Abstract
Populations of mitochondria reside within individuals. Among angiosperms, these populations are rarely considered as genetically variable entities and typically are not found to be heteroplasmic in nature, leading to the widespread assumption that plant mitochondrial populations are homoplasmic. However, empirical studies of mitochondrial variation in angiosperms are relatively uncommon due to a paucity of sequence variation. Recent greenhouse studies of Silene vulgaris suggested that heteroplasmy might occur in this species at a level that it is biologically relevant. Here, we use established qualitative methods and a novel quantitative PCR method to study the intraindividual population genetics of mitochondria across two generations in natural populations of S. vulgaris. We show incidences of heteroplasmy for mitochondrial atpA and patterns of inheritance that are suggestive of more widespread heteroplasmy at both atpA and cox1. Further, our results demonstrate that quantitative levels of mitochondrial variation within individuals are high, constituting 26% of the total in one population. These findings are most consistent with a biparental model of mitochondrial inheritance. However, selection within individuals may be instrumental in the maintenance of variation because S. vulgaris is gynodioecious. Male sterility is, in part, regulated by the mitochondrial genome, and strong selection pressures appear to influence the frequency of females in these populations.
THE level of heterozygosity frequently considered in population genetic studies represents a measure of the within-individual component of genetic variation for biparentally inherited nuclear genes. In diploids within-individual nuclear variation is generally limited to two possible states, homozygote or heterozygote. In contrast, the intraindividual component of genetic variance with regard to the mitochondrial genome has the potential to be far more complex because entire populations of mitochondria reside within individuals and are further subdivided among cells (Mackenzie and McIntosh 1999; Birky 2001; Rand 2001). Because of this, the magnitude of within-individual variation could potentially be considered a continuous trait. However, in flowering plants within-individual populations of mitochondria are widely assumed to be genetically invariant (homoplasmic), largely as a consequence of strict maternal inheritance.
With uniparental inheritance, mutation represents the main source of novel genetic variation within evolutionarily independent lineages because there is no opportunity for admixture. The thinking is that homoplasmy is enforced by the successive cell divisions that represent sequential founder events or bottlenecks for the mitochondrial population (Birky 2001; Rand 2001). As a result, co-occurrence of genetically distinct mitochondrial genomes within an individual (heteroplasmy) should be rare. Further, these rare and brief periods of heteroplasmy should be difficult to detect with sequence data, as new mutants should be nearly identical to the primary matrilineal haplotype (Avise et al. 1985; Takahata and Palumbi 1985). Thus, documentation of mitochondrial heteroplasmy in natural plant populations would suggest that either inheritance is not strictly maternal, with the heteroplasmic state maintained by at least occasional biparental inheritance, or that heteroplasmy is actively maintained by some within-individual process, such as selection favoring heteroplasmic cells or different mitochondrial haplotypes in different tissues.
The question of whether homoplasmy and strict maternal inheritance of the mitochondrial genome are the norm in flowering plants is relevant to several issues in plant evolutionary biology, including the utility of mitochondrial markers for studying gene flow by seeds (Petit et al. 2005), the impact of mitochondrial maternal inheritance and heteroplasmy on cytonuclear coevolution and conflict (Wade and McCauley 2005), the utility of cytoplasmic genes for phylogenetic studies (Barkman et al. 2000; Wolfe and Randle 2004), maintenance and measurement of genetic diversity (Birky et al. 1983, 1989; Rand and Harrison 1989), and issues related to the role of recombination in the evolution of the plant mitochondrial genome (Städler and Delph 2002). Should heteroplasmy occur in natural plant populations at levels greater than currently thought, it would be important to document its origins, maintenance, and mode of transmission.
While there are a small number of examples in angiosperms of biparental inheritance of the mitochondrial genome and/or mitochondrial heteroplasmy derived from experimental crosses (Reboud and Zeyl 1994; Mogensen 1996; Korpelainen 2004), direct evidence for heteroplasmy in natural populations of angiosperms is virtually nonexistent (Barr et al. 2005). Lack of such documentation may be due, in part, to the lack of polymorphic genetic markers that results from low levels of sequence variation observed in the mitochondrial genomes of many plant species (Wolfe et al. 1987; Palmer et al. 2000).
Two gynodioecious species in the genus Silene (Caryophyllaceae) provide notable exceptions to the general lack of mitochondrial sequence variation observed in most taxa (Städler and Delph 2002; McCauley et al. 2005). Gynodioecy is a mating system in which hermaphrodites and females co-occur, and gender is typically regulated by a cytonuclear interaction involving cytoplasmic male sterility (CMS) factors associated with the mitochondrial genome (Schnable and Wise 1998; Dudle et al. 2001). The relative number of females within gynodioecious populations is thought to be under frequency-dependent selection (Frank 1989; McCauley and Brock 1998), and it is generally believed that this selective force is responsible for maintaining mitochondrial variation that would otherwise be lost in these gynodioecious populations (McCauley and Taylor 1997; Laporte et al. 2001; Bailey et al. 2003; McCauley and Olson 2003; Byers et al. 2005).
Several recent studies have indicated that mitochondrial heteroplasmy could be relatively common in this genus. Observations consistent with intragenic recombination of the mitochondrial genome in Silene acaulis were considered by Städler and Delph (2002) to be indirect evidence for heteroplasmy in that species. Andersson (1999) has provided evidence that offspring sex ratios in S. vulgaris differ significantly even when generated by crosses involving a common pollen donor and pollen recipients that consisted of female and hermaphrodite flowers co-occurring on the same plant. One explanation for this finding is that different parts of the same maternal plant carry significantly different mitochondrial populations that include different forms of CMS.
Experimental crosses of S. vulgaris conducted by McCauley et al. (2005) utilized PCR/RFLPs of the mitochondrial genes atpA and cox1 to document rare cases in which offspring carried different alleles for these genes from their respective mothers. In some instances the offspring had the same allele as that of its father, suggesting paternal inheritance, although in others it did not match either parent. An explanation for the latter observation was that the mother was heteroplasmic, carrying a secondary mitochondrial haplotype at a level too low to visualize PCR/RFLPs on agarose gels. It was hypothesized that occasionally a rare secondary mitochondrial haplotype inherited by one of her offspring could become the dominant haplotype as a result of drift and/or selection during the series of cell divisions leading from the zygote to maturity, causing that individual to be scored for an allele that differed from its mother.
More direct evidence for heteroplasmy came from instances in which PCR/RFLP banding patterns appeared to be overlays of two distinct alleles. In other cases, readily distinguishable allelic designations of an individual could be changed by a “knockback” method, where whole genomic DNA is digested with a restriction enzyme that targets allele-specific sequence prior to PCR/RFLP genotyping. These latter observations were said to be cases of “cryptic heteroplasmy” because the level of the putative secondary haplotype within that individual was too low to be detected visually by the standard PCR/RFLP method (McCauley et al. 2005).
Despite strong evidence for mitochondrial heteroplasmy from these artificial crosses, the frequency of heteroplasmy in natural populations of S. vulgaris is unknown, the level of quantitative genetic variance of mitochondrial populations within individuals is unmeasured, and transmission rates of heteroplasmy from mother to offspring have never been described. In this study, we utilize mother–offspring arrays collected from three natural populations of S. vulgaris to document inheritance patterns of atpA and cox1 PCR/RFLP markers that would suggest heteroplasmy and/or biparental inheritance. We then quantify the magnitude of heteroplasmy in terms of relative frequencies of atpA alleles within individuals, using quantitative PCR (Q-PCR), which allows us to conduct within-individual population genetic analyses of allele frequencies and their impact on within-population structure, as well as their transmission rates across generations. Our findings of widespread heteroplasmy suggest that paternal leakage of mitochondria is common, that selection plays a role in regulating within-individual allele frequencies, or that both factors contribute to levels of heteroplasmy in natural populations. The case for selection is especially intriguing given that S. vulgaris is gynodioecious, a condition expected to generate strong frequency-dependent selection on the mitochondrial genome (Ingvarsson and Taylor 2002; Wade and McCauley 2005).
MATERIALS AND METHODS
Genetic material:
In the summer of 2004, collections of leaves and ripe fruit were taken from a number of individuals from each of three S. vulgaris populations in Virginia. Precise locations are available upon request. DNA was extracted from leaves of maternal plants and leaf material from greenhouse-reared seedlings, using a variation of the CTAB method (Doyle and Doyle 1997).
PCR/RFLPs:
Previous work on S. vulgaris described PCR/RFLPs in atpA and cox1, using universal primers (McCauley et al. 2005). All individuals in this study were genotyped for these PCR/RFLPs, using new species-specific primers that we have developed to enhance amplification (Table 1). PCR amplifications were conducted in 50-μl reactions that contained ∼5 ng of whole genomic DNA, 2 units of Taq polymerase, 125 nm of each primer, 72 mm tricine, 120 mm KCl, and 4.8 mm MgCl2. Mitochondrial fragments were amplified using a “touchdown” PCR protocol designed to reduce nonspecific primer associations and subsequent arbitrary fragment amplification (Don et al. 1991). An initial denaturing cycle of 3 min at 95° was followed by 10 touchdown cycles (the annealing temperature drops 1° each cycle) of 30 sec at 94°, 30 sec at the annealing temperature of 55°, and 45 or 90 sec, for atpA and cox1 respectively, at 72° for elongation. The touchdown cycles were followed by an additional 29 cycles of 30 sec at 94°, 30 sec at the final annealing temperature, and 45 or 90 sec at 72°, which in turn were followed by a 20-min final elongation period at 72°. Duration of elongation was determined by the length of the fragment being amplified. For digestions, 5 μl of PCR product were digested with restriction endonucleases and were visualized on 4% metaphor gels. Specifically, atpA amplicons were digested with AluI and MspI in two separate reactions, and cox1 amplicons underwent a double digest with DdeI and MspI. Representative sequences of each allele are available in GenBank (accession nos. DQ422872–DQ422877).
TABLE 1.
Oligonucleotide sequences
| Oligo | |
|---|---|
| atpA forward | 5′-GTC TGG TCC GAT CGT TTA GC-3′ |
| atpA reverse | 5′-TAA AGA GGG CGA TCT TGT CA-3′ |
| cox1 forward | 5′-TGG GCA CAT GCT TCT CAG TA-3′ |
| cox1 reverse | 5′-CTC TTC AAC AGC CCA AGG AC-3′ |
| Q forward | 5′-ACG TCG CCT GGG AAA GC-3′ |
| Q reverse | 5′-TCC GCG ATA ATG GAA TGC A-3′ |
| Probe X | 5′-6-FAM-CGG CCT GGT GGT CGG CGT AAC-BHQ1-3′ |
| Probe A | 5′-6-FAM-TGA CAT TTG TCG ATA TGC CAC CG-BHQ1-3′ |
| Probe D | 5′-HEX-AGA CAT TTG CCG GTA TGC CAC C-BHQ1-3′ |
Primers and probes for PCR and quantitative PCR of mitochondrial atpA and cox1 that are specific to Silene vulgaris are shown. Sequences underlined denote cut sites for restriction enzymes TaqαI, which cuts allele A, and MspI, which cuts allele D. Bases shown in italics are allele specific. Probe sequences also include fluorophores with which they are labeled (i.e., 6-FAM and HEX).
Quantitative PCR:
Copy numbers of two distinct atpA lineages within individuals were estimated using the TaqMan method of Q-PCR (Holland et al. 1991; Lie and Petropoulos 1998). These two lineages are very distinct, differing at 16 of the 665 bp registered with GenBank. Briefly, TaqMan Q-PCR involves three oligos, a forward primer, a reverse primer, and a probe. The primers are unmodified and typical of PCR primers. The probe is designed to target a region within the sequence to be amplified. It is modified on the 5′ end with a fluorophore and on the 3′ end with a quencher. When the fluorophore and quencher are near to one another, the majority of the fluorescence from the fluorophore is captured. However, once the fluorophore is separated from the probe by the exonucleic activity of Taq polymerase, its fluorescence can escape and be quantified. The intensity of fluorescence is standardized relative to a passive reference dye. This standardized value is referred to as 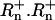 -values from the first 15 cycles, prior to any real accumulation of unquenched fluorophore are used to calculate a mean value termed
-values from the first 15 cycles, prior to any real accumulation of unquenched fluorophore are used to calculate a mean value termed 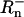 . The data collected for each cycle of a Q-PCR reaction are ΔRn (
. The data collected for each cycle of a Q-PCR reaction are ΔRn (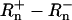 ). The value used to quantify the initial copy number in a sample is the critical cycle number (CT). The CT is the estimated number of cycles required for a significant increase in ΔRn.
). The value used to quantify the initial copy number in a sample is the critical cycle number (CT). The CT is the estimated number of cycles required for a significant increase in ΔRn.
TaqMan probes specific to different S. vulgaris atpA sequences were designed to work with the same primers and were labeled with different fluorophores such that they could be used together in a single Q-PCR reaction: the “A probe” for both the A and B alleles and the “D probe” for alleles D and E. Of the six atpA alleles previously identified (McCauley et al. 2005), only alleles A, B, and D were found among individuals in this study. A single-nucleotide difference within the sequence targeted by a TaqMan probe is generally considered sufficient for genotyping purposes (Livak 1999). In this study, allele-specific probes were distinguished by three nucleotide substitutions, including one on the 5′ end to ensure that probes function only with their intended alleles (Table 1).
Because several factors influence the rate at which Q-PCR reactions progress, and because of possible variation in the sensitivity of hardware to different fluorophores, we designed a third allele-neutral TaqMan assay, the allele X assay, to standardize results collected using the A/D assay, Q-PCR using the A and D probes. The “X probe” is designed to work with the same primers from the A/D assay; however, the X probe targets sequence that is invariant across all alleles in this study, and hence it should function equally well with any allele. The allele X assay allowed for the standardization of critical cycle (CT) values obtained using the A and D probes. Both the X assay and the A/D assay were conducted on 16 individuals for which allelic identity had previously been established using the PCR/RFLPs. Eight of these carried the A allele and 8 carried the D allele. At least five different concentrations of DNA template were used for both assays. CT was calculated for each individual at each concentration and standard curves were derived. These data were then utilized to generate Equations 1 and 2 that convert CT-values from alleles A (CTA) and D (CTD) into comparable copy number estimates:
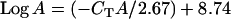 |
(1) |
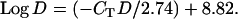 |
(2) |
All Q-PCR assays were designed using the software Primer 3 (Rozen and Skaletsky 2000) and Applied Biosystem's (Foster City, CA) software Primer Express v. 2.0. All primer and probe sequences are listed in Table 1. All Q-PCR assays were conducted on an ABI Prism 7000 (Applied Biosystems). All reaction conditions were optimized according to the protocol for TaqMan Universal PCR Master Mix (Applied Biosystems) with only minor adjustments. Total reaction volume was 25 μl including 12.5 μl of TaqMan Universal PCR Master Mix (Applied Biosystems). Forward and reverse primer concentrations were 900 nm each, and probe concentrations were 200 nm in all reactions. DNA template concentrations range from ∼0.1 ng/μl to ∼0.8 ng/μl, depending on assay and replicate. CT, the number of cycles required for a significant increase in fluorescence, was estimated with a threshold value of 0.2 using ABI Prism 7000 SDS software (Applied Biosystems).
Primary data collection:
Three different Q-PCR experiments were conducted. The first involved running the A/D assay on all individuals in the study at three different concentrations. The other two experiments were knockbacks that involve running the A/D assay on three different concentrations of genomic DNA that had each been digested with one of two specific restriction enzymes. MspI has a single cut site in the Q-PCR amplicon sequence of allele D, and hence it prevents its amplification, but not the amplification of the target sequence in allele A. TaqαI has the opposite effect of preventing the amplification of the Q-PCR amplicon sequence of allele A (see Table 1 for specifics). The knockback method was originally described as a means of revealing cryptic heteroplasmy, using traditional PCR methods, such as might exist when very unequal copy number limits the PCR amplification of a minority allele (McCauley et al. 2005). In any PCR, the exponential rate of the reaction slows and terminates with increasing product. The knockbacks allow for the necessary number of functional Q-PCR cycles required for quantifying minority alleles.
CT-values, estimates of the number of cycles required prior to a significant increase in PCR product to be detected, were collected and converted to relative copy numbers, which were then used to calculate the frequency of allele A within all individuals. Since a preliminary analysis showed no consistent difference in allele frequency among the three DNA concentrations that were used in the assays, these were treated as replicates and averaged. Allele scores were calculated for each individual using the copy number calculated for A when the D allele was knocked back and the copy number for D when the A allele was knocked back.
Analysis of population structure:
Under strict maternal inheritance most or all mitochondrial diversity should be partitioned among maternal lines, with little diversity expected within sibships or individuals. The magnitude of the within- and among-family components of diversity was estimated first from the PCR/RFLP data. The general principles for calculating genetic diversity indexes are those of Nei (1987) with a few exceptions that allowed for estimates of genetic variance attributable to family and individual. First, genetic diversity within families was calculated as 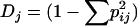 , where pij represents the frequency of allele i within family j. DF, genetic diversity attributable to family, was then obtained by averaging Dj over all families. Second, DF was used to calculate the proportion of genetic variance attributable to differences among families (Fft = [(DT − DF)/DT], where DT is the genetic diversity found in the entire population), a quantity similar to the more well-known Fst that quantifies genetic structure among potentially interbreeding populations. For similar methodologies of estimating diversity indexes at multiple levels that take heteroplasmy into account, see Birky et al. (1989) and Rand and Harrison (1989).
, where pij represents the frequency of allele i within family j. DF, genetic diversity attributable to family, was then obtained by averaging Dj over all families. Second, DF was used to calculate the proportion of genetic variance attributable to differences among families (Fft = [(DT − DF)/DT], where DT is the genetic diversity found in the entire population), a quantity similar to the more well-known Fst that quantifies genetic structure among potentially interbreeding populations. For similar methodologies of estimating diversity indexes at multiple levels that take heteroplasmy into account, see Birky et al. (1989) and Rand and Harrison (1989).
Quantification of mitochondrial heteroplasmy using Q-PCR allows for the analysis of genetic diversity within and among individuals in addition to that within and among families and populations. Variation within individuals was calculated as 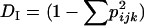 , where pijk represents the frequency p of allele i in individual k in family j. DI is then averaged for all individuals. Recall that the Q-PCR method was applied only to atpA and that the probes could distinguish only two classes of alleles, the A group and the D group. Note also that the individual allele scores represent the relative frequency of the A group (pA) such that qD = (1 − pA). Fft was calculated from the Q-PCR data. Fif (diversity within individuals, relative to families) was calculated as (DF − DI)/DF and Fit (diversity within individuals relative to the population) was calculated as (DT − DI)/DT.
, where pijk represents the frequency p of allele i in individual k in family j. DI is then averaged for all individuals. Recall that the Q-PCR method was applied only to atpA and that the probes could distinguish only two classes of alleles, the A group and the D group. Note also that the individual allele scores represent the relative frequency of the A group (pA) such that qD = (1 − pA). Fft was calculated from the Q-PCR data. Fif (diversity within individuals, relative to families) was calculated as (DF − DI)/DF and Fit (diversity within individuals relative to the population) was calculated as (DT − DI)/DT.
RESULTS
PCR/RFLP study:
The results of the PCR/RFLP survey are presented in Table 2, with a representative gel illustrated in Figure 1. A primary allele was readily scored for all individuals at the atpA locus and all but four individuals at the cox1 locus. All 4 individuals, 1 mother and 3 offspring from different families, failed to amplify during PCR using our primers after several attempts, and all are from population 1. However, other cox1 primers for invariant regions of the gene amplify product for these individuals, suggesting that variation in the original primer site prevents amplification in these individuals and preventing us from scoring two diagnostic restriction sites for this cox1 allele or class of alleles. We refer to this cox1 allele as a “null” allele throughout the remainder of the text. A total of five atpA/cox1 haplotypes, six if a null allele is accepted, were distinguished among the 18 mothers and 106 offspring surveyed. Overall, 91 of the 106 offspring studied (85.8%) resembled their respective mothers. All 15 of the individuals that did not resemble their respective mothers were found in population 1, the only population that was polymorphic for either atpA or cox1. Given that populations 2 and 3 expressed no PCR/RFLP genetic variation using these genetic markers, our techniques should not be able to detect nonmaternal inheritance. When only population 1 is considered, 61.5% of the offspring resembled their mother if we score the null allele. Of the 15 individuals that did not match their mothers, 13 differed at cox1 alone, and 2 individuals differed from their respective mothers at both loci.
TABLE 2.
Haplotypes and allele scores
| Population mother | Maternal haplotype | pi | No. offspring haplotype(s) | Range, p (SD) |
|---|---|---|---|---|
| 1-1 | D2 | 0.03 | 5 D2, 1 D1 | 0.05–0.44, 0.23 (0.17) |
| 1-2 | D* | 0.01 | 4 D2, 2 D3 | 0.01–0.84, 0.17 (0.31) |
| 1-3 | D2 | 0.00 | 6 D2 | 0.00–0.06, 0.02 (0.02) |
| 1-4 | D2 | 0.29 | 3 D2, 1 D3, 1 D1 | 0.00–0.98, 0.30 (0.41) |
| 1-5 | D2 | 0.02 | 2 D1, 1 D* | 0.03–0.13, 0.06 (0.06) |
| 1-6 | D2 | 0.00 | 1 D2, 1 B1, 1 D* | 0.22–1.00, 0.65 (0.40) |
| 1-7 | A3 | 1.00 | 9 A3, 1 D* | 0.01–1.00, 0.89 (0.33) |
| 2-1 | A3 | 1.00 | 6 A3 | 0.97–1.00, 0.99 (0.01) |
| 2-2 | A3 | 1.00 | 7 A3 | 1.00 |
| 2-3 | A3 | 1.00 | 7 A3 | 1.00 |
| 2-4 | A3 | 1.00 | 8 A3 | 1.00 |
| 2-5 | A3 | 1.00 | 5 A3 | 1.00 |
| 3-1 | B1 | 1.00 | 4 B1 | 0.94–1.00, 0.98 (0.03) |
| 3-2 | B1 | 1.00 | 6 B1 | 0.94–1.00, 0.99 (0.03) |
| 3-3 | B1 | 1.00 | 6 B1 | 0.85–1.00, 0.97 (0.07) |
| 3-4 | B1 | 1.00 | 8 B1 | 1.00 |
| 3-5 | B1 | 1.00 | 6 B1 | 1.00 |
| 3-6 | B1 | 1.00 | 4 B1 | 1.00 |
PCR/RFLP haplotypes and quantitative PCR allele scores for 18 families of Silene vulgaris collected from three populations (designated 1–3) in Virginia. The atpA allele is designated by letter (A, B, or D) and the cox1 allele is designated by number (1, 2, or 3). Individuals for which no cox1 allele could be ascribed are denoted with an *. Offspring not resembling their respective mothers are underlined for emphasis. Q-PCR allele scores for mothers (pi) and the ranges, averages, and standard deviations of offspring allele scores are also presented.
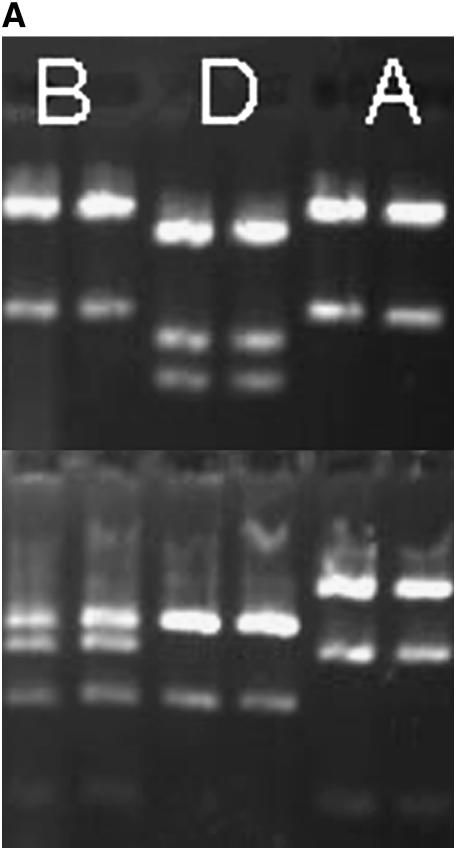
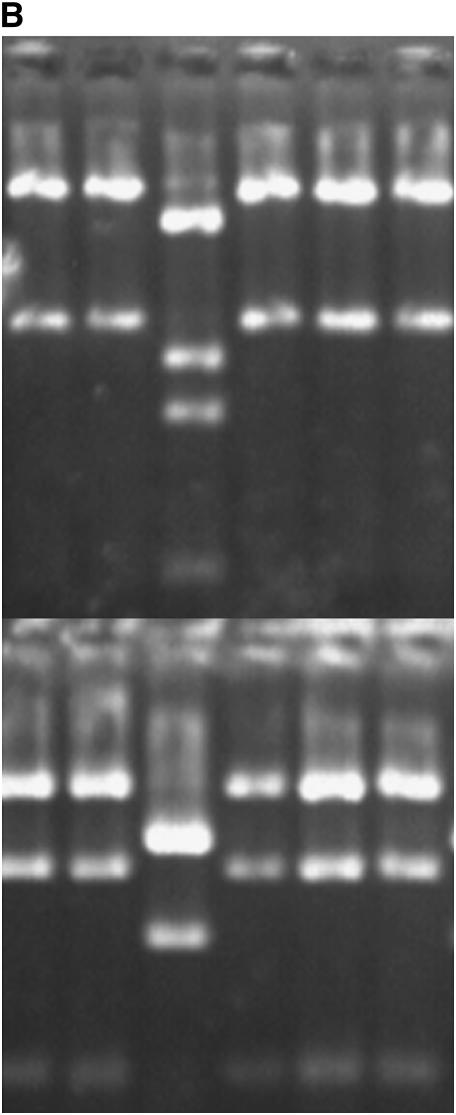
Figure 1.—
Agarose gel images of atpA PCR/RFLPs. Top frames depict atpA amplicons digested with MspI, and the bottom frames are digests with AluI. The three alleles, A, B, and D, found in this study are represented on the left (A). Six individuals from family 1-7 are shown on the right (B). Note that the third individual from the left carries allele D while its siblings carry allele A. The mother of family 1-7 also carries allele A.
Quantification of atpA heteroplasmy:
By the Q-PCR method of atpA genotyping all 18 mothers yielded results in complete agreement with qualitative results of the PCR/RFLP study when the template DNA was not subject to the knockback or restriction enzyme treatment and recognizing that the Q-PCR method cannot distinguish between atpA alleles A and B. The offspring atpA allele scores from the nonknockback Q-PCR assay were in agreement with the PCR/RFLP method with six exceptions. Five individuals, one each from families 1-1, 1-2, and 1-5, and 2 offspring from family 1-6, were found to be heteroplasmic (allele scores of 0.02, 0.02, 0.01, 0.41, and 0.73, respectively). Thus, by using the Q-PCR method, 6 of 106 offspring individuals (5.7%) were shown to have an atpA allele that differed from the mother, either because it was scored as entirely distinct from that of the mother or because it showed evidence of heteroplasmy when the mother did not. All six cases were found among the 39 offspring scored from population 1 (i.e., nonstrict maternal inheritance was found in 15.4% of offspring from population 1). Failing to find heteroplasmy in populations 2 and 3 using this method may simply reflect lack of genetic variation in these populations. Additionally, one of the apparently heteroplasmic individuals from family 1-6 and one individual from family 1-4 were scored as allele D by the PCR/RFLP method, but appear to be predominantly allele A when genotyped using the Q-PCR method. This dependency of allele on primers used during PCR was shown to be repeatable.
Application of the knockback technique yielded rather different results, as illustrated in Figure 2. The results obtained by calculating an allele score from reciprocal Q-PCR knockback experiments are summarized in Figure 3. Briefly, mothers 1-1, 1-2, 1-4, and 1-5 were classified as heteroplasmic by this method. All other mothers were scored as homoplasmic. Twenty-six offspring were scored as heteroplasmic, including the 5 offspring identified as heteroplasmic by the nonknockback method. Twenty-one of the heteroplasmic offspring were from population 1, 1 was from population 2, and 4 were from population 3.
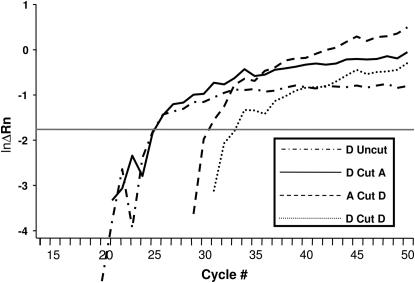
Figure 2.—
Graphical representation of quantitative PCR (Q-PCR) data. Presented are Q-PCR results from three treatments of the mitochondrial atpA gene for a single offspring from family 1-1. The y-axis, ln ΔRn, is a measure of relative fluorescence, and the x-axis is number of thermal cycles completed. Copy number is estimated from CT, the cycle number at which a significant increase in fluorescence is detected, approximately the cycle at which curves intersect the shaded line. Note that copy number for allele A in this individual cannot be estimated when template DNA is not treated with restriction enzymes (Uncut) and that CT for allele D changes very little when template DNA is undigested (Uncut) and when template is digested with TaqαI (Cut A) prior to amplification. However, when whole-genomic DNA is treated with MspI (Cut D), CT for allele A can readily be estimated. Presumably, this occurs because knocking allele D back with MspI allows for amplification of allele A to continue at an exponential rate for a sufficient number of cycles to be detected. Also noteworthy is that measurable quantities of allele D persist after treatment with MspI.
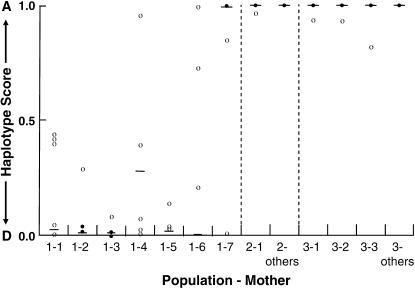
Figure 3.—
atpA allele scores calculated using the Q-PCR knockback method. Allele scores for 18 mothers (dashes) and their respective offspring (circles) collected from three natural populations of Silene vulgaris are shown. Solid circles represent cases in which two or more individuals from the same sibship display the same allele score.
Within-population structure:
Since no variation was observed within populations 2 and 3 in the PCR/RFLP study, and little variation was observed in these populations in the Q-PCR study, only population 1 was considered in the following analyses of population structure. Again, lack of variation in populations 2 and 3 does not necessarily indicate a lower frequency of heteroplasmy in these populations, but rather could reflect an issue of detectability with the markers at hand. From the PCR/RFLP data presented in Table 2, DT is estimated to be 0.69, DF = 0.37, and Fft = 0.46, suggesting that on average ∼54% (or 1 − Fft) of the PCR/RFLP diversity found in population 1 is contained within families. See materials and methods for a full description of summary statistics.
Using the Q-PCR data we get a notably lower index of total diversity, DT = 0.46, perhaps because the Q-PCR method distinguished only between two haplotypes instead of six. Diversity within families was comparable to that of the PCR/RFLP data, DF = 0.27, and diversity within individuals (DI) was calculated to be 0.12. These values obtained from the Q-PCR study yield an Fft of 0.41, an Fif of 0.55, and Fit of 0.74. Thus, on average, the proportion of mitochondrial genetic diversity found within families that resides within the individuals that compose those families is 0.45 (or 1 − Fif), and, on average, the proportion of variation found in the total population of offspring that can be found within the individuals that compose that population is 0.26 (or 1 − Fit).
DISCUSSION
Heteroplasmy in plants, especially mitochondrial heteroplasmy, is poorly studied (Korpelainen 2004; Barr et al. 2005). Extant cases generally fall into two categories: evidence for recombination has been used to suggest historic occurrences of heteroplasmy (e.g., Städler and Delph 2002; Lin et al. 2003), and experimental hybrids between divergent lineages and species have provided direct evidence of paternal leakage and heteroplasmy (e.g., Hattori et al. 2002). Mitochondrial heteroplasmy has apparently never been documented directly in natural plant populations (Barr et al. 2005) nor has the level of intraindividual variation been quantified in artificial or natural populations. The only attempt to quantify intraindividual cytoplasmic genetic variation in plants focused on the chloroplast genome and did not address issues of transmission (Frey et al. 2005).
The results presented here confirm findings of an earlier greenhouse crossing study of mitochondrial heteroplasmy in S. vulgaris (McCauley et al. 2005), extend them to natural populations, allow heteroplasmy to be viewed as a quantitative character, and consider the transmission of heteroplasmy from mother to offspring. The PCR/RFLP data indicate at least 15 incidences of mismatch between maternal and offspring mitochondrial alleles, an observation made earlier in the experimental crosses of McCauley et al. (2005). These mismatches were all from population 1 and were distributed among six of the seven families sampled. It is worth noting that the mismatches were observed using both atpA and cox1 markers, suggesting that this is a genomewide phenomenon rather than a locus-specific one.
Similarly, in the Q-PCR study most observations of heteroplasmy were in population 1. In that population 26% of the total mitochondrial diversity occurred within individuals, whereas variation among families accounted only for 41%, a result not at all consistent with the very low level of within-individual and within-family diversity expected to be enforced by strict maternal inheritance. It is clear that heteroplasmy is a common feature of at least one natural population of S. vulgaris, but the mechanism by which it is generated is unknown.
Evidence of heteroplasmy, historical or otherwise, is most frequently attributed to some degree of biparental inheritance (Mogensen 1996; Korpelainen 2004; Barr et al. 2005), and this is the most parsimonious explanation for our findings as well. Several possible explanations not requiring paternal leakage seem unlikely. The possibility that mutations can accumulate within individuals faster than they are eliminated or fixed by maternal inheritance can be dismissed because 16 nucleotide substitutions distinguish alleles A and D, yet these alleles co-occur within the same individuals. Selective maintenance of heteroplasmy within strictly maternal lineages also seems an unlikely explanation for our data. There are at least nine incidences of mothers yielding offspring with an alternative haplotype in population 1. Two of these incidences involve mothers that we scored as homoplasmic, giving rise to offspring that are predominantly of an alternative haplotype. Only very strong selection favoring the maintenance of multiple distinct mitochondrial lineages could account for these results given strict maternal inheritance, and this is in conflict with the largely invariant populations 2 and 3.
Two alternatives involving paralogous loci might also be invoked to explain some or all of our findings. First, plant mitochondrial genes are commonly transferred to the nucleus (Palmer et al. 2000; Richly and Leister 2004). However, in a recent survey of 280 plant genera, no incidents of loss of mitochondrial atpA or cox1 were uncovered, suggesting that these genes are not commonly transferred to the nucleus (Adams et al. 2002). Further, the experimental crosses conducted by McCauley et al. (2005) did not find a pattern of inheritance consistent with nuclear transmission of either of these mitochondrial genes.
Second, the plant mitochondrial genome is complex and often composed of multiple molecules, and some secondary molecules, or sublimons, can carry alternate copies of mitochondrial genes (Backert et al. 1997). Strong evidence supports the notion that sublimons can replicate independently of the rest of the mitochondrial genome and greatly increase or decrease in copy number in a single generation, resulting in substoichiometric shifts that could cause an apparent dissimilarity between mother and offspring (Janska et al. 1998). This is not the most parsimonious explanation for our results. Shifts in PCR/RFLP alleles occur at both atpA and cox1, suggesting that shifts are genomewide and not gene specific. However, this finding could also be explained by the co-occurrence of both genes on a single sublimon. Further, some of our shifts in relative frequency are dramatic, minimally five to six orders of magnitude, much greater than the three orders of magnitude that have previously been documented (Arrieta-Montiel et al. 2001). However, given the strong associations between substoichiometric shifting and CMS in other species (Janska et al. 1998), and the potential for this phenomenon to be regulated by nuclear genes (Abdelnoor et al. 2003), it seems unwise to discount this explanation entirely.
The results presented here have implications for other aspects of the population biology of S. vulgaris. The study of mitochondrial variation in this species was originally motivated by the fact that it is gynodioecious and displays cytonuclear sex determination (Charlesworth and Laporte 1998; Taylor et al. 2001), as do many gynodioecious systems (Frank 1989; Dudle et al. 2001). Much of the theory addressing the evolution and maintenance of cytonuclear gynodioecy focuses on the evolutionary conflict between maternally inherited mitochondrial CMS genes and biparentally inherited nuclear genes. This intergenomic conflict could be mitigated by occasional paternal leakage. Wade and McCauley (2005) have shown that paternal leakage can have considerable influence on the evolutionary dynamics of cytonuclear gynodioecious systems and can result in a type of frequency-dependent fitness that contributes to the maintenance of cytoplasmic polymorphism. Crossing studies have shown that S. vulgaris populations can carry several different CMS elements (Charlesworth and Laporte 1998; Taylor et al. 2001), an observation that, combined with the results presented here, suggests the possibility that CMS heteroplasmy could be a factor in the maintenance of gynodioecy in this system.
The occurrence of heteroplasmy also has implications for the evolution of the mitochondrial genome. In plants the mitochondrial genome is thought to be structurally quite fluid, owing to a propensity for both intra- and intermolecular recombination (Mackenzie and McIntosh 1999; Palmer et al. 2000). It would seem that generation of novel recombinants would be facilitated by heteroplasmy, since it would allow for the pairing of dissimilar DNA molecules during mitochondrial fusion events (Städler and Delph 2002). Indeed, McCauley et al. (2005) presented evidence for both inter- and intralocus mtDNA recombination in S. vulgaris, a finding consistent with our observations of frequent heteroplasmy.
The generality of these results to other angiosperm species is unclear, although the results of the study of S. acalis by Städler and Delph (2002) suggest that heteroplasmy might be found more generally in gynodioecious members of the genus Silene. The growing availability of PCR-based molecular markers and Q-PCR methods for assaying intraindividual genetic diversity should make investigations of heteroplasmy in a broad diversity of plant taxa increasingly feasible.
Acknowledgments
We thank Jonathan Ertelt who provided plant care in the Vanderbilt University greenhouses. We are extremely appreciative of Douglas McMahon of the Biological Sciences Department at Vanderbilt University. He allowed us the use of his ABI Prism 7000 for quantitative PCR, and he and his research group were extremely helpful when we were designing our Q-PCR assays. This work was funded by a Vanderbilt University Discovery grant to D.E.M.
References
- Abdelnoor, R. V., R. Yule, A. Elo, A. C. Christensen, G. Meyer-Gauen et al., 2003. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 100: 5968–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K. L., Y.-L. Qiu, M. Stoutemyer and J. D. Palmer, 2002. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 99: 9905–9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, H., 1999. Female and hermaphrodite flowers on a chimeric gynomonoecious Silene vulgaris plant produce offspring with different genders: a case of heteroplasmic sex determination? J. Hered. 90: 563–565. [Google Scholar]
- Arrieta-Montiel, M., A. Lyznik, M. Woloszynska, H. Janska, J. Tohme et al., 2001. Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics 158: 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise, J. C., J. E. Neigel and J. Arnold, 1985. Demographic influences on mitochondrial DNA lineage survivorship in animal populations. J. Mol. Evol. 20: 99–105. [DOI] [PubMed] [Google Scholar]
- Backert, S., B. L. Nielsen and T. Börner, 1997. The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 2: 477–483. [Google Scholar]
- Bailey, M. F., L. F. Delph and C. M. Lively, 2003. Modeling gynodioecy: novel scenarios for maintaining polymorphism. Am. Nat. 161: 762–776. [DOI] [PubMed] [Google Scholar]
- Barkman, T. J., G. Chenery, J. R. McNeal, J. Lyons-Weiler, W. J. Ellisens et al., 2000. Independent and combined analyses of sequences from all three genomic compartments converge on the root of flowering plant phylogeny. Proc. Natl. Acad. Sci. USA 97: 13166–13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, C. M., M. Neiman and D. R. Taylor, 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168: 39–50. [DOI] [PubMed] [Google Scholar]
- Birky, Jr., C. W., 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35: 125–148. [DOI] [PubMed] [Google Scholar]
- Birky, Jr., C. W., T. Maruyama and P. Fuerst, 1983. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics 103: 513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky, Jr., C. W., P. Fuerst and T. Maruyama, 1989. Organelle gene diversity under migration, mutation, and drift: equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparison to nuclear genes. Genetics 121: 613–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, D. L., A. Warsaw and T. R. Meagher, 2005. Consequences of prarie fragmentation on the progeny sex ratio of a gynodioecious species, Lobelia spicata (Campanulaceae). Heredity 95: 69–75. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., and V. Laporte, 1998. The male-sterility polymorphism of Silene vulgaris: analysis of genetic data from two populations and comparison with Thymus vulgaris. Genetics 150: 1267–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J. J., and J. L. Doyle, 1997. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 19: 11–15. [Google Scholar]
- Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker and J. S. Mattick, 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19: 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudle, D. A., P. Mutikainen and L. F. Delph, 2001. Genetics of sex determination in the gynodioecious species Lobelia siphilitica: evidence from two populations. Heredity 86: 265–276. [DOI] [PubMed] [Google Scholar]
- Frank, S. A., 1989. The evolutionary dynamics of cytoplasmic male-sterility. Am. Nat. 62: 345–376. [Google Scholar]
- Frey, J. E., B. Frey and D. Forcioli, 2005. Quantitative assessment of heteroplasmy levels in Senecio vulgaris chloroplast DNA. Genetica 123: 255–261. [DOI] [PubMed] [Google Scholar]
- Hattori, N., K. Kitagawa, S. Takumi and C. Nakamura, 2002. Mitochondrial DNA heteroplasmy in wheat, Aegilops and their nucleus-cytoplasm hybrids. Genetics 160: 1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland, P. M., R. D. Abramson, R. Watson and D. H. Gelfand, 1991. Detection of specific polymerase chain reaction product by utilizing the 5′ → 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88: 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson, P. K., and D. R. Taylor, 2002. Genealogical evidence for epidemics of selfish genes. Proc. Natl. Acad. Sci. USA 99: 11265–11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janska, H., R. Sarria, M. Woloszynska, M. Arrieta-Montiel and S. Mackenzie, 1998. Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10: 1163–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpelainen, H., 2004. The evolutionary processes of mitochondrial and chloroplast genomes differ from those of nuclear genomes. Naturwissenschaften 91: 505–518. [DOI] [PubMed] [Google Scholar]
- Laporte, V., F. Viard, F. Bena, M. Valero and J. Cugean, 2001. The spatial structure of sexual and cytonuclear polymorphism in the gynodioecious species Beta vulgaris ssp. maritima: I/ at a local scale. Genetics 157: 1699–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie, Y. S., and C. J. Petropoulos, 1998. Advances in quantitative PCR technology: 5′ nuclease assays. Curr. Opin. Biotechnol. 9: 43–48. [DOI] [PubMed] [Google Scholar]
- Lin, T.-P., W.-J. Chuang, S. S. F. Huang and S.-Y. Hwang, 2003. Evidence for the existence of some dissociation in an otherwise strong linkage disequilibrium between mitochondrial and chloroplastic genomes in Cyclobalanopsis glauca. Mol. Ecol. 12: 2661–2668. [DOI] [PubMed] [Google Scholar]
- Livak, K. J., 1999. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet. Anal. 14: 143–149. [DOI] [PubMed] [Google Scholar]
- Mackenzie, S., and L. McIntosh, 1999. Higher plant mitochondria. Plant Cell 11: 571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley, D. E., and M. T. Brock, 1998. Frequency-dependent fitness in Silene vulgaris, a gynodioecious plant. Evolution 52: 30–36. [DOI] [PubMed] [Google Scholar]
- McCauley, D. E., and M. S. Olson, 2003. Associations among cytoplasmic molecular markers, gender, and components of fitness in Silene vulgaris, a gynodioecious plant. Mol. Ecol. 12: 777–787. [DOI] [PubMed] [Google Scholar]
- McCauley, D. E., and D. R. Taylor, 1997. Local population structure and sex ratio: evolution in gynodioecious plants. Am. Nat. 150: 406–419. [DOI] [PubMed] [Google Scholar]
- McCauley, D. E., M. F. Bailey, N. A. Sherman and M. Z. Darnell, 2005. Evidence of paternal transmission and heteroplasmy in the mitochondrial genome of Silene vulgaris, a gynodioecious plant. Heredity 95: 50–58. [DOI] [PubMed] [Google Scholar]
- Mogensen, H. L., 1996. The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot. 83: 383–404. [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Palmer, J. D., K. L. Adams, Y. Cho, C. L. Parkinson, Y.-L. Qiu et al., 2000. Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc. Natl. Acad. Sci. USA 97: 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit, R. J., J. Duminil, S. Fineschi, A. Hampe, D. Salvini et al., 2005. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plants. Mol. Ecol. 14: 689–701. [DOI] [PubMed] [Google Scholar]
- Rand, D. M., 2001. The units of selection on mitochondrial DNA. Annu. Rev. Ecol. Syst. 32: 415–448. [Google Scholar]
- Rand, D. M., and R. G. Harrison, 2001. Molecular population genetics of mtDNA size variation in crickets. Genetics 121: 551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboud, X., and C. Zeyl, 1994. Organelle inheritance in plants. Heredity 72: 132–140. [Google Scholar]
- Richly, E., and D. Leister, 2004. NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol. 21: 1081–1084. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Schnable, P. S., and R. P. Wise, 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3: 175–180. [Google Scholar]
- Städler, T., and L. F. Delph, 2002. Ancient mitochondrial haplotypes and evidence for intragenic recombination in a gynodioecious plant. Proc. Natl. Acad. Sci. USA 99: 11730–11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., and S. R. Palumbi, 1985. Extranuclear differentiation and gene flow in the finite island model. Genetics 109: 441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D. R., M. S. Olson and D. E. McCauley, 2001. A quantitative genetic analysis of nuclear-cytoplasmic male sterility in structured populations of Silene vulgaris. Genetics 158: 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, M. J., and D. E. McCauley, 2005. Paternal leakage sustains the cytoplasmic polymorphism underlying gynodioecy but remains invasible by nuclear restorers. Am. Nat. 166: 592–602. [DOI] [PubMed] [Google Scholar]
- Wolfe, A. D., and C. P. Randle, 2004. Recombination, heteroplasmy, haplotype polymorphism, and paralogy in plastid genes: implications for plant molecular systematics. Syst. Bot. 29: 1011–1020. [Google Scholar]
- Wolfe, K. H., W.-H. Li and P. M. Sharp, 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 84: 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]