Abstract
Expression of KIT tyrosine kinase is critical for normal germ cell development and is observed in the majority of seminomas. Activating mutations in KIT are common in gastrointestinal stromal tumors and mastocytosis. In this study we examined the frequency and spectrum of KIT mutations in 54 testicular seminomas, 1 ovarian dysgerminoma and 37 non-seminomatous germ cell tumors (NSGCT). Fourteen seminomas (25.9%) contained exon 17 point mutations including D816V (6 cases), D816H (3 cases), Y823D (2 cases), and single examples of Y823C, N822K, and T801I. No KIT mutations were found in the ovarian dysgerminoma or the NSGCTs. In transient transfection assays, mutant isoforms D816V, D816H, Y823D, and N822K were constitutively phosphorylated in the absence of the natural ligand for KIT, stem cell factor (SCF). In contrast, activation of T801I and wild-type KIT required SCF. Mutants N822K and Y823D were inhibited by imatinib mesylate (Gleevec, previously STI571) whereas D816V and D816H were both resistant to imatinib mesylate. Biochemical evidence of KIT activation, as assessed by KIT phosphorylation and KIT association with phosphatidylinositol (PI) 3-kinase in tumor cell lysates, was largely confined to seminomas with a genomic KIT mutation. These findings suggest that activating KIT mutations may contribute to tumorigenesis in a subset of seminomas, but are not involved in NSGCT.
KIT is a 145-kd transmembrane glycoprotein that is the product of the KIT gene, the normal cellular homologue of the feline sarcoma virus oncogene v-kit. A member of the subclass III family of receptor tyrosine kinases, KIT is closely related to the receptors for PDGF, M-CSF, and FLT3 ligand.1–3 The ligand for KIT, stem cell factor (SCF; also known as steel factor), promotes the dimerization and autophosphorylation of KIT receptors. The resulting phosphorylated tyrosine residues provide binding sites for signaling molecules that contain SH2 domains, including phosphatidylinositol (PI) 3-kinase.4 In turn, these signaling molecules activate a variety of downstream targets. KIT signaling is critical for the normal development and survival of hematopoietic progenitor cells, mast cells, melanocytes, interstitial cells of Cajal (intestinal pacemaker cells), and germ cells.5
Activating mutations of KIT tyrosine kinase are common in gastrointestinal stromal tumors (GISTs)6–8 and mastocytosis/mast cell leukemia.9–11 Rarely, such mutations occur in acute myelogenous leukemia12,13 and sinonasal natural killer/T cell lymphoma.14 The mutations in GISTs are found primarily in exons 11 and 9 of the KIT gene, and the resulting mutant isoforms are sensitive to inhibition by imatinib mesylate (Gleevec, formerly STI571) in vitro.1 Correspondingly, many patients with malignant GIST respond to treatment with this drug.15,16 In contrast, nearly all cases of mastocytosis/mast cell leukemia are related to a mutation in exon 17 of the KIT gene (D816V), which yields an isoform that is resistant to imatinib mesylate.17
To date there have been two reports of KIT mutations in germ cell tumors. Tian and colleagues18 examined 23 cases of seminoma/dysgerminoma and found 2 tumors (1 seminoma [primary site unspecified], 1 ovarian dysgerminoma; 2 of 23 = 8.7%) with an activating mutation in exon 17 (D816H). There were no mutations in 10 non-seminomatous germ cell tumors evaluated. More recently, Przygodzki et al19 found KIT exon 17 mutations in 3 of 8 (37.5%) mediastinal seminomas (K818R, D820V, and N822K). While all three were novel mutations, one of them (K818R) was a conservative change for which the biochemical significance was not established.
To further examine the frequency and spectrum of KIT gene mutations in germ cell tumors, we screened a series of germ cell tumors tumors using the highly sensitive combination of denaturing high performance liquid chromatography (HPLC) and direct sequencing. Selected KIT mutant protein isoforms were profiled biochemically for constitutive KIT kinase activity and sensitivity to imatinib mesylate. In addition, we compared activation of intracellular signaling pathways in seminomas with and without KIT gene mutations, as well as in non-seminomatous germ cell tumors (NSGCT).
Materials and Methods
Tumor Specimens
Forty-six samples of paraffin-embedded germ cell tumor (32 testicular seminomas and 1 ovarian dysgerminoma, 13 NSGCT [5 also had corresponding frozen tumor]) were obtained from the archives of the Departments of Pathology at Oregon Health and Science University and the Portland VA Medical Center. An additional 8 fresh frozen testicular seminoma samples were from the Department of Pathology of Brigham & Women’s Hospital (Dr. Jonathan Fletcher). Twenty-four samples of paraffin-embedded germ cell tumor NSGCT were from the Department of Pathology, Indiana University (Dr. Oscar Cummings). Fourteen additional samples of fresh-frozen testicular seminoma were obtained from the Germ Cell Tumor Bank of Indiana University (Dr. Cummings). Sections of all paraffin-embedded tumor samples were reviewed by one of the authors (C.L.C.) to verify the diagnosis. Overall, our series included 54 seminomas of testicular origin (46 primary lesions, 8 metastases) and 1 primary ovarian dysgerminoma. The 37 NSGCT included 32 cases with a testicular primary site. In 5 cases of NSGCT the location of the primary site was not available to us. The majority of the NSGCT tumors examined were from recurrent, metastatic lesions. All samples were acquired in accordance with the regulations of the Institutional Review Boards for each institution.
Immunohistochemistry for KIT
All available samples of paraffin-embedded tumor were examined for KIT expression by immunohistochemistry using a rabbit antiserum from Dako (A4502, Dako Corp., Carpinteria, CA) as described previously.20
Genomic DNA Extraction and Analysis
Hematoxylin and eosin (H&E)-stained sections (5 μm) were reviewed under a microscope and areas rich in tumor (>50% cellularity) were marked. Corresponding areas on unstained sections were scraped from the slides with a sterile scalpel blade. In the case of NSGCT, we purposefully selected tumor regions that were rich in high-grade elements (embryonal carcinoma and yolk sac tumor). For lesions with elements of teratoma, we selected regions that were dominated by epithelial-lined cysts and nodules of cartilage rather than pure stroma. In cases of seminoma that had heavy lymphocytic infiltrates (≥50% cellularity), 7 μm sections were prepared on non-coated glass slides, deparaffinized, and stained with methyl green. Tumor cells were then microdissected using a Pixcell II Laser Capture Microscope (LCM) (Arcturus Engineering, Mountain View, CA). None of our cases of NSGCT were subjected to LCM. DNA was extracted from scraped tissue or the nuclear pellet of lysed fresh-frozen tumor (detailed below) using the QIAamp DNA Mini Kit and 500 ng was used for PCR (Qiagen, Valencia, CA). Caps bearing tumor cells from laser capture microdissection were incubated overnight in 50 μl of 10 mmol/L Tris-HCL (pH 8.0), 1 mmol/L EDTA, 1% Tween-20, and proteinase K (0.1 mg/ml) at 56°C. The mixture was then heated to 90°C to inactivate the proteinase K and 2 to 5 μl were used for PCR.
PCR was performed on all specimens for KIT exon 17 (High Fidelity System, Roche No. 1732078, Roche Diagnostics, Indianapolis, IN). PCR for KIT exons 9, 11, and 13 was completed in 20 of the seminoma cases that were known to be wild-type at exon 17. The following primer pairs were used:
KIT exon 17 forward 81318 5′-TGTATTCACAGAGACTTGGC-3′
KIT exon 17 reverse 81534 5′-GGATTTACATTATGAAAGTCACAGG-3′
KIT exon 9 forward 5′-ATGCTCTGCTTCTGTACTGCC-3′
KIT exon 9 reverse 5′-AGAGCCTAAACATCCCCTTA-3′
KIT exon 11 forward 5′-CCAGAGTGCTCTAATGACTG-3′
KIT exon 11 reverse 5′-ACCCAAAAAGGTGACATGGA-3′
KIT exon 13 forward 5′-CATCAGTTTGCCAGTTGTGC-3′
KIT exon 13 reverse 5′-ACACGGCTTTACCTCCAATG-3′
DNAs were amplified in 25 to 50 μl PCR reactions of 1 minute at 94°C, 1 minute at 56°C, and 1 minute at 72°C for 45 cycles (GeneAmp PCR System 9700, Applied Biosystems, Foster City, CA). Negative controls (no DNA) were included in every set of amplifications.
DNA from the laser-captured cells usually required nested PCR; 1 μl of the first reaction was used for the nested reaction. The nested PCR reaction was amplified for 25 cycles using the above PCR cycling conditions. Negative controls for the nested PCR reaction included a reaction using 1 μl of negative control (no template) specimen from the first round of amplification as a “template.”
Additional exon 17 primers for nested PCR:
KIT exon 17 forward 81289 (before nesting) 5′-GGTTTTCTTTTCTCCTCCAA-3′
KIT exon 17 reverse 81488 (nested) 5′-AGGACGTTTCCTAAAAATCAAAG-3′
Amplicons were analyzed by D-HPLC using a Transgenomic WAVE instrument (Transgenomics, Omaha, NE) as previously described.20 Denaturing temperatures for detection of point mutations were optimized for each primer pair (exon 9, 58°C; exon 11, 57°C; exon 13, 59°C; and exon 17, 56.5°C). Amplicons with an abnormal D-HPLC elution profile at denaturing temperature were subjected to bidirectional sequence analysis on an ABI 377 or 310 sequencer using the ABI Big Dye terminator kit (Applied Biosystems). All mutations were confirmed by repeat HPLC and sequencing analysis of an independently amplified specimen.
KIT Expression Vectors and Cell Transfection
Site-directed mutagenesis of wild-type KIT plasmid cDNA (cloned in pcDNA3.1 [Invitrogen, Carlsbad,CA]) was performed to produce the mutant KIT isoforms (Stratagene QuikChange-XL Site-Directed Mutagenesis Kit). The introduced mutations were confirmed by bi-directional sequencing. Chinese hamster ovary (CHO) cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in F12 (HAM) Nutrient Mixture (Gibco-BRL), + 10% fetal bovine serum. At 70% confluency in T25 flasks, the cells were transiently transfected with wild-type or mutant KIT expression plasmid using Lipofectamine PLUS (Invitrogen Life Technologies) according to the manufacturer’s suggested protocol. The next day, the transfected cells were exposed to concentrations of imatinib mesylate ranging from 0 to 5 μmol/L for 90 minutes (drug generously provided by Dr. Elisabeth Buchdunger, Novartis Pharma, Switzerland). Fresh stock solutions of drug (10 mmol/L) were made before each experiment by dissolving 5 mg STI571 in 1 ml Dulbecco’s phosphate-buffered saline (PBS; Gibco-BRL). When indicated, cells were stimulated with human SCF (final concentration 100 ng/ml) for 10 minutes before their removal from the flask. A 10-minute stimulation with human SCF was found to result in maximal receptor autophosphorylation, consistent with prior results (data not shown).20
Protein Lysates
After the 90-minute incubation period with drug, the transfected CHO cells were then scraped from the flasks and spun (1000 RPM). The cell pellets were lysed with 100 to 150 μl of protein lysis buffer (50 mmol/L Tris, 150 mmol/L sodium chloride, 5 mmol/L EDTA, 1% NP-40, and 0.25% deoxycholate), with addition of the inhibitors aprotinin, leupeptin, pepstatin, 4-(2-aminoethyl)-benzenesulfonyl fluoride, sodium pyrophosphate, and sodium orthovanadate [Sigma, St. Louis, MO]), as previously described.21 Protein concentration of homogenates was determined with Bio-Rad Protein Assay Reagent.
Fresh-frozen tumor specimens were ground in liquid nitrogen with mortar and pestle and solubilized with protein lysis buffer. Lysate was spun at 13,000 RPM for 15 minutes to pellet nuclei. Supernatant was saved for KIT and PI3-kinase immunoprecipitation studies and Western blots.
Immunoprecipitation and Western Blotting
Cellular lysates (250 to 500 μg of protein from CHO cell transfection or 4 mg of fresh-frozen tumor) were pre-cleared with 5 μl (1:100) of normal rabbit IgG agarose conjugate (Santa Cruz Biotech, Santa Cruz, CA) for 1 hour at 4°C. Next, they were immunoprecipitated with 5 μg of anti-KIT antibody agarose conjugate (Santa Cruz Biotech) overnight at 4°C. Separate samples of fresh-frozen tumor lysates were immunoprecipitated with 10 μg of anti-PI3-kinase antibody (N-SH3 domain p85 subunit) (Upstate Biotechnology, Lake Placid, NY). Immunoprecipitates were washed three times with PBS plus inhibitors, resuspended in 20 μl of 4X-Laemlli SDS-PAGE loading buffer, heated at 95°C for 10 minutes, and then separated by 10% SDS-PAGE and transferred to nitrocellulose. The membranes were blocked for one hour with TBS-Tween containing 5% BSA for anti-phosphotyrosine blotting or 5% nonfat milk for anti-KIT blotting. The following primary antibodies were used on the membranes: mouse monoclonal anti-phosphotyrosine antibody (PY20) at 1:1000 dilution (Transduction Laboratories, Lexington, KY) and rabbit anti-KIT polyclonal antibody at 1:500 (Oncogene, Cambridge, MA). Secondary antibodies were peroxidase-conjugated goat anti-mouse antibody at 1:5000 (Biorad, Hercules, CA) and goat-anti-rabbit antibody at 1:10000 (Biorad), respectively. Immunoblots were developed by enhanced chemiluminescence (Amersham Biosciences).
For Western blots of tumor lysates, 25 μg of lysate was combined with 5 μl of 4X-Laemlli SDS-PAGE loading buffer, heated at 95°C for 10 minutes, and then separated by 10% SDS-PAGE and transferred to nitrocellulose. The membranes were blocked for 1 hour with TBS-Tween containing 5% BSA for anti-phospho-blotting or 5% nonfat milk for all other blotting. The following primary antibodies were used on the membranes: anti-phospho-AKT (Ser473) antibody at 1:500 dilution, anti-total AKT 1:500, anti-phospho-MAPK 1:1000, anti-phospho-STAT3 1:666, and anti-STAT3 1:666 (all from Cell Signaling, Beverly, MA); anti-MAPK 1:1000 and anti-PI3-kinase (N-SH3 domain) 1:666 (Upstate Biotechnology, Lake Placid, NY), anti-placental alkaline phosphatase (PLAP) 1:100 (Biomeda Corp, Hayward, CA), and anti-actin 1:500 (Sigma). Secondary antibodies were the same as described above.
Results
Frequency of KIT Mutations in Germ Cell Tumors
The study was performed on 92 germ cells tumors, including 54 pure testicular seminomas, 1 ovarian dysgerminoma, 4 mixed seminoma/NSGCT, and 33 NSGCT. KIT expression was assessed in a subset of the seminomas by immunohistochemistry, which revealed strong surface membrane staining in 82% (31 of 38) of the pure and mixed seminoma tumors. KIT expression in the NSGCT samples was examined in a previous study, which showed patchy cytoplasmic staining in 48% (11 of 23).22 These results are consistent with other reports of KIT expression in testicular germ cell tumors.23–25
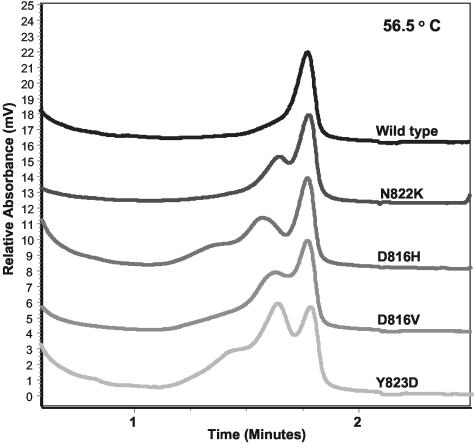
For 42 of the 54 cases of seminoma and the one case of ovarian dysgerminoma, the degree of lymphocytic infiltration was <50% and we were able to directly prepare genomic DNA from paraffin slides and/or fresh frozen samples. Amplicons of KIT exon 17 were screened for mutations using D-HPLC (Figure 1). Mutations of KIT exon 17 were found in 10 of the 42 cases of testicular seminoma (23.8%), including 3 cases with the D816H mutation originally described by Tian et al (Table 1).18 The most common mutation, found in 4 cases, was D816V. Two cases had Y823D, 1 case had Y823C, and 1 case had N822K (Table 1). None of these 43 cases required nested PCR for detection of mutations.
Figure 1.
Examples of high performance liquid chromatography (Transgenomic WAVE) elution profiles for KIT wild-type and representative exon 17 point mutants in seminoma tumors demonstrated at denaturing temperature (56.5°C). The estimated percentage of mutant allele ranges from approximately 25% (N822K, D816V, D816H) to 50% (Y823D) in these samples.
Table 1.
Activating KIT Exon 17 Point Mutations in Pure Testicular Seminomas (n = 54)
| Mutation (nucleotide substitution) | D816V (G → C @81401) | D816H (A → T @81402) | N822K (T → A @81421) | Y823D (T → G @81422) | Y823C (A → G @81423) |
|---|---|---|---|---|---|
| No. of cases (%) | 6 (11.1%) | 3 (5.6%) | 1 (1.9%) | 2 (3.7%) | 1 (1.9%) |
| Imatinib-sensitive | No | No | Yes | Yes | Not tested |
The frequency of activating KIT mutations in pure seminomas of testicular origin is tabulated. One case of seminoma had a somatic T801I mutation. This mutation is not associated with constitutive activation in our biochemical assays (Figure 2) and is therefore not included in this table.
For 12 of the 54 testicular seminoma specimens, the degree of lymphocytic infiltration (>50%) was deemed likely to interfere with detection of a possible KIT mutation. We therefore performed LCM to enrich for tumor cells before proceeding to KIT genotyping. The LCM-PCR assay was validated using five of the above cases for which genotyping had already been completed (1 case D816V, 4 wild-type cases). Only limited amounts of genomic DNA could be obtained using LCM and in 2 of the 5 validation cases we were not able to obtain an amplification product using single-step PCR. Therefore, we developed a nested PCR procedure in which 1 μl of the first-stage PCR product was used as template for a nested PCR reaction and the resultant PCR product as analyzed by D-HPLC. In all five of our validation cases, we obtained the identical mutational results using either our standard assay or the LCM procedure (with or without nesting). Genomic DNA was prepared using LCM from 12 seminomas with an unknown KIT mutational status and with >50% lymphocytic infiltration. Nested PCR was required to obtain an amplification product for 10 of these 11 LCM cases. Using the LCM-nested PCR assay, four additional cases with a KIT mutation were identified: two cases with D816V, one case with D816H, and one case with T801I (Table 1).
Overall, fourteen of the pure testicular seminomas (14 of 54, 25.9%) contained a KIT exon 17 point mutation. In five of the seminomas harboring an exon 17 mutation (1 case each of D816V, D816H, Y823C, Y823D, and T801I), DNA extracted from the adjacent normal tissue was wild-type, indicating that the mutations were somatic rather than germline. Twenty pure seminomas that were wild-type for exon 17 were screened for mutations in exons 9, 11, and 13 (all commonly mutated in GISTs); none were found. No KIT mutation was found in the solitary ovarian dysgerminoma. In contrast to our results from pure seminoma/dysgerminoma germ cell tumors, no exon 17 mutations were found in the 33 NSGCT or the 4 mixed seminoma/NSGCT (P = 0.0015, Fisher’s exact test).
KIT Kinase Activity in Transiently Transfected CHO Cells
There are only a few reports of human testicular seminoma cell lines, and these cell lines have not been fully characterized as to their usefulness in modeling seminoma biology in vitro.26–28 In particular, it has not been reported if these cell lines express KIT. Therefore, we used an alternative in vitro eukaryotic system to assess the biochemical consequences of the observed KIT mutations on KIT kinase activity. Plasmid expression vectors encoding each mutation were generated by site-directed mutagenesis of wild-type KIT cDNA. CHO cells were transiently transfected with mutant or wild-type KIT vector. Transfected cells were then exposed to concentrations of imatinib mesylate ranging from 0 to 5 μmol/L for 90 minutes. Cell lysates were then immunoprecipitated with anti-KIT antibodies and analyzed by Western blotting for evidence of KIT kinase activity, as evidenced by phosphorylation of tyrosine residues. As expected, wild-type KIT required ligand stimulation for phosphorylation, while D816V and D816H did not (Figure 2).18 The N822K and Y823D mutant isoforms were also constitutively phosphorylated. The T801I mutant isoform was phosphorylated only in the presence of SCF. The kinase activities of mutant isoforms N822K and Y823D were inhibited by imatinib mesylate with an approximate IC50 of 0.5 μmol/L and 0.1 μmol/L, respectively (Figure 2). Mutant isoform D816H was much less sensitive to imatinib mesylate, with an IC50 of 5.0 μmol/L. Mutant D816V was resistant to 5.0 μmol/L imatinib mesylate, consistent with previous reports (see Table 1 for summary).17
Figure 2.
CHO cell temporary transfections with wild-type (WT) or mutant KIT expression vectors and sensitivity to imatinib mesylate. Transfected cells were treated with varying concentrations of imatinib mesylate (STI571) for 90 minutes. Cell lysates were immunoprecipitated with anti-KIT antibodies and phosphorylation of KIT tyrosine residues was demonstrated by anti-phosphotyrosine immunoblotting (P-TYR). The membrane was then stripped and reprobed with an antibody for total KIT. Phosphorylation of WT KIT and T801I were dependent on SCF. Phosphorylation of Y823D, N822K, D816H, and D816V was constitutive. Imatinib mesylate inhibited WT KIT, Y823D, and N822K with IC50s of 0.1 μmol/L, <0.1 μmol/L, and 0.5 μmol/L, respectively. D816H and D816V were resistant to drug, IC50 >5 μmol/L.
KIT Kinase Activity in Fresh-Frozen Tumor Specimens
To determine whether KIT kinase is active in seminomas, total KIT protein was immunoprecipitated from 13 fresh-frozen seminoma lysates and analyzed by Western blotting with a phospho-tyrosine-specific monoclonal antibody (PY20). Two of the 13 frozen samples contained the D816V mutation; the remaining 11 were wild-type. While total KIT protein was immunoprecipitated in all of the samples, phosphorylated KIT was detectable only in the specimens containing the D816V mutation (Figure 3A). Notably, in two wild-type cases (lanes 6 and 13) in which an equivalent amount of total KIT protein was immunoprecipitated relative to the D816V seminomas, there was no evidence of phosphorylated KIT. Since it has been demonstrated that PI3-kinase is required for SCF-induced cell proliferation and survival in vitro and for male mouse fertility in vivo, we also performed successive immunoblotting of the KIT-immunoprecipitated protein complexes with an antibody to the p85 subunit of PI3-kinase.29–32 PI3-kinase was co-precipitated with KIT in two of the tumors, most strongly in a D816V mutant, but also in a wild-type tumor (Figure 3A, lanes 5 and 11). Conversely, immunoprecipitation of PI3-kinase protein from the seminoma lysates yielded strong signals for co-precipitated total and phospho-KIT in both of the D816V mutants (Figure 3B). In contrast, very little total or phospho-KIT was co-precipitated from the wild-type seminoma samples even though equivalent amounts of p85 subunit of PI3-kinase were immunoprecipitated relative to the D816V seminomas. Thus, evidence of strong KIT activation could be demonstrated only in the seminomas with KIT-activating mutations.
Figure 3.
Coimmunoprecipitation of KIT and PI3-kinase in seminoma tumor lysates. A: Total KIT protein was immunoprecipitated from 13 fresh-frozen seminoma lysates (4 mg) and analyzed by Western blotting with a phospho-tyrosine-specific antibody (P-TYR). The membrane was then stripped and reprobed with an antibody for total KIT. Successive stripping and immunoblotting was done with an antibody for PI3-kinase (N-SH3 domain p85 subunit). B: Total PI3-kinase protein was immunoprecipitated from the seminoma lysates (specimens 2–13) and analyzed by Western blotting with P-TYR. The membrane was then sequentially stripped and reprobed with antibodies for total KIT and PI3-kinase (N-SH3 domain p85 subunit).
To further evaluate intracellular signaling in seminomas, Western blots of seminoma lysates were probed with antibodies to proteins involved in cellular proliferation and survival (Figure 4). The signal for placental alkaline phosphatase (PLAP) was compared across all lanes to ensure that comparable amounts of seminoma lysate were loaded even in the one case (lane 13) for which LCM had been used for the genotyping analyses. Notably, total KIT was expressed at much higher levels in the two D816V tumors than in any of the KIT wild-type specimens, even though the PLAP staining suggested that these samples contained slightly less seminoma-derived protein than most of the other lanes. In reviewing the KIT IHC staining, we also noted that the mutant KIT seminomas had much stronger KIT staining than the non-mutant cases (data not shown).
Figure 4.
Seminoma whole cell lysate immunoblotting. Whole cell lysates (25 μg) from frozen seminoma tumor samples were immunoblotted with antibodies for total KIT, phospho-MAPK, total MAPK, phospho-AKT, total AKT, PI3-kinase (N-SH3 domain p85 subunit), phospho-STAT3, and total STAT3. Placental alkaline phosphatase (PLAP) immunoblotting was performed to assess equivalency of lane loading with seminoma tissue.
We analyzed the expression of total forms of PI3-kinase, AKT, mitogen-activated protein (MAP)-kinase, and STAT3 by immunoblotting. We also analyzed the seminoma lyates for expression of activated forms of AKT, MAP-kinase, and STAT3. The expression and activation of the various signaling intermediates varied between tumors, with no apparent difference between the KIT-mutant and wild-type tumors. These differences in expression and activation of signaling proteins are not explained by variations in the amount of seminoma protein analyzed as the PLAP staining was comparable among the seminoma cases.
Five frozen samples of NSGCT were available for comparison of intracellular signaling. Total KIT expression was not detected in any of the NSGCT, as predicted by their paucity of KIT staining on immunohistochemistry (3 of 5 tumors had <5%, patchy KIT staining; the remaining 2 were negative). There was tumor to tumor variation in the expression of total and activated MAPK, AKT, and STAT3, but the overall signal transduction profile of these tumors was not distinguishable from the seminoma tumors (Figure 5). It should be noted that normalization of the protein loading on these blots was problematic due to the inherent heterogeneity of the NSGCTs. Actin was added as an additional loading control because PLAP expression is not as uniform in NSGCT as it is in seminoma. The actin staining would suggest some unequal loading across lanes (Figure 5), but PLAP staining was quite similar among all of the NSGCT cases. Given that PLAP expression is more characteristic of embryonal carcinoma and yolk sac tumor than teratoma, the findings in Figure 5 may be somewhat biased toward these high-grade NSGCT elements.33
Figure 5.
Non-seminoma whole cell lysate immunoblotting. Whole cell lysates (25 μg) from frozen non-seminoma tumor samples were immunoblotted with antibodies for total KIT, phospho-MAPK, total MAPK, phospho-AKT, total AKT, PI3-kinase (N-SH3 domain p85 subunit), phospho-STAT3, and total STAT3. Placental alkaline phosphatase (PLAP) and actin immunoblotting was performed to assess equivalency of lane loading. The histology of these tumors was as follows: pure embryonal carcinoma (A), embryonal and teratoma (B), predominantly teratoma with 10% embryonal and microscopic yolk sac (C), pure yolk sac (D), and embryonal carcinoma (E).
Discussion
The receptor tyrosine kinase KIT and its ligand SCF are essential for germ cell development and spermatogenesis, as well as for the normal development of blood cells (especially erythrocytes), melanocytes, mast cells, and the interstitial cells of Cajal.34 Activating mutations of KIT are most commonly associated with tumors that arise from cells developmentally dependent on an intact SCF/KIT axis, most notably GIST and mastocytosis and, much less frequently, acute myelogeous leukemia (AML). Using a denaturing HPLC method to screen for mutations, we found activating point mutations in exon 17 in 24.1% (13 of 54) of the pure testicular seminomas examined. The majority of the KIT mutations affected aspartic acid 816 (D816V, n = 6; D816H, n = 3), which is in the activation loop located between the ATP-binding and substrate phosphostransferase domains and is highly conserved among type III tyrosine kinases. The D816V mutation has previously been reported only in mastocytosis and AML.9,11,12,35–38 Two other exon 17 codons were mutated in seminomas, codon 822 in one case (N822K) and codon 823 in three cases (Y823D, n = 2; Y823C, n = 1). The N822K mutation was recently identified in a mediastinal seminoma, and has been found in GISTs and in one AML cell line.19,39,40 The Y823D and Y823C mutations have not previously been reported in seminoma, although we recently identified rare cases of AML harboring the Y823D mutation.41 Both of these latter mutations (N822K and Y823D) were found to result in ligand-independent activation when expressed in CHO cells in vitro. A novel somatic mutation (T801I) of KIT was also identified in a single seminoma. In our assay, this variant of KIT was dependent on SCF for kinase activation. The biological significance of T801I could not be demonstrated because no frozen tissue was available to examine KIT phosphorylation and downstream signaling. Additional regions of the KIT gene were screened for mutations in 20 cases that lacked an exon 17 mutation. No mutations in exons 9, 11, or 13 were identified. Thus, activating mutations of KIT in testicular seminoma appear to be localized to exon 17.
The activating mutation frequency in our series (24.1%) is higher than that reported by Tian et al, 18 probably due to methodological differences. Seminomas are frequently triploid or near-tetraploid, and therefore a KIT allele with a gain-of-function mutation may be outnumbered by two or more wild-type alleles.42 This ratio of mutant to wild-type alleles approaches the limit of detection by direct sequencing, a problem that may be compounded by concomitant contamination of the tumor by infiltrating lymphocytes. We believe that D-HPLC screening, along with application of laser capture microscopy to selected cases, allowed more point mutations to be identified in our series. It remains possible that some KIT mutations were missed and could perhaps be detected through the use of even more sensitive methodologies (eg, allele-specific PCR).
Przygodzki et al19 recently assayed for KIT mutations in primary mediastinal seminomas using direct sequencing of exon 17 amplicons. Three of eight studied tumors had protein sequence altering mutations: D820V, N822K, and K818R. Asp-820 and Asn-822 mutations have been previously identified as activating in GISTs, mast cell tumors, and in seminomas as presented here, but the biological significance of the conservative change K818R mutation is unknown.39,43,44 The frequency of KIT mutations in our series of seminomas of testicular origin does not appear different from the frequency reported in the smaller series of mediastinal seminomas.
Interestingly, all 33 NSGCT cases in our study were negative for KIT gene mutations. This observation confirms the trend noted by Tian et al, 18 who found no mutations among 10 NSGCT. The extent to which KIT mutations are present in mixed seminoma/NSGCT is less clear. One of the two D816H mutations identified by Tian et al18 was found in a combined dysgerminoma/yolk sac tumor of the ovary. In contrast, we found no KIT mutations in 4 mixed seminoma/NSGCT originating in the testis.
It is possible that stromal cells present in the NSGCT samples may have contributed wild-type DNA that interfered with KIT mutation detection. This is certainly a concern, but there are several reasons why we believe that our results were not significantly influenced in this regard. First, we purposefully selected areas of NSGCT that were rich in high-grade elements (embryonal carcinoma and yolk sac tumor). Second, the areas of teratoma that were analyzed were dominated by epithelial-lined cysts and nodules of cartilage rather than pure stroma. Third, there is recent evidence that at least some of the stroma present in teratomas is tumor-derived rather than a host reaction.45 For these reasons, it is unlikely that the complete absence of detectable KIT gene mutations in all our NSGCT can be explained by contamination from reactive tissue elements.
To explore the role of KIT activation, whether resulting from mutation or ligand stimulation, we examined the phosphorylation status of KIT in seminoma lysates. KIT tyrosyl-phosphorylation, indicative of kinase activation, was readily detected only in seminomas with the D816V mutation. Another measure of KIT activation is the association of KIT with PI3K; we detected co-immunprecipitated PI3K in 1 of 2 cases with D816V and in 1 of 11 wild-type KIT seminomas. On reciprocal immunoprecipitation with a PI3K antibody there was much more phosphorylated and total D816V KIT associated with PI3K than was detected in wild-type KIT seminomas. Thus, were able to find compelling evidence of KIT kinase activation only in the D816V seminomas and not in any of the wild-type KIT cases. These findings indicate that KIT is not hyperactivated in seminomas lacking KIT mutations in the same manner observed for several KIT mutant seminomas. It is likely that KIT can be activated by ligand, or by other mechanisms, in wild-type tumors. However, such mechanisms presumably result in levels of KIT activation that are below the detection threshold for our assays (estimated to be <1% of the activation seen in seminomas with D816 KIT mutation).
There is ample precedent of KIT expression in human cancers in the absence of substantial or pathogenetically significant KIT activation (eg, melanoma, adenoid cystic carcinoma, small cell lung cancer). Indeed, in the case of melanoma, loss of KIT expression correlates directly with increasing clinical stage.1 We believe, based on our results, that in the majority of seminomas KIT expression may be a differentiation marker without playing a central pathogenetic role in the initiation or progression of these tumors. Indeed, not all seminomas even express KIT as assessed by immunohistochemistry.46 In contrast, seminomas with KIT mutations have biochemical evidence of strong KIT activation and it is likely that KIT plays a role in the initiation or maintenance of the seminoma tumor. Further studies are required to confirm this hypothesis.
If oncogenic activation of KIT is responsible for tumorigenesis and/or the maintenance of tumor phenotype in a subset of seminomas, then these tumors might have a distinct pattern of KIT-mediated intracellular signals that differs from wild-type tumors; however, this was not the case. PI3-kinase, AKT, phospho-AKT, MAP-kinase, phospho-MAP-kinase, STAT3, and phospho-STAT3 were all found at similar levels in 11 wild-type and 2 mutant (both D816V) seminoma lysates. This finding suggests that there might be alternate receptor tyrosine kinase oncogenes or redundant growth signaling pathways in germ cell tumors that have yet to be identified. Indeed, our finding of similar intracellular signal patterns in NSGCT lysates supports this hypothesis. Further proteomic- and/or genomic-based studies will be needed to identify alternative mechanisms for signal transduction pathway activation in seminomas lacking a KIT mutation.
What role activated KIT may play in the biology of seminomas remains unclear. As there is abundant evidence that seminomas and NSGCT share a common neoplastic germ cell of origin, it is intriguing that KIT mutations appear to be limited to seminomas.47 Seminomas likely deviate from NSGCT very early in the oncogenesis of germ cell tumors, and acquired KIT gene mutations may favor the seminoma pathway. From studies in mice it is known that signaling from KIT kinase through the PI3K pathway is essential for normal spermatogenesis.29,48 Insofar as seminomas retain morphological and immunophenotypic features that closely resemble primary germ cells, KIT activity may be important in maintaining this germ cell-like character. “Atypical seminomas,” which show evidence of dedifferentiation (eg, expression of cytokeratins or CD30), have a significantly lower frequency of KIT expression than “usual seminomas” (57% vs. 95%, respectively).46 These “atypical seminomas” have higher mitotic counts and tend to present at a higher clinical stage.46 Thus, loss of KIT expression in seminomas may correlate with more aggressive clinical behavior.
To the extent that KIT is important in the biology of seminomas, KIT kinase inhibitors may have a role in their treatment, particularly for those seminomas with activating mutations. Imatinib mesylate has proven very effective in the treatment of advanced GI stromal tumors with KIT mutations.15,16 Unlike the case with GISTs, however, most seminomas in our study harbored activating KIT mutations (9 of 13) that are biochemically resistant to imatinib mesylate in vitro. Thus, only 4 of 54 testicular seminomas in our series had an imatinib mesylate-sensitive KIT mutation. Targeting the D816V and D816H mutations found in seminomas will require different kinase inhibitors that can inhibit the specific KIT conformation induced by these mutations.49
In contemplating the treatment of seminomas with a KIT kinase inhibitor, it must be noted that current medical treatment of seminoma results in a cure in the vast majority of patients. Indeed, even patients with advanced metastatic disease do well with platinum-based chemotherapy. A clinical trial sponsored by Cancer and Leukemia Group B (CALGB) is currently underway to examine the efficacy of imatinib mesylate in the treatment of advanced metastatic seminoma that is refractory to chemotherapy. KIT inhibitors might also be tested in a “window of opportunity” neoadjuvant setting in those patients with low volume retroperitoneal disease before their radiotherapy. If active in these settings, future development could include use of KIT inhibitors as an adjuvant therapy for patients with stage I disease, with the goal of decreasing the need for subsequent radiation treatment or chemotherapy and perhaps reducing consequent toxicities, eg, infertility. Rational design and interpretation of such trials will require molecular assessment of KIT activation, at both the protein and nucleic acid levels, in individual patient tumor specimens.
Footnotes
Address reprint requests to Michael C. Heinrich, M.D., R&D19, Portland VA Medical Center, 3710 U.S. Veterans Hospital Road, Portland, OR 97239. E-mail heinrich@ohsu.edu.
Supported in part with federal funds from the National Cancer Institute, National Institutes of Health (Contract N01-CO-12400 [to M.C.H.), and a Department of Veterans Affairs Merit Review Grant (to M.C.H.).
References
- Heinrich MC, Blanke CD, Druker BJ, Corless CL. Inhibition of KIT tyrosine kinase activity: a novel molecular approach to the treatment of KIT-positive malignancies. J Clin Oncol. 2002;20:1692–1703. doi: 10.1200/JCO.2002.20.6.1692. [DOI] [PubMed] [Google Scholar]
- Besmer P, Murphy JE, George PC, Qiu FH, Bergold PJ, Lederman L, Snyder HW, Jr, Brodeur D, Zuckerman EE, Hardy WD. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature. 1986;320:415–421. doi: 10.1038/320415a0. [DOI] [PubMed] [Google Scholar]
- Qiu FH, Ray P, Brown K, Barker PE, Jhanwar S, Ruddle FH, Besmer P. Primary structure of c-kit: relationship with the CSF-1/PDGF receptor kinase family: oncogenic activation of v-kit involves deletion of extracellular domain and C terminus. EMBO J. 1988;7:1003–1011. doi: 10.1002/j.1460-2075.1988.tb02907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Vliagoftis H, Worobec AS, Metcalfe DD. The protooncogene c-kit and c-kit ligand in human disease. J Allergy Clin Immunol. 1997;100:435–440. doi: 10.1016/s0091-6749(97)70131-3. [DOI] [PubMed] [Google Scholar]
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Muhammad TG, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- Beghini A, Tibiletti MG, Roversi G, Chiaravalli AM, Serio G, Capella C, Larizza L. Germline mutation in the juxtamembrane domain of the kit gene in a family with gastrointestinal stromal tumors and urticaria pigmentosa. Cancer. 2001;92:657–662. doi: 10.1002/1097-0142(20010801)92:3<657::aid-cncr1367>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Moskaluk CA, Tian Q, Marshall CR, Rumpel CA, Franquemont DW, Frierson HF., Jr Mutations of c-kit JM domain are found in a minority of human gastrointestinal stromal tumors. Oncogene. 1999;18:1897–1902. doi: 10.1038/sj.onc.1202496. [DOI] [PubMed] [Google Scholar]
- Nagata H, Worobec AS, Oh CK, Chowdhury BA, Tannenbaum S, Suzuki Y, Metcalfe DD. Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci USA. 1995;92:10560–10564. doi: 10.1073/pnas.92.23.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley BJ, Jr, Metcalfe DD, Tharp M, Wang X, Tyrrell L, Lu SZ, Heitjan D, Ma Y. Activating and dominant inactivating c-KIT catalytic domain mutations in distinct clinical forms of human mastocytosis. Proc Natl Acad Sci USA. 1999;96:1609–1614. doi: 10.1073/pnas.96.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakura Y, Furitsu T, Tsujimura T, Butterfield JH, Ashman LK, Ikeda H, Kitayama H, Kanayama Y, Matsuzawa Y, Kitamura Y. Activating mutations of the c-kit proto-oncogene in a human mast cell leukemia cell line. Leukemia. 1994;8(Suppl 1):S18–S22. [PubMed] [Google Scholar]
- Ning ZQ, Li J, Arceci RJ. Activating mutations of c-kit at codon 816 confer drug resistance in human leukemia cells. Leuk Lymphoma. 2001;41:513–522. doi: 10.3109/10428190109060342. [DOI] [PubMed] [Google Scholar]
- Gari M, Goodeve A, Wilson G, Winship P, Langabeer S, Linch D, Vandenberghe E, Peake I, Reilly J. c-kit proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br J Haematol. 1999;105:894–900. doi: 10.1046/j.1365-2141.1999.01449.x. [DOI] [PubMed] [Google Scholar]
- Hongyo T, Li T, Syaifudin M, Baskar R, Ikeda H, Kanakura Y, Aozasa K, Nomura T. Specific c-kit mutations in sinonasal natural killer/T-cell lymphoma in China and Japan. Cancer Res. 2000;60:2345–2347. [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, Fletcher JA, Silverman SG, Silberman SL, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker BJ, Corless C, Fletcher CD, Joensuu H. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- van Oosterom AT, Judson I, Verweij J, Stroobants S, Donato DP, Dimitrijevic S, Martens M, Webb A, Sciot R, Van Glabbeke M, Silberman S, Nielsen OS. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a phase I study. Lancet. 2001;358:1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- Ma Y, Zeng S, Metcalfe DD, Akin C, Dimitrijevic S, Butterfield JH, McMahon G, Longley BJ. The c-KIT mutation causing human mastocytosis is resistant to STI571 and other KIT kinase inhibitors; kinases with enzymatic site mutations show different inhibitor sensitivity profiles than wild-type kinases and those with regulatory-type mutations. Blood. 2002;99:1741–1744. doi: 10.1182/blood.v99.5.1741. [DOI] [PubMed] [Google Scholar]
- Tian Q, Frierson HF, Jr, Krystal GW, Moskaluk CA. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol. 1999;154:1643–1647. doi: 10.1016/S0002-9440(10)65419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przygodzki RM, Hubbs AE, Zhao FQ, O’Leary TJ. Primary mediastinal seminomas: evidence of single and multiple KIT mutations. Lab Invest. 2002;82:1369–1375. doi: 10.1097/01.lab.0000032410.46986.7b. [DOI] [PubMed] [Google Scholar]
- Corless CL, McGreevey L, Haley A, Town A, Heinrich MC. KIT mutations are common in incidental gastrointestinal stromal tumors one centimeter or less in size. Am J Pathol. 2002;160:1567–1572. doi: 10.1016/S0002-9440(10)61103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich MC, Griffith DJ, Druker BJ, Wait CL, Ott KA, Zigler AJ. Inhibition of c-kit receptor tyrosine kinase activity by STI 571, a selective tyrosine kinase inhibitor. Blood. 2000;96:925–932. [PubMed] [Google Scholar]
- Madani A, Kemmer K, Sweeney C, Corless C, Ulbright T, Heinrich M, Einhorn L. Expression of KIT and epidermal growth factor receptor in chemotherapy refractory non-seminomatous germ-cell tumors. Ann Oncol. 2003;14:873–880. doi: 10.1093/annonc/mdg244. [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Kuczyk MA, Dunn T, Serth J, Hartmann K, Jonasson J, Pietsch T, Jonas U, Schmoll HJ. Expression of stem-cell factor and its receptor c-kit protein in normal testicular tissue and malignant germ-cell tumours. J Cancer Res Clin Oncol. 1996;122:301–306. doi: 10.1007/BF01261407. [DOI] [PubMed] [Google Scholar]
- Izquierdo MA, Van der Valk P, Ark-Otte J, Rubio G, Germa-Lluch JR, Ueda R, Scheper RJ, Takahashi T, Giaccone G. Differential expression of the c-kit proto-oncogene in germ cell tumours. J Pathol. 1995;177:253–258. doi: 10.1002/path.1711770307. [DOI] [PubMed] [Google Scholar]
- Rajpert-De Meyts E, Skakkebaek NE. Expression of the c-kit protein product in carcinoma-in-situ and invasive testicular germ cell tumours. Int J Androl. 1994;17:85–92. doi: 10.1111/j.1365-2605.1994.tb01225.x. [DOI] [PubMed] [Google Scholar]
- Kinugawa K, Hyodo F, Matsuki T, Jo Y, Furukawa Y, Ueki A, Tanaka H. Establishment and characterization of a new human testicular seminoma cell line, JKT-1. Int J Urol. 1998;5:282–287. doi: 10.1111/j.1442-2042.1998.tb00604.x. [DOI] [PubMed] [Google Scholar]
- von Keitz AT, Riedmiller H, Neumann K, Gutschank W, Fonatsch C. Establishment and characterization of a seminoma cell-line (S2). Invest Urol (Berl) 1994;15:28–31. [PubMed] [Google Scholar]
- Fujii T, Otsuki T, Moriya T, Sakaguchi H, Kurebayashi J, Yata K, Uno M, Kobayashi T, Kimura T, Jo Y, Kinugawa K, Furukawa Y, Morioka M, Ueki A, Tanaka H. Effect of hypoxia on human seminoma cells. Int J Oncol. 2002;20:955–962. [PubMed] [Google Scholar]
- Blume-Jensen P, Jiang G, Hyman R, Lee KF, O’Gorman S, Hunter T. Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3′-kinase is essential for male fertility. Nat Genet. 2000;24:157–162. doi: 10.1038/72814. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Ronnstrand L, Gout I, Waterfield MD, Heldin CH. Modulation of Kit/stem cell factor receptor-induced signaling by protein kinase C. J Biol Chem. 1994;269:21793–21802. [PubMed] [Google Scholar]
- Blume-Jensen P, Janknecht R, Hunter T. The kit receptor promotes cell survival via activation of PI 3-kinase and subsequent Akt-mediated phosphorylation of Bad on Ser136. Curr Biol. 1998;8:779–782. doi: 10.1016/s0960-9822(98)70302-1. [DOI] [PubMed] [Google Scholar]
- Kozawa O, Blume-Jensen P, Heldin CH, Ronnstrand L. Involvement of phosphatidylinositol 3′-kinase in stem-cell-factor- induced phospholipase D activation and arachidonic acid release. Eur J Biochem. 1997;248:149–155. doi: 10.1111/j.1432-1033.1997.00149.x. [DOI] [PubMed] [Google Scholar]
- Manivel JC, Jessurun J, Wick MR, Dehner LP. Placental alkaline phosphatase immunoreactivity in testicular germ-cell neoplasms. Am J Surg Pathol. 1987;11:21–29. doi: 10.1097/00000478-198701000-00003. [DOI] [PubMed] [Google Scholar]
- Geissler EN, Liao M, Brook JD, Martin FH, Zsebo KM, Housman DE, Galli SJ. Stem cell factor (SCF), a novel hematopoietic growth factor and ligand for c-kit tyrosine kinase receptor, maps on human chromosome 12 between 12q14.3 and 12qter. Somat Cell Mol Genet. 1991;17:207–214. doi: 10.1007/BF01232978. [DOI] [PubMed] [Google Scholar]
- Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, Sugahara H, Butterfield JH, Ashman LK, Kanayama Y. Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest. 1993;92:1736–1744. doi: 10.1172/JCI116761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Bain BJ, Knight CL. Cytogenetic and molecular genetic abnormalities in systemic mastocytosis. Acta Haematol. 2002;107:123–128. doi: 10.1159/000046642. [DOI] [PubMed] [Google Scholar]
- Longley BJ, Tyrrell L, Lu SZ, Ma YS, Langley K, Ding TG, Duffy T, Jacobs P, Tang LH, Modlin I. Somatic c-KIT activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishment of clonality in a human mast cell neoplasm. Nat Genet. 1996;12:312–314. doi: 10.1038/ng0396-312. [DOI] [PubMed] [Google Scholar]
- Beghini A, Peterlongo P, Ripamonti CB, Larizza L, Cairoli R, Morra E, Mecucci C. C-kit mutations in core binding factor leukemias. Blood. 2000;95:726–727. [PubMed] [Google Scholar]
- Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA, Demetri GD, Fletcher CD, Fletcher JA. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- Beghini A, Magnani I, Ripamonti CB, Larizza L. Amplification of a novel c-Kit activating mutation Asn(822)-Lys in the Kasumi-1 cell line: a t(8;21)-Kit mutant model for acute myeloid leukemia. Hematol J. 2002;3:157–163. doi: 10.1038/sj.thj.6200168. [DOI] [PubMed] [Google Scholar]
- Zwaan C, Miller M, Goemans B, Hahlen K, van Wering E, Meshinchi S, Zimmermann M, Creutzig U, Kaspers G, Heinrich M. Frequency and clinical significance of c-kit exon 17 mutations in childhood acute myeloid leukemia. Blood. 2002;100:746a. [Google Scholar]
- Chaganti RS, Houldsworth J. Genetics and biology of adult human male germ cell tumors. Cancer Res. 2000;60:1475–1482. [PubMed] [Google Scholar]
- Hirota S, Nishida T, Isozaki K, Taniguchi M, Nishikawa K, Ohashi A, Takabayashi A, Obayashi T, Okuno T, Kinoshita K, Chen H, Shinomura Y, Kitamura Y. Familial gastrointestinal stromal tumors associated with dysphagia and novel type germline mutation of KIT gene. Gastroenterology. 2002;122:1493–1499. doi: 10.1053/gast.2002.33024. [DOI] [PubMed] [Google Scholar]
- Pignon JM, Giraudier S, Duquesnoy P, Jouault H, Imbert M, Vainchenker W, Vernant JP, Tulliez M. A new c-kit mutation in a case of aggressive mast cell disease. Br J Haematol. 1997;96:374–376. doi: 10.1046/j.1365-2141.1997.d01-2042.x. [DOI] [PubMed] [Google Scholar]
- Brandli DW, Ulbright TM, Foster RS, Cummings OW, Zhang S, Sweeney CJ, Eble JN, Cheng L. Stroma adjacent to metastatic mature teratoma after chemotherapy for testicular germ cell tumors is derived from the same progenitor cells as the teratoma. Cancer Res. 2003;63:6063–6068. [PubMed] [Google Scholar]
- Tickoo SK, Hutchinson B, Bacik J, Mazumdar M, Motzer RJ, Bajorin DF, Bosl GJ, Reuter VE. Testicular seminoma: a clinicopathologic and immunohistochemical study of 105 cases with special reference to seminomas with atypical features. Int J Surg Pathol. 2002;10:23–32. doi: 10.1177/106689690201000105. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Oosterhuis JW. Pathogenesis of testicular germ cell tumours. Rev Reprod. 1999;4:90–100. doi: 10.1530/ror.0.0040090. [DOI] [PubMed] [Google Scholar]
- Kissel H, Timokhina I, Hardy MP, Rothschild G, Tajima Y, Soares V, Angeles M, Whitlow SR, Manova K, Besmer P. Point mutation in kit receptor tyrosine kinase reveals essential roles for kit signaling in spermatogenesis and oogenesis without affecting other kit responses. EMBO J. 2000;19:1312–1326. doi: 10.1093/emboj/19.6.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton L, Morley GM, Chopra R, Khwaja A. The Src-selective kinase inhibitor PP1 also inhibits Kit and Bcr-Abl tyrosine kinases. J Biol Chem. 2003;278:4847–4853. doi: 10.1074/jbc.M209321200. [DOI] [PubMed] [Google Scholar]