Abstract
DNA damage response proteins identify sites of DNA damage and signal to downstream effectors that orchestrate either apoptosis or arrest of the cell cycle and DNA repair. The C. elegans DNA damage response mutants mrt-2, hus-1, and clk-2(mn159) displayed 8- to 15-fold increases in the frequency of spontaneous mutation in their germlines. Many of these mutations were small- to medium-sized deletions, some of which had unusual sequences at their breakpoints such as purine-rich tracts or direct or inverted repeats. Although DNA-damage-induced apoptosis is abrogated in the mrt-2, hus-1, and clk-2 mutant backgrounds, lack of the apoptotic branch of the DNA damage response pathway in cep-1/p53, ced-3, and ced-4 mutants did not result in a Mutator phenotype. Thus, DNA damage checkpoint proteins suppress the frequency of mutation by ensuring that spontaneous DNA damage is accurately repaired in C. elegans germ cells. Although DNA damage response defects that predispose humans to cancer are known to result in large-scale chromosome aberrations, our results suggest that small- to medium-sized deletions may also play roles in the development of cancer.
THE genome is under continuous surveillance for DNA damage by checkpoint sensor proteins, which, if activated, can elicit temporary arrest of the cell cycle and repair of the damaged lesion. In multicellular eukaryotes, an alternative fate for cells that reach a critical threshold of DNA damage is apoptosis. Malfunction of DNA damage response proteins can stimulate tumorogenesis in humans (Venkitaraman 2002), probably as a consequence of improper processing of endogenous or exogenous forms of DNA damage that results in alterations to the genome. An understanding of the precise relationship between genome instability and the development of cancer is currently a topic of intense study and includes analysis of gross chromosomal rearrangements (GCRs) for many forms of cancer. While GCRs are commonly observed in cancer cells and may contribute to tumorogenesis, it is unclear if they herald the presence of other forms of DNA damage that are also relevant. We have chosen to address this issue by studying DNA damage response mutants in the nematode Caenorhabditis elegans.
Proteins that respond to DNA damage are thought to function via a tiered network of biochemical interactions. Putative sensors of DNA damage such as the phosphatidyl inositol-3 (PI-3)-like kinases ATR and ATM and their respective partners ATRIP and the MRE11/RAD50/NBS1 complex help to initiate a DNA damage response (DDR) (Sancar et al. 2004). A complex of checkpoint proteins that is independently recruited to sites of DNA damage is the RAD9/RAD1/HUS1 (9-1-1) proliferating cell nuclear antigen (PCNA)-like sliding clamp, which is loaded onto single-stranded DNA by the RAD17 clamp loader and its four replication factor C subunits (Griffiths et al. 1995; Kostrub et al. 1998; Burtelow et al. 2000; Caspari et al. 2000; Zou et al. 2002; Bermudez et al. 2003). The ATR and ATM checkpoint kinases respond to DNA damage by phosphorylating members of the 9-1-1 damage sensor complex as well as downstream “mediators” such as BRCA1, CLASPIN, 53BP1, the signaling kinases CHK1 and CHK2, and effectors such as p53 (Sancar et al. 2004).
Genetic studies in mice and yeast suggest that DNA damage response proteins may suppress genome instability, in part, by facilitating the repair of endogenous DNA double-strand breaks (DSBs) (Patel et al. 1998; Moynahan et al. 1999, 2001; Deng and Scott 2000; Myung et al. 2001a,b; Howlett et al. 2002; Kraakman-Van Der Zwet et al. 2002; Venkitaraman 2002; D'Andrea and Grompe 2003; Pennaneach and Kolodner 2004). Examination of genome stability in haploid yeast mutants defective for the 9-1-1 checkpoint complex has revealed modestly elevated levels of GCRs (Myung et al. 2001b). Mutation of vertebrate 9-1-1 complex subunits results in genome instability and lethality (Weiss et al. 2000; Budzowska et al. 2004; Hopkins et al. 2004; Kobayashi et al. 2004). Deficiency for mediators of the DNA damage response such as BRCA1 or BRCA2 also results in lethality accompanied by translocations, loss of chromosome arms, and aneuploidy (Venkitaraman 2002). In contrast, mutation of p53 in mice does not lead to genome instability or a Mutator phenotype (Nishino et al. 1995; Buettner et al. 1997; Reliene and Schiestl 2003). Rather, p53 is thought to suppress cancer by acting to control cell proliferation via either apoptosis or senescence (Lowe et al. 2004).
C. elegans displays tissue-specific responses to DNA damage. Checkpoint proteins such as the 9-1-1 complex or ATM/ATR can initiate apoptosis in meiotic germ cells at the pachytene stage or cell cycle arrest in the mitotic germ cells (Gartner et al. 2004; Garcia-Muse and Boulton 2005), whereas somatic cells are refractory to these cellular responses to DNA damage (Gartner et al. 2000). Mammalian p53 responds to DNA damage by eliciting either apoptosis or cell cycle arrest (Attardi 2005). However, C. elegans and Drosophila orthologs of p53 affect only the induction of DNA-damage-induced apoptosis (Derry et al. 2001; Schumacher et al. 2001; Brodsky et al. 2004), suggesting that the mammalian cell cycle arrest function may be derived.
Deficiency for the C. elegans 9-1-1 complex subunits HUS-1 or MRT-2 results in defective responses to both ionizing radiation (IR)-induced apoptosis and cell cycle arrest in germ cells (Ahmed and Hodgkin 2000; Gartner et al. 2000; Hofmann et al. 2002). Mutations in a third C. elegans DNA damage checkpoint gene, clk-2, also abrogate the apoptotic and cell cycle arrest responses to IR and confer an additional defect in the S-phase DNA replication checkpoint (Ahmed et al. 2001). clk-2 has recently been shown to function downstream of atl-1, a homolog of the PI-3-like DNA-damage-signaling kinase ATR that interacts with DSBs (Garcia-Muse and Boulton 2005). Both atl-1 and clk-2 are essential, and a strong defect in either gene results in the accumulation of single-stranded DNA and mitotic failure (Garcia-Muse and Boulton 2005). Two conditional mutations of clk-2 have been identified: mn159 was recovered in a screen for radiation-hypersensitive mutants of C. elegans (hence its former gene name “rad-5”) (Hartman and Herman 1982), and qm37 was identified in a screen for mutations that confer a maternally rescued Slow Growth phenotype (Lakowski and Hekimi 1996).
Here we show that mutations in the C. elegans DDR genes mrt-2, hus-1, and clk-2 result in an elevation in the frequency of spontaneous mutation, whereas defects in genes required exclusively for DNA-damage-induced apoptosis do not. Mutations that result from DDR defects are often small- to medium-sized deletions, suggesting a failure to repair spontaneous lesions that result in strand breaks.
MATERIALS AND METHODS
Strains:
All strains were cultured at 20° as described previously (Sulston and Hodgkin 1988). Strains used in this study were Bristol N2 wild type, hus-1(op241) I, dpy-5(e61) I, cep-1(lg12501) I, cep-1(gk138) I, dpy-17(e164) III, ced-4(n1162) III, clk-2(mn159) III, clk-2(qm37) III, unc-32(e191) III, mrt-2(e2663) III, unc-17(e2754) IV, ced-3(n717) IV, unc-60(e2763) V, him-7(e1480) V, unc-46(e177) V, vab-8(e2764) V, lon-2(e2775) X, unc-58(e665) X, 18 unc-58(e665) suppressor mutants numbered unc-58(e2815) X to unc-58(e2832) X, and ypIs1 [rol-6(su1006) mrt-2(+)].
Genetics:
All marker strains were outcrossed five times vs. Bristol N2 wild type before use. The presence of mrt-2 or hus-1 was assessed by scoring for sterility following gamma irradiation of six L1 larvae per strain at a dose of 60 Gy. The presence of clk-2(mn159) was assessed on the basis of its temperature-sensitive sterile phenotype (Hartman and Herman 1982). The mutation hus-1(op241), which lies on chromosome I, was originally identified in the strain CB1480 him-7(e1480) V (Gartner et al. 2000). The hus-1(op241) and him-7(e1480) mutations were separated by crossing them into and out of unc-46 V and dpy-5 I backgrounds, respectively. Two F2 lines each of the hus-1(op241);unc-58(e665) and him-7(e1480);unc-58(e665) lines were established. Two mrt-2 F2 lines were established after six and seven outcrosses with N2 wild type, both were crossed with unc-58(e665), and two mrt-2;unc-58(e665) F2 lines each were established for the mrt-2 parental lines. clk-2(mn159) was outcrossed four times with N2 wild type, crossed into and out of backgrounds containing the flanking genetic markers dpy-17 and unc-32, and crossed an additional three times vs. dpy-17,unc-32; two clk-2(mn159) strains, each used to generate independent clk-2(mn159);unc-58(e665) strains, were established. unc-30;unc-58(e665) and dpy-17;unc-58(e665) were used to generate ced-3;unc-58(e665) and ced-4;unc-58(e665) doubles, respectively. For rescue of mrt-2, an extrachromosomal array containing a wild-type copy of the mrt-2 gene and a rol-6(su1006) Dm marker was integrated into a single locus in the genome by irradiating L4 larvae with 20 Gy IR to create the insertion ypIs1. This rescuing transgene was crossed with three independent lines of mrt-2;unc-58(e665) to generate mrt-2;unc-58(e665);ypIs1 strains.
Mutator assays:
To confirm the presence of spontaneous visible mutants that appeared in the mrt-2 mutant background, L4 larvae with unusual phenotypes were singled and their progeny were scored for transmission of the parental phenotype. For sterile mutants, nonsterile siblings were picked and their progeny were scored for segregation of the sterile phenotype.
For unc-58(e665) assays, OP50 bacteria was spread over the entire surface of 50 or 60 mm NGM plates and allowed to dry for 2 days. For each assay, a total of 40 plates were seeded with five unc-58(e665) L4 larvae each per genotype. One to eight additional trials were conducted using independently derived strains to assess mutation frequency (Hodgkin 1974). Plates were allowed to grow for a month until starvation, and the agar from each plate was then transferred to a 90-mm NGM plate with a streak of bacteria to one side. The presence of suppressor mutations was assessed 3 and 4 days later, and a single L4 larva was picked for each suppressor.
The five unc-58(e665) larvae used to seed each plate gave rise to ∼150 F1 progeny apiece, corresponding to 1500 haploid genomes. However, unc-58(e665) moves very slowly and brood size subsequently drops as a consequence of starvation and overcrowding, so 750 F1 might give rise to 30 F2 progeny each, on average, for a total of 22,500 F2 animals. The mean number of animals on a starved unc-58(e665) plate was 57,000 ± 7000 for 50-mm plates (Table 2) or 82,200 ± 10,500 for 60-mm plates (Table 3) (n = 16). For 50-mm plates, an additional 34,500 progeny would arise from F2, F3, and F4 parents, each of which were estimated to produce 15 progeny apiece, on average, accounting for 2300 parents or an additional 4600 haploid genomes. Thus, the total number of haploid genomes sampled per 50-mm plate was estimated as 6100. For parents that produce 15 progeny apiece, there is a 1.8% probability that none of the progeny of a heterozygous parent will be homozygous for the mutation, which lowers the estimated number of haploid genomes sampled per 50-mm plate to 6020. Similar calculations for the 60-mm plates used in Table 2 predict 9320 haploid genomes per plate.
TABLE 2.
unc-58(e665) spontaneous revertant frequencies
| unc-58(e665) background | Trial | Plates with revertants/ total plates | f(0) | Mutation frequency (revertants/gamete) | Spontaneous mutations/gamete |
|---|---|---|---|---|---|
| Wild type | 1 | 2/40 | 0.95 | 8.56 × 10−6 | |
| 2 | 0/40 | 1 | 0 | ||
| 3 | 0/40 | 1 | 0 | ||
| 4 | 0/40 | 1 | 0 | ||
| 5 | 1/40 | 0.975 | 8.88 × 10−6 | ||
| 6 | 1/40 | 0.975 | 8.88 × 10−6 | ||
| 7 | 0/40 | 1 | 0 | ||
| 8 | 0/40 | 1 | 0 | ||
| 9 | 1/40 | 0.975 | 8.88 × 10−6 | 3.9 ± 4.4 × 10−6 | |
| clk-2(mn159) | 1 | 6/40 | 0.85 | 5.7 × 10−5 | |
| 2 | 5/40 | 0.875 | 4.69 × 10−5 | ||
| 3 | 5/40 | 0.875 | 4.69 × 10−5 | ||
| 4 | 3/40 | 0.925 | 2.74 × 10−5 | ||
| 5 | 3/40 | 0.925 | 2.74 × 10−5 | ||
| 6 | 3/40 | 0.925 | 2.74 × 10−5 | ||
| 7 | 3/40 | 0.925 | 2.74 × 10−5 | ||
| 8 | 4/40 | 0.9 | 3.7 × 10−5 | ||
| 9 | 3/40 | 0.925 | 2.74 × 10−5 | 3.6 ± 1.1 × 10−5 | |
| clk-2(qm37) | 1 | 0/40 | 1 | 0 | |
| 2 | 0/40 | 1 | 0 | ||
| 3 | 0/40 | 1 | 0 | ||
| 4 | 0/40 | 1 | 0 | ||
| 5 | 0/40 | 1 | 0 | ||
| 6 | 1/40 | 0.975 | 8.88 × 10−6 | 1.5 ± 3.3 × 10−6 | |
| hus-1 | 1 | 7/40 | 0.825 | 6.75 × 10−5 | |
| 2 | 6/40 | 0.85 | 5.7 × 10−5 | ||
| 3 | 4/40 | 0.9 | 3.7 × 10−5 | ||
| 4 | 2/40 | 0.95 | 1.8 × 10−5 | ||
| 5 | 3/40 | 0.925 | 2.74 × 10−5 | ||
| 6 | 6/40 | 0.85 | 5.7 × 10−5 | 4.4 ± 1.8 × 10−5 | |
| mrt-2 | 1 | 12/40 | 0.7 | 1.25 × 10−4 | |
| 2 | 6/40 | 0.85 | 5.7 × 10−5 | ||
| 3 | 8/40 | 0.8 | 3.7 × 10−5 | ||
| 4 | 5/40 | 0.875 | 1.8 × 10−5 | ||
| 5 | 9/40 | 0.775 | 2.74 × 10−5 | ||
| 6 | 10/40 | 0.75 | 1.0 × 10−4 | 6.1 ± 3.9 × 10−5 | |
| mrt-2;ypIs1 | 1 | 0/40 | 1 | 0 | |
| 2 | 1/40 | 0.975 | 8.88 × 10−6 | 4.4 ± 4.4 × 10−6 | |
| ced-3 | 1 | 1/40 | 0.975 | 8.88 × 10−6 | |
| 2 | 0/40 | 1 | 0 | 4.4 ± 4.4 × 10−6 | |
| ced-4 | 1 | 1/40 | 0.975 | 8.88 × 10−6 | |
| 2 | 1/40 | 0.975 | 8.88 × 10−6 | 8.8 ± 0.0 × 10−6 | |
| him-7 | 1 | 1/40 | 0.975 | 8.88 × 10−6 | |
| 2 | 1/40 | 0.975 | 8.88 × 10−6 | ||
| 3 | 0/40 | 1 | 0 | 5.9 ± 4.2 × 10−6 |
The frequency of spontaneous germline mutations conferring suppression of unc-58(e665) was assessed using the Poisson equation to account for the possibility that more than one reversion event may have occurred on a single plate on the basis of the number of plates that lacked reversion events. The general Poisson equation for n (not necessarily) = 0 is p(n) = (mn)e−m/n!, which reduces to f(0) = e−m for n = 0, where f(0) is the fraction of plates from a given trial that did not contain an unc-58(e665) revertant. Mutation frequencies were calculated as: (m, the mean number of spontaneous mutations per plate)/6020 (= number of haploid genomes per plate) (Zalevsky et al. 1999). See materials and methods for an estimation of the number of haploid genomes per 50-mm plate used in Table 2. Standard deviations are shown for mean mutation frequencies.
TABLE 3.
unc-58(e665) spontaneous revertant frequencies for cep-1 mutants
| unc-58(e665) background | Trial | Plates with revertants/ total plates | f(0) | Mutation frequency (revertants/gamete) | Spontaneous mutations/gamete |
|---|---|---|---|---|---|
| Wild type | 1 | 2/40 | 0.95 | 5.52 × 10−6 | |
| 2 | 0/40 | 1 | 0 | ||
| 3 | 0/40 | 1 | 0 | ||
| 4 | 0/40 | 1 | 0 | ||
| 5 | 1/40 | 0.975 | 2.73 × 10−6 | ||
| 6 | 0/40 | 1 | 2.73 × 10−6 | ||
| 7 | 1/40 | 0.975 | 0 | ||
| 8 | 0/40 | 1 | 0 | 1.4 ± 1.9 × 10−6 | |
| cep-1(lg12501) | 1 | 1/40 | 0.975 | 2.73 × 10−6 | |
| 2 | 0/40 | 1 | 0 | ||
| 3 | 1/40 | 0.975 | 2.73 × 10−6 | ||
| 4 | 0/40 | 1 | 0 | ||
| 5 | 0/40 | 1 | 0 | ||
| 6 | 0/40 | 1 | 0 | ||
| 7 | 0/40 | 1 | 0 | ||
| 8 | 0/40 | 1 | 0 | 0.68 ± 1.2 × 10−6 | |
| cep-1(gk138) | 1 | 2/40 | 0.95 | 5.52 × 10−6 | |
| 2 | 0/40 | 1 | 0 | ||
| 3 | 1/40 | 0.975 | 2.73 × 10−6 | ||
| 4 | 2/40 | 0.95 | 5.52 × 10−6 | ||
| 5 | 0/40 | 1 | 0 | ||
| 6 | 0/40 | 1 | 0 | 2.3 ± 2.5 × 10−6 |
Calculations were made as for Table 2, except that 60-mm plates were used, yielding an estimated 9320 haploid genomes sampled per plate.
Deletion analysis:
Genomic DNA was prepared from suppressors using a Puregene cell and tissue kit (Gentra, Minneapolis), as recommended. Primers spanning each mutant locus were spaced ∼1 kb apart and amplified using the polymerase chain reaction (PCR). Primers were designed such that the ends of adjacent PCR products overlapped, except for the lin-15 locus, where nonoverlapping primers spaced 10 kb apart were also used. Deletions were identified on the basis of the change in size of a PCR product or on the absence of one or more PCR products and the ability to generate a novel PCR product from mutant, but not wild-type, template DNA using primers flanking predicted breakpoints.
G-tract PCR:
For template DNA, 10 C. elegans adults were mixed with 10 μl of lysis buffer (50 mm KCL, 10 mm, Tris pH 8.3, 2.5 mm MgCl2, 0.45% NP-40, 0.45% Tween-20, 0.01% gelatin) and 2 μl of 10 mg/ml proteinase K, incubated at 60° for 60 min and then at 95° for 25 min. One microliter of template DNA was then added to 24 μl of PCR reaction containing 50 mm LiCl, 10 mm Tris–HCl, pH 9.0, 0.1% Triton X-100, 0.1 mm MgCl2, 2 mm dNTPs, 5–10 μm G-tract primers, and 5 units/μl Taq DNA polymerase and incubated at 94° for 2 min, followed by 34 cycles of 94° for 45 sec, 60° for 30 sec, 72° for 90 sec and finally a 72° 5-min extension. Li+ was used instead of K+ as the monovalent cation in the 10× PCR buffer, as stable G-quartets that can inhibit DNA polymerases may form at long G-tracts in the presence of K+ (Woodford et al. 1994). Primer sets were F55F3 (GCCAATCTATCAAAACTCTGACTG and CGACCAAGGTTCTATCATACGAA), F38A6 (GAACCACTTCTGGGACCTCCG and TGGCTGGGCTGATGTAGTTCG), and Y41E3 (ACCGTTTCGAAACATGTTGCCAA and TGTATGCCATTTACTAAACTCTCC). Template DNA was prepared from strains grown at 20° and 25° to determine if growth temperature might affect the frequency of deletion, but identical results were obtained for all primer sets.
Life-span analysis:
A total of 100 L4 larvae of each genotype were used to initiate life-span assays. Adults were aged in groups of six, as described (Meier et al. 2006).
RESULTS
Elevated levels of spontaneous mutation in C. elegans DDR mutants:
Several spontaneous visible mutations were noted when propagating the C. elegans DNA damage response mutant mrt-2 (Ahmed and Hodgkin 2000). To confirm this observation, 40 freshly outcrossed F3 mrt-2 founders were singled, most of which failed to segregate visible mutations in the F4 generation. However, when progeny of 680 F4 mrt-2 hermaphrodites that had F3 siblings with no visible phenotypes were examined, 11 visible mutants were identified: 4 Uncoordinated (Unc), 2 Dumpy (Dpy), 2 Multivulva, 1 Long, and 1 Variable (Vab), whereas no spontaneous visible mutants were observed for progeny of 940 control mrt-1 hermaphrodites.
Spontaneous mutations occur rarely in the wild-type C. elegans strain Bristol N2 (Anderson 1995). To determine if mrt-2 might confer a Mutator phenotype, a mrt-2 strain carrying a rescuing extrachromosomal array containing the wild-type mrt-2 gene was compared with siblings from the same strain that had lost the rescuing array and were therefore checkpoint defective (Ahmed and Hodgkin 2000). The progeny of 780 mrt-2 mutants carrying the rescuing array failed to segregate visible mutations, whereas a total of 10 visible mutations were identified among the progeny of 780 nonrescued sibling mrt-2 individuals: 2 Unc, 1 Vab, 3 sterile Dpy, 2 egg-laying defective, 1 protruding Vulva, and 1 gonad development abnormal. Thus, the mrt-2 DDR gene suppresses the frequency of spontaneous mutation.
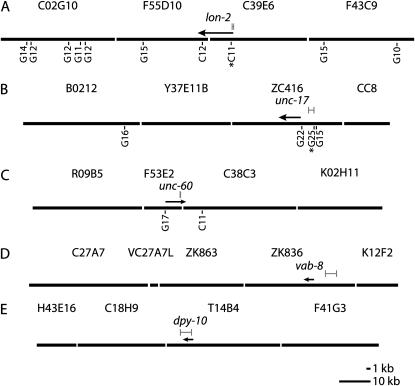
Inspection of the spontaneous mrt-2-induced visible mutants suggested candidate genes, and linkage and complementation tests demonstrated that alleles of dpy-31 (Novelli et al. 2004), lon-2, unc-17, unc-60, vab-8, and lin-15 had been isolated (Brenner 1974; Horvitz et al. 1983). A lin-15 Multivulva phenotype arises only when the functions of two neighboring genes, lin-15a and lin-15b, have been eliminated (Huang et al. 1994). Thus, isolation of a lin-15 mutation conferring a Multivulva phenotype from a mrt-2 background suggested that a genome rearrangement may have occurred. PCR analysis of the lin-15 locus amplified the expected products from N2 wild-type genomic DNA but not from that of the lin-15 mutant, whereas control loci on a different chromosome were intact for both strains. Further experimentation revealed that a deletion extending 165 kb to the right and 67 kb to the left of the lin-15 locus had occurred. Nested PCR reactions using primers flanking the putative breakpoints failed to yield a PCR product, suggesting a complex genome rearrangement. Analysis of four other mrt-2-induced mutations revealed that small- to medium-sized deletions had occurred: 0.15 kb for unc-60(e2763) (McKim et al. 1994), 0.435 kb for lon-2(e2775) (http://www.wormbase.org), 1.813 kb for unc-17(e2754) (Rand 1989), and 3.436 kb for vab-8(e2764) (Wolf et al. 1998). For two of these four deletions, a tract of G-rich sequence was adjacent to one breakpoint (Table 1, Figure 1). Such sequences occur rarely in the C. elegans genome (Figures 1 and 4), suggesting the involvement of the 9-1-1 DDR complex in resolving G-rich secondary structures (Cheung et al. 2002). The sixth visible mutant isolated from mrt-2 was the founding dpy-31 allele, e2770, which was sequenced and shown to result from a T-to-C transition (Novelli et al. 2004).
TABLE 1.
Breakpoints of spontaneous mrt-2 and clk-2(mn159) deletion products
| lon-2(e2755) (mrt-2) |
| AAACATAAATCTTTTTGTCATGCCATTTTTGGAAA..7073..gtcgtaaatttcaattacacacgcggcttcttttt |
| AT (insert) |
| caggtgagttaaaaatatttgggaggggggggggg..6623..ATCATAGAAAAACGTTTTTGTAATTTTTTTAAATT |
| (polypurine tract) |
| unc-17(e2754) (mrt-2) |
| TTTAAAATTTAAAAAAAGGTCTGAACTATTTTTTT..24834..ggatttttttttggattttttacaacggaaaaatt |
| cgcggggagtggggggggggggggggggggggggg..26647..GGATTTGCTACCGGTTGTCGAAAAAGGAGTTCAAA |
| (polypurine tract) |
| unc-60(e2763) (mrt-2) |
| AAAAATTTCCATGTATTTCCACATTCCACTTCCCA..11548..aactcctattttcagctgcccagacaacgccccag |
| AAgtacaggcctccgagatgtcggacctcgacgag..11690..AAGAGCGTGAAGAGCGACCTGATGTCCAACCAAAG |
| (hairpin) |
| vab-8(e2764) (mrt-2) |
| CACACCGTTTTCTTCTCGAGACCACCCATAAAAGT..26399..cgtgggtggcgcaaccagaaatactgtcggcccgt |
| (hairpin) |
| tcaatcaaatactgatatggatgtttctgaaatgg..29834..GTTTACCGGAGAGCAAGCCATTTGGAAGAATATGC |
| (hairpin) |
| dpy-10(e2760) (clk-2 mn159) |
| TTAATGAAAAATTAAAACCGCTAGAGCTTGAAAAT..4183..ccaacaaacgcagcttcttgagaaaaattaattgg |
| CTAAACAAATAAACAATAAAAAAACTAAAAAATAAAAAAAAAAA (insert) |
| acaatgtagatgagaagagaagtactacggtaggt..7799..ATGAGTGAGTGTGTGGATCGAAAAGTGAAATTTCAAAGTGAGC GGGGGGCATAGGGTGGTGGCGGCGCCCGCCGCGCGCGC |
| (direct repeat and G/C-rich tract) |
Sequences present at the breakpoints are in uppercase. Deleted sequences are in lowercase. Positions of breakpoints relative to the cosmid sequence for each gene are indicated (http://www.wormbase.org). Microhomology at breakpoints is underlined and in italics. Novel DNA sequence inserted between each breakpoint during NHEJ is indicated. DNA sequences with unusual qualities are underlined, in boldface, and defined in parentheses beneath the sequence. A direct repeat at the e2760 breakpoint is italicized, underlined, and in boldface.
Figure 1.—
Models for genome regions surrounding five spontaneous visible mutants. (A–D) Mutants from the mrt-2 genetic background. (E) Mutant from the clk-2(mn159) background. Genome sequence is presented as contiguous cosmids (uppercase letters). Wild-type position of mutant genes is indicated by arrows. Sequence deleted for each mutant is indicated by shaded bars. Note that deletions affecting unc-17 and vab-8 occur in the promoters of these genes. G-tracts longer than nine nucleotides are indicated below the genome sequence and are labeled “G” if oriented 5′–3′ or “C” if oriented 3′–5′. The length of each G-tract is indicated and G-tracts at deletion breakpoints have an asterisk.
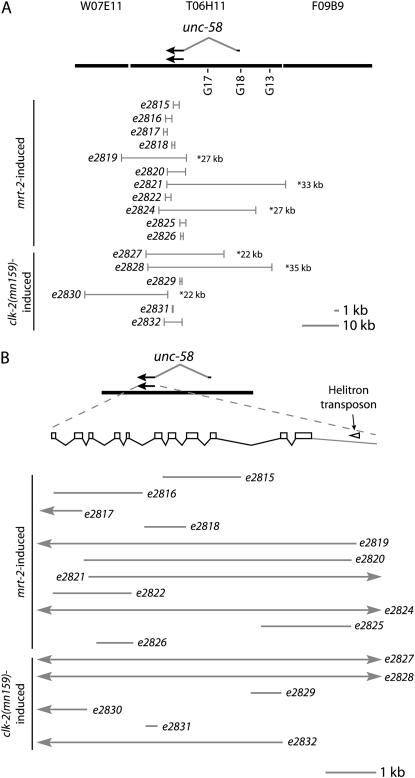
Figure 4.—
Models of unc-58 deletions induced in checkpoint-defective backgrounds. (A) Large-scale model of the genome region surrounding unc-58 (http://www.wormbase.org). Alternative unc-58 transcripts and G-tracts are shown above or below the cosmid sequence, respectively. Deletions are indicated as shaded bars with allele designations to the left and approximate lengths of large deletions to the right. (B) Enlargement of the smaller unc-58 transcription unit. The nonautonomous Helitron transposon is indicated as an open arrowhead. Deletions are indicated as shaded bars if their breakpoints fell within the enlarged region or as shaded bars with arrowheads for breakpoints that occurred upstream or downstream of the area shown.
The clk-2 checkpoint gene functions in the mrt-2 DDR pathway and also in a parallel DDR pathway (Ahmed et al. 2001). A spontaneous mutation in dpy-10 was isolated from the clk-2(mn159) mutant background, and analysis of the dpy-10 locus (Levy et al. 1993) revealed a 3.616-kb deletion and a long 42-bp G/C-rich sequence located near one deletion breakpoint (Table 1, Figure 1). Thus, five of six spontaneous mrt-2-induced mutations and one spontaneous clk-2-induced mutation resulted from deletions, three of which displayed G- or G/C-rich sequences near one deletion breakpoint.
IR hypersensitivity of checkpoint mutants:
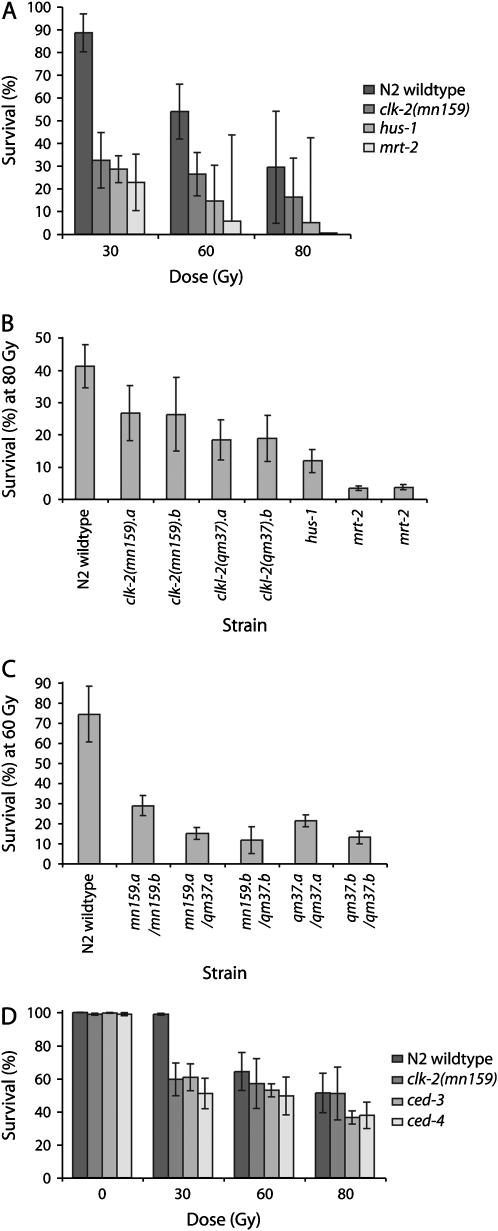
Given that both mrt-2 and clk-2(mn159) appeared to be Mutators on the basis of the isolation of visible mutations, multiple outcrosses were performed to remove extraneous mutations from strains used in this study. Prior to quantifying their effects on the frequency of spontaneous mutation, the severity of the checkpoint defects of the DDR mutations was assessed. Although mrt-2 and hus-1 physically interact and encode members of the heterotrimeric 9-1-1 PCNA-like sliding clamp (Boulton et al. 2002), the mrt-2(e2663) mutation is a null allele that is fully defective for the G2/M DNA damage checkpoint and for telomere replication, whereas hus-1(op241) is a partial loss-of-function mutation that has weaker checkpoint defects and is proficient for telomere replication (Ahmed and Hodgkin 2000; Hofmann et al. 2002). The clk-2(mn159) and clk-2(qm37) mutations confer radiation response defects that are somewhat weaker than those of hus-1(op241), but both clk-2 alleles have additional S-phase checkpoint defects and fail to respond to DNA replication forks that stall as a consequence of hydroxyurea-mediated nucleotide depletion (Ahmed et al. 2001). The magnitude of the DNA damage response defects of outcrossed DDR mutant strains was mrt-2 > hus-1 > clk-2(mn159), as assessed by measuring relative survival of progeny when L4 larvae were treated with gamma irradiation (Figure 2A). Previous results indicated that the germlines of clk-2(qm37) display a mildly enhanced radiation hypersensitivity phenotype in comparison with those of clk-2(mn159) (Ahmed et al. 2001). These results were recapitulated with isogenic versions of the clk-2 alleles, and both clk-2(qm37) homozygotes and clk-2(mn159)/clk-2(qm37) trans-heterozygotes were more sensitive to IR than clk-2(mn159) homozygotes (Figure 2, B and C). The radiation hypersensitivity of the germlines of the above mutants provided a measure of the relative magnitude of their DDR defects, and it seemed plausible that their frequencies of spontaneous mutation might scale accordingly.
Figure 2.—
Radiation hypersensitivity of checkpoint mutants. (A) Dose-response analysis of radiation hypersensitivity. Comparisons of IR hypersensitivity of clk-2 mutant strains that were independently outcrossed (a and b) at doses of (B) 80 Gy and (C) 60 Gy. (D) Apoptosis removes germ cells with lethal levels of DNA damage. For all panels, progeny derived from irradiated L4 larvae were scored for embryonic lethality (n = 5 broods scored/strain). Percentage of survival was normalized relative to unirradiated controls and standard deviations are shown.
unc-58 Mutator assays:
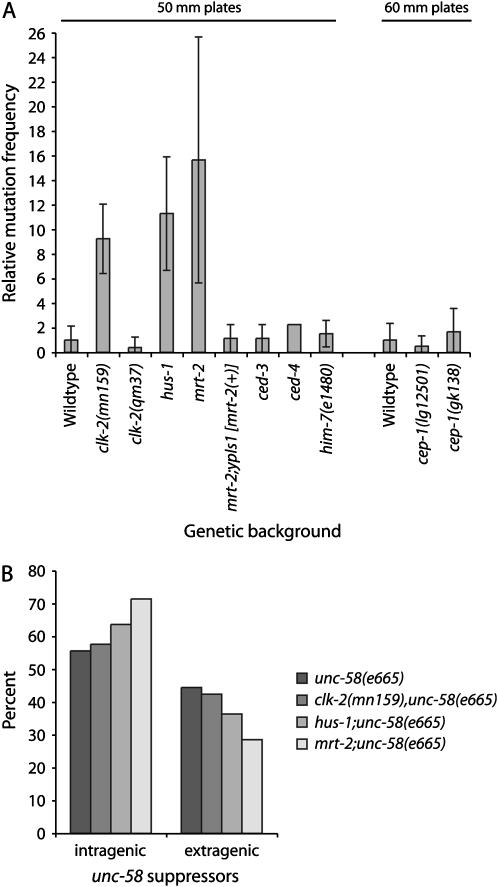
To quantify the Mutator phenotypes of these DDR mutants, an assay was developed based on the dominant uncoordinated mutation unc-58(e665), which results in almost paralyzed, small hermaphrodites with a distinctive “shaker” phenotype (Brenner 1974). unc-58(e665) can be suppressed by either intra- or extragenic mutations (Hodgkin 1974). Double mutants were made between unc-58(e665) and the DNA damage response mutations mrt-2, hus-1(op241), clk-2(mn159), and clk-2(qm37). The frequency of spontaneous mutation was elevated 8- to 15-fold for each of the checkpoint-defective unc-58(e665) doubles in comparison with the unc-58(e665) control, with the exception of clk-2(qm37), which did not display a Mutator phenotype (Figure 3A, Table 2). A transgene containing the wild-type mrt-2 gene rescued the Mutator phenotype of mrt-2 in the unc-58(e665) background (Figure 3A, Table 2). These unc-58 assays confirmed the presence of spontaneous visible mutants in mrt-2 and clk-2(mn159) genetic backgrounds (Figure 1) and agreed with previous observations suggesting that clk-2(mn159) and hus-1 may be Mutators (Hartman and Herman 1982; Hofmann et al. 2002).
Figure 3.—
Frequencies of spontaneous mutation as determined using unc-58(e665). (A) Relative levels of spontaneous mutation in various unc-58(e665) genetic backgrounds. For each experiment, 40 50-mm plates were scored for the strains unc-58(e665), clk-2(mn159);unc-58(e665), clk-2(qm37);unc-58(e665), hus-1(op241);unc-58(e665), mrt-2;unc-58(e665), mrt-2;unc-58(e665);ypIs1[rol-6(su1006) mrt-2(+)], ced-3;unc-58(e665), ced-4;unc-58(e665), and him-7(e1480);unc-58(e665). Additional results obtained using 60-mm petri dishes are shown for cep-1/p53 mutants. unc-58(e665) assays were repeated for control, mrt-2, hus-1(op241), and clk-2(mn159) genetic backgrounds with similar results. (B) Relative percentage of total intragenic vs. extragenic unc-58 suppressors. Note that wild-type suppressors were rare, which violates some assumptions of the z-test for proportions.
Loss-of-function mutations in unc-58 result in adult hermaphrodites that display a weak Uncoordinated phenotype, whereas extragenic suppressors are medium-sized twitchers that move well. Intragenic unc-58(e665) suppressor mutations were identified by crossing each suppressor with N2 wild-type males and scoring for absence of the dominant unc-58(e665) phenotype in F1 cross-progeny. Despite the increase in the frequency of spontaneous mutation in most DDR mutant backgrounds, the relative frequency of intra- vs. extragenic suppressors for clk-2(mn159), hus-1, and mrt-2 was not significantly different from that of wild type (P > 0.05 in all cases; z-test for proportions) (Figure 3B).
hus-1, mrt-2, and clk-2 checkpoint mutants are defective for DNA-damage-induced apoptosis (Gartner et al. 2000; Ahmed et al. 2001). Meiotic germ cells of C. elegans hermaphrodites display a continuous low level of physiological apoptosis, such that half of all germ cells die (Gumienny et al. 1999). Although levels of physiological apoptosis are not significantly altered in hus-1, mrt-2, or clk-2 mutant backgrounds (Gartner et al. 2000; Schumacher et al. 2001), apoptosis may normally occur at low levels in response to endogenous DNA damage and could be absent from checkpoint-defective mutant backgrounds. To determine if the elevated spontaneous mutation frequencies of checkpoint mutants occurred as a consequence of abrogation of DNA-damage-induced apoptosis, we examined cep-1/p53 mutants, which are defective for the apoptotic branch of the DNA damage response pathway (Derry et al. 2001; Schumacher et al. 2001), and ced-3 and ced-4 mutants, which are defective for core components of the apoptotic machinery and therefore lack both DNA-damage-induced and physiological forms of germ-cell apoptosis (Gartner et al. 2000). At a low dose of IR (30 Gy), N2 wild-type germlines produced zygotes with nearly normal levels of survival, whereas progeny of ced-3 or ced-4 mutants displayed moderate levels of embryonic lethality (Figure 2D). At higher doses of irradiation, similar levels of IR-induced lethality were observed for wild type and for ced mutants. Given that the hypersensitivity of clk-2 mutant germlines was similar to that of ced-3 and ced-4 (Figure 2D), the strong radiation-induced apoptosis defects of the clk-2 alleles may account for their hypersensitivity to IR (Ahmed et al. 2001). While apoptosis did clear the germline of some nuclei with critical levels of DNA damage (Figure 2D) (Bhalla and Dernburg 2005), deficiency for cep-1/p53, ced-3, or ced-4 had no effect on the frequency of spontaneous unc-58(e665) revertants (Figure 3A, Tables 2 and 3). These results agree with the lack of a Mutator phenotype for clk-2(qm37), which is deficient for DNA-damage-induced apoptosis (Figure 3A, Table 2). Similarly, studies of p53 mouse mutants have indicated that defects in DNA-damage-induced apoptosis do not affect the frequency of spontaneous mutation in mammals (Nishino et al. 1995; Buettner et al. 1997; Reliene and Schiestl 2003).
hus-1(op241) was originally identified in a genetic background containing him-7(e1480), which confers a weak chromosome-loss phenotype suggestive of a defect in genome stability (Hodgkin et al. 1979; Gartner et al. 2000). As a control, we determined the effect of him-7 on the frequency of spontaneous mutation. him-7 was separated from the hus-1(op241) mutation, and him-7;unc-58(e665) double mutants were constructed. him-7 displayed a frequency of spontaneous mutation similar to that of wild type (Figure 3A, Table 2), indicating that it plays little or no role in the repair of endogenous DNA damage. In agreement with this possibility, him-7 germlines displayed normal sensitivity to ionizing radiation (data not shown).
Molecular analysis of unc-58 mutations:
Given that most of the spontaneous visible mutations isolated from the mrt-2 and clk-2(mn159) backgrounds were deletions, the molecular nature of intragenic unc-58 mutations from the Mutator assays was determined. Eleven/twelve, 3/5, and 12/19 intragenic unc-58 revertants derived from mrt-2, hus-1, and clk-2(mn159) backgrounds, respectively, were deletions (Figure 4; data not shown). In contrast, clk-2(qm37) and wild-type backgrounds did not give rise to deletions (n = 5 and 6, respectively), as expected from numerous spontaneous C. elegans mutations that have been previously isolated and sequenced (Pulak and Anderson 1988). Thus, checkpoint defects can trigger an increase in the frequency of spontaneous deletions. cDNA sequencing has defined long and short transcripts from the unc-58 locus, where the longer transcription product contains two additional 5′ exons (Figure 4A) (http://www.wormbase.org). However, all of the intragenic unc-58(e665) deletions that were recovered eliminated segments of the short unc-58 transcript, suggesting that it may be responsible for the dominant Unc phenotype of unc-58(e665).
Deletions can result from DNA double-strand breaks that are healed either by homology-driven recombination or by nonhomologous end-joining (NHEJ). Canonical NHEJ seals breaks that have little or no homology via proteins that include DNA ligase IV, XRCC4/Lif1, and the Ku heterodimer (Lieber et al. 2003). An alternative noncanonical NHEJ pathway known as microhomology-mediated end-joining relies either on a short patch of microhomology or on insertion of short DNA sequences to seal a DSB (Feldmann et al. 2000; Ma et al. 2003; Yu and Gabriel 2003; McVey et al. 2004b). The breakpoints of 11 mrt-2 and 6 clk-2(mn159) deletions of the unc-58 locus were mapped using PCR and sequenced. Deletions from both mutant backgrounds primarily consisted of either small deletions between 0.5 and 2 kb or large deletions between 10 and 35 kb (Figure 4). Molecular analysis of the breakpoints of these unc-58 deletions revealed 7- to 47-bp insertions at breakpoints for 3/6 deletions from the clk-2(mn159) background and 1- to 199-bp insertions at breakpoints for 4/11 deletions from the mrt-2 background. In addition, short patches of microhomology >1 nucleotide were apparent at breakpoints for 3/6 clk-2(mn159) deletions and for 4/11 mrt-2 deletions (Tables 4 and 5). Thus, microhomology-mediated end-joining may be responsible for many of the recovered deletions, which would be consistent with observations that lig-4-mediated NHEJ is repressed in the C. elegans germline (Martin et al. 2005; Clejan et al. 2006).
TABLE 4.
Breakpoints of unc-58 deletions from a clk-2(mn159) background
| unc-58(e2827) |
| CCGGAAAGATATCGAAAGGATCAGGATCCAAACCA..3993..gaatcttcgctcggctatgaagcaagttgcgtctg |
| agattccactaattatttgtttttaatgtaaacgt..25682.GAATGGTTTTTGTTATCAATTGAAACAGTCTCCGT |
| unc-58(e2828) |
| TTCGGGACACGATGGTGGAACTGGAGCTTCGAGTT..4523..ggactggtattaaacacgcaggattgccatggga |
| TTTTTAGT (insert) |
| tagatagagcattccgtgataattttcattccgtg..39000.AAAATTTTCATTCCGTGATAATTTTCATTCCGTGA |
| (direct repeats) |
| unc-58(e2829) |
| GTGTGAACAAGAGTCTTCAAAATTCTGAATTATTC..13421..accagaatacttctcgagattcttcaatttatttt |
| acagaagaaacaggtgaagcacatagctccaagaa..14020..ACCGGTAGGCGGTTGTGATCCGGTCCATAGTTCTT |
| unc-58(e2830) |
| CTCGAACCCATTTTTTTGATGGGATACTGTAACTT.-12374..tgtgacatcaactctcacacagcccaccgccaatt |
| ATTCCTAAAGTAGTGAATTCTGGTGAAAGTAGTGAATTCTGTGAATT (insert) |
| accgaaaactttcaattaactaccgttttgctcga..10051..CCAAAGTAGTGAATTCTGGTGAAGTACGCCTTGAG |
| unc-58(e2831) |
| GTTGAATCAATAAACCAAAATAGGTACCTATTGGG..11302..aaccaaagtttctcttcagttccgtcgacaccaag |
| TGCTACT (insert) |
| tcggcagctgttatgaacataagtggtataccaac..11520..AACGCGAATGCTACTGAGAAAACCTTTCCGACGTC |
| unc-58(e2832) |
| AATAACTGGCAGGCAAGTTTTTGTTTCCTTATTGT..9055..gaggtacattttcaacagttctatgatttcaattg |
| caagaaaccggtaggcggttgtgatccggtccata..14048.GTTCTTGATTCTCATCTATTATGAAACGGAGCTGT |
Sequence is annotated as for Table 1. Positions relative to the cosmid T06H11 are indicated.
TABLE 5.
Breakpoints of unc-58 deletions from a mrt-2 background
| unc-58(e2815) |
| AACCTGACAATAATAAATTTTAATTATTTCTGAAT..11665..tatgaatcaacaataccgcaagtagttaacacagt |
| cgaaggcaatgttgcgaaaacggcaagagtcagaa..13229..TATTTTGAGAGCGCTGATCATGCGTTCTTTTCTAA |
| unc-58(e2816) |
| ATTCAAAAATTATAATGAAAGATAAAAGAGAAAAA..9418..gttgttcgcatgagtcgcaaacaagattgggcaaa |
| (polypurine tract) |
| TTT (insert) |
| aaaccataatttcgttcgttcaccatttggaactg..11192..TTAGGATACGATATTTCAATTATGATTTAAATAA |
| (hairpin) |
| unc-58(e2817) |
| CTGTTAGGATACGATATTTCAATTATGATTTAAAT..8919..gtaatttgttaccagctcttactatcaagtaggct |
| atcgcccccagtaaagcaacgatttctttgagttc..9976..CTCACTCATTCCTAAGAATCTGTAACATTCGAAAT |
| (hairpin) |
| unc-58(e2818) |
| TCCTTACCGATCGGAAACCATAATTTCGTTCGTTC..11177..accatttggaactgttaggatacgatatttcaatt |
| (hairpin) |
| taattcaggtaaaagaatgttactataagatgttt..11996..CCCTGAAAGAAATCAAACACACAGTTACATTATAA |
| unc-58(e2819) |
| ATTCCGCGTCCATCATTAAAGAACACTCTTTGATA..-2818..atatcaaagacacgtgatatgcgtatggcaaacat |
| T (insert) |
| ctacagtaccccgactatatccctacactaagccc..15255..AACTCACATTCCCTCCAGAAGCCAAAACTTCATAG |
| unc-58(e2820) |
| AAACTTTCAATTAACTACCGTTTTGCTCGACCAAA..10055..gtagtgaattctggtgaagtacgccttgagatgtg |
| (hairpin) |
| atttagatgcgtaaaactaatataagtaaccagcc..15117..GTGGACCGCACCTCCGGCGCGGCCCCGACTTCTGG |
| (hairpin) |
| unc-58(e2821) |
| TAGAATTACATACTAGTTACATAACCTTTGCGGCC..9791..ccatcagtaaaaacgtaccagaagatgtttcgtca |
| T (insert) |
| gtgttattattttctatcaaactcactgctccttg..42818..ATATATATTTCCAGCCAGCGAAACTCCCACCATTT |
| unc-58(e2822) |
| AAAGCAGGTTTTTGTTGTTCCCGTTTTCAAGTTGT..9378..cagaaattcaaaaattataatgaaagataaaagag |
| (hairpin) |
| gacagaaacggaaaaatactttaataaaacagcaa..10966..GATAGATGACATTGATAGACAAGAGTTGTATGTTA |
| unc-58(e2824) |
| TGGGAAGGACTTGACCCAGCCAACAAGAAGGTTAT..7656..cattctgggaggcggagacacggctactgactgta |
| ataaatattattgtgaaaaaaaaatcaaatgtgca..34517..GAAAAAATGCTCAAAGAAACCCTTTGTTTAAAGAT |
| (direct repeat) |
| unc-58(e2825) |
| TATGTGTGAACAAGAGTCTTCAAAATTCTGAATTA..13421..tccaccagaatacttctcgagattcttcaatttat |
| TCCAATGAACCAAATTTGTAGAGTATTTTGCTAAAGATAGTCTAACTGGAATTCGATCTGCTATTAAAAACCAAAAAATAGAAACA AAACAGAATGGTCCCATGGAATCCAAATATTAAAAAACCAGCCGTGGACCGCACCTCCGGCGCGGCCCCGCGACTTCTGAGGCTGA AAACTAATTTTTCTGAAACTACCGTAA (insert) |
| (hairpin) |
| ggggttgaaaactaatttttctgaaactaccgtaa..15109..TCATACAGCACTCCTACCGTAACCCTATTGTACCA |
| unc-58(e2826) |
| TTAAATCTTATTTTAAGCCTTGACAATCTGAATAT..10280..AAAtaaactctaaaatattgcacaaatatttgccg |
| gatgacattgatagacaagagttgtatgttaattg..11005..aaaCAAACAACACTCAAAGAAGACATAATCATTGT |
| (hairpin) |
Studies of deletions that result from NHEJ of DSBs induced at specific sites in the genome have revealed that one end of the DSB can be protected from degradation while the other end is resected, which may aid the search for regions of microhomology that can be used for end-joining (Yu and Gabriel 2003). Therefore, the breakpoints of unc-58 deletions were examined for sequences that might explain spontaneous DSB induction. Several deletions that occurred in strains deficient for either mrt-2 or clk-2 displayed unusual qualities at their breakpoints. The clk-2(mn159)-induced e2828 deletion contained three direct AATTTTCATTCCGTGA repeats at one breakpoint, where the breakpoint occurred between the first and second repeats (Table 4). Similarly, the mrt-2-induced e2824 deletion had two direct GAAAAAAAAATCAAA repeats at one breakpoint, which occurred between the direct repeats, and the mrt-2-induced e2816 deletion had a 30-bp polypurine-rich tract adjacent to one breakpoint (Table 5). Such sequences have been proposed to form unusual structures during DNA replication that may trigger replication fork collapse and DSB formation (Bacolla et al. 2004).
Two groups of deletions had breakpoints that occurred near one another. The first group involved e2816, e2818, e2822, e2826, and e2831, all of which had one breakpoint that occurred within a 350-bp interval. However, no obvious sequence anomalies were recognized in this area, and the e2816 deletion had an unusual and potentially unstable polypurine sequence located at its other breakpoint. The second group of deletions, e2819, e2820, and e2825, all had one breakpoint located within a 240-bp repetitive element that corresponds to a nonautonomous Helitron transposon, which is thought to transpose via rolling circle replication (Figure 4B) (Kapitonov and Jurka 2001). One breakpoint of e2820 corresponded precisely to the terminus of a small 12-bp hairpin that is diagnostic of the 3′-ends of Helitron transposons (Arkhipova and Meselson 2005), whereas one breakpoint of e2825 was just upstream of this hairpin (Table 5). The e2825 deletion breakpoint contained an insertion that was identical to the 3′-end of a homologous Helitron transposon and 121 bp of adjacent unique sequence found on cosmid C11G10, which is located 20 MU to the right of unc-58 on chromosome X. Although the Helitron transposons that lie within C11G10 and the unc-58 locus share identical hairpin and 3′-end sequences, polymorphisms exist immediately 5′ to the hairpin that suggest that the genesis of e2825 involved a recombination event that occurred between 15 and 45 bp upstream from the 3′ hairpin. Thus, five unc-58 deletions that were identified in checkpoint-defective backgrounds contained either a polypurine tract or direct or inverted repeats at or near their breakpoints.
Given the unusual sequences observed at some deletion breakpoints, genome sequence corresponding to 35 bp upstream and downstream of each deletion breakpoint identified in this study was assessed for its propensity to form secondary structures using the Mfold program (Zuker 2003). Of 44 sequences corresponding to 22 deletion breakpoints, 10 were predicted to form hairpins >8 bp in length. In comparison, of 68 contiguous 70-bp segments of sequence corresponding to the genomic DNA of the short unc-58 transcript, 4 were predicted to fold into hairpins >8 bp (P = 0.018, Fisher exact test). Of the 20 unc-58 breakpoints that fell within the genomic region of the short unc-58 transcript, 7 could fold into hairpins >8 bp in length, which was also significantly different from the frequency observed for 68 contiguous segments of sequence from the same interval (P = 0.002, Fisher exact test). Thus, breakpoints of spontaneous mutations that occurred in checkpoint-defective backgrounds were enriched for sequences that could form hairpins.
Sequencing of spontaneous mrt-2-induced visible mutations revealed that G-tracts flanked two of the four breakpoints (Table 1). Three G-tracts span the 5′-end of the unc-58 gene, but no G-tract-induced deletions were recovered among 17 unc-58 deletions examined, even though several of these deletion events removed one or more of the G-tracts (Figure 4A, Tables 2 and 3). Thus, the G-tracts adjacent to breakpoints of deletions that occurred in the mrt-2 background may have been fortuitous, although G-tracts greater than nine nucleotides in length are rare in the C. elegans genome (Figures 1 and 4; S. Ahmed, unpublished data). C. elegans dog-1 (deletion of guanine tract-1) mutants display small deletions where a G-tract always flanks one of the breakpoints, and previous PCR-based analysis revealed that some, but not all, G-tracts >22 bp in length are capable of generating deletions (Cheung et al. 2002). Therefore, the G-tracts in the unc-58 gene may be of the “stable” variety, and some caution may be warranted when assessing the relevance of the mutation spectra obtained at this locus.
To determine if mrt-2 displays a general defect in G-tract stability, genomic DNA from mrt-2 mutants was examined at two G-tract loci that were previously shown to be highly unstable in the dog-1 background: F55F3 (tract length of C26) and F38A6 (tract length of C29). In addition, a third locus, Y41E3 (tract length of C17), for which no deletions were previously observed, was examined. PCR-based deletion products were not observed for any of these loci using either mrt-2 or dog-1 genomic DNA as a template (n = 30 for each genotype) (data not shown), despite extensive optimization of PCR conditions and robust wild-type-sized PCR products. Thus, this PCR-based assay may vary from lab to lab (M. Tijsterman and R. Plasterk, personal communication), and we were unable to confirm that the mrt-2 mutant has a general defect in G-tract stability using this PCR-based approach.
clk-2(qm37) but not clk-2(mn159) has an extended life span:
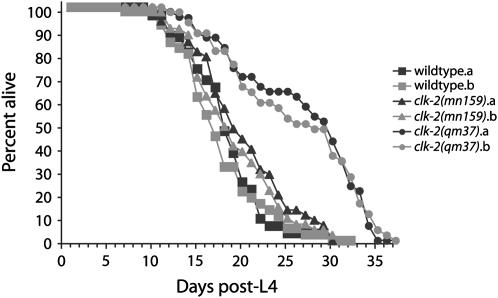
unc-58(e665) Mutator assays suggested that clk-2(mn159) had a deletion-based genome stability defect whereas clk-2(qm37) did not (Figure 3A). Interestingly, clk-2(qm37) has checkpoint defects that are modestly more severe than those of clk-2(mn159) (Figure 2), and the clk-2(qm37) mutant has a stronger Slow Growth phenotype than clk-2(mn159) (Ahmed et al. 2001). Given the phenotypic disparity between the clk-2 alleles, and given that clk-2(qm37) has been reported to confer an extended life span (Lakowski and Hekimi 1996), we sought to determine how strains of clk-2(qm37) and clk-2(mn159) compared with respect to their effects on longevity. Life spans of independently outcrossed clk-2(qm37) strains (a = 30.4 ± 1.4 days; b = 29.4 ± 1.6 days) were significantly greater than either clk-2(mn159) (a = 21.1 ± 0.8 days; b = 20.3 ± 0.8 days) or N2 wild type (a = 20.7 ± 0.9 days; b = 21.4 ± 1.0 days) strains (P < 0.002 in all cases; log-rank Mantel Cox test), whereas clk-2(mn159) strains were not significantly longer lived than wild-type strains (P = 0.67, 0.84, 0.62, and 0.21 for comparisons with a and b wild-type strains, respectively), revealing a fourth difference between these two clk-2 alleles (Figure 5). Thus, the clk-2 mutations qm37 and mn159 have distinct properties with respect to all phenotypes examined, as might be expected for hypomorphic alleles of an essential gene.
Figure 5.—
clk-2(qm37) but not clk-2(mn159) is long lived. Life spans of strains that were independently crossed nine times vs. a dpy-17;unc-32 marker strain (n = 100 nematodes scored for each strain).
DISCUSSION
Our results indicate that DNA damage response genes are required to suppress the frequency of spontaneous mutation in C. elegans. Of six spontaneous mutations isolated from the mrt-2 background and one spontaneous mutation isolated from the clk-2(mn159) background, all but one occurred as a consequence of deletions (Figure 1, Table 1). In addition, 26/36 unc-58 mutations identified in mrt-2, hus-1, or clk-2(mn159) genetic backgrounds were deletions, whereas no deletions were observed for 11 unc-58 mutations in wild-type or clk-2(qm37) backgrounds. In agreement with these observations, a previous study revealed that 11% of spontaneous mutations in the C. elegans unc-54 gene were found to be deletions, indicating that deletions are rare events (Pulak and Anderson 1988). Similarly, the frequency of spontaneous deletions segregating in human families corresponds to 5–10% of all mutations (Verhoog et al. 1998; Puget et al. 1999; Cooper 2002).
A recent study in Schizosaccharomyces pombe indicated that the frequency of spontaneous deletions that occurred as a consequence of DNA polymerase α-mediated replication stress were suppressed by hypomorphic alleles of rad17 (Kai and Wang 2003). These results are surprising as rad17 encodes a component of the replication factor C complex that loads the 9-1-1 complex onto DNA damage, so mutation of 9-1-1 complex subunits in C. elegans might be expected to suppress rather than promote the formation of spontaneous deletions. Note, however, that deficiency for Cds1/Chk2, an effector kinase that acts downstream of the 9-1-1 complex, dramatically enhanced the frequency of deletions, but not of point mutations, in response to DNA polymerase α-mediated replication stress (Kai and Wang 2003). Therefore, the deletions observed in our study may ultimately reflect malfunction of downstream signaling components of the DDR pathway such as Chk1 or Chk2.
A genomewide RNA interference (RNAi) screen recently identified mrt-2 as a gene that may suppress small deletions or insertions of one or more nucleotides in a repetitive array of an out-of-frame LacZ reporter construct in a manner that is independent of mismatch repair (Pothof et al. 2003). However, the majority of mutations identified in the checkpoint-defective backgrounds used in our study were spontaneous deletions large enough to be detected by agarose gel electrophoresis. The other mutations may have been point or frameshift mutations. The dpy-31 mutation isolated from the mrt-2 background resulted from a T-to-C point mutation (Novelli et al. 2004), and two additional mrt-2-induced spontaneous mutations at the unc-93 locus arose as a consequence of a T-to-C transition and a T-to-G transversion (data not shown). Thus, limited sequence analysis failed to reveal frameshifts arising from the mrt-2 background. The restoration of reading frame by mrt-2(RNAi) may therefore have occurred as a consequence of larger deletions within the repetitive LacZ array. However, a more thorough examination of point or frameshift mutations from mrt-2 or other checkpoint-defective backgrounds may reveal increases in the frequency of frameshifting. Note that deficiency for the 9-1-1 complex or downstream mediator checkpoint kinases such as Chk1/Chk2 has been shown to affect the frequency of frameshifting in response to replication stress in S. pombe (Kai and Wang 2003).
Although both clk-2(mn159) and clk-2(qm37) have strong defects for the S-phase DNA replication checkpoint that responds to stalled replication forks (Ahmed et al. 2001), only clk-2(mn159) displayed a Mutator phenotype (Figure 3A). This result was surprising because S-phase checkpoint defects in Saccharomyces cerevisiae typically confer high frequencies of spontaneous GCRs relative to defects in the 9-1-1 complex (Myung et al. 2001a,b). The pleiotropic nature of clk-2 mutations extends to additional phenotypes, such as growth rate and life span, where only the qm37 allele has pronounced effects (Figure 5) (Lakowski and Hekimi 1996; Ahmed et al. 2001). The life span of clk-2(mn159) was not significantly different from that of wild type (Figure 5), indicating that neither a strong S-phase checkpoint defect (Ahmed et al. 2001) nor an enhanced frequency of endogenous mutation is sufficient to affect C. elegans life span. Note, however, that deficiency for several additional S-phase checkpoint genes has recently been shown to confer an extended life span in C. elegans (Olsen et al. 2006) and that the differential effects of the mn159 and qm37 alleles of clk-2 on longevity may provide an opportunity to more precisely determine the relationship between the S-phase checkpoint and life span.
Frequencies of spontaneous mutation were 8- to 15-fold higher for hus-1 and mrt-2 mutants than for wild type (Figure 3). These mutation frequencies mirror the 7- to 10-fold increases in GCRs observed for S. cerevisiae rad9 and rad17 mutants (homologs of hus-1 and mrt-2, respectively) and for mutations that affect the S. cerevisiae rad51 homologous recombination (HR) pathway (Myung et al. 2001b). If endogenous DSBs are not accurately repaired by checkpoint-orchestrated HR, which usually relies on a sister chromatid for repair, then resection and end-joining of DSBs may occur (Yu and Gabriel 2003), as observed for many mutations that occurred in checkpoint-defective backgrounds in this study. One unc-58 deletion breakpoint, e2825, occurred near the 3′-end of a nonautonomous Helitron transposable element and was unique because sequence from a homologous repetitive element elsewhere on chromosome X was inserted at the e2825 breakpoint (Table 5). Thus, the e2825 deletion may have been created by a break in the Helitron transposable element at the unc-58 locus, strand invasion of a homologous transposon on C11G10, and dissociation and end-joining at the unc-58 locus to seal the break. These results suggest that C. elegans checkpoint-defective mutants may be deficient for a model of HR-dependent break repair known as synthesis-dependent strand annealing, which may involve processive cycles of template-directed DNA synthesis followed by dissociation to search for homology with the other end of the break (Nassif et al. 1994; McVey et al. 2004a). Recombination events similar to that of the e2825 deletion have been observed for DNA sequences flanking P elements in Drosophila (Tsubota and Huong 1991; Heslip et al. 1992). In addition, studies in S. cerevisiae have indicated that DSBs in Ty retrotransposons can result in gene conversion (Parket et al. 1995) and translocations (Lemoine et al. 2005), and recombination between Ty elements represents a major source of genome rearrangements (Dunham et al. 2002). Roughly 2% of the C. elegans genome is composed of autonomous and nonautonomous Helitron elements (Kapitonov and Jurka 2001), although their present activity is currently uncertain.
For two of five spontaneous deletions that gave rise to visible mutant phenotypes in the mrt-2 background, a G-tract was present immediately adjacent to one of the breakpoints. The DOG-1 helicase has been shown to suppress deletions at such G-tracts, and one model suggests that this helicase may resolve unusual non-Watson–Crick structures formed by G-tracts during replication of the lagging DNA strand prior to DSB induction (Cheung et al. 2002). Given that similar deletions were recovered from a mrt-2 background (Table 1), the MRT-2 protein may help to recruit DOG-1 to resolve unusual G-rich structures prior to DSB induction. Alternatively, both mrt-2 and dog-1 may act downstream of DSB induction to facilitate synthesis-dependent strand-annealing-mediated DSB repair. Note that the mammalian DOG-1 homolog BRIP1/BACH1 has been implicated in the response to interstrand crosslinks that are processed via a DSB intermediate (Magana-Schwencke et al. 1982; Bridge et al. 2005). The sizes of the deletion events observed in the absence of dog-1 were only several hundred base pairs in length (Cheung et al. 2002), whereas the events observed at mrt-2 G-tract deletions were several kilobases and most deletions in the mrt-2, hus-1, and clk-2(qm37) backgrounds were >300 bp. Thus, it is possible that the DDR mutants used in this study have an additional defect that renders DSBs more susceptible to exonuclease-mediated degradation prior to end-joining. Alternatively, the initial visible mutants isolated from the dog-1 background may have represented the short end of the dog-1 deletion spectrum (Cheung et al. 2002). In summary, our genetic results suggest that dog-1 and mrt-2 may act to repress genome instability caused by G-rich non-Watson–Crick DNA structures such as G-quadruplexes.
Several unc-58 deletions had unusual sequences at or near their breakpoints, including short direct repeats, short hairpins, and long G/A-rich polypurine tracts. Thus, the initiation of a spontaneous deletion event may involve stretches of nucleotides with propensities to form slipped intermediates, which may be vulnerable to endonucleolytic attack and result in replication fork stalling and collapse. Stalled replication forks have been shown to result in DSB formation in Escherichia coli (Michel et al. 1997), which may lead to exonuclease-mediated DSB resection and end-joining if checkpoint-mediated homologous recombination fails to resolve the lesion (Moore and Haber 1996; Yu and Gabriel 2003; Guirouilh-Barbat et al. 2004). Note that polypurine tracts and direct or inverted repeats have been observed at genome rearrangement breakpoints in bacteria, yeast, and humans (Abeysinghe et al. 2003; Chuzhanova et al. 2003; Bacolla et al. 2004). The biological relevance of such structures in vivo is emphasized by the two unc-58 deletions with breakpoints at or near a stem loop at the 3′-end of a Helitron transposable element. This 3′ stem loop is conserved in other Helitron transposons, is essential for transposition, and may facilitate termination of rolling circle replication by direct inhibition of the DNA replication machinery (Mendiola et al. 1994; Kapitonov and Jurka 2001; Ton-Hoang et al. 2005). This suggests a model where the Helitron stem loop may be sufficient to induce spontaneous DSBs at a low frequency. As hairpins were significantly enriched for near deletion breakpoints, such sequences may be relevant to the genesis of DSBs or may aid in sealing resected breaks. Although some deletion breakpoints that we identified were close to short direct repeats, short hairpins, and long G/A-rich polypurine tracts, potential roles for these sequences in the genesis of chromosome rearrangements in C. elegans may be bolstered by analysis of many more spontaneous unc-58 deletion events or by transgene reporter assays (Pothof et al. 2003).
Our data suggest that DNA damage response proteins help to maintain genome stability by coordinating proper HR-mediated repair of spontaneous DSBs, some of which are likely to occur as a consequence of unusual nucleic acid structures that form during DNA replication (Bacolla et al. 2004; Lovett 2004). Defects in DNA damage response proteins such as ATM, ATR, BRCA1, BRCA2, MRE11, and NBS1 result in familial cancer syndromes and confer GCRs such as inversions, translocations, or large deletions suggestive of DSB repair defects (Venkitaraman 2002). A recent study of BRCA1 and BRCA2 tumors revealed a number of large hemizygous deletions based on comparative genome hybridization using microarrays with an average resolution of 1 Mb (Jonsson et al. 2005). Further, the second allele of BRCA1 or BRCA2 is almost always inactivated by genomic rearrangements or deletions in cancers that develop in patients who are heterozygous at one of these loci (Welcsh and King 2001; Hendrickson et al. 2005). Although we recovered one visible mutant from a mrt-2 background with an ∼230-kb deletion, mrt-2 and clk-2(mn159) defects resulted in 22 additional spontaneous deletions that ranged from 0.15 to 35 kb. Note that the unc-58 target gene is located on the X chromosome, which harbors few essential genes, and that the closest genes annotated to possess sterile or lethal phenotypes were located 71 kb to the left of and 101 kb to the right of unc-58 (http://www.wormbase.org). Thus, small- to medium-sized deletions occur more frequently than large chromosomal aberrations in checkpoint-defective C. elegans mutants. Gross changes to chromosomes that are visible by microscopy and microsatellite analyses may contribute to the development of cancer. However, our results suggest that genomes of cancer cells that evolve in backgrounds with DNA damage response defects may be peppered with small- to medium-sized deletions and that the resulting aberrant gene products or hemizygosity may be a driving force in the development of cancer.
Acknowledgments
We thank M. de Bono for assistance with sequencing lon-2, A. Gartner for strains, P. Lansdorp for technical advice, A. Carr for encouragement, J. Stough for discussion, members of the Ahmed lab for discussion and critical reading of the manuscript, B. Johnson, T. Zucchero, D. Walker, and J. Boerckel for technical assistance, and the C. elegans Genetics Center for providing strains. I.C. is the recipient of a University of North Carolina Lineberger Comprehensive Cancer Center postdoctoral fellowship. This work was supported in part by the United Kingdom Medical Research Council. J.H., M.L., and S.A. were supported by National Institutes of Health grant GM066228.
References
- Abeysinghe, S. S., N. Chuzhanova, M. Krawczak, E. V. Ball and D. N. Cooper, 2003. Translocation and gross deletion breakpoints in human inherited disease and cancer I: nucleotide composition and recombination-associated motifs. Hum. Mutat. 22: 229–244. [DOI] [PubMed] [Google Scholar]
- Ahmed, S., and J. Hodgkin, 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403: 159–164. [DOI] [PubMed] [Google Scholar]
- Ahmed, S., A. Alpi, M. O. Hengartner and A. Gartner, 2001. C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 11: 1934–1944. [DOI] [PubMed] [Google Scholar]
- Anderson, P., 1995. Mutagenesis. Methods Cell Biol. 48: 31–58. [PubMed] [Google Scholar]
- Arkhipova, I. R., and M. Meselson, 2005. Diverse DNA transposons in rotifers of the class Bdelloidea. Proc. Natl. Acad. Sci. USA 102: 11781–11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi, L. D., 2005. The role of p53-mediated apoptosis as a crucial anti-tumor response to genomic instability: lessons from mouse models. Mutat. Res. 569: 145–157. [DOI] [PubMed] [Google Scholar]
- Bacolla, A., A. Jaworski, J. E. Larson, J. P. Jakupciak, N. Chuzhanova et al., 2004. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. USA 101: 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudez, V. P., L. A. Lindsey-Boltz, A. J. Cesare, Y. Maniwa, J. D. Griffith et al., 2003. Loading of the human 9–1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc. Natl. Acad. Sci. USA 100: 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla, N., and A. F. Dernburg, 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310: 1683–1686. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., A. Gartner, J. Reboul, P. Vaglio, N. Dyson et al., 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127–131. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge, W. L., C. J. Vandenberg, R. J. Franklin and K. Hiom, 2005. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 37: 953–957. [DOI] [PubMed] [Google Scholar]
- Brodsky, M. H., B. T. Weinert, G. Tsang, Y. S. Rong, N. M. McGinnis et al., 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 24: 1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzowska, M., I. Jaspers, J. Essers, H. De Waard, E. Van Drunen et al., 2004. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 23: 3548–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner, V. L., H. Nishino, J. Haavik, A. Knoll, K. Hill et al., 1997. Spontaneous mutation frequencies and spectra in p53 (+/+) and p53 (−/−) mice: a test of the ‘guardian of the genome’ hypothesis in the Big Blue transgenic mouse mutation detection system. Mutat. Res. 379: 13–20. [DOI] [PubMed] [Google Scholar]
- Burtelow, M. A., S. H. Kaufmann and L. M. Karnitz, 2000. Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J. Biol. Chem. 275: 26343–26348. [DOI] [PubMed] [Google Scholar]
- Caspari, T., M. Dahlen, G. Kanter-Smoler, H. D. Lindsay, K. Hofmann et al., 2000. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20: 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, I., M. Schertzer, A. Rose and P. M. Lansdorp, 2002. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 31: 405–409. [DOI] [PubMed] [Google Scholar]
- Chuzhanova, N., S. S. Abeysinghe, M. Krawczak and D. N. Cooper, 2003. Translocation and gross deletion breakpoints in human inherited disease and cancer II: potential involvement of repetitive sequence elements in secondary structure formation between DNA ends. Hum. Mutat. 22: 245–251. [DOI] [PubMed] [Google Scholar]
- Clejan, I., J. Boerckel and S. Ahmed, 2006. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics 173: 1301–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, D. N., 2002. Human gene mutation in pathology and evolution. J. Inherit. Metab. Dis. 25: 157–182. [DOI] [PubMed] [Google Scholar]
- D'Andrea, A. D., and M. Grompe, 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3: 23–34. [DOI] [PubMed] [Google Scholar]
- Deng, C. X., and F. Scott, 2000. Role of the tumor suppressor gene Brca1 in genetic stability and mammary gland tumor formation. Oncogene 19: 1059–1064. [DOI] [PubMed] [Google Scholar]
- Derry, W. B., A. P. Putzke and J. H. Rothman, 2001. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294: 591–595. [DOI] [PubMed] [Google Scholar]
- Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown et al., 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, E., V. Schmiemann, W. Goedecke, S. Reichenberger and P. Pfeiffer, 2000. DNA double-strand break repair in cell-free extracts from Ku80-deficient cells: implications for Ku serving as an alignment factor in non-homologous DNA end joining. Nucleic Acids Res. 28: 2585–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse, T., and S. J. Boulton, 2005. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 24: 4345–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner, A., S. Milstein, S. Ahmed, J. Hodgkin and M. O. Hengartner, 2000. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- Gartner, A., A. J. MacQueen and A. M. Villeneuve, 2004. Methods for analyzing checkpoint responses in Caenorhabditis elegans. Methods Mol. Biol. 280: 257–274. [DOI] [PubMed] [Google Scholar]
- Griffiths, D. J., N. C. Barbet, S. McCready, A. R. Lehmann and A. M. Carr, 1995. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 14: 5812–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat, J., S. Huck, P. Bertrand, L. Pirzio, C. Desmaze et al., 2004. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14: 611–623. [DOI] [PubMed] [Google Scholar]
- Gumienny, T. L., E. Lambie, E. Hartwieg, H. R. Horvitz and M. O. Hengartner, 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Hartman, P. S., and R. K. Herman, 1982. Radiation-sensitive mutants of Caenorhabditis elegans. Genetics 102: 159–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, B. C., T. Judkins, B. D. Ward, K. Eliason, A. E. Deffenbaugh et al., 2005. Prevalence of five previously reported and recurrent BRCA1 genetic rearrangement mutations in 20,000 patients from hereditary breast/ovarian cancer families. Genes Chromosomes Cancer 43: 309–313. [DOI] [PubMed] [Google Scholar]
- Heslip, T. R., J. A. Williams, J. B. Bell and R. B. Hodgetts, 1992. A P element chimera containing captured genomic sequences was recovered at the vestigial locus in Drosophila following targeted transposition. Genetics 131: 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., 1974. Genetic and anatomical aspects of the Caenorhabditis elegans male. Ph.D. Thesis, Darwin College, Cambridge, UK.
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, E. R., S. Milstein, S. J. Boulton, M. Ye, J. J. Hofmann et al., 2002. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12: 1908–1918. [DOI] [PubMed] [Google Scholar]
- Hopkins, K. M., W. Auerbach, X. Y. Wang, M. P. Hande, H. Hang et al., 2004. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol. 24: 7235–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz, H. R., P. W. Sternberg, I. S. Greenwald, W. Fixsen and H. M. Ellis, 1983. Mutations that affect neural cell lineages and cell fates during the development of the nematode Caenorhabditis elegans. Cold Spring Harbor Symp. Quant. Biol. 48 (Pt. 2): 453–463. [DOI] [PubMed] [Google Scholar]
- Howlett, N. G., T. Taniguchi, S. Olson, B. Cox, Q. Waisfisz et al., 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297: 606–609. [DOI] [PubMed] [Google Scholar]
- Huang, L. S., P. Tzou and P. W. Sternberg, 1994. The lin-15 locus encodes two negative regulators of Caenorhabditis elegans vulval development. Mol. Biol. Cell 5: 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson, G., T. L. Naylor, J. Vallon-Christersson, J. Staaf, J. Huang et al., 2005. Distinct genomic profiles in hereditary breast tumors identified by array-based comparative genomic hybridization. Cancer Res. 65: 7612–7621. [DOI] [PubMed] [Google Scholar]
- Kai, M., and T. S. Wang, 2003. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 17: 64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov, V. V., and J. Jurka, 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98: 8714–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, M., A. Hirano, T. Kumano, S. L. Xiang, K. Mihara et al., 2004. Critical role for chicken Rad17 and Rad9 in the cellular response to DNA damage and stalled DNA replication. Genes Cells 9: 291–303. [DOI] [PubMed] [Google Scholar]
- Kostrub, C. F., K. Knudsen, S. Subramani and T. Enoch, 1998. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 17: 2055–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman-Van Der Zwet, M., W. J. Overkamp, R. E. Van Lange, J. Essers, A. Van Duijn-Goedhart et al., 2002. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 22: 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski, B., and S. Hekimi, 1996. Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272: 1010–1013. [DOI] [PubMed] [Google Scholar]
- Lemoine, F. J., N. P. Degtyareva, K. Lobachev and T. D. Petes, 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell 120: 587–598. [DOI] [PubMed] [Google Scholar]
- Levy, A. D., J. Yang and J. M. Kramer, 1993. Molecular and genetic analyses of the Caenorhabditis elegans dpy-2 and dpy-10 collagen genes: a variety of molecular alterations affect organismal morphology. Mol. Biol. Cell 4: 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber, M. R., Y. Ma, U. Pannicke and K. Schwarz, 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 4: 712–720. [DOI] [PubMed] [Google Scholar]
- Lovett, S. T., 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52: 1243–1253. [DOI] [PubMed] [Google Scholar]
- Lowe, S. W., E. Cepero and G. Evan, 2004. Intrinsic tumour suppression. Nature 432: 307–315. [DOI] [PubMed] [Google Scholar]
- Ma, J. L., E. M. Kim, J. E. Haber and S. E. Lee, 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23: 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magana-Schwencke, N., J. A. Henriques, R. Chanet and E. Moustacchi, 1982. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc. Natl. Acad. Sci. USA 79: 1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, J. S., N. Winkelmann, M. I. Petalcorin, M. J. Mcilwraith and S. J. Boulton, 2005. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol. Cell. Biol. 25: 3127–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K. S., C. Matheson, M. A. Marra, M. F. Wakarchuk and D. L. Baillie, 1994. The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol. Gen. Genet. 242: 346–357. [DOI] [PubMed] [Google Scholar]
- McVey, M., M. Adams, E. Staeva-Vieira and J. J. Sekelsky, 2004. a Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey, M., D. Radut and J. J. Sekelsky, 2004. b End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 168: 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, B., I. Clejan, Y. Liu, M. Lowden, A. Gartner et al., 2006. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PLoS Genet. 2: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola, M. V., I. Bernales and F. De La Cruz, 1994. Differential roles of the transposon termini in IS91 transposition. Proc. Natl. Acad. Sci. USA 91: 1922–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, B., S. D. Ehrlich and M. Uzest, 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16: 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J. K., and J. E. Haber, 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 2164–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan, M. E., J. W. Chiu, B. H. Koller and M. Jasin, 1999. BRCA1 controls homology-directed DNA repair. Mol. Cell 4: 511–518. [DOI] [PubMed] [Google Scholar]
- Moynahan, M. E., A. J. Pierce and M. Jasin, 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7: 263–272. [DOI] [PubMed] [Google Scholar]
- Myung, K., C. Chen and R. D. Kolodner, 2001. a Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076. [DOI] [PubMed] [Google Scholar]
- Myung, K., A. Datta and R. D. Kolodner, 2001. b Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408. [DOI] [PubMed] [Google Scholar]
- Nassif, N., J. Penney, S. Pal, W. R. Engels and G. B. Gloor, 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino, H., A. Knoll, V. L. Buettner, C. S. Frisk, Y. Maruta et al., 1995. p53 wild-type and p53 nullizygous Big Blue transgenic mice have similar frequencies and patterns of observed mutation in liver, spleen and brain. Oncogene 11: 263–270. [PubMed] [Google Scholar]
- Novelli, J., S. Ahmed and J. Hodgkin, 2004. Gene interactions in Caenorhabditis elegans define DPY-31 as a candidate procollagen C-proteinase and SQT-3/ROL-4 as its predicted major target. Genetics 168: 1259–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A., M. C. Vantipalli and G. J. Lithgow, 2006. Lifespan extension of Caenorhabditis elegans following repeated mild hormetic heat treatments. Biogerontology 7: 221–238. [DOI] [PubMed] [Google Scholar]
- Parket, A., O. Inbar and M. Kupiec, 1995. Recombination of Ty elements in yeast can be induced by a double-strand break. Genetics 140: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, K. J., V. P. Yu, H. Lee, A. Corcoran, F. C. Thistlethwaite et al., 1998. Involvement of Brca2 in DNA repair. Mol. Cell 1: 347–357. [DOI] [PubMed] [Google Scholar]
- Pennaneach, V., and R. D. Kolodner, 2004. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast. Nat. Genet. 36: 612–617. [DOI] [PubMed] [Google Scholar]
- Pothof, J., G. Van Haaften, K. Thijssen, R. S. Kamath, A. G. Fraser et al., 2003. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 17: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puget, N., D. Stoppa-Lyonnet, O. M. Sinilnikova, S. Pages, H. T. Lynch et al., 1999. Screening for germ-line rearrangements and regulatory mutations in BRCA1 led to the identification of four new deletions. Cancer Res. 59: 455–461. [PubMed] [Google Scholar]
- Pulak, R. A., and P. Anderson, 1988. Structures of spontaneous deletions in Caenorhabditis elegans. Mol. Cell. Biol. 8: 3748–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, J. B., 1989. Genetic analysis of the cha-1-unc-17 gene complex in Caenorhabditis. Genetics 122: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reliene, R., and R. H. Schiestl, 2003. Mouse models for induced genetic instability at endogenous loci. Oncogene 22: 7000–7010. [DOI] [PubMed] [Google Scholar]
- Sancar, A., L. A. Lindsey-Boltz, K. Unsal-Kacmaz and S. Linn, 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73: 39–85. [DOI] [PubMed] [Google Scholar]
- Schumacher, B., K. Hofmann, S. Boulton and A. Gartner, 2001. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11: 1722–1727. [DOI] [PubMed] [Google Scholar]
- Sulston, J., and J. Hodgkin, 1988. Methods, pp. 587–606 in The Nematode Caenorhabditis elegans, edited by W. B. Wood. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Ton-Hoang, B., C. Guynet, D. R. Ronning, B. Cointin-Marty, F. Dyda et al., 2005. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J. 24: 3325–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota, S. I., and D. V. Huong, 1991. Capture of flanking DNA by a P element in Drosophila melanogaster: creation of a transposable element. Proc. Natl. Acad. Sci. USA 88: 693–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman, A. R., 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108: 171–182. [DOI] [PubMed] [Google Scholar]
- Verhoog, L. C., C. T. Brekelmans, C. Seynaeve, L. M. Van Den Bosch, G. Dahmen et al., 1998. Survival and tumour characteristics of breast-cancer patients with germline mutations of BRCA1. Lancet 351: 316–321. [DOI] [PubMed] [Google Scholar]
- Weiss, R. S., T. Enoch and P. Leder, 2000. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 14: 1886–1898. [PMC free article] [PubMed] [Google Scholar]
- Welcsh, P. L., and M. C. King, 2001. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 10: 705–713. [DOI] [PubMed] [Google Scholar]
- Wolf, F. W., M. S. Hung, B. Wightman, J. Way and G. Garriga, 1998. vab-8 is a key regulator of posteriorly directed migrations in C. elegans and encodes a novel protein with kinesin motor similarity. Neuron 20: 655–666. [DOI] [PubMed] [Google Scholar]
- Woodford, K. J., R. M. Howell and K. Usdin, 1994. A novel K(+)-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J. Biol. Chem. 269: 27029–27035. [PubMed] [Google Scholar]
- Yu, X., and A. Gabriel, 2003. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics 163: 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky, J., A. J. MacQueen, J. B. Duffy, K. J. Kemphues and A. M. Villeneuve, 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L., D. Cortez and S. J. Elledge, 2002. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M., 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]