Abstract
Olfactory receptors (ORs) located in the cell membrane of olfactory sensory neurons of the nasal epithelium are responsible for odor detection by binding specific odorant ligands. Primates are thought to have a reduced sense of smell (microsmatic) with respect to other mammals such as dogs or rodents. We have previously demonstrated that over 70% of the human OR genes have become nonfunctional pseudogenes, leading us to hypothesize that the reduced sense of smell could correlate with the loss of functional genes. To extend these results, we sampled the OR gene repertoire of 10 primate species, from prosimian lemur to human, in addition to mouse. About 221 previously unidentified primate sequences and 33 mouse sequences were analyzed. These sequences encode ORs distributed in seven families and 56 subfamilies. Analysis showed a high fraction (≈50% on average) of pseudogenes in hominoids. In contrast, only ≈27% of OR genes are pseudogenes in Old World monkeys, and New World monkeys are almost free of pseudogenes. The prosimian branch seems to have evolved differently from the other primates and has ≈37% pseudogene content. No pseudogenes were found in mouse. With the exception of New World monkeys, we demonstrate that primates have a high fraction of OR pseudogenes compared with mouse. We hypothesize that under relaxed selective constraints, primates would have progressively accumulated pseudogenes with the highest level seen in hominoids. The fraction of pseudogenes in the OR gene repertoire could parallel the evolution of the olfactory sensory function.
Keywords: olfaction, pseudogenes, evolution
Mammals are able to discriminate between thousands of odor molecules. This capacity relies on a multigene family encoding 500–1,000 olfactory receptors (ORs; ref. 1). These receptors are expressed mainly in the olfactory epithelium and have been found in a number of species including mammals (1–5), birds (6, 7), amphibians (8), and fish (9). All these receptors belong to the G protein-coupled receptor superfamily and share features of sequence and structure, such as seven hydrophobic transmembrane domains.
The sense of smell plays an important role in mammalian social behavior, location of food, and detection of predators. However, mammals vary in their olfactory ability (10, 11). The sense of smell in primates is greatly reduced (microsmatic) with respect to other mammals such as dogs (12) or rodents (10, 11). Various explanations for the differences in olfactory performance have been hypothesized. Differences in the anatomical structures (e.g., size and location) devoted to olfaction could partly explain these differences. For example, dogs, which have an olfactory sensitivity up to 100 times greater than humans, have on average ≈100 cm2 of olfactory epithelium, whereas humans have only 10 cm2 (see ref. 4 and references therein). Variations in the size and diversity of the expressed OR gene family could also account for these differences. We recently demonstrated that the human OR gene repertoire is distributed in over 25 chromosomal sites, and over 70% of these OR genes are pseudogenes, i.e., the sequences have accumulated deleterious mutations such as in-frame stop codons and/or indel frameshifts (3). This finding led us to hypothesize that the reduction of the sense of smell observed in primates could parallel the reduction of the number of functional OR genes.
To test this hypothesis, we wished to characterize the evolution of the OR gene family in other primates. We performed a random survey of OR genes from primate hominoids to prosimians. In parallel, we constructed a mouse OR-enriched library from genomic DNA to sequence a number of OR. The comparison of the OR gene repertoire from macrosmatic mouse and primates provides insight into the evolution of this multigene family and could reflect the evolution of a sensory function in mammals in response to selective constraints.
Materials and Methods
Cloning and Analysis of OR-Like Sequences in Primates and Mouse.
The isolation of OR-related sequences has been described elsewhere (3, 13). Briefly, 100 ng of genomic DNA from each species was subjected to PCR by using consensus OR primers OR5B-OR3B [OR5B (TM2), 5′-CCCATGTA(T/C)TT-(G/C/T)TT(C/T)CTC(A/G/T)(G/C)(C/T)AA(C/T)-(T/C)T(G/A)TC-3′; PMY(F/L)FL(S/A/T/G/C)NLS; OR3B (TM7), 5′-AG(A/G)C(A/T)(A/G)TAIATGAAIGG-(A/G)TTCAICAT-3′; M(L/F/V/I)NPF(I/M)Y(S/C)L; ref. 14]. A second pair of consensus primers, OR3.1-OR7.1 [OR3.1 (TM3), 5′-GCIATGGCITA(C/T)GA(C/T)(A/C)GITA-3′; AMAYD(S/R)Y; OR7.1 (TM7), 5′-A(A/G)I(G/C)(A/T)(A/G)TA(A/G/T)AT(A/G)AAIGG(A/G)TT-3′; NPFIY-(S/R/T/C/W)(L/F); refs. 15 and 16], was also used to amplify primate OR sequences. PCR products were subcloned in the TA vector (Invitrogen), and recombinant clones were identified by PCR. Sequencing of the OR sequences was performed, and sequences were assembled and analyzed as detailed elsewhere (3). The following species were studied: human (Homo sapiens, HSA), chimpanzee (Pan troglodytes, PTR), gorilla (Gorilla gorilla, GGO), orangutan (Pongo pygmaeus, PPY), gibbon (Hylobates lar, HLA), macaque (Macaca sylvanus, MSY), baboon (Papio papio, PPA), marmoset (Callithrix jacchus, CJA), squirrel monkey (Saimiri sciureus, SSC, and Saimiri boliviensis, SBO), lemur (Eulemur fulvus, EFU, and Eulemur rubriventer, ERU), and mouse (Mus musculus domesticus, MMU). In addition, a few zebrafish (Danio rerio, DRE) sequences were characterized with primers OR3.1-OR7.1.
Pairwise sequence comparisons and multiple alignments were performed with gap and pileup from the GCG package (Wisconsin Package, version 8).
Construction and Screening of an OR-Specific Mouse Sublibrary.
Mouse OR clones obtained by PCR as described above were gridded in 96-well microtiter dishes (1,536 clones in eight plates). For hybridization screening, the clones were robot-spotted in duplicate on high-density filters as described elsewhere (17). Approximately 90% of the clones were identified as OR genes. This library was screened to identify clones hybridizing to human OR pseudogene sequences. Human plasmid DNA probes were radiolabeled to a specific activity of 108–109 cpm/μg by random hexamer priming (18) by using [α-32P]dCTP (Amersham Pharmacia). Filter hybridizations were carried out under standard hybridization conditions (19) and exposed to Kodak x-ray film at −80°C. Three human OR probes were used: OR1-72, OR912-47, and OR15-71 (GenBank accession nos. U86218, U86230, and U86296, respectively).
Results
Isolation and Analysis of Primate OR Sequences.
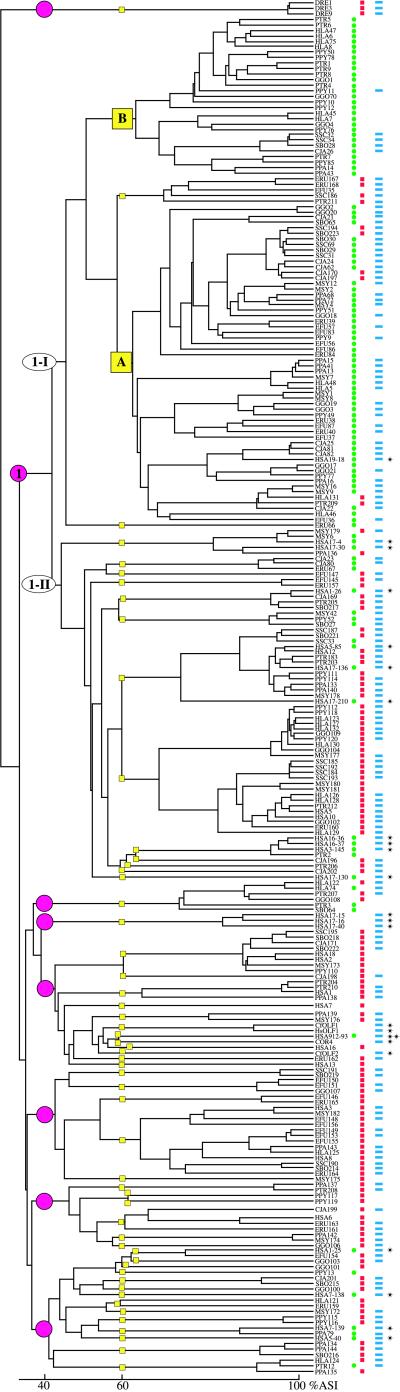
To sample the OR genes in primate species, we randomly sequenced OR genes from anthropoids and prosimians (Fig. 1). OR genes were obtained by PCR on genomic DNA from the different species by using consensus OR primer pairs OR5B-OR3B and OR3.1-OR7.1 chosen, respectively, in the transmembrane domains TM2 and TM7 as well as TM3 and TM7. Except human, 18–35 individual OR clones were sequenced per taxon. A total of 221 OR sequences, representing 10 species, was analyzed. These sequences are distributed in different groups whose percentages of nucleotide sequence identity range from ≈35 to >99% (not shown). The corresponding amino acid sequences were compared with a variety of OR sequences from the public databases and previous studies (3). All sequences have the characteristic features of ORs, with a heptahelical structure and conserved motifs as defined (1, 3, 14). The use of two pairs of consensus primers made our sampling representative of the OR gene repertoire. Primate sequences are distributed in seven families [sequences that share >40% amino acid identity (ASI) define a family] and 56 subfamilies (ASI>60%), with group 1-II of family 1 representing the zone of overlap of sequences derived by using the two primer pairs (Fig. 2). Nonhuman primate OR genes are represented in six families and about 45 subfamilies. Numerous sequences are grouped in family 1 (≈66%) comprising subfamily 1A, the largest subfamily (57 of 221 or 26%). Subfamily 1B is almost devoid of coding human OR sequences (Fig. 2). Subfamily 1A contains only human pseudogenes originating from chromosomes 14 and 19 (not shown), whereas subfamily 1B contains human pseudogenes lying on various chromosomes (not shown; ref. 3). As we found previously for human (3), the amino acid sequences deduced from the nonhuman primate sequences identified many pseudogenes (Fig. 2 and Table 1). Table 1 provides information about the evolution of the pseudogene fraction along with the evolution of primates. Hominoids present the highest fraction of pseudogenes (39 to >70%; average ≈50%). Old World monkeys (macaque and baboon) have a lower pseudogene fraction (20 to 35%; average 27%), whereas even fewer pseudogenes were found among the sequences derived from New World monkeys. Only one pseudogene (SBO64) was identified among the 49 sequences obtained from marmoset and two species of squirrel monkey. In contrast, 37% of the prosimian lemur OR sequences were pseudogenes.
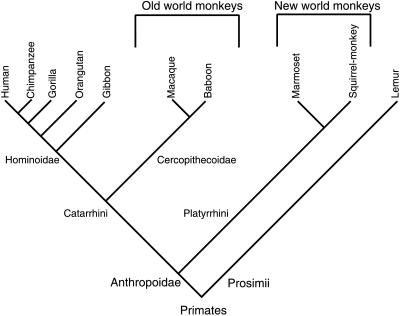
Figure 1.
Schematic phylogeny tree of the primate species used in the present study (adapted from ref. 32).
Figure 2.
Comparison of the deduced protein OR sequences obtained from the different primate species characterized in this study. The dendogram was established with the pileup program from the GCG Package. Percentage of ASI was determined by pairwise sequence comparisons with the gap program and is indicated along the abscissa of the tree. Sequences from the literature are indicated by asterisks. Human OR sequences derived from the use of the OR5B-OR3B primers and representing the main OR families were selected from refs. 3 and 13. Dog (CfOLF1, its human counterpart HsOLF1, and CfOLF2) and chicken (COR4) sequences were selected from refs. 4 and 7, respectively. OR families (greater than 40% ASI) are indicated by pink circles, and subfamilies (greater than 60% ASI) are indicated by yellow squares. The main family was arbitrarily named family 1 and subdivided into two subfamily groups, 1-I and 1-II (ovals). Group 1-II comprises subfamilies A and B. Beside the sequence names, green dots indicate sequences derived from the use of the OR5B-OR3B consensus primers; red squares indicate sequences derived from the OR3.1–7.1 consensus primers; and blue rectangles indicate potentially functional genes (uninterrupted ORFs). In the case of HSA 912–93 (blue rectangle and double asterisk), this sequence contains only one nonsense point mutation in human but is potentially coding in other primates (ref. 13; see also Table 1).
Table 1.
Fraction of pseudogenes in the OR gene repertoire of primate species and mouse
| Family/species | No. sequences analyzed | Percentage ORF | Percentage pseudogenes | Average percentage pseudogenes by family |
|---|---|---|---|---|
| Hominoids | 50 | |||
| Human (HSA) | 99 | 30 | 70 | |
| Chimpanzee (PTR) | 21 | 52 | 48 | |
| Gorilla (GGO) | 18 | 50 | 50 | |
| Orangutan (PPY) | 23 | 61 | 39 | |
| Gibbon (HLA) | 22 | 59 | 41 | |
| Old World monkeys | 27 | |||
| Macaque (MSY) | 20 | 65 | 35 | |
| Baboon (PPA) | 21 | 81 | 19 | |
| New World monkeys | 2 | |||
| Marmoset (CJA) | 19 | 100 | 0 | |
| Squirrel monkey (SSC) | 15 | 100 | 0 | |
| Squirrel monkey (SBO) | 15 | 93 | 7 | |
| Prosimians | 37 | |||
| Lemur (EFU) | 19 | 58 | 42 | |
| Lemur (ERU) | 16 | 69 | 31 | |
| Rodents | 0 | |||
| Mouse (MMU) | 33 | 100 | 0 | |
| Fish | 0 | |||
| Zebrafish (DRE) | 3 | 100 | 0 |
Sequence Analysis of Mouse OR Sequences.
To test whether mammals thought to be microsmatic or macrosmatic differ in the fraction of pseudogenes in their OR repertoire, we surveyed OR sequences in the mouse genome. We constructed a mouse sublibrary enriched for OR-related sequences amplified by PCR from the mouse genome (see Materials and Methods).
Randomly selected mouse OR clones (n = 19) were sequenced. All 19 have an uninterrupted ORF and are potentially functional. These sequences group primarily in family 1 (not shown) and vary from ≈52 to >99% nucleotide sequence identity (not shown). In addition, in an attempt to bias in favor of selecting mouse OR pseudogenes, we searched for mouse OR sequences homologous to human pseudogenes. One member was chosen from three different OR pseudogene families: clones 1–72, 15–71, and 912-47 from chromosomes 1, 15, and 11, respectively (3). Each of these genes belongs to one of the three main groups of human OR sequences and has accumulated a number of mutations such as stop codons and indel frameshifts (3). The amino acid sequence identity between these three ranges from 31% to 41%.
High-density filters from the mouse OR sublibrary were then hybridized separately with the three human pseudogene probes at a high stringency; 14 clones were sequenced on both strands. These sequences showed 38% to 53% ASI to the human sequences used to select them, indicating that they are not the orthologs of the human pseudogenes. All have an uninterrupted ORF from TM2 to TM7 (not shown). Together, we sequenced 33 mouse OR sequences, none of which contained characteristic features of pseudogenes.
Discussion
What is the basis for the differences in olfactory ability observed among mammals? Diverse reasons have been suggested, i.e., the size of the anatomical structures devoted to olfaction (e.g., olfactory epithelium, olfactory bulb, and cortical structures), the number of OR families/subfamilies, and the total number and diversity of expressed OR genes. The olfactory epithelial surface of macrosmatic animals such as dogs is larger than that in microsmatic humans (see ref. 4 and references therein). On the other hand, by using unique dog sequence probes that represent specific OR subfamilies that will not cross-hybridize with other subfamilies, comparative analyses have been performed by Southern blot analysis among a panel of mammals including dog and human. This study indicates that the number of OR sequences per subfamily is similar in microsmatic and macrosmatic animals (4). Recently, we demonstrated that a high fraction (>70%) of the human OR genes have lost function during evolution and are represented as pseudogenes (3). We found that chromosomes 7, 16, and 17 contain a high fraction of potentially coding OR sequences, whereas other chromosomes, such as chromosome 3 (3, 20) or 11 (3), contained primarily pseudogenes. Other studies on chromosome 17 (14, 21) and on chromosome 11 (22) in which 75% of the OR sequences identified were pseudogenes support these observations. These findings led us to hypothesize that the number of functional OR genes could be correlated to the olfactory capability of a given animal. In an attempt to test this hypothesis, we sampled the OR gene repertoire of the main primate species, thought to be microsmatic, and of mouse, thought to be macrosmatic. Our analysis clearly indicates that (see Fig. 1 and Table 1), from New World monkeys to hominoids, there is an increase in the percentage of OR pseudogenes from ≈0 to ≈70%, with the highest pseudogene content observed in gorilla, chimpanzee, and human. Supporting this observation, during the course of this work, Sharon and colleagues (23) published a study about the evolution in primates of the OR genes orthologous to the human OR gene cluster located on chromosome 17p13.3 (14, 21). The authors conclude that a rapid decline (≈10 million years ago, corresponding to the radiation of hominoids) of the functional OR repertoire occurred in mammals. In addition, we showed in a recent study that the pool of pseudogenes is still growing in human, which will probably evolve toward a minimal set of functional OR genes (13). It is therefore likely that there is a selective advantage for New World monkeys to retain a high proportion of functional OR genes, whereas this advantage seems to be reduced in Old World monkeys. The prosimian branch represented by two lemur species does not follow this rule and has accumulated a high fraction of OR pseudogenes (Table 1). This particular taxon is localized in Madagascar and is composed of diurnal (as EFU) and nocturnal (as ERU) animals, both of which are thought to have evolved from a common ancestral species. The two species showed no striking difference in the OR pseudogene fraction, suggesting that loss of functional OR genes preceded their divergence. Nevertheless, despite the number of OR families and subfamilies presented in this study (Fig. 2), the collection of sequences of the present work represents only a subset of OR genes, and it is still possible that the fraction of functional OR genes and OR pseudogenes could be different in other segments of the OR family in different species. However, although some studies indicate that mice and dogs have increased olfactory abilities compared with humans (10, 24), accurate experiments to compare the olfactory ability between the different primates species remain to be conducted to support our hypothesis.
All OR sequences we derived from mouse are potentially coding. No pseudogenes were detected either by sequencing randomly selected OR sequences or by deliberately screening with human OR pseudogene probes. This result indicates that the OR pseudogene content is either zero or restricted to rare examples in mouse (25).
Taken together, this study led us to hypothesize that the reduction of the sense of smell could correlate with the fraction of functional OR genes in the genome. This phenomenon would probably result from the relaxation of the selective pressure exerted on the different species, i.e., as soon as the function becomes nonessential for the survival or the social behavior of a particular species, the genes responsible for that function tend to accumulate deleterious mutations. Actually, it is likely that the different hypotheses evoked to explain the loss of function would be not exclusive but that a parallel might exist between the reduction of the anatomical structures devoted to olfaction, the decrease in the number of functional OR genes, and the reduction of the sense of smell. This hypothesis is strongly supported by the fact that aquatic mammals such as dolphin, which has a reduced olfactory apparatus, have only OR pseudogenes (15). These animals live in water and do not need to smell volatile odorants. Therefore, a parallel degeneration of the olfactory organs and the OR gene repertoire has occurred probably because of the relaxation of the selective constraints. This sort of observation has also been made in the blind marsupial mole, in which degeneration of the eyes is accompanied by mutation of the interphotoreceptor retinoid binding protein gene, which is involved in the regeneration of rhodopsin in the visual cycle (26).
However, it is difficult to measure and compare the olfactory efficiency of different animal species. Various parameters such as the threshold of detection of odorants (sensitivity), the range of odors detectable, and the discriminatory power (acuity) are key parts of the olfactory ability. Thus, it is difficult to determine precisely which of these parameters are taken in account when comparing two species, and therefore the origin of the olfactory deficiency of primates remains a controversial and difficult point to address. Furthermore, there are no data available in the literature comparing the olfactory ability between the different primate species studied in this work, and most of the few studies conducted to compare the performances of mouse or dog versus human concern either the threshold of detection of very few odorants (for example n-amyl acetate; ref. 27) or the observation that trained dogs are far better than humans in detecting hidden objects such as mines, drugs, or people buried after natural disasters (28, 29). Nonetheless, although the fraction of OR pseudogenes has been estimated in mouse, it remains to be determined in dog.
The chromosomal distribution of the OR gene repertoire arose through multiple duplication rounds (3, 5, 20, 30, 31) giving rise to paralogous regions. Even though the number of duplication events may be different among the mammals, overall it seems that the number of OR genes was established before the divergence of mammals (4). This observation explains why, as determined by Southern analysis, there is no striking difference in the number of OR genes of four different subfamilies between the sea lion, which has an underdeveloped olfactory apparatus, and other mammals (4). On the other hand, the Southern blot approach does not indicate the functionality of the OR sequences, and we predict that a large fraction of the sea lion OR genes could be pseudogenes as described for the dolphin (23). Similarly striking differences have been observed in the olfactory abilities of different breeds of dogs (12). Despite the variations in the size of the olfactory epithelium of the different breeds (ref. 12 and references therein), it would be interesting to determine the biological basis for the differences in performances between sight and scent hounds. One obvious possibility is loss of functional OR genes; however, given the recent origin of all modern dogs, this explanation seems unlikely. Other explanations could be changes in behavior, in expression brought about by the modification of a key master transcription factor, or in the unusual mechanism that allows only one OR gene allele or the other to be expressed exclusively in any one epithelium cell.
Finally, we hypothesize that the study of the evolution of the OR gene repertoire through the determination of the pseudogene fraction could mirror the evolution of the olfactory sensory function in microsmatic and macrosmatic mammals.
Acknowledgments
We are grateful to Drs. Barbara Trask and Andrew Goldsborough for critical comments on manuscript, Dr. Joachim Freitag for technical advice, Dr. Armand Renucci for kindly providing zebrafish DNA, Prof. Jacques Demaille for his interest to the work, and Dr. Concepcion Ferraz for technical support. This research was supported by a grant from the Programme Génome du Centre National de la Recherche Scientifique.
Abbreviations
- OR
olfactory receptors
- HSA
Homo sapiens
- PTR
Pan troglodytes
- GGO
Gorilla gorilla
- PPY
Pongo pygmaeus
- HLA
Hylobates lar
- MSY
Macaca sylvanus
- PPA
Papio papio
- CJA
Callithrix jacchus
- SSC
Saimiri sciureus
- SBO
Saimiri boliviensis
- EFU
Eulemur fulvus
- ERU
Eulemur rubriventer
- MMU
Mus musculus domesticus
- DRE
Danio rerio
- ASI
amino acid identity
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF022649, AF073959–AF073989, AF127814–AF127907, and AF179716–AF179846).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040580197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040580197
References
- 1.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Selbie L A, Townsend-Nicholson A, Iismaa T I, Shine J. Mol Brain Res. 1992;13:159–163. doi: 10.1016/0169-328x(92)90057-i. [DOI] [PubMed] [Google Scholar]
- 3.Rouquier S, Taviaux S, Trask B, Brand-Arpon V, van den Engh G, Demaille J, Giorgi D. Nat Genet. 1998;18:243–250. doi: 10.1038/ng0398-243. [DOI] [PubMed] [Google Scholar]
- 4.Issel-Tarver L, Rine J. Genetics. 1997;145:185–195. doi: 10.1093/genetics/145.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan S L, Adamson M C, Ressler K J, Kozak C A, Buck L B. Proc Natl Acad Sci USA. 1996;93:884–888. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nef P, Hermans-Borgmeyer I, Artières-Pin H, Beasley L, Dionne V E, Heinemann S F. Proc Natl Acad Sci USA. 1992;89:8948–8952. doi: 10.1073/pnas.89.19.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leibovici M, Lapointe F, Aletta P, Ayer-Le Lièvre C. Dev Biol. 1996;175:118–131. doi: 10.1006/dbio.1996.0100. [DOI] [PubMed] [Google Scholar]
- 8.Freitag J, Krieger J, Strotman J, Breer H. Neuron. 1995;15:1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 9.Ngai J, Dowling M M, Buck L, Axel R, Chess A. Cell. 1993;72:657–666. doi: 10.1016/0092-8674(93)90395-7. [DOI] [PubMed] [Google Scholar]
- 10.Moulton D G. Am Zool. 1967;7:421–429. doi: 10.1093/icb/7.3.421. [DOI] [PubMed] [Google Scholar]
- 11.Stoddart D. The Ecology of Vertebrate Olfaction. New York: Chapman and Hall; 1980. [Google Scholar]
- 12.Issel-Tarver L, Rine J. Proc Natl Acad Sci USA. 1996;93:10897–10902. doi: 10.1073/pnas.93.20.10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouquier S, Friedman C, Delettre C, van den Engh G, Blancher A, Crouau-Roy B, Trask B, Giorgi D. Hum Mol Genet. 1998;7:1337–1345. doi: 10.1093/hmg/7.9.1337. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Arie N, Lancet D, Taylor C, Khen M, Walker N, Ledbetter D H, Carrozzo R, Patel K, Sheer D, Lehrach H, et al. Hum Mol Genet. 1994;3:229–235. doi: 10.1093/hmg/3.2.229. [DOI] [PubMed] [Google Scholar]
- 15.Freitag J, Ludwig G, Andreini P, Roessler P, Breer H. J Comp Physiol. 1998;183:635–650. doi: 10.1007/s003590050287. [DOI] [PubMed] [Google Scholar]
- 16.Freitag J, Beck A, Ludwig G, von Buchholtz L, Breer H. Gene. 1999;226:165–174. doi: 10.1016/s0378-1119(98)00575-7. [DOI] [PubMed] [Google Scholar]
- 17.Rouquier S, Stubbs L, Gaillard-Sanchez I, Giorgi D. Mamm Genome. 1999;10:1172–1174. doi: 10.1007/s003359901185. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg A, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 19.Rouquier S, Giorgi D, Trask B, Bergmann A, Phillips M, MacLennan D, de Jong P J. Genomics. 1993;17:330–340. doi: 10.1006/geno.1993.1329. [DOI] [PubMed] [Google Scholar]
- 20.Brand-Arpon V, Rouquier S, Massa H, de Jong P, Ferraz C, Ioannou P, Demaille J, Trask B, Giorgi D. Genomics. 1999;56:98–110. doi: 10.1006/geno.1998.5690. [DOI] [PubMed] [Google Scholar]
- 21.Glusman G, Clifton S, Roe B, Lancet D. Genomics. 1996;37:147–160. doi: 10.1006/geno.1996.0536. [DOI] [PubMed] [Google Scholar]
- 22.Buettner J, Glusman G, Ben-Arie N, Ramos P, Lancet D, Evans G. Genomics. 1998;53:56–68. doi: 10.1006/geno.1998.5422. [DOI] [PubMed] [Google Scholar]
- 23.Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D. Genomics. 1999;61:24–36. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- 24.Krestel D, Passe D, Smith J C, Jonsson L. Neurosci Biobehav Rev. 1984;8:169–174. doi: 10.1016/0149-7634(84)90037-x. [DOI] [PubMed] [Google Scholar]
- 25.Mombaerts P. Curr Opin Genet Dev. 1999;9:315–320. doi: 10.1016/s0959-437x(99)80047-1. [DOI] [PubMed] [Google Scholar]
- 26.Springer M, Burk A, Kavanagh J, Waddell V, Stanhope M. Proc Natl Acad Sci USA. 1997;94:13754–13759. doi: 10.1073/pnas.94.25.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moulton D G, Celebi G, Fink R P. In: Taste and Smell in Vertebrates. Wolstenholme G, Knight J, editors. London: Churchill; 1970. pp. 238–250. [Google Scholar]
- 28.Komar D. J Forensic Sci. 1999;44:405–408. [PubMed] [Google Scholar]
- 29.Ashton E H, Eayrs J T. In: Taste and Smell in Vertebrates. Wolstenholme G, Knight J, editors. London: Churchill; 1970. pp. 251–263. [Google Scholar]
- 30.Trask B, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Hum Mol Genet. 1998;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- 31.Trask B J, Massa H, Brand-Arpon V, Chan K, Friedman C, Nguyen O T, Eichler E, van den Engh G, Rouquier S, Shizuya H, et al. Hum Mol Genet. 1998;7:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- 32.O'Brien S, Seuanez H, Womack J. Annu Rev Genet. 1988;22:323–351. doi: 10.1146/annurev.ge.22.120188.001543. [DOI] [PubMed] [Google Scholar]