Abstract
The Arabidopsis short integuments 2-1 (sin2-1) mutant produces ovules with short integuments due to early cessation of cell division in these structures. SIN2 was isolated and encodes a putative GTPase sharing features found in the novel DAR GTPase family. DAR proteins share a signature DAR motif and a unique arrangement of the four conserved GTPase G motifs. We found that DAR GTPases are present in all examined prokaryotes and eukaryotes and that they have diversified into four paralogous lineages in higher eukaryotes. Eukaryotic members of the SIN2 clade of DAR GTPases have been found to localize to mitochondria and are related to eubacterial proteins that facilitate essential steps in biogenesis of the large ribosomal subunit. We propose a similar role for SIN2 in mitochondria. A sin2 insertional allele has ovule effects similar to sin2-1, but more pronounced pleiotropic effects on vegetative and floral development. The diverse developmental effects of the mitochondrial SIN2 GTPase support a mitochondrial role in the regulation of multiple developmental pathways.
ARABIDOPSIS ovules are a useful model system for studying developmental mechanisms. Arabidopsis ovules initiate on the inner surface of immature carpels as relatively featureless finger-like primordia. As they elongate, three different regions become specialized and undergo morphogenesis and cellular differentiation (Robinson-Beers et al. 1992). The distal-most region, the nucellus, is the site of meiosis and embryo sac formation. The central chalazal region is the site of the most visible morphogenic changes as it gives rise to two appendages, the inner and outer integuments. The basal region elongates through division and coordinated expansion of cells forming the funiculus, a supporting stalk. During this process, the developing ovule becomes bilaterally symmetrical as a result of differential growth that causes the funiculus to curve toward the base of the carpel and the outer integument to curve toward the carpel apex. At maturity, both integuments have grown to enclose the nucellus and form a terminal micropylar opening (Figure 1A). Mutations altering ovule morphogenesis may also disrupt other plant developmental pathways and the relative morphological simplicity of ovules can facilitate overall understanding of the underlying biochemical or molecular processes.
Figure 1.—
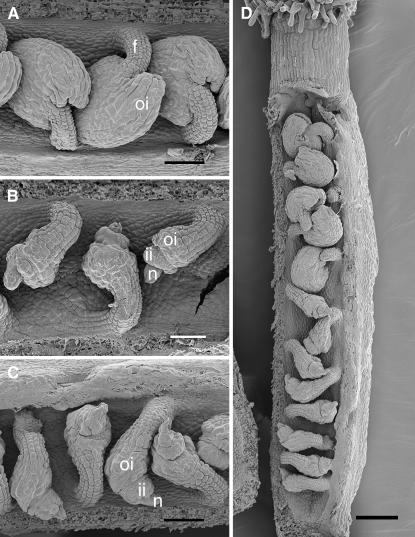
Scanning electron micrographs of stage 3-VI ovules (anthesis; stages from Schneitz et al. 1995). (A) Wild-type Ler. (B) sin2-1. (C and D) sin2-2 plants. In D, the gradation in severity of effects of sin2-2 from the base to the apex of the carpel is apparent. f, funiculus; ii, inner integument; n, nucellus; oi, outer integument. Bar, 50 μm (A–C) and 100 μm (D).
Numerous genes affecting growth and patterning of ovules have been identified via mutagenesis and cloning (Schneitz 1999; Skinner et al. 2004). Several of the genes regulating Arabidopsis ovule development manifest their effects through alterations in the pattern or progress of cell division. These genes encode proteins with a variety of biochemical functions (Skinner et al. 2004). For example, mutations in AINTEGUMENTA (ANT) and INNER NO OUTER (INO), encoding AP2 and YABBY-domain transcription factors, respectively, result in a complete absence of both integuments or of only the outer integument (Gaiser et al. 1995; Elliott et al. 1996; Klucher et al. 1996; Baker et al. 1997; Villanueva et al. 1999). TSO1 encodes a novel nuclear protein required for proper orientation of cell elongation and cytokinesis during floral organ and integument development (Hauser et al. 1998, 2000; Song et al. 2000). Severe mutations in the HUELLENLOS (HLL) gene, encoding a mitochondrial L14 ribosomal subunit, lead to an arrest in ovule growth and degeneration of the apical regions of the primordia, revealing a role for mitochondrial activity in the process of ovule growth (Schneitz et al. 1998; Skinner et al. 2001). Among other floral effects, reduced activity of the putative protein kinase TOUSLED (TSL) causes short outer and protruding inner integuments (Roe et al. 1997a,b, 1993). The variety of protein classes involved in ovule growth implies complex regulation of this process at the levels of transcription, signal transduction, and metabolism.
SHORT INTEGUMENTS 2 (SIN2) is required for sustaining cell divisions during integument development (Broadhvest et al. 2000). At anthesis, sin2-1 ovules have fewer integument cells than wild type, leaving their nucelli exposed (Figure 1B). sin2-1 also has subtle pleiotropic effects on flower development. The gynoecia of sin2-1 mutants generally have a cleft stigma and occasionally bear an outgrowth on the corresponding valve. Additionally, in sin2-1 sepals marginal cells appear to be absent or highly reduced in number. These pleiotropic effects suggest that SIN2 facilitates several morphogenic pathways.
Double mutants with sin2-1 revealed functional relationships between SIN2 and other ovule development genes (Broadhvest et al. 2000). ant-5 sin2-1 double mutants were indistinguishable from ant-5 mutants, but ant and sin2 had similar synergistic interactions with hll. Both ant hll and sin2-1 hll double mutants exhibited a reduced number of ovules and an earlier abortion of primordia development than hll single mutants. The ovule effects of sin2-1 were additive with the cell expansion defects of tso1-3. Thus SIN2 may act downstream of ANT in a common pathway with HLL, and in parallel to TSO1, to promote cell division during integument growth (Broadhvest et al. 2000).
To better understand regulation of cell division during organ formation, we have identified and characterized the SIN2 gene and isolated a second allele, sin2-2. SIN2 encodes a putative GTPase of a relatively uncharacterized subclass, termed the DAR GTPases, on the basis of a conserved aspartate–alanine–arginine (DAR) motif and other conserved features (Fu et al. 1998). Some DAR GTPases have been found to be important for cell division in bacteria, fungi, and human stem cell lines, where they are associated with assembly or subcellular transport of ribosomal subunits (Bassler et al. 2001; Saveanu et al. 2001; Bialkowska and Kurlandzka 2002; Morimoto et al. 2002; Tsai and McKay 2002; Kallstrom et al. 2003; Matsuo et al. 2006; Uicker et al. 2006). We found SIN2 to localize to mitochondria and hypothesize a function in mitochondrial ribosome assembly. In conjunction with hll, sin2 mutants provide an attractive system with which to study the role of mitochondrial function in the development of a multicellular organism.
MATERIALS AND METHODS
Plant material:
Plants were grown on soil as previously described (Kranz and Kirchheim 1987; Robinson-Beers et al. 1992). Some plants were germinated on 1% agar containing 1% sucrose, 1× Murashige and Skoog (MS) salts, and 1× Gamborg's B-5 vitamins (Murashige and Skoog 1962; Gamborg et al. 1968). The Student's t-test was used to determine if mutant plants were significantly different from wild-type plants in germination rate, number of days to flowering, number of rosette leaves produced, leaf production rates, and number of axillary meristem elongations. Plant transformation vectors and methods were as described (Meister et al. 2002).
Genetic mapping and complementation:
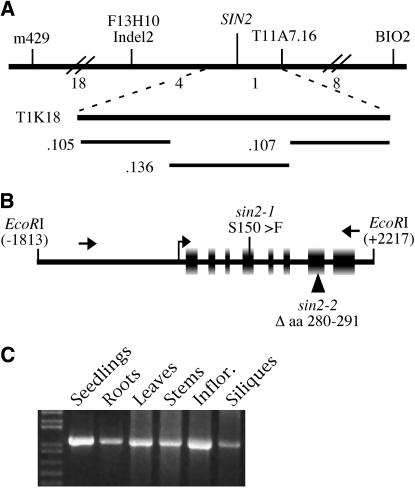
Using a mapping population generated by crossing sin2-1 (Landsberg erecta, Ler) with wild-type Columbia (Col-3), SIN2 was previously localized to chromosome 2 between the cleaved amplified polymorphic sequence (CAPS) m429 and the simple sequence length polymorphism (SSLP) AthBIO2 (Broadhvest et al. 2000). Of 31 chromosomes that had a recombination event between m429 and AthBIO2, four recombination points were between SIN2 and the SSLP marker F13H10 Indel2 [Cereon accession CER448978; The Arabidopsis Information Resource (TAIR) http://www.arabidopsis.org/Cereon/] and one recombination was between SIN2 and a ClaI RFLP within T11A7.16 (At2G41740). This region is spanned by three overlapping bacterial artificial chromosome (BAC) clones (T11A7, T1K18, and T32G6; Choi et al. 1995). Cosmid subclones were generated from BAC T1K18 by partial digestion with Sau3AI and insertion of 12- to 25-kb fragments in pOCA28-15 (Eshed et al. 1999). The overlapping cosmids cJBT1K18.136, cJBT1K18.105, and CJBT1K18.107 were identified and assembled into a contig using hybridization and fingerprint data. These cosmids spanned the annotated genes At2G41600.1–At2G41730.1 and were transformed into plants from the sin2-1 mapping population. The genotype of transgenic plants at the SIN2 locus was determined with the F13H10 Indel2 SSLP and the DdeI CAPS marker 6D20R in At2G42090.1.
Molecular methods:
Coding regions for genes within the complementing cosmid (cJBT1K18.136) were isolated from sin2-1 homozygous and Ler plants by reverse transcription polymerase chain reaction (RT–PCR) using gene-specific primers and SuperscriptII reverse transcriptase (Invitrogen, Carlsbad, CA). The cDNAs were sequenced by Davis Sequencing (Davis, CA). Sequences were compared using Sequencher (Gene Codes, Ann Arbor, MI). Genomic subclones, including each gene within the complementing cosmid along with the sequences 5′ and 3′ extending into the next closest genes, were isolated from cJBT1K18.136 and transformed into plants from the sin2-1 mapping population. For At2g41670 (SIN2), a 4.0-kb EcoRI fragment was isolated and inserted into these same sites in pBJ97 (Gleave 1992), creating pTH13. At2g42670 was removed from pTH13 on a NotI fragment and inserted into the same site in pMLBART (Gleave 1992) for plant transformation. The 5′-end of the SIN2 cDNA was determined by sequencing clones obtained with the GeneRacer 5′ RACE System (Invitrogen) using primers specific to SIN2. The full-length cDNA was inserted as an SpeI/BamHI fragment into XbaI/BamHI-digested pMON999 (Meister et al. 2002), creating a P35S∷SIN2∷NOS3′ transcriptional fusion (pTH52). The cDNA was also attached to a 5.6-kb SIN2 5′-flanking region using a common NruI site in the first exon to create pTH64. The gene fusions in pTH52 and pTH64 were transferred to pMLBART (Gleave 1992) on NotI fragments for plant transformation.
Primers designed to amplify the SIN2 genomic region, SIN2KOF2 (GAT GGG TTA TTA CGA TTT GGG CAG TTA TT) and SIN2KOR (CAG TTT CAG GGA CAT CGT CAA GGA TAA AG), were used to screen the Wisconsin α-population of insertion lines for additional sin2 alleles as described by Krysan et al. (1999). The insertion site in sin2-2 was determined by sequencing PCR products using SIN2 primers on either side of the insertion and the T-DNA JL202 primer. The SIN2KOR and JL202 primers (Krysan et al. 1999) were used to detect the sin2-2 insertion in segregating populations.
An RT–PCR product generated using synthetic primers T32G6.19F and SIN2GR on first-strand cDNA from Ler inflorescence poly(A)+ RNA was cloned into the pCR2.1 vector (Invitrogen), forming pTH19. The insert was sequenced and found to include the entire SIN2 coding region with no mutations except for those intentionally introduced to remove the stop codon. This cDNA was mobilized on an SpeI/BamHI fragment and inserted into pRJM86 (Meister et al. 2002) at the same sites to form a fusion with the green fluorescent protein (GFP) 1.1.15 (Schumacher et al. 1999) coding region. The SpeI/KpnI fragment containing the SIN2:GFP fusion protein-coding region was inserted between the CaMV 35S promoter and the nopaline synthase polyadenlyation region (NOS3′) in pMON999 (Meister et al. 2002). A NotI fragment consisting of P35S∷SIN2:GFP NOS3′ was inserted into the NotI site of the plant transformation vector pMLBART (Gleave 1992) and transformed into plants.
Microscopy:
Samples were prepared for scanning electron microscopy and images were acquired and edited as described (Broadhvest et al. 2000). Samples were fixed for light microscopy as described by Baum and Rost (1996). For viewing petal veins, following fixation, flowers were cleared overnight in a saturated aqueous chlorylhydrate solution. Petals were removed from the flowers and observed with a Zeiss Axioplan (Zeiss, Oberkochen, Germany) microscope using dark-field optics. GFP localization was determined using transgenic SIN2:GFP T2 seedlings, which were stained for 1 hr in a 1× MS solution containing the mitochondrial-specific stain MitoFluor Red589 (Molecular Probes, Eugene, OR) at 500 nm and were visualized on a Zeiss Axioscope microscope using fluorescein isothiocyanate and tetramethylrhodamine isothiocyanate filter sets, respectively. Images were recorded using the Openlab system (Improvision, Lexington, MA) and were adjusted for contrast in Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
Phylogenetic analysis:
BLAST (Altschul et al. 1997) searches were used to identify sequences in GenBank encoding proteins similar to SIN2 and other Arabidopsis DAR proteins. A large number of proteins containing G motifs were identified. DAR proteins (defined as those including a DAR-G4-G1-G2-G3 arrangement of motifs but which did not have two complete G domains in tandem; Fu et al. 1998) uniformly gave the best matches, followed by ENGA proteins. Parsimony analysis was performed with PAUP4.0b6 (Swofford 1999) on alignments created using ClustalX (Thompson et al. 1997). Three separate alignments were produced using gap opening:substitution costs of 2.5:0.5, 5:0.4, and 10:0.3, and the BLOSUM series of weightings (Henikoff and Henikoff 1992). ENGA sequences from the ENGA DAR motif to the second G3 motif were used as an outgroup. Regions unstable between alignments were detected using SOAP (Loytynoja and Milinkovitch 2001). Sequences <70% stable were not included in the phylogenetic analysis (culling). A second analysis was performed in which all three alignments, arranged in tandem, were included (elision; Wheeler et al. 1995). In parsimony analysis, characters were weighted using a BLOSUM45 amino acid substitution matrix, which was generated from the published BLOSUM45 matrix (Henikoff and Henikoff 1992) by substituting “15 − n” for each value “n” of the matrix. This results in all positive values, ranging from 0 to 20. An amino-acid-to-gap transition was given a cost equal to the highest value in the matrix, 20. Bootstrap values were calculated from 500 bootstrap replicates of heuristic searches on 10 random sequence additions per bootstrap replicate and tree bisection–reconnection procedure for branch swapping. Branches that were not consistent between the culling and elision methods of character weighting or those that had bootstrap values ≤50% were collapsed.
Accession numbers:
The GenBank/EMBL accession number for the SIN2 cDNA is AY254472. The accession numbers for other protein sequences used in the analysis are: Arabidopsis DGP2 (CAB40031); Arabidopsis DGP3 (AAD15335); Arabidopsis DGP4 (AAF27009); Arabidopsis DGP5 (AAG52287); Arabidopsis DGP6 (AAD26884); Arabidopsis DGP7 (AAF22888); Oryza DGP4 (BAB61154); Oryza DGP3 (AU077558); Oryza DGP1 (AAAA01011536); Oryza DGP2 (AC077693); Medicago DGP6 (BG452200); Hordeum DGP5 (BF619769); Zea DGP6 (AAD41267); Lycopersicon DGP1 (BG133077); Leishmania DGP3 (CAC32254); Leishmania DGP2 (CAC32261); Leishmania DGP1 (AAF73086); Bacillus ylqF (F69880); Borrelia DGP (AAC67000); Vibrio DGP (AAF96431); Streptococcus DGP (AAK75264); Synechocystis DGP (NP_441269); Nostoc DGP (NP_484788); Methanococcus DGP (G64482); Pyrococcus DGP (F75050); Pyrobaculum DGP (NP_558827); human MTG1 (AAH04409); human LSG1 (NP_060855); human nucleostemin (GNL3, NP_055181); human GNL2 (NGP1, AAH00107); human HSR1 (XP_041722); human GNL3L (NP_061940); Drosophila DGP2 (Dme1_CG3983, AAF55384); Drosophila DGP1 (Dmel_CG6501; AAF56060); Drosophila DGP4 (Dme1_CG14788, AAF45628); Drosophila DGP5 (Dme1_CG9320, AAF53920); Drosophila Ngp (AAF57834); Caenorhabditis DGP4 (C53H9.2, AAK68267); Caenorhabditis DGP3 (T24970); Caenorhabditis nst-1(CAA88860); Caenorhabditis DGP1 (Y67D2.4, NM_065016); Saccharomyces LSG1 (NP_011416); Saccharomyces MTG1 (S55083); Saccharomyces NUG1 (S50464); Saccharomyces NOG2 (NP_014451); Treponema ENGA (P96128); Aquifex ENGA (O67749); Borrelia ENGA (O51461); Rickettsia ENGA (NP_360667)
RESULTS
SIN2 encodes a putative GTPase:
The ethyl-methanesulfonate-induced sin2-1 allele was previously mapped to chromosome 2 using a sin2-1 Ler/Col segregating population (Broadhvest et al. 2000). Further analysis of this population localized SIN2 to a region spanned by three overlapping clones (T11A7, T1K18, and T32G6; Choi et al. 1995). Figure 2A illustrates the overlapping cosmid subclones of T1K18 that were transformed into sin2-1 heterozygous progeny from the mapping population. Eight of the nine identified homozygous sin2-1 plants (genotyped with mapping markers flanking the SIN2 locus) carrying cJBT1K18.136 exhibited a wild-type ovule phenotype, indicating complementation of the mutation. cJBT1K18.136 comprises four complete predicted coding regions (At2g41660, 41670, 41680, and 41690; TAIR, http://www.arabidopsis.org/). Subclones spanning these coding regions from Ler and sin2-1 plants were sequenced and a single gene, At2g41670, was found to contain a C-to-T mutation unique to sin2-1 plants. A 4.0-kb EcoRI fragment spanning At2g41670 (Figure 2B) was also able to complement the sin2-1 mutation, confirming this gene as SIN2.
Figure 2.—
Identification and expression of the SIN2 gene. (A) The chromosomal region surrounding the SIN2 locus with the molecular markers used to map SIN2 shown above, and the number of recombination events between markers found in 918 F2 plants indicated below the top horizontal line. The BAC T1K18 spans part of the region between the closest flanking markers. Cosmid subclones .105, .136, and .107, derived from T1K18 and used in the complementation test, are illustrated. (B) The 4.0-kb EcoRI fragment spanning a single gene, At2G41670, derived from cJBT1K18.136, which was able to complement the sin2-1 mutant phenotype. The exons in this region and the transcriptional start site as determined by RT–PCR are shown as boxes and a bent arrow, respectively. Arrows indicate the positions of primers used to screen T-DNA lines for additional sin2 alleles. The positions of the missense (sin2-1) and insertional (sin2-2) mutations are indicated. Numbering is relative to the translational start site. (C) RT–PCR with total RNA from several plant parts used as template shows that SIN2 RNA was present in all structures assayed.
RT–PCR generated a 1.25-kb SIN2 cDNA that derived from the eight exons spanning 2 kb of genomic sequence predicted for At2g41670 and encoded a 386-amino-acid protein (Figure 2B). The 5′-end of the SIN2 mRNA was determined by 5′-RACE PCR to be 34 bases upstream of the putative translation start codon. The complete coding region of the cDNA was fused to 5.6 kb of genomic sequence 5′ to the SIN2 coding region or to the CaMV35S promoter, and both constructs complemented the sin2-1 mutation. While we were unable to detect SIN2 transcript by in situ hybridization, transcript was detected by RT–PCR in all structures assayed (Figure 2C). Fusions of either the 5.6- or the 1.5-kb region 5′ of the SIN2 coding sequence to the GUS coding region failed to produce GUS activity sufficient to be detected by histochemical staining in transgenic plants. These observations indicate that SIN2 is likely expressed at low levels in a variety of plant organs.
The predicted SIN2 protein contains the four sequence motifs (G1–G4) typical of all members of the GTPase superfamily (Bourne et al. 1991). The sin2-1 lesion produces a change of a conserved serine to phenylalanine at position 150 within the P-loop or putative GTP binding pocket present in the G1 motif (Figure 2B). Because sin2-1 was the only mutant allele of this gene that we had identified, and since mutations in the G1 motif can have a gain-of-function effect (Bourne et al. 1991), it was not possible to assess if the effects of sin2-1 on plant development were typical of SIN2 loss of function. Determination of the SIN2 sequence enabled screening for an insertional allele. A transgenic line in which a T-DNA had inserted in the seventh exon of SIN2, and associated with deletion of the SIN2 nucleotides corresponding to amino acids 281–290 at the insertion site, was identified in the Wisconsin Knock-Out population (Krysan et al. 1999). This lesion was designated sin2-2 (Figure 2B).
Plants heterozygous for sin2-2 were not visibly different from wild-type siblings, indicating that sin2-2 is a recessive allele. sin2-2 plants produced ovules similar to those of sin2-1 mutants with a reduced number of ovule integument cells (Figure 1, B and C; Broadhvest et al. 2000). However, the extent of sin2-2 integument reduction was more variable, with some sin2-2 pistils including as many as six ovules with a nearly wild-type appearance in addition to those with short integuments (Figure 1D). These morphologically wild-type ovules were usually at the distal end of the carpel. Even with this variation in ovule phenotype, sin2-2 plants were completely female sterile but male fertile as were sin2-1 plants. The similarity of the most severe integument phenotype between the sin2-2 insertional mutant and the sin2-1 point mutant suggests that sin2-1 is a loss-of-function allele and that SIN2 acts to promote cell divisions in the developing integuments.
sin2-2 illuminates additional developmental roles for SIN2:
sin2-1 and sin2-2 plants displayed both overlapping and allele-specific phenotypes. However, it is possible that some of the specific phenotypic effects observed in sin2-1 and sin2-2 plants were linked to their respective genetic backgrounds [Ler and Wassilewskija (Ws), respectively]. Mutant phenotypes were always compared to those of wild-type siblings found in the same segregating population (unless specified otherwise). A reduction in the expected 1:3 (sin2-1:wild type) segregation ratio had been previously noted and was proposed to be due to reduced viability of sin2-1 plants (Broadhvest et al. 2000). A greater degree of reduction was observed in sin2-2 plants. When seeds from sin2-2 heterozygote plants were sown in soil, 15% failed to germinate, an additional 12% failed to survive to maturity, and sin2-2 mutants were underrepresented in the population of plants reaching maturity (3 of 143 total vs. 35 of 143 that would be expected). However, the sin2-2 allele was found at the expected frequency among 70 phenotypically wild-type plants (47 of 140 chromosomes, a 1:2.04 ratio of homozygotes to heterozygotes), consistent with Mendelian transmission of the allele. On media, the sin2-2 germination defect was partially rescued, with an overall germination efficiency of 98.5% (336 of 341) in a segregating population. The underrepresentation of sin2-2 mutants when grown in soil indicates that sin2-2 plants exhibit a greater reduction in viability in soil than sin2-1 plants.
Vegetative growth rates were also affected in sin2 mutants. Both sin2-1 and sin2-2 mutants exhibited an extended germination period. In a sin2-2 segregating population at day 13 of growth on media, 82 seedlings had only cotyledons while 257 had developed visible rosette leaves. Forty-eight of these seedlings with two leaves >1 mm and 48 bearing only cotyledons were transplanted to soil. All seedlings without leaves that subsequently matured to flowering were shown to be homozygous for the sin2-2 allele, demonstrating a slower germination rate for sin2-2 seeds compared to wild-type siblings. Detailed analyses showed that both sin2-1 and sin2-2 seedlings exhibited slower emergence of radicle, cotyledons, and secondary leaves than their wild-type siblings, with sin2-2 showing the strongest effects in regard to germination and seedling growth (Table 1). The leaf emergence rate of sin2-2 was reduced by half, but sin2-1 plants behaved as their wild-type siblings (Table 1). The sin2-2 plants also appeared darker green than their wild-type siblings (supplemental Figure 1A at http://www.genetics.org/supplemental/). These alterations in germination, leaf production, and greening suggest the involvement of SIN2 activity in a variety of growth processes during vegetative development.
TABLE 1.
Growth measurements
| Genotype | Days to radicle emergence | Days to cotyledon emergence | Days to two leaves | Leaf emergence (days/leaf) | Days to bolting | No. of rosette leaves | No. of active branches |
|---|---|---|---|---|---|---|---|
| SIN2 (Ler) | 2.2 ± 1.2 (57) | 4.7 ± 1.5 (57) | 10.7 ± 0.8 (57) | 2.8 R2 = 0.98 | 29.6 ± 3.4 (57) | 5.4 ± 1.0 (57) | 3.0 ± 0.6 (17) |
| sin2-1 (Ler) | 2.6 ± 0.7 (10) | 7.2 ± 2.3 (10)*** | 12.2 ± 2.2 (10) | 2.2 R2 = 0.99 | 34.6 ± 1.9 (14)**** | 7.6 ± 1.4 (14)**** | 3.2 ± 0.7 (12) |
| SIN2 (Ws) | 1.1 ± 0.2 (39) | 3.1 ± 0.6 (39) | 9.1 ± 0.4 (39) | 2.2 R2 = 0.98 | 24.5 ± 1.7 (36) | 6.1 ± 0.9 (36) | 4.1 ± 0.7 (16) |
| sin2-2 (Ws) | 3.7 ± 1.5 (23)**** | 9.1 ± 1.8 (23)**** | 19.2 ± 5.5 (23)**** | 5.1 R2 = 0.98 | 49.4 ± 9.2 (19)**** | 7.6 ± 2.3 (13)* | 10.4 ± 2.4 (11)*** |
Numbers represent the mean ± the standard deviation of the mean. Numbers of plants analyzed are in parentheses. Statistically significant differences between wt-1 and sin2-1 or wt-2 and sin2-2 are designated *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 where P-values are determined by the Student's t-test.
In addition to the reduction in integument growth, sin2 mutations affected other aspects of reproductive development. The number of rosette leaves produced prior to flowering was increased in both mutants relative to wild type when grown in continuous light (Table 1). This alteration of flowering time appeared specific to long-day conditions since sin2-1 mutants grown under short-day (12 hr light) conditions produced a wild-type number of rosette leaves (12.63 ± 1.39 vs. 12.85 ± 0.96). sin2-2 mutants also showed reduced apical dominance, characterized by a doubling in the number of secondary inflorescences, while sin2-1 did not show any obvious alterations in apical dominance (Table 1). In some cases, sin2-1 plants had multiple branches originating from the axil of a single cauline leaf, resulting in a bushy phenotype (supplemental Figure 1 at http://www.genetics.org/supplemental/). sin2-2 plants also exhibited a marked increase in vascular discontinuity within the petals relative to wild type, sometimes exhibiting a completely isolated vein region (supplemental Figures 1 and 2 at http://www.genetics.org/supplemental/). Taken together, the pleiotropic effects of both sin2 mutant alleles suggest that SIN2 may play a role in the production, perception, or response to a variety of developmental signals.
SIN2 is a member of a family of atypical GTPases:
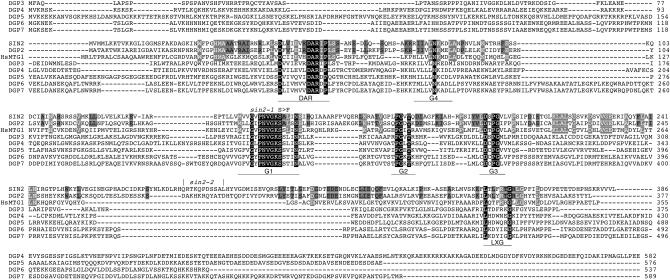
Database searches indicated that SIN2 shares several conserved features, including the unique conserved aspartate–alanine–arginine (DAR) motif, with a relatively uncharacterized family of GTPases, the DAR GTPases (Fu et al. 1998). We performed similarity searches of all publicly available sequence databases and identified DAR proteins from bacteria, archaea, yeast, plants, and animals. Sequence alignments generated from 61 representatives of the identified proteins revealed additional unreported conserved features. Figure 3 shows that sequence and order conservation was present within the four G motifs [in the noncanonical order: G4-G1-G2-G3 (Reynaud et al. 2005)], the DAR motif, and a previously unreported C-terminal motif. In addition to its unique position, the G4 motif showed the greatest intersequence variability among the G motifs. However, the typical G4 motif found in all GTPase superfamily proteins, (Z)4NKXD (where Z is any nonpolar or hydrophobic amino acid residue and X is any amino acid residue), was found in 80% of these DAR proteins, and 7 of the 10 deviations from this consensus consisted of conservative lysine-to-arginine or aspartate-to-glutamate substitutions. The DAR proteins were also found to invariably have a lysine or arginine one to three amino acids preceding the four hydrophobic or nonpolar amino acids of the G4 motif. Examination of the DAR motif region revealed two additional conserved amino acid residues, aspartate (D) and proline (P), flanking the DAR sequence, and several positions generally consisting of nonpolar or hydrophobic residues, expanding the DAR consensus to D(Z)3XZXDARXP. The DAR motif was always found N-terminal to the four G motifs in our sample. C-terminal to the G motifs, a loosely conserved sixth motif, L(X)5G, was also identified. This additional motif, as well as the DAR motif, is likely to be involved in specific protein–protein interactions.
Figure 3.—
DAR protein alignment. An alignment of the seven Arabidopsis DAR proteins, SIN2 and DGP2–7, and the human SIN2 ortholog, human DGP1, highlights the six conserved motifs of this family. Residues found in all of these proteins are shown in white on a black background. These include amino acids within the GTPase motifs G1–G4 and the DAR motif, labeled and underscored below each sequence. There is also a region C-terminal to the recognizable GTPase motifs, which appears loosely conserved and includes a L(X)5G motif. Amino acids conserved between SIN2 and the closely related proteins DGP2 (38% identity, 57% similarity) and human DGP1 (31% identity, 47% similarity) are shaded. Residues conserved between SIN2 and DGP2 or HsDGP1 are shown in black or white letters, respectively. The positions corresponding to the sin2-1 S150F transition and the sin2-2 insertion are indicated above the sequence.
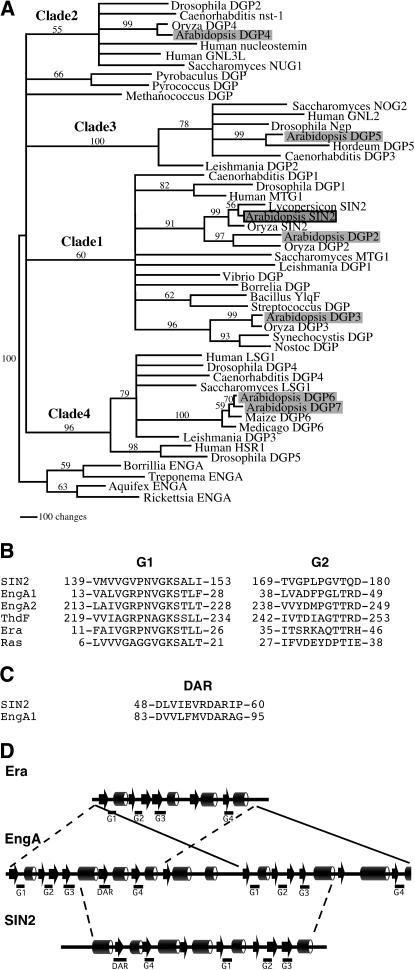
While each prokaryotic genome examined included only a single DAR gene, multiple DAR genes were observed in eukaryotes, with seven members found in Arabidopsis [SIN2 and DAR GTPases 2–7 (DGP2–7)]. Maximum-parsimony analysis was used to evaluate the relationships among 51 of the identified DAR proteins selected to represent the breadth of the entire class. The resulting tree indicated that eukaryotic and eubacterial DAR proteins fall into four distinct clades, which we have designated 1–4 in Figure 4A. Each clade was found to include representatives from all major groups of multicellular crown eukaryotes: plants, animals, and fungi. Each clade also included at least one Arabidopsis DGP. This analysis was not sufficient to resolve the relationships between the archeal proteins and the rest of the DAR proteins or among clades 1–4.
Figure 4.—
Phylogenetic analysis of DAR GTPases. (A) The single tree generated by parsimony analysis of a culled alignment was modified to collapse branches that were not supported in both culling and elision analyses by bootstrap values >50%. Four main clades are resolved, designated clades 1–4. Eukaryotes are represented in each clade. Prokaryotic sequences are found in clade 1 and two orphan branches (including Methanococcus and Pyrobaculus sequences). Arabidopsis proteins are shaded and SIN2 is outlined in black. Numbers on the branches indicate bootstrap values based on 500 bootstrap replicates. Branch lengths are relative to distance as calculated using the transformed BLOSUM45 matrix. Clade designations are shown above the respective branches. Proteins without previous names were given “DGP” designations. (B–D) DAR proteins share conserved sequences and secondary structure with Era family members. (B) An alignment of the G1 and G2 motifs of SIN2, E. coli Era, ThdF, EngA1, and EngA2 demonstrates that the SIN2 G1 motif is similar to that of Era family members (Era, ThdF, and EngA). (C) A region of EngA similar to a DAR motif was found between EngA1 G3 and EngA1 G4. (D) A comparison of the clade 1 consensus secondary structure predicted by the Jpred server (Cuff et al. 1998) with that derived from the E. coli EngA and E. coli Era crystal structures (Chen et al. 1999; Robinson et al. 2002). EngA structure (arrows and cylinders indicate β-sheet and α-helix regions, respectively) appears to derive from a direct repeat of two Era-like units. The DAR GTPases, represented by SIN2, resemble the central (G4–G3) region of the EngA proteins that include DAR motifs.
All eubacterial DAR proteins and SIN2 were included in clade 1. Also included in this clade were the Arabidopsis DGP2 (At4g10650) and DGP3 (At4g02790) proteins as well as proteins from rice, humans, Drosophila, Caenorhabditis, and Saccharomyces. Interestingly, some of the nonplant clade 1 proteins were shown to localize to mitochondria (Barrientos et al. 2003). SIN2 and DGP2, along with their respective rice orthologs Oryza SIN2 and Oryza DGP2, appeared to be more closely related to each other than to any other DAR proteins, suggesting that SIN2 and DGP2 resulted from a duplication postdating the divergence of plants from other eukaryotes. Thus, it is possible that these proteins share overlapping functions. DGP3, which includes a predicted chloroplast transit peptide (data not shown), was placed with high bootstrap support into a small subgroup within clade 1 containing the cyanobacterial DAR proteins. Accordingly, DGP3 may play a role in the chloroplast that is analogous to that of SIN2.
DGP4 (At3g07050), DGP5 (At1g52980), and DGP6/7 (At2g27200/At1g08410) were found in clades 2, 3, and 4, respectively, each of which also includes plant, animal, and fungal representatives, but no bacterial proteins. The clade 4 proteins DGP6 and DGP7 were 72% identical and likely resulted from a relatively recent duplication event. Only one clade 4 protein was found from each of the other plant species represented in the sequence databases. Proteins within a particular clade exhibited increased amino acid conservation among residues flanking each of the six described motifs. Clade 2–4 proteins were longer than the clade 1 proteins at both their N- and C-termini. The N-terminal extensions had conserved features within each group. However, the sequences C-terminal to the LXG motif were highly variable both among and within groups.
Families of GTPases have been defined by similarities among G1 and G2 sequence motifs (Caldon et al. 2001). Our database searches and a previous study (Reynaud et al. 2005) indicated that the DAR subfamily shares some sequence conservation with members of the Era family of GTPases, which include Era, EngA, and ThdF/TrmE and which are found in all eubacteria (Caldon et al. 2001). A comparison of the SIN2 G1 and G2 motifs with the same regions from Escherichia coli Era family members showed that the G1 sequence GXPNVGKS is conserved between SIN2 and Era family members (Figure 4B). The secondary structure predicted from the DAR consensus sequence was compared with the E. coli EngA and Era crystal structures (Chen et al. 1999; Robinson et al. 2002). All the G motifs of the DAR proteins were predicted to lie within similar structural regions of EngA and Era (Figure 4D). Within this family, the DAR GTPases share the greatest sequence similarity with EngA proteins. The G2 sequence PGXT was found in both SIN2 and EngA (Figure 4B; Caldon et al. 2001) and several EngA proteins were found of have a D(X)6DAR motif 29 amino acids N-terminal to a G4-G1-G2-G3 segment (Figure 4C). This is similar to the 22-amino-acid distance between the SIN2 DAR and G4 motifs. In addition, the predicted DAR protein consensus structure had features similar to the region of EngA from its DAR-like motif to the C-terminal G3 (Figure 4D). These traits suggest that SIN2 and other DAR GTPases may be derived from an EngA-like progenitor.
SIN2 localizes to mitochondria:
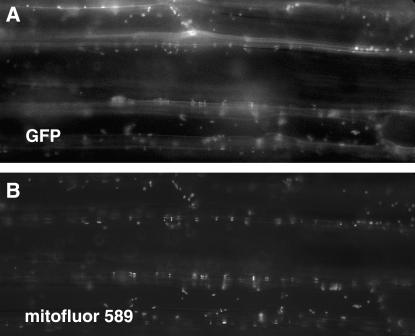
The Saccharomyces DAR proteins have been found to localize to different cellular compartments, including the nucleus, cytoplasm, and mitochondria (Bassler et al. 2001; Barrientos et al. 2003). The subcellular localization of SIN2 was evaluated in transgenic Arabidopsis plants expressing a SIN2:GFP fusion protein. The localization of the fused GFP moiety was determined in the roots, hypocotyls, cotyledons, and trichomes of T2 seedlings. Most cells showed a punctate GFP fluorescence pattern that colocalized with fluorescent-stained mitochondria indicative of a mitochondrial localization of the SIN2 protein in planta (Figure 5). This is consistent with the reported localization of the Saccharomyces and human DAR proteins that group within the SIN2 clade 1 (Barrientos et al. 2003).
Figure 5.—
Localization of SIN2:GFP in Arabidopsis. Arabidopsis hypocotyl cells transformed with 35S:SIN2:GFP. Fluorescence appears white. (A) Green fluorescent protein is localized to small particles within the cells that correspond to mitochondria stained with MitoFluor 589 dye, shown in B.
DISCUSSION
SIN2 is a member of a bacterial/organellar subclass of the DAR GTPase family:
Sequence similarity analyses show that SIN2 includes the DAR motif and reveal a noncanonical G-motif order in the DAR GTPases with the G4 motif preceding G1–G3 (Figure 3; Fu et al. 1998). Structural similarities were found between the G domains of DAR proteins and the E. coli Era GTPases, including the presence of conserved residues in the G1 and G2 motifs (Figure 4B), suggesting that the DAR family is closely related to the Era family (Caldon et al. 2001).
Interestingly, members of the EngA subclass of Era proteins have two canonical G1–G4 GTPase domains in tandem (Figure 4D; Mehr et al. 2000) and, therefore, contain the G4-G1-G2-G3 arrangement observed in DAR proteins. In addition, several EngA proteins include a recognizable D(X)6DAR sequence motif at a position relative to the first G4 motif that is similar to that seen in DAR proteins (Figure 4C). Notably, there is additional amino acid conservation between EngA and DAR proteins within the effector-binding G2 motif (Figure 4B). This level of conservation between EngA and DAR proteins suggests that the two classes derive from a common ancestor that included the duplication of the canonical G1–G4 GTPase domains and a DAR motif and that the DAR proteins derived from this common ancestor through truncation (Figure 4, B and D).
Eukaryotic DAR proteins resolve into four moderately to well-supported clades (1–4), suggesting an early expansion of the gene family (Figure 4A; Reynaud et al. 2005). In contrast, eubacterial DAR GTPases are confined to clade 1, along with SIN2, DPG2, DGP3, and other eukaryotic representatives. This association is supported by another recent phylogenetic analysis (Reynaud et al. 2005). These results suggest that the eukaryotic proteins in this clade could have been acquired in an early endosymbiotic event or other horizontal gene transfer. The first possibility is consistent with the mitochondrial localization of SIN2 (this work) and of its putative orthologs in yeast and mammals (Barrientos et al. 2003). The sequence similarity and close association among SIN2, DGP2, and plant orthologs in the phylogram indicates that DGP2 is also likely a mitochondrial protein that derived from a recent gene duplication event. In contrast, DGP3 and a rice ortholog associate with the DAR proteins from known bacterial relatives of chloroplasts (Figure 4) and include putative chloroplast transit peptides (data not shown). Thus, DGP3 likely represents the chloroplast DAR ortholog in higher plants, as has been previously hypothesized (Reynaud et al. 2005). The structure of clade 1 therefore suggests that each of the two endosymbiotic events related to plant evolution, acquisition of mitochondria and plastids, brought with them specific DAR GTPase lineages.
DAR proteins are important for ribosomal assembly in many subcellular compartments:
Recent reports shed light on the biological function of some of the clade 1 proteins that are putative orthologs of SIN2. Bacillus subtilis YlqF (Figure 4A) was shown to facilitate an essential step in the assembly of the large ribosomal subunit (Matsuo et al. 2006; Uicker et al. 2006). Cells deficient in YlqF accumulate incomplete 50S ribosomal subunits lacking the L16 and L25 protein components (Matsuo et al. 2006; Uicker et al. 2006). Evidence of participation of the GTPase activity of YlqF in the assembly process is provided by the stimulation of YlqF GTPase activity in the presence of 50S subunits and by an association of YlqF with 50S subunits upon addition of a nonhydrolyzable GTP analog (Matsuo et al. 2006). Functional studies on humans and Saccharomyces MTG1 proteins indicate conservation of the ribosome assembly function for these proteins in the eubacterial-derived mitochondria. Saccharomyces MTG1 was shown to be important for translation in mitochondria and a role in assembly of the mitochondrial large ribosomal subunit was hypothesized (Barrientos et al. 2003). It was further shown that the human MTG1 protein could partially complement the Saccharomyces mtg1 mutant, indicating that the ribosome biogenesis function is likely also conserved in animals (Barrientos et al. 2003).
Other diverse DAR proteins have also been found to participate in ribosome biogenesis in other subcellular compartments. The Saccharomyces clade 2 protein NUG1 appears to be involved in ribosome assembly in the nucleus and in transport through nuclear pores (Bassler et al. 2001; Nissan et al. 2002). The Saccharomyces clade 3 protein NOG2 has functions similar to NUG1 and in addition may be involved in mRNA processing (Bassler et al. 2001; Saveanu et al. 2001; Bialkowska and Kurlandzka 2002). LSG1, the Saccharomyces clade 4 protein, is located in the cytoplasm and is also important for ribosome transport through nuclear pores (Nissan et al. 2002; Kallstrom et al. 2003). This indicates conservation of a ribosome biogenesis function among divergent DAR GTPase in the subcellular compartments where they reside.
The reported functions of SIN2 orthologs as well as the mitochondrial localization of SIN2 and of its closest eukaryotic orthologs suggests strongly that SIN2 is a DAR GTPase that functions in mitochondrial ribosome assembly in Arabidopsis. SIN2 mRNA was present in all tested plant parts as would be expected for a mitochondrial protein.
Effects of SIN2-reduced activity in Arabidopsis are consistent with a role in supporting mitochondrial translation:
At the molecular level, the mutation found in the sin2-1 allele would generate a protein with a single amino acid substitution in the GTP loop, the GTP-binding motif of the GTPase. Mutations in this motif were identified in other GTPases and led to gain- or loss-of-function phenotypes (Bourne et al. 1991). However, the phenotypic overlap observed between the sin2-1 and the insertional sin2-2 allele suggests that both alleles cause loss of function or a decrease in SIN2 protein activity.
Mitochondria are involved in biochemical energy utilization and one would expect lower energy levels to translate into reduced or retarded growth. The reduced growth observed in the integuments of sin2 ovules resulted from a decrease in the number of cell divisions in these structures (Broadhvest et al. 2000; this study). This reduced growth could result from insufficient energy production due to compromised mitochondrial activity. Indeed, we have previously characterized the phenotypic effects of mutations in HLL, which encodes a mitochondrial ribosomal protein homologous to the L14 proteins of eubacteria (Skinner et al. 2001). hll mutations result in a dramatic reduction in ovule growth, apparently due to compromised translation in mitochondria (Schneitz 1999; Skinner et al. 2001). The combination of sin2-1 and hll-1 mutations is synergistic, leading to almost complete loss of ovule primordial growth (Broadhvest et al. 2000). This could be due to the combination of effects of the two mutations leading to an even more severe deficit in mitochondrial function, at least during ovule development. Interestingly, the ant mutation was found to be epistatic to sin2-1 but synergistic with hll, leading to a similar but somewhat less severe phenotype in comparison to the sin2-1 hll-1 double mutant (Broadhvest et al. 2000). This supports a hypothesis that ANT is needed to promote growth and HLL is needed for growth to actually occur. SIN2 would then facilitate the role of HLL in ovule and integument growth. Thus, this suggests that HLL is more important for mitochondrial activity than SIN2, at least during ovule development, consistent with the proposed structural and assembly roles for the two proteins with respect to the large ribosomal subunit.
Many of the other phenotypic effects observed for sin2 Arabidopsis plants such as slower growth, reduced pollen formation, and reduced organ size are consistent with a general decrease in energy metabolism that would be expected for plants with reduced mitochondrial activity. However, the more complex phenotypes observed also suggest a role for mitochondria in integration of signaling pathways. For example, both maintenance of axillary bud dormancy and continuity of vascular tissue are under control of auxin-related pathways (Hardtke and Berleth 1998; Chatfield et al. 2000). In addition, inhibition of auxin transport or mutation of the auxin response factor gene ETTIN causes differential effects along the length of the gynoecium similar to the gradation of effects observed for sin2-2 (Nemhauser et al. 2000). Thus, some aspects of SIN2 action may reflect an interaction between phytohormone signaling and mitochondrial function. The broad range of effects that sin2 mutations have on plant growth and architecture is consistent with the SIN2 GTPase regulating mitochondrial activity in a variety of developmental pathways within the plant.
The observation that available sin2 mutants are not more seriously growth deficient suggests that significant mitochondrial function likely persists in these mutant plants. This implies either that SIN2 activity is not always required for assembly of the mitochondrial translational apparatus or that a redundant activity exists. Such a redundant activity could be supplied by DGP2, which our sequence and phylogenic studies have identified as a late-branching paralog of SIN2. DGP2 could have overlapping function with SIN2 and could partially compensate for the reduction or loss of SIN2 activity in the sin2 mutants. Preliminary analyses of Arabidopsis dgp2 mutants show that this gene is functional and seems to affect whole-plant architecture (T. Hill, unpublished results). This would parallel the observed redundancy for HLL and the paralogous HULLENLOS PARALOG (HLP) gene (Skinner et al. 2001). More studies are needed to evaluate the role of DGP2 in mitochondria and putative paralogous interactions with SIN2.
Complexity in mitochondrial modulation of growth pathways:
In plants, mitochondria have been shown to be important for male fertility, appropriate expression of floral homeotic genes, and programmed cell death (Christensen et al. 2002; Hoeberichts and Woltering 2003; Leino et al. 2003; Linke et al. 2003). Work in our laboratory has shown that at least two ribosomal proteins targeted to the mitochondria, HLL and SIN2, are necessary for ovule development and that both also affect the regulation of other developmental pathways in Arabidopsis. Interestingly, both of these genes have functional paralogs in Arabidopsis: DGP2 and HLP, respectively. Together, regulation of these genes could allow for modulation of mitochondrial activity to facilitate the differential growth necessary for morphogenesis. The added complexity that cell mitochondrial number can vary in a tissue- and developmentally dependent manner provides additional plasticity for mitochondria-integrated pathways. SIN2 and HLL, along with their paralogs, offer an entry point to understanding the regulation and impact of mitochondrial protein synthesis in plant development.
Acknowledgments
The authors thank Chris Roxas and Roderick Kumimoto for dedicated assistance in plant care and genotyping, Yuval Eshed for the gift of pOCA28-15, the National Science Foundation Arabidopsis Biological Resource Center at Ohio State for Arabidopsis DNA clones, and members of the Gasser and Bowman Labs for helpful discussions. This work was supported by the National Science Foundation (grant IBN-0079434) and by a U. S. Department of Agriculture National Research Initiative Competitive Grants Program award (2001-35304-09989).
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. AY254472.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, S. C., K. Robinson-Beers, J. M. Villanueva, J. C. Gaiser and C. S. Gasser, 1997. Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 145: 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos, A., D. Korr, K. J. Barwell, C. Sjulsen, C. D. Gajewski et al., 2003. MTG1 codes for a conserved protein required for mitochondrial translation. Mol. Biol. Cell 14: 2292–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski et al., 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8: 517–529. [DOI] [PubMed] [Google Scholar]
- Baum, S. F., and T. L. Rost, 1996. Root apical organization in Arabidopsis thaliana. 1. Root cap and protoderm. Protoplasma 192: 178–188. [Google Scholar]
- Bialkowska, A., and A. Kurlandzka, 2002. Additional copies of the NOG2 and IST2 genes suppress the deficiency of cohesin Irr1p/Scc3p in Saccharomyces cerevisiae. Acta Biochim. Pol. 49: 421–425. [PubMed] [Google Scholar]
- Bourne, H. R., D. A. Sanders and F. McCormick, 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349: 117–127. [DOI] [PubMed] [Google Scholar]
- Broadhvest, J., S. C. Baker and C. S. Gasser, 2000. SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development. Genetics 155: 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldon, C. E., P. Yoong and P. E. March, 2001. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41: 289–297. [DOI] [PubMed] [Google Scholar]
- Chatfield, S. P., P. Stirnberg, B. G. Forde and O. Leyser, 2000. The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 24: 159–169. [DOI] [PubMed] [Google Scholar]
- Chen, X., D. L. Court and X. H. Ji, 1999. Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc. Natl. Acad. Sci. USA 96: 8396–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S., R. A. Creelman, J. E. Mullet and R. A. Wing, 1995. Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Weeds World 2: 17–20. [Google Scholar]
- Christensen, C. A., S. W. Gorsich, R. H. Brown, L. G. Jones, J. Brown et al., 2002. Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14: 2215–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay and G. J. Barton, 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14: 892–893. [DOI] [PubMed] [Google Scholar]
- Elliott, R. C., A. S. Betzner, E. Huttner, M. P. Oakes, W. Q. J. Tucker et al., 1996. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., S. F. Baum and J. L. Bowman, 1999. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99: 199–209. [DOI] [PubMed] [Google Scholar]
- Fu, G., S. Melville, S. Brewster, J. Warner and D. C. Barker, 1998. Analysis of the genomic organisation of a small chromosome of Leishmania braziliensis M2903 reveals two genes encoding GTP-binding proteins, one of which belongs to a new G-protein family and is an antigen. Gene 210: 325–333. [DOI] [PubMed] [Google Scholar]
- Gaiser, J. C., K. Robinson-Beers and C. S. Gasser, 1995. The Arabidopsis SUPERMAN gene mediates asymmetric growth of the outer integument of ovules. Plant Cell 7: 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg, O. L., R. A. Miller and K. Ojima, 1968. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50: 151–158. [DOI] [PubMed] [Google Scholar]
- Gleave, A. P., 1992. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207. [DOI] [PubMed] [Google Scholar]
- Hardtke, C. S., and T. Berleth, 1998. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, B. A., J. He, S. O. Park and C. S. Gasser, 2000. TSO1 is a novel protein regulating cell division and directional cell expansion in Arabidopsis. Development 127: 2219–2226. [DOI] [PubMed] [Google Scholar]
- Hauser, B. A., J. M. Villanueva and C. S. Gasser, 1998. Arabidopsis TSO1 regulates directional processes in cells during floral organogenesis. Genetics 150: 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and J. G. Henikoff, 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts, F. A., and E. J. Woltering, 2003. Multiple mediators of plant programmed cell death: interplay of conserved cell death mechanisms and plant-specific regulators. BioEssays 25: 47–57. [DOI] [PubMed] [Google Scholar]
- Kallstrom, G., J. Hedges and A. Johnson, 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23: 4344–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucher, K. M., H. Chow, L. Reiser and R. L. Fischer, 1996. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8: 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, A. R., and B. Kirchheim, 1987. Handling of Arabidopsis, pp. 4.1.1–4.2.7 in Arabidopsis Information Service, Vol. 24: Genetic Resources in Arabidopsis, edited by A. R. Kranz. Arabidopsis Information Service, Frankfurt, Germany.
- Krysan, P. J., J. C. Young and M. R. Sussman, 1999. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leino, M., R. Teixeira, M. Landgren and K. Glimelius, 2003. Brassica napus lines with rearranged Arabidopsis mitochondria display CMS and a range of developmental aberrations. Theor. Appl. Genet. 106: 1156–1163. [DOI] [PubMed] [Google Scholar]
- Linke, B., T. Nothnagel and T. Borner, 2003. Flower development in carrot CMS plants: mitochondria affect the expression of MADS box genes homologous to GLOBOSA and DEFICIENS. Plant J. 34: 27–37. [DOI] [PubMed] [Google Scholar]
- Loytynoja, A., and M. C. Milinkovitch, 2001. SOAP, cleaning multiple alignments from unstable blocks. Bioinformatics 17: 573–574. [DOI] [PubMed] [Google Scholar]
- Matsuo, Y., T. Morimoto, M. Kuwano, P. C. Loh, T. Oshima et al., 2006. The GTP-binding protein YlqF participates in the late step of 50S ribosomal subunit assembly in Bacillus subtilis. J. Biol. Chem. 281: 8110–8117. [DOI] [PubMed] [Google Scholar]
- Mehr, I. J., C. D. Long, C. D. Serkin and H. S. Seifert, 2000. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics 154: 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, R. J., L. M. Kotow and C. S. Gasser, 2002. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129: 4281–4289. [DOI] [PubMed] [Google Scholar]
- Morimoto, T., P. C. Loh, T. Hirai, K. Asai, K. Kobayashi et al., 2002. Six GTP-binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148: 3539–3552. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and F. Skoog, 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15: 473–479. [Google Scholar]
- Nemhauser, J. L., L. J. Feldman and P. C. Zambryski, 2000. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127: 3877–3888. [DOI] [PubMed] [Google Scholar]
- Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey and E. Hurt, 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21: 5539–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud, E. G., M. A. Andrade, F. Bonneau, T. B. Ly, M. Knop et al., 2005. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, V. L., J. Hwang, E. Fox, M. Inouye and A. M. Stock, 2002. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 10: 1649–1658. [DOI] [PubMed] [Google Scholar]
- Robinson-Beers, K., R. E. Pruitt and C. S. Gasser, 1992. Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4: 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe, J. L., C. J. Rivin, R. A. Sessions, K. A. Feldmann and P. C. Zambryski, 1993. The TOUSLED gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 75: 939–950. [DOI] [PubMed] [Google Scholar]
- Roe, J. L., T. Durfee, J. R. Zupan, P. P. Repetti, B. G. McLean et al., 1997. a TOUSLED is a nuclear serine-threonine protein kinase that requires a coiled-coil region for oligomerization and catalytic activity. J. Biol. Chem. 272: 5838–5845. [DOI] [PubMed] [Google Scholar]
- Roe, J. L., J. L. Nemhauser and P. C. Zambryski, 1997. b TOUSLED participates in apical tissue formation during gynoecium development in Arabidopsis. Plant Cell 9: 335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas et al., 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20: 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneitz, K., 1999. The molecular and genetic control of ovule development. Curr. Opin. Plant Biol. 2: 13–17. [DOI] [PubMed] [Google Scholar]
- Schneitz, K., M. Hulskamp and R. E. Pruitt, 1995. Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 7: 731–749. [Google Scholar]
- Schneitz, K., S. C. Baker, C. S. Gasser and A. Redweik, 1998. Pattern formation and growth during floral organogenesis: HUELLENLOS and AINTEGUMENTA are required for the formation of the proximal region of the ovule primordium in Arabidopsis thaliana. Development 125: 2555–2563. [DOI] [PubMed] [Google Scholar]
- Schumacher, K., D. Vafeados, M. McCarthy, H. Sze, T. Wilkins et al., 1999. The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 13: 3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner, D. J., S. C. Baker, R. J. Meister, J. Broadhvest, K. Schneitz et al., 2001. The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 13: 2719–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner, D. J., T. A. Hill and C. S. Gasser, 2004. Regulation of ovule development. Plant Cell 16: S32–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.-Y., T. Leung, L. K. Ehler, C. Wang and Z. Liu, 2000. Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development 127: 2207–2217. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 1999. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods), Version 4.0. Sinauer Associates, Sunderland, MA.
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL*X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, R. Y., and R. D. McKay, 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16: 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uicker, W. C., L. Schaefer and R. A. Britton, 2006. The essential GTPase RbgA (YlqF) is required for 50S ribosome assembly in Bacillus subtilis. Mol. Microbiol. 59: 528–540. [DOI] [PubMed] [Google Scholar]
- Villanueva, J. M., J. Broadhvest, B. A. Hauser, R. J. Meister, K. Schneitz et al., 1999. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 13: 3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, W. C., J. Gatesy and R. Desalle, 1995. Elision: a method for accommodating multiple molecular sequence alignments with alignment-ambiguous sites. Mol. Phylogent. Evol. 4: 1–9. [DOI] [PubMed] [Google Scholar]