-conserved 18S coding region. Interestingly, this common functional variant is scarcely amplified at all under standard PCR conditions. Group 2, on the other hand, occurs at very low copy numbers and therefore seems unlikely to have much functional significance.
Even within group 1, some of the lineages have been diverging for at least 3.7 million years. This result is in contrast to the expectation that functional arrays would be particularly prone to homogenization through concerted evolution (Kovarik et al. 2004). What could explain a low rate of homogenization in Podisma? Several studies strongly suggest that concerted evolution is much more potent for sequences on the same chromosome than for loci on different chromosomes (Schlötterer and Tautz 1994; Copenhaver and Pikaard 1996; Polanco et al. 1998; Parkin and Butlin 2004). The results of fluorescent in situ hybridization indeed show that multiple rDNA loci are present in P. pedestris. From two to seven loci have been detected, fused populations generally having two to three, whereas unfused populations vary from one population to another, usually having more (P. Veltsos, unpublished data). However, the sequencing results detected many more haplotypes than there are loci, implying that individual arrays must have more than one haplotype and indicating low rates of concerted evolution even at the intrachromosomal level. Individual CM1, for example, has six different rDNA loci but a minimum of 14 different haplotypes from group 1 alone (Figure 2). Furthermore, the different sequence variants persist in the P. pedestris genome for millions of years, while the number and chromosomal location of the arrays changes extraordinarily rapidly. Populations BL and CM, for example, have no rDNA loci in common but are remarkably similar at the sequence level (Figure 2).
This inefficiency of concerted evolution could be associated with the exceptionally large genome of P. pedestris (Westerman et al. 1987), a genome that is 100 times larger than that of Drosophila melanogaster (Hartl 2000). There are extensive heterochromatic blocks (Bella et al. 1991) and restriction digests (Figure 1b) show evidence of methylation, unlike Drosophila. Methylated sequences and DNA condensation, possibly associated with heterochromatinization, have been implicated in markedly reducing rates of concerted evolution (Lim et al. 2000).
It is possible that there is a causal link between genome size and the abundance of rDNA pseudogenes in this species. It has been demonstrated that the relative rate of DNA loss (per kilobases of sequence) is significantly lower in P. pedestris than in model organisms such as D. melanogaster or Caenorhabditis elegans with their compact genomes (Petrov et al. 1996; Robertson 2000; Bensasson et al. 2001). Pseudogenes in these small genomes are being removed rapidly relative to mutation (Petrov 2001), so that little genetic divergence accumulates over their lifetime before they are eroded away by deletion. The Drosophila genome, in particular, is characterized by a remarkable paucity of pseudogenes (Harrison et al. 2003). In Podisma, by contrast, the rate of removal is five hundred times lower than in Drosophila, and only 5% of the point substitution rate (Bensasson et al. 2001), which would help explain why the pseudogenes have accumulated such remarkable divergence.
It is, at first sight, surprising that the cDNA sequences obtained from transcribed rRNA include many potential pseudogenes. However, pseudogenes can still be transcribed, provided that their transcriptional control elements remain functional. Expressed pseudogenes have indeed been documented (Mighell et al. 2000; Hirotsune et al. 2003; Yamada et al. 2003). It follows that transcription is not an indicator of functionality (Bailey et al. 2003). Pseudogene transcripts can actually be more abundant than RNA from functional paralogues (Fujii et al. 1999). It is possible that the pool of standing rRNA in the cell has a disproportionately high frequency of pseudogenes. ITS1 is included in the primary transcript and is excised during the processing of the precursor rRNA (van Nues et al. 1995). If this process is less efficient for pseudogene transcripts, they could persist in the cell and hence be the more frequent template for our RT–PCR. One particular attribute of the cDNA sequences, which could interfere with efficient splicing, is that three quarters of them contain deletions in the ITS1. Our observation of very deeply diverged pseudogenes in cDNA indicates that at least some of these copies are accessible to the nucleus's transcriptional machinery, although not necessarily within active nucleoli.
Methodological issues associated with the study of multigene families:
Our study illustrates PCR drift and a particularly severe case of bias (Table 2), which can affect surveys of multigene families (Wagner et al. 1994; Kanagawa 2003). The results obtained under standard reaction conditions appear to provide a particularly inaccurate estimate of the actual genomic frequencies of different sequence variants. Group 2 sequences, for example, seem to be preferentially amplified and are consistently present in standard PCR products even though their genomic frequency is very low (as demonstrated by three approaches: qPCR, direct sequencing, and digestion of PCR products). The most highly repeated functional group, group 1, on the other hand, is often not amplified at all under standard PCR conditions. Fenton et al. (1998) and Hartmann et al. (2001) also found that particular rDNA types were readily amplified by PCR even though Southern blots showed that their genomic frequency was very low.
The addition of DMSO to the reaction dramatically alters the frequency of the different sequence variants in the pool of PCR products. Group 1 sequences consistently predominate while group 2 sequences become undetectable in the restriction digests. Buckler et al. (1997) have suggested that DMSO may facilitate the amplification of sequences with a more stable secondary structure. At first glance, this would seem consistent with our results, since the ITS1 of group 1 units tends to have higher predicted secondary-structure stability than the other variants (data not shown). However, a follow-up experiment indicated that PCR efficiency was little affected by these sequence differences. Using standard conditions, we amplified mixtures containing varying proportions of group 1 and group 2 clones. In contrast to the results from the genomic DNA extracts, group 1 sequences were clearly detectable in restriction digests of the PCR product whenever they were present in the template mixture. Further, more group 1 sequences were recovered from the genomic DNA extracts when the annealing temperature was increased to 68°, consistent with the proposal that the effect of the DMSO had been to increase stringency.
Using the hybrid zone to date the dynamics of the rDNA loci:
The biogeography of the Alps indicates that the hybrid zone must be ∼10,000 years old and that the initial separation of the chromosomal races occurred at least 100,000 years ago and possibly several times longer ago than that (Barton and Hewitt 1981). The existence of the different rDNA haplotypes on both sides of the zone is therefore consistent with our much older estimates for their age of origin. Even though Dallas et al. (1988) found very strong linkage disequilibrium between the presence of a particular IGS restriction site and the chromosomal fusion in one locality, they did not find the association elsewhere. One possible mechanism that could both make and break such associations is the movement of the rDNA variants around the genome. This process does indeed seem to be occurring, as reflected in the differing number and location of rDNA loci on either side of the zone, and indeed between populations on the same side of the zone. It follows, then, that the ancient rDNA lineages within Podisma are persisting despite this substantial intrachromosomal mobility having occurred since they arose.
Acknowledgments
We thank Lee Crainey, Thomas Giger, and Raj Joseph for substantial help with molecular work and Hussna Jahan and Joe Nichols for assistance in the field. We are very grateful to Andrew Leitch, Carlo Largiadèr, Douda Bensasson, and Steve Rossiter for helpful discussions and to anonymous reviewers for valuable comments on earlier versions of this manuscript. This work was financially supported by fellowships from the Swiss National Science Foundation (grant PBBEA-104447) and the Roche Research Foundation to I.K. and a Natural Environment Research Council grant to R.A.N. (NER-B-S-2003-00859).
References
- Akaike, H., 1974. A new look at the statistical model identification. IEEE Trans. Autom. Control 19: 716–723. [Google Scholar]
- Aljanabi, S., and I. Martinez, 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25: 4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez, I., and J. F. Wendel, 2003. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 29: 417–434. [DOI] [PubMed] [Google Scholar]
- Amedegnato, C., W. Chapco and G. Litzenberger, 2003. Out of South America? Additional evidence for a southern origin of melanopline grasshoppers. Mol. Phylogenet. Evol. 29: 115–119. [DOI] [PubMed] [Google Scholar]
- Bailey, C. D., T. G. Carr, S. A. Harris and C. E. Hughes, 2003. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Mol. Phylogenet. Evol. 29: 435–455. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and G. M. Hewitt, 1981. A chromosomal cline in the grasshopper Podisma pedestris. Evolution 35: 1008–1018. [DOI] [PubMed] [Google Scholar]
- Bella, J. L., M. Westerman, C. Lopezfernandez, J. Delatorre, J. M. Rubio et al., 1991. Sex-chromosome and autosome divergence in Podisma (Orthoptera) in Western-Europe. Genet. Sel. Evol. 23: 5–13. [Google Scholar]
- Bensasson, D., D. A. Petrov, D.-X. Zhang, D. L. Hartl and G. M. Hewitt, 2001. Genomic gigantism: DNA loss is slow in mountain grasshoppers. Mol. Biol. Evol. 18: 246–253. [DOI] [PubMed] [Google Scholar]
- Buckler, E. S., A. Ippolito and T. P. Holtsford, 1997. The evolution of ribosomal DNA: Divergent paralogues and phylogenetic implications. Genetics 145: 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrero, J., F. Perfectti, R. Gomez, J. P. M. Camacho and M. D. Lopez-Leon, 2003. Population variation in the A chromosome distribution of satellite DNA and ribosomal DNA in the grasshopper Eyprepocnemis plorans. Chromosome Res. 11: 375–381. [DOI] [PubMed] [Google Scholar]
- Chapco, W., G. Litzenberger and W. R. Kuperus, 2001. A molecular biogeographic analysis of the relationship between North American melanoploid grasshoppers and their Eurasian and South American relatives. Mol. Phylogenet. Evol. 18: 460–466. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G. P., and C. S. Pikaard, 1996. Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant J. 9: 273–282. [DOI] [PubMed] [Google Scholar]
- Crease, T. J., 1995. Ribosomal DNA evolution at the population level: nucleotide variation in intergenic spacer arrays of Daphnia pulex. Genetics 141: 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas, J. F., N. H. Barton and G. A. Dover, 1988. Interracial rDNA variation in the grasshopper, Podisma pedestris. Mol. Biol. Evol. 5: 660–674. [Google Scholar]
- DeBry, R. W., and R. G. Olmstead, 2000. A simulation study of reduced tree-search effort in bootstrap resampling analysis. Syst. Biol. 49: 171–179. [DOI] [PubMed] [Google Scholar]
- Denduangboripant, J., and Q. C. B. Cronk, 2000. High intraindividual variation in internal transcibed spacer sequences in Aeschynanthus (Gesneriaceae): implications for phylogenetics. Proc. Biol. Soc. 267: 1407–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder, J. F., and B. J. Turner, 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 70: 297–320. [DOI] [PubMed] [Google Scholar]
- Farris, J. S., 1969. A successive approximations approach to character weighting. Syst. Zool. 18: 374–385. [Google Scholar]
- Felsenstein, J., 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17: 368–376. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1985. Confidence limits on phylogenies—an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Fenton, B., G. Malloch and F. Germa, 1998. A study of variation in rDNA ITS regions shows that two haplotypes coexist within a single aphid genome. Genome 41: 337–345. [PubMed] [Google Scholar]
- Flanagan, N. S., P. L. Mason, J. Gosalvez and G. M. Hewitt, 1999. Chromosomal differentiation through an alpine hybrid zone in the grasshopper Chorthippus parallelus. J. Evol. Biol. 12: 577–585. [Google Scholar]
- Fujii, G. H., A. M. Morimoto, A. E. Berson and J. B. Bolen, 1999. Transcriptional analysis of the PTEN/MMAC1 pseudogene, (phi)PTEN. Oncogene 18: 1765–1769. [DOI] [PubMed] [Google Scholar]
- Gernandt, D. S., and A. Liston, 1999. Internal transcribed spacer region evolution in Larix and Pseudotsuga (Pinaceae). Am. J. Bot. 86: 711–723. [PubMed] [Google Scholar]
- Goldman, N., 1993. a Simple diagnostic statistical tests of models for DNA substitutions. J. Mol. Evol. 37: 650–661. [DOI] [PubMed] [Google Scholar]
- Goldman, N., 1993. b Statistical tests of models of DNA substitutions. J. Mol. Evol. 36: 182–198. [DOI] [PubMed] [Google Scholar]
- Graur, D., and W. H. Li, 2000. Fundamentals of Molecular Evolution. Sinauer Associates, Sunderland, MA.
- Hall, T. A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95. [Google Scholar]
- Harris, D. J., and K. A. Crandall, 2000. Intragenomic variation within ITS1 and ITS2 of freshwater crayfishes (Decapoda: Cambaridae): implications for phylogenetic and microsatellite studies. Mol. Biol. Evol. 17: 284–291. [DOI] [PubMed] [Google Scholar]
- Harrison, P. M., D. Milburn, Z. Zhang, P. Bertone and M. Gerstein, 2003. Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Res. 31: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, D. L., 2000. Molecular melodies in high and low C. Nat. Rev. Genet. 1: 145–149. [DOI] [PubMed] [Google Scholar]
- Hartmann, S., J. D. Nason and D. Bhattacharya, 2001. Extensive ribosomal DNA genic variation in the columnar cactus Lophocereus. J. Mol. Evol. 53: 124–134. [DOI] [PubMed] [Google Scholar]
- Hillis, D. M., and J. J. Bull, 1993. An empirical-test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42: 182–192. [Google Scholar]
- Hirotsune, S., N. Yoshida, A. Chen, L. Garrett, F. Sugiyama et al., 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423: 91–96. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and J. P. Bollback, 2001. Empirical and hierarchical Bayesian estimation of ancestral states. Syst. Biol. 50: 351–366. [PubMed] [Google Scholar]
- Huelsenbeck, J. P., and F. Ronquist, 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., B. Larget, R. E. Miller and F. Ronquist, 2002. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst. Biol. 51: 673–688. [DOI] [PubMed] [Google Scholar]
- Hugall, A., J. Stanton and C. Moritz, 1999. Reticulate evolution and the origins of ribosomal internal transcribed spacer diversity in apomictic Meloidogyne. Mol. Biol. Evol. 16: 157–164. [DOI] [PubMed] [Google Scholar]
- John, B., and G. M. Hewitt, 1970. Inter-population sex chromosome polymorphism in the grasshopper Podisma pedestris—I. Fundamental facts. Chromosoma 31: 291–308. [DOI] [PubMed] [Google Scholar]
- Jordan, S., C. Simon and D. Polhemus, 2003. Molecular systematics and adaptive radiation of Hawaii's endemic damselfly genus Megalagrion (Odonata: Caenagrionidae). Syst. Biol. 52: 89–109. [DOI] [PubMed] [Google Scholar]
- Kanagawa, T., 2003. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96: 317–323. [DOI] [PubMed] [Google Scholar]
- Kovarik, A., R. Matyasek, K. Y. Lim, K. Skalicka, B. Koukalova et al., 2004. Concerted evolution of 18–5.8–26S rDNA repeats in Nicotiana allotetraploids. Biol. J. Linn. Soc. 82: 615–625. [Google Scholar]
- Larget, B., and D. L. Simon, 1999. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol. Biol. Evol. 16: 750–759. [Google Scholar]
- Lim, K. Y., A. Kovarik, R. Matyasek, M. Bezdek, C. P. Lichtenstein et al., 2000. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma 109: 161–172. [DOI] [PubMed] [Google Scholar]
- Litzenberger, G., and W. Chapco, 2001. Molecular phylogeny of selected Eurasian Podismine grasshoppers (Orthoptera: Acrididae). Ann. Entomol. Soc. Am. 94: 505–511. [Google Scholar]
- Márquez, L. M., D. J. Miller, J. B. MacKenzie and M. J. H. van Oppen, 2003. Pseudogenes contribute to the extreme diversity of nuclear ribosomal DNA in the hard coral Acropora. Mol. Biol. Evol. 20: 1077–1086. [DOI] [PubMed] [Google Scholar]
- Martin, D. P., C. Williamson and D. Posada, 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21: 260–262. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34: 126–129. [DOI] [PubMed] [Google Scholar]
- Mayol, M., and J. A. Rossello, 2001. Why nuclear ribosomal DNA spacers (ITS) tell different stories in Quercus. Mol. Phylogenet. Evol. 19: 167–176. [DOI] [PubMed] [Google Scholar]
- Mighell, A. J., N. R. Smith, P. A. Robinson and A. F. Markham, 2000. Vertebrate pseudogenes. FEBS Lett. 468: 109–114. [DOI] [PubMed] [Google Scholar]
- Muir, G., C. C. Fleming and C. Schlötterer, 2001. Three divergent rDNA clusters predate the species divergence in Quercus petraea (Matt.) Liebl. and Quercus robur L. Mol. Biol. Evol. 18: 112–119. [DOI] [PubMed] [Google Scholar]
- Nichols, R. A., and G. M. Hewitt, 1986. Population structure and the shape of a chromosomal cline between two races of Podisma pedestris (Orthoptera: Acrididae). Biol. J. Linn. Soc. 29: 301–316. [Google Scholar]
- O'Donnell, K., and E. Cigelnik, 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7: 103–116. [DOI] [PubMed] [Google Scholar]
- Parkin, E. J., and R. K. Butlin, 2004. Within- and between-individual sequence variation among ITS1 copies in the meadow grasshopper Chorthippus parallelus indicates frequent intrachromosomal gene conversion. Mol. Biol. Evol. 21: 1595–1601. [DOI] [PubMed] [Google Scholar]
- Pereira, S. L., A. L. Baker and A. Wajntal, 2002. Combined nuclear and mitochondrial DNA sequences resolve generic relationships within the Cracidae (Galliformes, Aves). Syst. Biol. 51: 946–958. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., 2001. Evolution of genome size: new approaches to an old problem. Trends Genet. 17: 23–28. [DOI] [PubMed] [Google Scholar]
- Petrov, D. A., E. R. Lozovskaya and D. L. Hartl, 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384: 346–349. [DOI] [PubMed] [Google Scholar]
- Polanco, C., A. I. Gonzalez, A. de la Fuente and G. A. Dover, 1998. Multigene family of ribosomal DNA in Drosophila melanogaster reveals contrasting patterns of homogenization for IGS and ITS spacer regions: a possible mechanism to resolve this paradox. Genetics 149: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- Posada, D., and K. A. Crandall, 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98: 13757–13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razafimandimbison, S. G., E. A. Kellogg and B. Bremer, 2004. Recent origin and phylogenetic utility of divergent ITS putative pseudogenes: a case study from Naucleeae (Rubiaceae). Syst. Biol. 53: 177–192. [DOI] [PubMed] [Google Scholar]
- Robertson, H. M., 2000. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 10: 192–203. [DOI] [PubMed] [Google Scholar]
- Ruggiero, M. V., and G. Procaccini, 2004. The rDNA ITS region in the Lessepsian marine angiosperm Halophila stipulacea (Forssk.) Aschers. (Hydrocharitaceae): intragenomic variability and putative pseudogenic sequences. Mol. Biol. Evol. 58: 115–121. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schlötterer, C., and D. Tautz, 1994. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr. Biol. 4: 777–783. [DOI] [PubMed] [Google Scholar]
- Sharpe, R. G., R. E. Harbach and R. Butlin, 2000. Molecular variation and phylogeny of members of the Minimus group of Anopheles subgenus Cellia (Diptera: Culicidae). Syst. Entomol. 25: 263–272. [Google Scholar]
- Swofford, D. L., 2002. PAUP* Version 4.0b10 for Macintosh. Sinauer Associates, Sunderland, MA.
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues, R. W., J. Venema, J. M. J. Rientjes, A. Dirks-Mulder and H. A. Raué, 1995. Processing of eukaryotic pre-rRNA: the role of the transcribed spacers. Biochem. Cell Biol. 73: 789–801. [DOI] [PubMed] [Google Scholar]
- Waddell, P. J., Y. Cao, J. Hauf and M. Hasegawa, 1999. Using novel phylogenetic methods to evaluate mammalian mtDNA, including amino acid invariant sites LogDet plus site stripping, to detect internal conflicts in the data, with special reference to the positions of hedgehog, armadillo, and elephant. Syst. Biol. 48: 31–53. [DOI] [PubMed] [Google Scholar]
- Wagner, A., N. Blackstone, P. Cartwright, M. Dick, B. Misof et al., 1994. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst. Biol. 43: 250–261. [Google Scholar]
- Westerman, M., N. H. Barton and G. M. Hewitt, 1987. Differences in DNA content between two chromosomal races of the grasshopper Podisma pedestris. Heredity 58: 221–228. [Google Scholar]
- Whang, I.-J., J. Jung, J.-K. Park, G.-S. Min and W. Kim, 2002. Intragenomic length variation of the ribosomal DNA intergenic spacer in a malaria vector, Anopheles sinensis. Mol. Cells 14: 158–162. [PubMed] [Google Scholar]
- Yamada, K., J. Lim, J. M. Dale, H. Chen, P. Shinn et al., 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and D. Roberts, 1995. On the use of nucleic acid sequences to infer early branchings in the tree of life. Mol. Biol. Evol. 12: 451–458. [DOI] [PubMed] [Google Scholar]
- Yoder, A. D., and Z. H. Yang, 2000. Estimation of primate speciation dates using local molecular clocks. Mol. Biol. Evol. 17: 1081–1090. [DOI] [PubMed] [Google Scholar]