Abstract
pp32 (ANP32A) is a nuclear phosphoprotein expressed as a nonmutated form in self-renewing cell populations and neoplastic cells. Mechanistically, pp32 may regulate pathways important in the process of differentiation as part of separate complexes inhibiting histone acetylation and regulating immediate-early and cytokine mRNA stability. Prostatic adenocarcinomas express pp32 in a differentiation related manner—well-differentiated tumors express lower levels of pp32 than poorly differentiated tumors. In benign prostate, pp32 is expressed in basal cells but not in terminally differentiated glandular cells. Based on these observations, we hypothesized that reduction of pp32 expression might be an important differentiation signal. We used anti-sense pp32 and RNAi transfection to study the effects of reduced pp32 expression in the TSU-Pr1 carcinoma cell line. pp32 reduction induced TSU-Pr1 cells to differentiate into neuronal-like cells with associated inhibition of growth. Reduction of pp32 and consequent differentiation were accompanied by a marked reduction in expression of SET, which complexes with pp32, by a marked change in acetylation status of histone H4, and by further differential expression of genes in differentiation pathways. Thus, reduction of pp32 in the undifferentiated TSU-Pr1 neoplastic cell line induces differentiation and thus may be an element of a differentiation control pathway in both normal and neoplastic cells.
Numerous signaling pathways converge on self-renewing cell populations and cause them to differentiate. Recent work on chromatin structure and the histone code suggests that the conformational and covalent modification of chromatin leads to specific programs and patterns of gene expression that may underlie differentiation. In support of this, agents that inhibit chromatin-remodeling enzymes such as histone deacetylases lead to differentiation.1 Here, we report that modulation of the expression of a single gene, pp32, a key component of an inhibitor of histone acetylation, leads to differentiation and diminished proliferation in the carcinoma cell line, TSU-Pr1.
pp32 is a member of the ANP32 gene family and is the most ubiquitously and highly expressed of the four known and two predicted members (JR Brody and SS Kadkol, unpublished data). pp32 mRNA and protein are found in self-renewing and stem cell-like populations in a broad array of benign tissues and neoplastic cells including prostate adenocarcinomas.2–4 Neoplastic and benign tissues differ significantly in their pp32 expression patterns.4 By in situ hybridization analysis, pp32 expression in benign prostatic glands is restricted to the basal cells, a compartment that contains prostatic epithelial stem cells.5 In contrast, terminally differentiated luminal secretory cells are negative. Restriction of pp32 expression to self-renewing compartments in the normal prostatic glands has also been inferred from animal studies involving castration in rats.6 In contrast, high-level pp32 expression occurs in prostatic adenocarcinomas of Gleason scores ≥5, whereas a low level of expression is seen in tumors with a Gleason score <5, indicating that the intensity of pp32 expression correlates with the differentiation status of prostate tumors.2
The pattern of expression suggests that pp32 expression is high in relatively undifferentiated cells, whether benign or malignant, but is low to absent in more fully differentiated cells. This notion is supported by additional data in hematopoietic cell lines. K562 and ML-1 leukemic cell lines express high levels of pp32 in an undifferentiated state, but show a significant decrease in expression on differentiation with 12-O-tetraoccanolyphorbol B-acetate (TPA). Likewise, CD34+ hematopoietic stem cells express high levels of pp32 (JR Brody and SS Kadkol, unpublished data).
From a biological standpoint, pp32 is a nuclear phosphoprotein that isolates in a complex with SET, which is both structurally and functionally significant.3,7 Similar to pp32 expression, others have reported that SET expression correlates with the state of differentiation.8 Because pp32 and SET are physically associated with each other, SET and pp32 might cooperate in regulating the process of cellular differentiation. As part of a complex known as inhibitor of histone acetyl transferase (INHAT), pp32 and SET inhibit histone acetylation by masking specific regions of the chromatin with consequent effects on transcription.9 As part of the HuR complex, pp32 and SET stabilize messages containing AU-rich elements located in the 3′- and 5′-untranslated regions of cytokines, oncogenes, and other immediate-early mRNA species.10 Recently, it has also been shown that pp32 interacts with the adenovirus type 5, E1B-55k.11 Hence, it is possible pp32 affects the stabilization of p53 and/or of late messages produced by this adenovirus.
We have previously shown that pp32 overexpression possesses many features of a tumor suppressor in a number of in vitro and in vivo assays; although it is neither mutated nor subject to loss of heterozygosity and therefore does not meet the strict definition of tumor suppressor, the abnormal expression in cancer suggests that it may play a suppressor-like role in certain settings. Specifically, pp32 inhibits transformation mediated by a broad range of oncogenes. Taken together with the above data, these models suggest that pp32 overexpression in stem cell-like populations may protect cells from the transformation process.
Based on the above observations, we hypothesized that a reduction of pp32 expression might lead to differentiation and cessation of proliferation of cancer cells; we further hypothesized that this might occur through pp32’s effects on histone acetylation or its other activities that can modulate gene expression. To test this hypothesis, we reduced pp32 expression in the TSU-Pr1 cell line and followed the effects on cell growth and differentiation. TSU-Pr1, a malignant cell line with p53 and ras mutations, was originally thought to be derived from a prostatic adenocarcinoma and is now believed to be more consistent with bladder carcinoma origin.12,13 TSU-Pr1 cells are competent to differentiate into neuroendocrine or microglial cells depending on whether they are treated with staurosporine or TPA.14,15 Because of their phenotypic plasticity and high level of pp32 expression, TSU-Pr1 cells were chosen as a model system to study the effects of pp32 reduction. Here we report that a reduction in pp32 expression causes TSU-Pr1 cells to undergo neuronal differentiation accompanied by a marked decrease in growth potential. We also show that pp32 reduction is accompanied by gene expression changes in pathways that either collaborate or cross-talk during differentiation.16–18
Materials and Methods
Transfection of TSU-Pr1 Cell Lines with pp32 Constructs
Either anti-sense pp32 or sense pp32 sequences were subcloned into an expression vector under pCMV promoter control. Equal numbers of TSU-Pr1 cells were maintained in RPMI 1640 with 10% fetal bovine serum and antibiotics (complete medium) in separate T-75 flasks in a humidified atmosphere containing 5% CO2. Cells were transfected with the anti-sense pp32, sense pp32, or vector-only constructs at ∼50% confluency with Lipofectin reagent (Invitrogen, Carlsbad, CA) for 5 hours. The medium was then changed to complete medium and cells were allowed to recover for 48 to 72 hours. After this interval, the cells were trypsinized and replated on to 100-mm dishes and the selection process was started by addition of 500 μg/ml of G418 (Invitrogen) to the culture medium. After several weeks of culture and selection to allow stable integration of the plasmids to occur, the resultant clones were counted and single clones picked for evaluation or pooled clones created to negate a position effect and clonal variation. The AS-32 cells used in this study consist of a pool of ∼25 randomly selected G418-resistant clones.
Transfection with RNAi pp32 Construct into TSU-Pr1 Cells
One day before transfection TSU-Pr1 cells were counted and plated at 1 × 105 cells per well in a six-well plate. After seeding overnight, OPTIMEM plus Oligofectamine and OPTIMEM plus RNA (Invitrogen) were incubated separately for 10 minutes at room temperature and then combined into one polystyrene tube. The combined Oligofectamine-RNA-OPTIMEM solution was incubated in polystyrene tubes for 20 minutes at room temperature. The solution was then mixed gently and pipetted onto the TSU-Pr1 cells. The cells were checked for pp32 and neuron-specific enolase (NSE) expression after 3 to 5 days of incubation as described in the Western blot method. TSU-Pr1 cells were transfected with 250 nmol/L of RNAi construct and retransfected once more during a 5-day period. The following RNAi oligonucleotide is specific for pp32 cDNA and does not target pp32r1 or pp32r2: pp32 sense-1—modified to 19 nucleotides, GC 52%, 5′-CGATAACAGAGTCTCAGGG-3′; sense and anti-sense RNA oligos, 5′-CGAUAACAGAGUCUCAGGGdTdT-3′ and 3′-dTdTGCUAUUGUCUCAGAGUCCC-5′. We also used scramble-II sequence (target sequence: 5′-GCG CGC TTT GTA GGA TTC G-3′) from Dharmacon.com as a control that should not target any known functional gene (www.dharmacon.com).
Analysis of Growth Properties
Cells (5 × 104) were plated in T-75 flasks and maintained in complete medium. In addition, AS-32 and vector only control cells were maintained in medium containing G418 (Invitrogen). Cells were washed twice in phosphate-buffered saline, trypsinized, and viable cells were counted on indicated days by Trypan blue exclusion in a hemocytometer. Each data point represents the mean number of viable cells from duplicate flasks.
Western Blot Analysis
Cells were lysed in M-per reagent (Pierce, Rockford, IL) containing one tablet of Complete Mini Protease Inhibitor (Roche Molecular Biochemicals, Indianapolis, IN) per 5 ml mol/L-per on ice for 10 minutes in Petri dishes. The lysate was scraped into an Eppendorf tube, vortexed, and then centrifuged for 10 minutes at maximum speed. The supernatant was then transferred to a fresh tube and used for Western blot analysis. Protein amounts were measured using the BCA protein assay (Pierce) and 7.5 μg of protein lysate were loaded in each lane of a 10% Bis-Tris polyacrylamide gel (Invitrogen). After electrophoresis, proteins were transferred onto polyvinylidene difluoride membranes. The membranes were then blocked in 5% nonfat milk in TTBS (20 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl) for 1 hour at room temperature. The membranes were then incubated with the indicated primary antibodies diluted in 5% nonfat milk in TTBS overnight at 4°C: anti-pp32 1:200, anti-NSE 1:200 (Chemicon, Temecula, CA), anti-AChE 1:200 (Santa Cruz Biotechnology, Santa Cruz, CA), or anti-acetyl H4 2 μg/ml (Upstate Biotechnology, Waltham, MA). Subsequently, the membranes were washed and incubated with a 1:5000 dilution of the appropriate secondary antibodies for 1 hour (biotin-labeled anti-rabbit IgG, Amersham Biosciences, Inc., Piscataway, NJ; biotin-labeled anti-mouse IgG, Sigma Chemical Co., St. Louis, MO). Membranes were washed extensively and chemiluminescent detection was performed with Supersignal West (Pierce). Immunoblots were stripped with Restore Western Blot Stripping Buffer (Pierce) and reprobed as necessary.
RNA Extraction
Total RNA was extracted using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA) as per the protocol. Contaminating DNA was eliminated by extensively treating the RNA samples with RNase-free DNase I (Amersham Biosciences, Inc., Piscataway, NJ). Absence of DNA contamination was confirmed by 40 cycles of polymerase chain reaction (PCR) amplification omitting the reverse transcriptase step using the same primer set used for reverse transcriptase (RT)-PCR.
Quantitative RT-PCR
Quantitation of pp32-specific mRNA was performed by competitive RT-PCR. Sense and anti-sense primer sequences corresponding to pp32 were incorporated at either end of the competitor DNA sequence by PCR so that both the competitor and the endogenous pp32 transcripts could be amplified using the same primer set in a single tube. The competitor DNA was subcloned into the expression vector, pCR II TOPO (Invitrogen) and sense strand RNA was generated by in vitro transcription using the MegaScript Kit (Ambion Inc., Austin, TX). The template DNA used for in vitro transcription was eliminated by DNase I treatment.
Twenty ng of DNA-free total RNA were reverse-transcribed and PCR-amplified in the presence of three different concentrations of competitor RNA in three separate tubes using the One-Step RT-PCR kit (Qiagen). For each sample, three tubes each containing 20 ng of total RNA and either 4.63 × 105, 4.63 × 104, or 4.63 × 103 copies of competitor RNA were used. Reverse transcription was performed with the anti-sense primer only (5′-TAGTCGTTCAGGTTGGTTAC-3′) at 52°C for 45 minutes followed by inactivation of the enzyme at 95°C for 15 minutes. This step also activated the HotStartaq polymerase in the enzyme mix. At this point, the sense primer (5′-TAAGAAGCTTGAACTAAGCGA-3′) was added to each tube and PCR amplification was performed for 24 cycles consisting of 92°C for 30 seconds, 57°C for 35 seconds, and 72°C for 35 seconds followed by a final extension step at 72°C for 10 minutes. Ten μl of the product were stained with 1 μl of 1:1000 dilution of SYBR GOLD (Molecular Probes, Inc., Eugene, OR) and electrophoresed on a 2% agarose gel in Tris borate-ethylenediaminetetraacetic acid. The product generated by the competitor (280 bp) was easily distinguishable from the product of the endogenous pp32 transcript (195 bp).
Image Analysis
The image of the gel under UV light was captured digitally and analyzed by the TotalLab software v. 1.10 (www.phoretix.com) to determine the densities of the bands from the competitor and the endogenous pp32 transcript. Because incorporation of dye is dependent on mass, the intensity of the smaller pp32 band was multiplied by the ratio (competitor band mass/pp32 band mass). The result was designated as the corrected pp32 band intensity.
The ratio of the intensities of the competitor to the corrected pp32 band density was obtained for each of the three tubes from every sample. A linear regression analysis was performed with the log copy number of the competitor on the x axis and the log of the above ratio on the y axis. The point at which the regression line intersected the x axis indicated an equal amount of starting endogenous pp32 mRNA and exogenously added competitor.
Image analysis of Western blots and of other amplifications was performed in a similar manner. The percent reduction or fold induction was determined as the quotient of the pixel volumes of the bands corresponding to the indicated species and samples.
RT-PCR to Confirm Anti-Sense pp32 Expression
Anti-sense expression was confirmed in TSU-Pr1 transfectants by qualitative RT-PCR. Twenty ng of DNA-free RNA were reverse-transcribed with the sense primer at 52°C for 45 minutes followed by inactivation of the RT enzyme at 95°C for 15 minutes. The anti-sense primer was then added and PCR amplification was performed for 30 cycles under the conditions described above.
Proliferating Cell Nuclear Antigen (PCNA), Interleukin (IL)-6, CDK4, and SET Expression Analysis by RT-PCR
PCNA, IL-6, and SET expression were analyzed by RT-PCR with the Qiagen One-Step RT-PCR system with the following primers: PCNA, sense 5′- ATGGGCGTGAACCTCACCAGTAT-3′ and anti-sense 5′- TCCGTCTTTTGCACAGGAAATTACAA-3′; IL-6, sense 5′- TTGCCTGGTGAAAATCATC-3′ and anti-sense 5′- GGTGCCCATGCTACATTT-3′; SET, sense 5′-GAAGAGGCACTGCATTATTTG-3′ and anti-sense 5′-CTCTCCTGCTGGCTTTATTCT-3′; CDK4, sense 5′-GAGGCG ACTGGAGGCTTTTG-3′ and anti-sense 5′-CTGGCGCATCAGATCCTTGA-3′.
For PCNA and IL-6 analysis, 60 ng of DNA-free total RNA were reverse-transcribed with the anti-sense primer at 52°C for 45 minutes followed by inactivation of the RT enzyme at 95°C for 15 minutes. Subsequently 25 cycles of PCR, each cycle consisting of 92°C for 30 seconds, 57°C for 30 seconds, and 72°C for 35 seconds and a final extension step of 72°C for 10 minutes were performed. For SET analysis, 60 ng of DNA-free total RNA were reverse-transcribed with the anti-sense primer at 50°C for 45 minutes followed by inactivation of the RT enzyme at 95°C for 15 minutes. Subsequently, 21 cycles of PCR, each cycle consisting of 92°C for 30 seconds, 52°C for 30 seconds, and 72°C for 35 seconds and a final extension step of 72°C for 10 minutes, were performed. Ten μl of the product from each sample were stained with 1 μl of 1:1000 solution of SYBR GOLD and electrophoresed on a 2% agarose gel in Tris borate-ethylenediaminetetraacetic acid. For CDK4 analysis, 60 ng of DNA-free total RNA were reverse-transcribed with the anti-sense primer at 50°C for 45 minutes, followed by inactivation of the RT enzyme at 95° for 15 minutes. Subsequently, 27 cycles of PCR were performed with each cycle consisting of 92°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, followed by one cycle of 92°C for 30 seconds, 60°C for 30 seconds, and 72°C for 10 minutes. Ten μl of the product was stained with SYBR GOLD and electrophoresed as described above.
Microarray Analysis of TSU-Pr1 Cell Lines
This procedure was performed at The Johns Hopkins University Oncology Microarray facility by using a human glass 12K cDNA chip. RNA was prepared as the same preparation for RT-PCR analysis and a total of 10 μg were used. RNA was extracted from the TSU-Pr1 cell line and TSU-Pr1 cells reduced in pp32 expression. cDNA is transcribed using reverse transcriptase and then Cy3 and Cy5 monofunctional, N-hydroxysuccinimide-activated fluorescent dye is labeled to the prepared cDNA. After labeling with the either fluorescent dye, cDNA probes are then purified to extract the unreacted dye. Then the microarray chip, which is prepared by the Microarray core facility, 1) is prehybridized, 2) hybridized with the coupled two cDNA samples, and 3) then washed several times for stringency. Once data were read off the chip we used the following program to quantify the fold changes of gene expression: http://www.nhgri.nih.gov/DIR/LCG/15K/HTML/img_analysis.html.
Results
pp32 Is Expressed in Basal and Neoplastic Cells in the Prostate and in Other Neoplastic Cell Lines
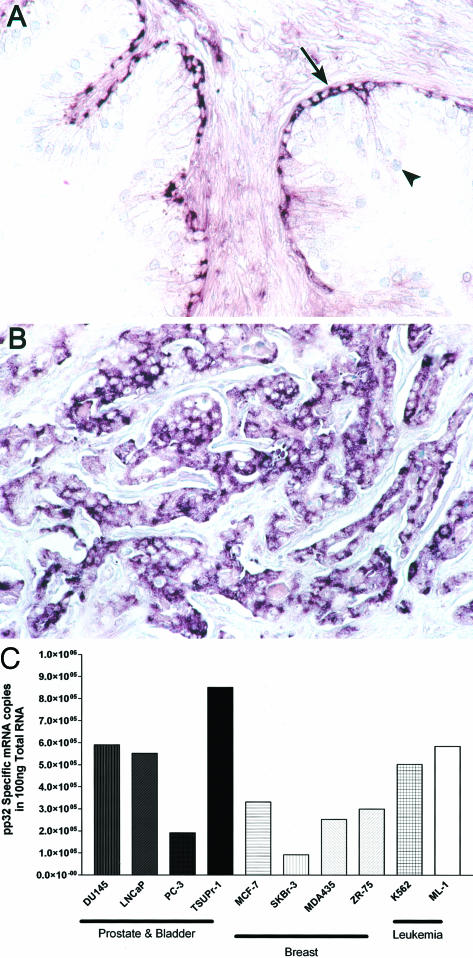
pp32’s expression pattern in benign tissues is both informative and striking. Basal cells strongly hybridize with a probe that detects pp32, whereas the differentiated luminal secretory cells are negative (Figure 1A). The basal cells of the prostatic glands are believed to contain the epithelial stem cell compartment in the prostate.5 Like basal cells, prostatic adenocarcinomas also hybridize strongly to a pp32 probe; this is consistent with our previous findings (Figure 1B).2 Prostatic adenocarcinoma cell lines DU 145, PC 3, LNCAP, and TSU-Pr1 cells, all express pp32. Consistent with our previous findings that pp32 is expressed in cancers from different origins, we show that pp32 is also expressed in breast carcinoma and leukemia cell lines. The TSU-Pr1 cell lines showed the highest level of specific pp32 expression and hence were chosen for this study (Figure 1C).
Figure 1.
pp32 mRNA expression analyzed by in situ hybridization. A: Benign prostatic hyperplasia. Basal cells of the benign prostatic gland express pp32 message (arrow) whereas the differentiated secretory cells (arrowhead) are negative. B: Poorly differentiated prostatic adenocarcinoma (Gleason score 9) showing abundant pp32 mRNA expression. C: pp32 is expressed in a number of prostatic carcinoma cell lines (DU 145, PC 3, LNCAP, and TSU-Pr1 cells), breast carcinoma cell lines (MCF-7, SKBr-3, MDA435, ZR-75 cells), and leukemia cell lines (K562 and ML-1 cells). Expression was measured by quantitative RT-PCR. Original magnifications, ×250 (A, B).
Reduction of pp32 mRNA Expression Consistently Reduces Expression of pp32 Protein
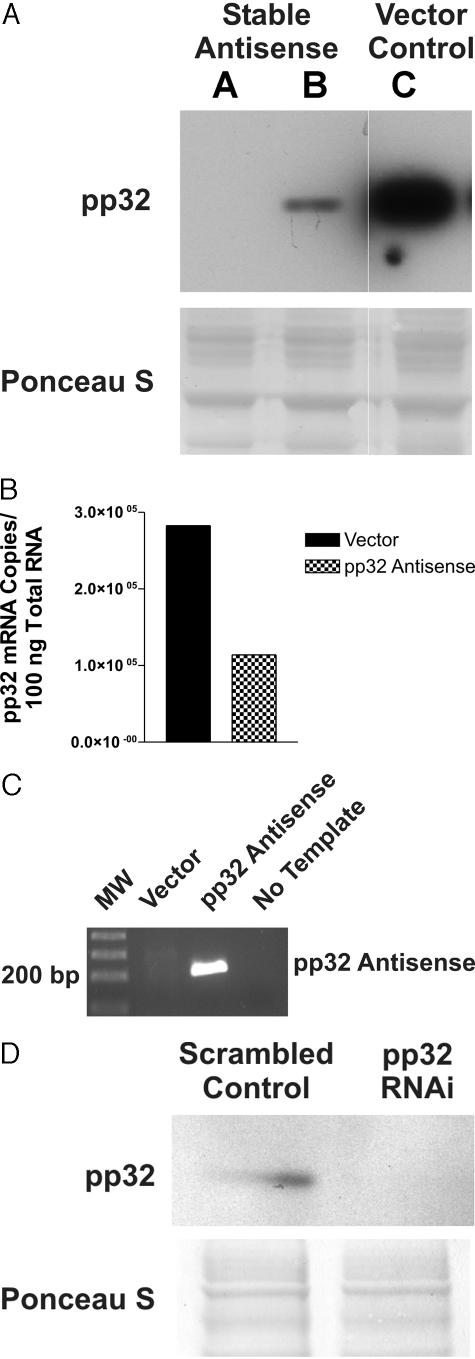
As a first step in studying the effects of pp32 anti-sense, we demonstrated that stable transfection of pp32 anti-sense reduces the expression of pp32 in TSU-Pr1 cells. Figure 2A shows that anti-sense pp32 expression decreased pp32 protein levels in pooled transfected cells (AS-32 cells) surviving selection as compared to the vector-only cells. This progressive reduction in pp32 protein correlated with a reduction of sense pp32 mRNA by quantitative pp32-specific RT-PCR (Figure 2B). To confirm that the anti-sense construct was responsible for the reduction of pp32 expression, we determined by RT-PCR that AS-32 cells indeed produced the desired anti-sense RNA species. The AS-32 cell line showed significant expression of the anti-sense pp32 strand when compared with vector-only cells (Figure 2C). Because the process of stable transfection required selection and took place throughout a relatively extended period of time, we also transfected TSU-Pr1 cells with an RNA interference (RNAi) construct directed specifically against pp32 to determine whether a more immediate, transient reduction of pp32 expression could result in comparable biological effects and thereby validate the anti-sense method. Figure 2D shows that a pp32 RNAi construct transfected into TSU-Pr1 cells substantially reduced pp32 protein expression within 5 days.
Figure 2.
Characterization of stable TSU-Pr1 transfectants. A: Immunoblot analysis with anti-pp32 antibody. Lanes A and B, AS-32; lane C, vector control. The Ponceau S-stained membrane shown below the blot demonstrates that equal amounts of protein were loaded onto each lane. Lanes A and B represent two different lines of the AS-32 cells. B: Quantitative analysis of pp32-specific mRNA by competitive RT-PCR. The graph shows the actual copy numbers computed from densitometric calibration curves as described in Material and Methods. C: The transfected anti-sense construct is expressed by RT-PCR analysis. AS-32 cells stably transfected with pp32 anti-sense express the anti-sense pp32 construct mRNA whereas vector control transfectants do not express the anti-sense strand. D: pp32 RNAi abolishes expression of pp32. Top: A Western blot demonstrating that no pp32 is detected in TSU-Pr1 cells treated with pp32 RNAi, whereas pp32 continues to be expressed in cells treated with the scrambled control RNAi construct. Bottom: A Ponceau S stain of the membrane, demonstrating loading of equal amounts of protein.
Reduction of pp32 Expression Inhibits Growth of TSU-Pr1 Cells
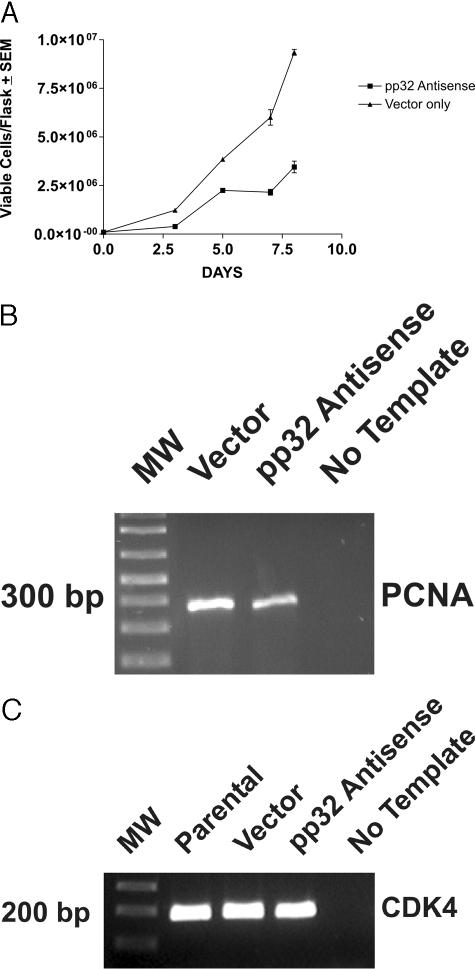
Because RNAi transient transfection is inherently unstable and incomplete, affecting only a fraction of cells, growth properties are better analyzed in stable transfectants. TSU-Pr1 cells stably transfected to express pp32 anti-sense grew at a substantially slower rate than did cells transfected with vector alone (Figure 3A). By RT-PCR analysis, the growth inhibition was associated with a significant reduction of PCNA expression, a well-known marker for cycling cells (Figure 3A).19 Quantitation by image analysis showed that PCNA was reduced to 67% of its control level in anti-sense-transfected cells. These data show that reduction of pp32 expression inhibits both growth of TSU-Pr1 cells and the expression of a marker associated with proliferation. Figure 3C, which shows similar amounts of CDK4 amplified from the same RNA samples, serves as a loading control for Figure 3B and for subsequent figures using the same RNA samples (see Figures 7 and 8). Image analysis revealed less than 5% band intensity variation.
Figure 3.
Anti-sense pp32 expression decreases the growth of TSU-Pr1 cells. A: Analysis of cell proliferation. Growth analysis was performed by seeding equal numbers of cells in T-75 flasks. Viable cells were counted on a hemocytometer on the indicated days and the results were plotted. B: Analysis of PCNA expression. The gel shows the results of PCNA expression by RT-PCR analysis on RNA that was extracted on day 4. MW indicates the ladder, with the size of the marker nearest the PCNA amplicon indicated. C: Analysis of CDK4 expression. CDK4 expression was analyzed as described in Materials and Methods in each of the same RNA samples used in B, and in Figures 7 and 8. CDK4 levels were comparable in all samples.
Figure 7.
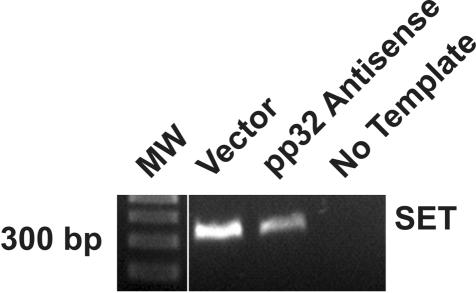
Analysis of SET expression after reduction of pp32 in TSU-Pr1 transfectants. The figure shows analysis of SET by RT-PCR in cells stably transfected with pp32 anti-sense or control cells. SET expression is down-regulated in anti-sense-transfected cells.
Figure 8.
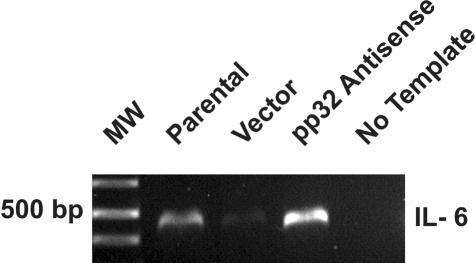
Expression of IL-6 in TSU-Pr1 cells. TSU-Pr1 cells stably transfected with pp32 anti-sense express higher levels of IL-6 message as compared to parental TSU-Pr1 cells and vector-only control by RT-PCR analysis, which validates the cDNA microarray analysis (see Figure 6).
Reduction of pp32 Expression Causes TSU-Pr1 Cells to Acquire Neuronal Morphological Features and to Express Neuronal Markers
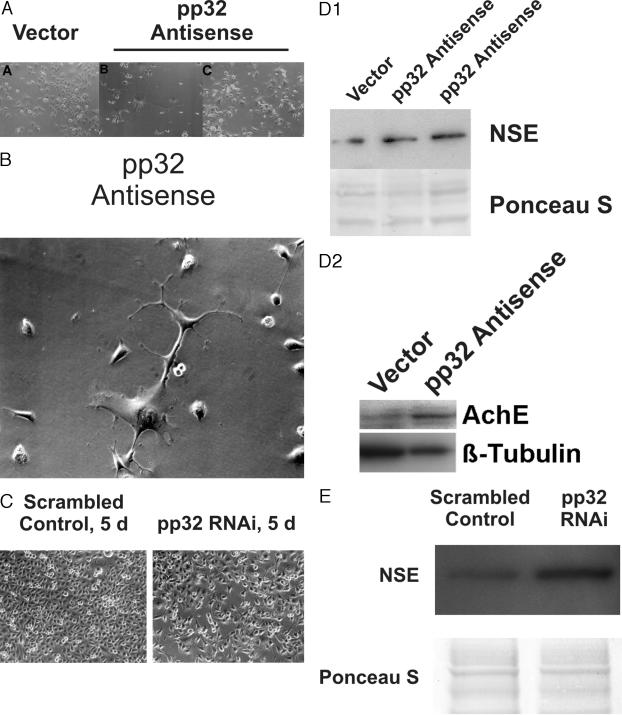
Both pp32 anti-sense and RNAi affect the state of cellular differentiation seen both in morphology (Figure 4; A to C) and in expression of differentiation markers (Figure 4, D and E). In culture, AS-32 cells displayed long arborizing processes whose lengths were greater than two to three times the maximum dimensions of the cell bodies (Figure 4B). The processes from different cells appeared to touch each other at several places. Approximately 80 to 90% of the cells showed such morphology in any individual flask. These morphological features are consistent with previous reports of neuronal differentiation in TSU-Pr1 cells induced by staurosporine.14 In marked contrast, only a few (<10%) of vector control cells elaborated elongated cellular processes. Reduction of pp32 by RNAi caused similar morphological changes in treated TSU-Pr1 cells, whereas cells transfected with the scrambled control were unchanged (Figure 4C).
Figure 4.
Effect of pp32 reduction on TSU-Pr1 cell morphology. A: Low-power view. A, Vector control—a representative focus of polygonal TSU-Pr1 cells devoid of neuron-like features; B and C, AS-32 cells showing distinct neuronal features characterized by outgrowth of cellular processes. B: High-power view. AS-32 cells displaying neuron-like features including outgrowth of processes greater than two to three times the maximum cell body dimension. C: Effects of pp32 RNAi on TSU-Pr1 morphology. The figure shows that pp32 RNAi induces morphological changes in TSU-Pr1 cells comparable to those produced by stable pp32 anti-sense transfection. Note that the scrambled control is equivalent to the TSU-Pr1 parental line. D: Immunoblot analysis of NSE (top) and AchE (bottom) expression. Lysates of the indicated cells were analyzed for neuron-specific enolase or for acetyl cholinesterase. Equal loading of protein on each lane was verified by Ponceau S staining or by anti-β-tubulin analysis by immunoblot. E: Induction of NSE expression by pp32 RNAi. The figure shows Western blots demonstrating decreased levels of pp32 and increased levels of NSE expression in cells treated with pp32 RNAi as compared to the scrambled RNAi control. Equal loading is verified by the Ponceau S-stained membrane. Original magnifications: ×200 (A); × 400 (B).
Depletion of pp32 in TSU-Pr1 cells by stable pp32 anti-sense transfection led to an induction of NSE of 1.9-fold and of acetyl cholinesterase (AChE) of 4.4-fold normalized to β-tubulin, respectively (Figure 4D), as determined by image analysis. Transfection of the RNAi construct also led to comparable induction of NSE (Figure 4E), as compared to the scrambled oligonucleotide, accompanied by an increase in AChE (data not shown) similar to that seen in the stably transfected cells. The induced differentiation of TSU-Pr1 cells is thus independent of the method used to achieve a reduction of pp32, because two related but independent methods, transient transfection of an RNAi construct and stable transfection of an anti-sense construct led to the same results and conclusions.
Differentiation of TSU-Pr1 Cells Induced by a Reduction of pp32 Expression Is Accompanied by a Reduction in Histone Acetylation
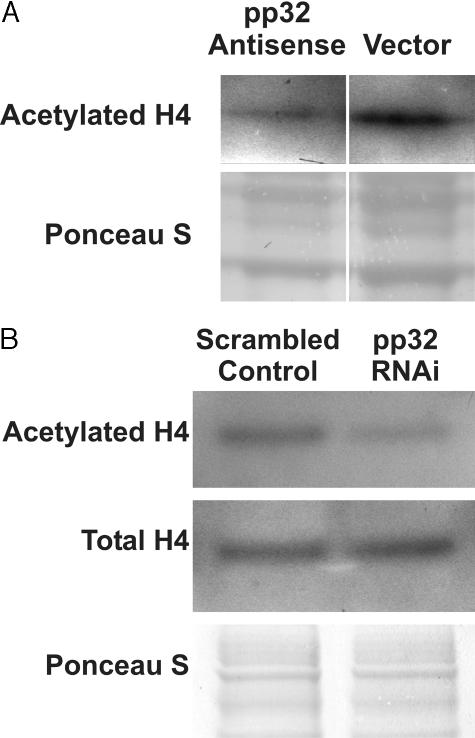
pp32 is part of a complex that inhibits histone acetyl transferase activity and transcription of certain genes. We hypothesized that high levels of pp32 might serve to maintain TSU-Pr1 cells in an undifferentiated and proliferative state by inhibiting histone acetylation. To test this hypothesis, we analyzed histone H4 acetylation status20 in both the anti-sense and RNAi systems. Rather than the predicted elevation, histone acetylation was reduced to 32% of the vector control value in TSU-Pr1 cells stably transfected with anti-sense as determined by image analysis of the Western blot (Figure 5A). Likewise, in RNAi-treated cells, histone H4 acetylation was decreased to 43% of the scrambled oligonucleotide control (Figure 5B) at 72 hours after transfection. Note that the total level of histone H4 remained constant and that only the acetylated fraction was reduced in the RNAi-treated cells. As discussed below, this reduction is consistent with other systems and reflects the transient nature of elevations of histone acetylation in differentiating cells.1
Figure 5.
Analysis of histone acetylation. A: Immunoblot analysis of histone acetylation. AS-32 cells stably transfected with pp32 anti-sense showed a reduced expression of hyperacetylated histone H4 compared to cells transfected with vector only. B: Effect of pp32 RNAi on histone acetylation. The figure shows Western blots demonstrating that pp32 RNAi induces decreased levels of acetylated histone H4 in treated TSU-Pr1 cells as compared to the scrambled control. Only acetylation was affected because analysis of total histone H4 protein demonstrated comparable levels in both control and RNAi-treated cells, and the Ponceau S stain confirmed that equal amounts of protein were loaded.
Reduction in pp32 Expression in TSU-Pr1 Cells Changes the Gene Expression Pattern
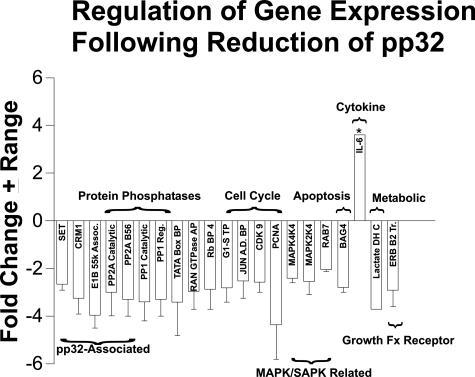
Because reduction of pp32 changed the acetylation of histone H4, we wanted to define the pathways affected by changes in pp32 expression. Figure 6 summarizes the fold change of known genes affected by a pp32 reduction in the TSU-Pr1 cell line. Note that expression of the vast majority of genes on the 12K cDNA chip was unaffected by pp32 anti-sense transfection, eliminating general, nonspecific effects on gene expression. Because the selection process lowered the pp32 expression slightly in vector-only cells, we chose to compare gene expression patterns in the TSU-Pr1 parental cell line to the AS-32 cells. All of the genes that reduced in expression on pp32 reduction were validated in two separate array experiments. In one experiment, the RNA was extracted from cells sampled early after selection, in the other, the RNA derived from cells sampled late in the postselection process after neuronal differentiation and expression of neuronal markers had been confirmed. Hence, we believe that these are early and consistent expression events that are needed in the process of differentiation of these cells. We found that levels of pp32 regulate genes that have been described previously to physically and functionally interact with pp32: SET, CRM1, and PP2A. pp32 also affects the expression of genes involved in differentiation and/or proliferation: cell cycle, MAP kinase, apoptosis, cytokine, metabolic, PP2A, p53 stabilization, and growth factor receptor pathways (Figure 6). Interestingly, pp32 also inhibits PP2A activity at the biochemical level.21 The PP2A-B56 subunit confers specificity on PP2A’s enzymatic function, and has been shown to regulate apoptotic and wnt signaling pathways.22,23 Here we show that in addition to the above effects, pp32 also regulates the steady-state mRNA levels of the catalytic subunit of PP2A in cancer cells.21 Hence, Figure 6 shows that a down-regulation of pp32 affects the expression of a number of pathways that collaborate in differentiation.
Figure 6.
Pathways displaying altered gene expression after pp32 reduction in TSU-Pr1 cells. The figure shows the fold change in gene expression in pathways involved in differentiation and/or proliferation after reduction of pp32 expression in TSU-Pr1 cells and their subsequent differentiation into neuronal-like cells. Genes shown were positive in two separate array experiments as described, and the error bars reflect the range of the fold change in expression. Expression of genes marked with an asterisk was validated by semiquantitative RT-PCR. The full names of the genes illustrated are: SET, CRM1, E1B-55-kd-associated protein 1, protein phosphatase 2A catalytic subunit, protein phosphatase 2A B56 subunit, protein phosphatase 1 catalytic subunit, protein phosphatase 1 regulatory subunit, TATA box-binding protine, RAN GTPase-activating protein, RB-binding protein 4, G1-S transition protein, Jun AD-binding protein, CDK9, proliferating cell nuclear antigen (PCNA), MAP kinase 4K4, MAP kinase 2K4, RAB7, BAG4, lactate dehydrogenase C, transducer of ERB B2.
Because pp32 is physically associated with SET in the INHAT complex, we analyzed SET mRNA expression as a function of pp32 to determine whether there is coordinate regulation of expression of the two molecules.24 AS-32 cells expressed 47% of the level of SET message of vector control cells as determined by image analysis (Figure 7), indicating that reduction of pp32 expression in turn leads to reduced SET expression. This data validates our cDNA microarray data that SET expression is co-regulated by pp32 expression in the TSU-Pr1 cell line (Figure 6). These results suggest the novel concept that pp32 not only binds SET physically, but also regulates SET expression.
pp32 Down-Regulation Induces IL-6 Expression
Because TSU-Pr1 cells were previously shown to differentiate with staurosporine through its effects on p21 expression, we sought to study p21 expression changes in our model. We found that p21 expression does not significantly change on differentiation through a reduction on pp32 expression (JR Brody, data not shown). However, Figure 8 demonstrates that differentiation induced by reduction of pp32 expression is associated with a 10-fold up-regulation of IL-6 message as determined by an image analysis comparison of vector-transfected cells with cells transfected with anti-sense. Although IL-6 message was up-regulated only in the first array, after selection but before confirmation of neuronal marker expression, the up-regulation was validated independently by RT-PCR (Figures 6 and 8), suggesting that it is required for induction but not maintenance of differentiation. We based these findings on cDNA microarray analysis of TSU-Pr1 cells with reduction of pp32 as compared to the TSU-Pr1 parental cells (Figure 6). These findings showed that pp32 reduction in TSU-Pr1 cells down-regulated the exportin, CRM1, that has been proven to be part of the HuR complex, which stabilizes certain cytokine mRNAs such as IL-6 (Figure 6).10 Recently, it has been shown that IL-6 can differentiate prostate adenocarcinoma cell lines into cells displaying neuroendocrine-like features.25
Discussion
We report that reduction of pp32 expression by two different methodologies induces differentiation and reduces proliferation in vitro of the poorly differentiated TSU–Pr1 carcinoma cell line. These results suggest a possible mechanistic explanation for the low levels of pp32 observed in well-differentiated prostatic adenocarcinomas and the high levels in poorly differentiated tumors. It is conceivable that persistent pp32 expression and an inability to reduce pp32 expression might contribute to a blockade of a critical differentiation pathway and result phenotypically in poorly differentiated prostatic tumors. The self-renewal capacity of cells in the basal cell compartment of normal or benign prostatic glands might also depend on pp32 expression. Extrapolating the results from this TSU-Pr1 model, a reduction of such pp32 expression might lead to the differentiation of basal cells into secretory cells in the prostate. This possibility will need to be tested in future experimentation.
As an inhibitor of histone acetylation, pp32 plays a role in a critical cellular mechanism that is likely to be involved in the control of differentiation. pp32 inhibits histone acetyl transferase activity through its interaction with SET, leading to a decrease in histone acetylation in vitro.24 We show that pp32 expression levels affect the acetylation status of histone H4 in vivo in two independent systems. Our initial supposition had been that the increased transcriptional requirements of differentiation would be accompanied by a concomitant increase in histone acetylation, rather than the observed decrease. Several possibilities may account for the variance of the results from this supposition. In these experiments, histone acetylation was assessed after terminal differentiation had occurred, and not during the process of differentiation. It is possible that histone acetylation would have shown an increase had it been assessed during, rather than after, the process of differentiation; this possibility is quite likely because acetylation induced by histone deacetylase inhibitors produces only rapid, transient elevations of acetylation in TSU-Pr1 cells.26,27 Likewise, the transient transfection used in the RNAi studies precluded examination of a homogeneous, synchronous population. Finally, aberrant histone H4 acetylation in TSU-Pr1 cells could also account for these findings. These observations may also suggest that pp32 expression may correlate with the acetylation status of core histones in cancer, which may in the future translate into an informative prognostic and therapeutic marker.
The observation that pp32 reduction leads to concomitant reduction of SET expression bears comment. The data indicate that pp32 acts not only as a physical subunit of the INHAT complex, but also as a regulator of the activity of the complex through regulation of expression of at least one additional subunit of the complex, namely SET (Figure 6). Like pp32, SET is highly expressed in a variety of tumor types8 and is highly expressed in more poorly differentiated tumors.2,4 pp32 regulation of SET expression, with a consequent maintenance of an undifferentiated state, might explain why SET is also highly expressed in cancers.9 For example, SET expression was high in developing rat and human kidney in comparison to terminally differentiated kidney cells, suggesting a role in differentiation.8 Functionally, the combination of pp32 and SET leads to more potent INHAT activity than does either subunit alone.20 Moreover, simultaneous overexpression of SET and pp32 block histone acetyl transferase-mediated transcription.20,24 Hence, coordinated expression of both SET and pp32 may be required to obtain physiological levels of histone acetyl transferase inhibition as well as inhibition of demethylation of certain genes that we found change expression on manipulation of pp32 expression.9 Because pp32 and SET are both expressed at high levels in stem cell-like undifferentiated cells, we suggest that SET and pp32 may be jointly expressed to provide a coordinated control mechanism for proliferation and differentiation that serves to keep cells in an undifferentiated state. The data thus support the notion that pp32 is a key regulator of the INHAT complex. Studies are in progress to determine the precise mechanism whereby pp32 regulates SET expression.
The microarray data, although restricted to a single cell line, suggest that pp32 may regulate additional molecules associated with chromatin modulation, control of gene expression, and control of proliferation. Future experiments will need to determine their generality and full significance.
Although presently restricted to a single cell line, the theme of pp32-mediated regulation of components of complexes in which it participates is echoed in the effect of pp32 reduction on several other molecules with which pp32 interacts. pp32 expression reduces the expression of CRM1, a member of the HuR complex along with pp32. pp32 is part of a complex that stabilizes specific messages with the exportin, CRM1, and HuR, a member of the Elav family of RNA-binding proteins.10 Messages affected by this mechanism contain AU-rich elements and consist predominantly of cytokine and proto-oncogene mRNAs. Central nervous system tumors, regardless of cell origin, express high levels of HuR, where it may stabilize messages involved in tumor progression such as vascular endothelial growth factor and IL-6.28 IL-6 plays a critical role in cell differentiation.25
The hypothesis that pp32 reduction up-regulates IL-6- mediated differentiation of the TSU-Pr1 cell line is consistent with previously published reports that IL-6 mediates differentiation of prostate cancer lines displaying phenotypic plasticity.17,29 Up-regulation of IL-6 mRNA is likely to occur at a transcriptional level. Reduction of pp32 could lead to increased acetylation of the IL-6 gene; this could be confirmed by chromosome immunoprecipitation experiments that are beyond the scope of the present study.
A second mechanism could also serve to increase IL-6 protein expression. It is interesting to postulate that pp32, HuR, SET, and CRM1, acting as described above, might act to stabilize and/or sequester mRNAs encoding important cellular differentiation signals such as IL-6, interferons, and transforming growth factor-β. pp32 down-regulation could control the HuR complex’s ability to stabilize specific cytokine messages directly and through regulation of expression of its constituents. According to this scheme, inhibition of pp32 would release IL-6 and possibly other messages for translation and subsequent differentiation by effecting the transcription of mRNA nucleocytoplasmic shuttling molecule, CRM1 and pp32’s partner SET.10
Our model suggests that a primary reduction of pp32 expression may be necessary for differentiation in both neoplastic and normal tissues. Underscoring the importance of pp32 expression in these settings, we have not found a mutation in pp32 in either undifferentiated or cancer cells to date.3 Hence, pp32 appears to be intact in all cancers thus far examined.3,4 This concept has important therapeutic implications in treating prostate cancer, because differentiating agents are already in use in promyelocytic leukemia treatment, and inhibitors of histone deacetylase are showing great promise.30 Treatment of TSU-Pr1 cells with a histone deacetylase inhibitor causes growth suppression in the absence of cell death that is substantially equivalent to the effects of pp32 reduction.30 As future experimentation extends the present findings and provides support for hypotheses suggested by the data, it may become reasonable to identify and explore small molecule inhibitors of pp32 function or expression as potential differentiating agents with therapeutic value, that preserve the inherent highly selective target specificity of pp32 acting through chromatin modulation. Our studies are consistent with the notions that divergent pathways collaborate in a selective manner to induce differentiation and that these specific pathways might, in some instances, be coordinated by pp32.
Footnotes
Address reprint requests to Gary R. Pasternack, M.D., Ph.D., Department of Pathology, The Johns Hopkins University School of Medicine, 418 N. Bond St., Room B300, Baltimore, MD 21231. E-mail: gpastern@jhmi.edu.
Supported in part by the National Institutes of Health (R01 CA 54404).
J. R. B. and S. S. K. share first authorship of this article based on their distinct and equal contributions to the research reported herein.
Disclosure: Under a licensing agreement between Marligen, Inc. and The Johns Hopkins University, J. R. B., S. S. K., and G. R. P. are entitled to a share of royalty received by The Johns Hopkins University on sales of products described in this article. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
References
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- Kadkol SS, Brody JR, Epstein JI, Kuhajda FP, Pasternack GR. Novel nuclear phosphoprotein pp32 is highly expressed in intermediate- and high-grade prostate cancer. Prostate. 1998;34:231–237. doi: 10.1002/(sici)1097-0045(19980215)34:3<231::aid-pros11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Brody JR, Kadkol SS, Mahmoud MA, Rebel JM, Pasternack GR. Identification of sequences required for inhibition of oncogene-mediated transformation by pp32. J Biol Chem. 1999;274:20053–20055. doi: 10.1074/jbc.274.29.20053. [DOI] [PubMed] [Google Scholar]
- Bai J, Brody JR, Kadkol SS, Pasternack GR. Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene. 2001;20:2153–2160. doi: 10.1038/sj.onc.1204294. [DOI] [PubMed] [Google Scholar]
- De Marzo AM, Nelson WG, Meeker AK, Coffey DS. Stem cell features of benign and malignant prostate epithelial cells. J Urol. 1998;160:2381–2392. doi: 10.1097/00005392-199812020-00004. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Coffey DS, Chen TH, Wu TC, Pasternack GR. A novel M(r) 32,000 nuclear phosphoprotein is selectively expressed in cells competent for self-renewal. Cancer Res. 1993;53:4720–4726. [PubMed] [Google Scholar]
- Mencinger M, Panagopoulos I, Contreras JA, Mitelman F, Aman P. Expression analysis and chromosomal mapping of a novel human gene, APRIL, encoding an acidic protein rich in leucines. Biochim Biophys Acta. 1998;1395:176–180. doi: 10.1016/s0167-4781(97)00165-6. [DOI] [PubMed] [Google Scholar]
- Carlson SG, Eng E, Kim EG, Perlman EJ, Copeland TD, Ballermann BJ. Expression of SET, an inhibitor of protein phosphatase 2A, in renal development and Wilms’ tumor. J Am Soc Nephrol. 1998;9:1873–1880. doi: 10.1681/ASN.V9101873. [DOI] [PubMed] [Google Scholar]
- Cervoni N, Detich N, Seo SB, Chakravarti D, Szyf M. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J Biol Chem. 2002;277:25026–25031. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151:1–14. doi: 10.1083/jcb.151.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada JN, Shevchenko A, Pallas DC, Berk AJ. Analysis of the adenovirus E1B–55K-anchored proteome reveals its link to ubiquitination machinery. J Virol. 2002;76:9194–9206. doi: 10.1128/JVI.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter BS, Epstein JI, Isaacs WB. Ras gene mutations in human prostate cancer. Cancer Res. 1990;50:6830–6832. [PubMed] [Google Scholar]
- van Bokhoven A, Varella-Garcia M, Korch C, Miller GJ. TSU-Pr1 and JCA-1 cells are derivatives of T24 bladder carcinoma cells and are not of prostatic origin. Cancer Res. 2001;61:6340–6344. [PubMed] [Google Scholar]
- Takahashi N, Shimizu T, Takeda K. Low-dose staurosporine suppresses proliferation and induces neurites in human prostatic cancer TSU-Pr1 cells. Prostate. 2000;44:328–333. doi: 10.1002/1097-0045(20000901)44:4<328::aid-pros10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Sugibayashi R, Shimizu T, Suzuki T, Yamamoto N, Hamada H, Takeda K. Upregulation of p21(WAF1/CIP1) leads to morphologic changes and esterase activity in TPA-mediated differentiation of human prostate cancer cell line TSU-Pr1. Oncogene. 2001;20:1220–1228. doi: 10.1038/sj.onc.1204206. [DOI] [PubMed] [Google Scholar]
- Bonkhoff H. Neuroendocrine differentiation in human prostate cancer. Morphogenesis, proliferation and androgen receptor status. Ann Oncol. 2001;12(Suppl 2):S141–S144. doi: 10.1093/annonc/12.suppl_2.s141. [DOI] [PubMed] [Google Scholar]
- Cox ME, Deeble PD, Lakhani S, Parsons SJ. Acquisition of neuroendocrine characteristics by prostate tumor cells is reversible: implications for prostate cancer progression. Cancer Res. 1999;59:3821–3830. [PubMed] [Google Scholar]
- Bostwick DG, Qian J, Pacelli A, Zincke H, Blute M, Bergstralh EJ, Slezak JM, Cheng L. Neuroendocrine expression in node positive prostate cancer: correlation with systemic progression and patient survival. J Urol. 2002;168:1204–1211. doi: 10.1016/S0022-5347(05)64626-5. [DOI] [PubMed] [Google Scholar]
- Carroll PR, Waldman FM, Rosenau W, Cohen MB, Vapnek JM, Fong P, Narayan P, Mayall BH. Cell proliferation in prostatic adenocarcinoma: in vitro measurement by 5-bromodeoxyuridine incorporation and proliferating cell nuclear antigen expression. J Urol. 1993;149:403–407. doi: 10.1016/s0022-5347(17)36104-9. [DOI] [PubMed] [Google Scholar]
- Seo SB, Macfarlan T, McNamara P, Hong R, Mukai Y, Heo S, Chakravarti D. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J Biol Chem. 2002;277:14005–14010. doi: 10.1074/jbc.M112455200. [DOI] [PubMed] [Google Scholar]
- Li M, Makkinje A, Damuni Z. Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry. 1996;35:6998–7002. doi: 10.1021/bi960581y. [DOI] [PubMed] [Google Scholar]
- Li X, Yost HJ, Virshup DM, Seeling JM. Protein phosphatase 2A and its B56 regulatory subunit inhibit Wnt signaling in Xenopus. EMBO J. 2001;20:4122–4131. doi: 10.1093/emboj/20.15.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeling JM, Miller JR, Gil R, Moon RT, White R, Virshup DM. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science. 1999;283:2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Deeble PD, Murphy DJ, Parsons SJ, Cox ME. Interleukin-6- and cyclic AMP-mediated signaling potentiates neuroendocrine differentiation of LNCaP prostate tumor cells. Mol Cell Biol. 2001;21:8471–8482. doi: 10.1128/MCB.21.24.8471-8482.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler LM, Agus DB, Scher HI, Higgins B, Rose A, Cordon-Cardo C, Thaler HT, Rifkind RA, Marks PA, Richon VM. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 2000;60:5165–5170. [PubMed] [Google Scholar]
- Munster PN, Troso-Sandoval T, Rosen N, Rifkind R, Marks PA, Richon VM. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces differentiation of human breast cancer cells. Cancer Res. 2001;61:8492–8497. [PubMed] [Google Scholar]
- Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- Mori S, Murakami-Mori K, Bonavida B. Interleukin-6 induces G1 arrest through induction of p27(Kip1), a cyclin-dependent kinase inhibitor, and neuron-like morphology in LNCaP prostate tumor cells. Biochem Biophys Res Commun. 1999;257:609–614. doi: 10.1006/bbrc.1999.0515. [DOI] [PubMed] [Google Scholar]
- He LZ, Tolentino T, Grayson P, Zhong S, Warrell RP, Jr, Rifkind RA, Marks PA, Richon VM, Pandolfi PP. Histone deacetylase inhibitors induce remission in transgenic models of therapy-resistant acute promyelocytic leukemia. J Clin Invest. 2001;108:1321–1330. doi: 10.1172/JCI11537. [DOI] [PMC free article] [PubMed] [Google Scholar]