Abstract
The introduction of gene expression profiling has resulted in the production of rich human data sets with potential for deciphering tumor diagnosis, prognosis, and therapy. Here we demonstrate how artificial neural networks (ANNs) can be applied to two completely different microarray platforms (cDNA and oligonucleotide), or a combination of both, to build tumor classifiers capable of deciphering the identity of most human cancers. First, 78 tumors representing eight different types of histologically similar adenocarcinoma, were evaluated with a 32k cDNA microarray and correctly classified by a cDNA-based ANN, using independent training and test sets, with a mean accuracy of 83%. To expand our approach, oligonucleotide data derived from six independent performance sites, representing 463 tumors and 21 tumor types, were assembled, normalized, and scaled. An oligonucleotide-based ANN, trained on a random fraction of the tumors (n = 343), was 88% accurate in predicting known pathological origin of the remaining fraction of tumors (n = 120) not exposed to the training algorithm. Finally, a mixed-platform classifier using a combination of both cDNA and oligonucleotide microarray data from seven performance sites, normalized and scaled from a large and diverse tumor set (n = 539), produced similar results (85% accuracy) on independent test sets. Further validation of our classifiers was achieved by accurately (84%) predicting the known primary site of origin for an independent set of metastatic lesions (n = 50), resected from brain, lung, and liver, potentially addressing the vexing classification problems imposed by unknown primary cancers. These cDNA- and oligonucleotide-based classifiers provide a first proof of principle that data derived from multiple platforms and performance sites can be exploited to build multi-tissue tumor classifiers.
Making the correct pathological diagnosis is always preferred before the initiation of treatment of the cancer patient because cancer therapy is primarily directed by tumor origin. Using standard pathological techniques, it is estimated that up to 5 to 10% of all tumors may actually be misclassified.1,2 Furthermore, current pathological techniques still find the differential diagnosis of a number of cancers problematic. In fact, the diagnosis of “unknown primary” is applied to nearly 5 to 10% of all tumors because the origin of the lesion cannot be identified or predicted.3 Pathologists must often apply their best estimate of the correct tissue of origin for any given metastatic lesion, based primarily on histological and morphological features, and secondarily on semiquantitative immunohistochemical stains. Unfortunately, this approach is not error-free, particularly in the circumstance in which no obvious primary tumor source has been identified. Accurate diagnosis of these and other tumors may lead to more effective treatment and better outcome.
The recent development of gene expression profiling technology has permitted the development of prototypical clinical classifiers that demonstrate the feasibility of this molecular approach to diagnosis.4–13 Some of these classifiers have potential clinical application in narrowly defined diagnostic applications, however, none of these classifiers are suitable for broad based clinical application because of limitations in the numbers and types of tumors included in the analyses. Moreover, levels of accuracy in diagnosis on test sets of data are generally too low for clinical application in which high degrees of accuracy are necessary. The most comprehensive approach to classification published to date involved 14 common tumor types and was able to achieve a 78% success rate using support vector machines for classification,9 a rate below what is routinely considered clinically acceptable.
The goal of this study was to develop a prototype classifier with sufficient accuracy and breadth of coverage that would demonstrate the feasibility for genome-wide microarray analysis in clinical application. To accomplish this, we realized that large data sets from multiple sources and types of human microarray data would have to be exploited. Both cDNA- and oligonucleotide-based microarray platforms are currently being used to produce the majority of human tumor data. It was our belief that, by using meticulous normalization and scaling techniques, both cDNA- and oligonucleotide-based microarray platforms could be assessed independently, or even jointly, if a relatively robust analysis tool was applied. For this purpose, we investigated a number of analytical tools including hierarchical clustering, principal components analysis, and artificial neural networks (ANNs). Ultimately, we discovered that our ANN-based classification technique was capable of classifying cDNA-, oligonucleotide-, or even mixed-platform data, derived from multiple tumor types and multiple performance sites, with high levels of accuracy suitable for clinical application.
Materials and Methods
Sources of Human Microarray Data
Data used to develop diagnostic classifiers were derived from both cDNA and oligonucleotide microarray platforms from up to seven different performance sites, including the Moffitt Cancer Center, as outlined in Table 1. Metastatic tumors of mixed origin and primary colon tumors derived from brain, liver, and lung (Moffitt Cancer Center Tumor Bank) and interrogated by cDNA microarrays, served as blinded, independent validation sets to test the classifiers (see Table 3). All tumor samples derived from the Moffitt Cancer Center Tumor Bank underwent independent pathological review by a single pathologist (DC) as well as frozen section guided microdissection before RNA extraction.
Table 1.
Description of Tumors and Their Sources Used in Classifier Construction
| Tumor type | Number of samples | Array platform† | Website‡ | Reference§ |
|---|---|---|---|---|
| Bladder | 19 | U95, HU68 | A, B | 9, β |
| Breast | 42 | U95, HU68, T32 | A, B, F | 9, β, β |
| Central-nervous atypical teratoid/rhabdoid | 10 | HU68 | C | 1 |
| Central-nervous glioma | 10 | HU68 | C | 1 |
| Central-nervous meduloblastoma | 70 | HU68 | B, C | β, 1 |
| Colon | 41 | U95, HU68, T32 | A, B, F | 9, β, β |
| Stomach/EG junction | 30 | U95, T32* | A, F | 9, β |
| Kidney | 31 | U95, HU68, T32 | A, B, F | 9, β, β |
| Leukemia-acute lymphocytic B cell | 10 | HU68 | B | β |
| Leukemia-acute lymphocytic T cell | 10 | HU68 | B | β |
| Leukemia-acute myelogenous | 10 | HU68 | B | β |
| Lung-adenocarcinoma | 71 | U95, HU68, T32 | A, B, D, E, F | 9, β, 1, α, β |
| Lung-squamous cell carcinoma | 21 | U95 | A, D, E | 9, 1, α |
| Lymphoma-follicular | 11 | HU68 | B | β |
| Lymphoma-large B cell | 11 | HU68 | B | β |
| Melanoma | 10 | HU68 | B | β |
| Mesothelioma | 10 | HU68 | B | β |
| Ovary | 44 | U95, HU68, T32 | A, B, F | 9, β, β |
| Pancreas | 26 | U95, HU68, T32 | A, B, F | 9, β, β |
| Prostate | 42 | U95, HU68 | A, B, E | 9, β, α |
| Uterus | 10 | HU68 | B | β |
Array legend: U95, affymetrix U95A; HU68, affymetrix HU6800FL; T32, 32K TIGR cDNA array;
EG junction and stomach were distinct tumor types in the T32 classifier.
Data source URL legend: A, http://carrier.gnf.org/welsh/epican/1; B, http://www-genome.wi.mit.edu/mpr/GCM.html2; C, http://www-genome.wi.mit.edu/mpr/CNS/3; D, http://www.pnas.org/cgi/content/full/191502998/DC14; E, http://www.moffitt.usf.edu/research/classifier Moffitt5; F, http://cancer.tigr.org/classifier.html TIGR/Moffitt.6
Publication legend: In addition to the references to published reports, additional data were provided by: α, Jove/Bepler, personal communication; β, TIGR 〈http://cancer.tigr.org/data/classifier.html〉 (from this study).
Table 3.
Classification of Site of Origin of 50 Adenocarcinomas Metastatic to Brain, Lung, and Liver
| Primary site | Met site | Classifier used | Result |
|---|---|---|---|
| Colon | Liver | cDNA | 9/10 |
| Breast | CNS | cDNA | 2/2 |
| Breast | Lung | cDNA | 1/1 |
| Lung | Liver | cDNA | 0/1 |
| Lung | CNS | cDNA | 1/1 |
| Ovary | Lung | cDNA | 1/2 |
| Pancreas | Liver | cDNA | 1/1 |
| Renal | Lung | cDNA | 9/11 |
| Renal | CNS | cDNA | 2/2 |
| Breast | Liver | Oligonucleotide | 4/5 |
| Colon | Ovary | Oligonucleotide | 1/1 |
| Colon | Liver | Oligonucleotide | 1/1 |
| Gastric | NA | Oligonucleotide | 0/1 |
| Renal | Colon | Oligonucleotide | 0/1 |
| Lung adenocarcinoma | NA | Oligonucleotide | 2/2 |
| Lung squamous carcinoma | NA | Oligonucleotide | 2/2 |
| Ovary | Omentum | Oligonucleotide | 4/4 |
| Prostate | Lymph node | Oligonucleotide | 2/2 |
cDNA Microarray Chip Construction and Analysis
To assess tumor-associated gene expression, we first used a spotted cDNA microarray containing 32,448 elements (ten exogenous controls printed 36 times, four negative controls printed 36 to 72 times, 31,872 human cDNAs representing 30,849 distinct transcripts, 23,936 unique Institute for Genomic Research (TIGR) Tentative Consensus (TCs), and 6913 expressed sequence tags) to profile multiple tumor types of nearly identical histological appearance. A description of this array along with allprimary and filtered data are available at http://cancer.tigr.org/data/classifier.html. Total RNA was prepared from adenocarcinomas (n = 10) derived from eight different primary sites of origin (breast, pancreas, lung, ovary, kidney, colon metastases, stomach, and esophagogastric junction). Tumors were selected such that all appeared nearly identical on histological inspection, despite originating from different organ sites. Labeled first-strand cDNA was prepared and co-hybridized with labeled samples prepared from a universal reference RNA consisting of equimolar quantities of total RNA derived from three cell lines, CaCO2 (colon), KM12L4A (colon), and U118MG (brain), as described previously14 using standard protocols (http://cancer.tigr.org/protocols.shtml).15 All hybridizations were replicated with a dye-reversal to eliminate any fluor-specific effects. Data from each hybridization were normalized using total intensity normalization.14 Dye-reversed hybridizations were subjected to replicate flip-dye trimming to eliminate inconsistent data, and the geometric mean was calculated for the remaining array elements.14 Data were further filtered to select genes that could suitably distinguish tissues and the resulting gene expression vectors were subjected to average linkage hierarchical clustering using a Pearson correlation coefficient distance measure.
Normalization and Scaling of Data
Our cDNA classifiers use a comparison of expression measured in a tumor relative to that in a reference RNA sample. For application to the Affymetrix (Santa Clara, CA) oligonucleotide-based platform, we first labeled and hybridized our reference RNA source to the HU6800 and U95A GeneChips and measured the expression for each gene. For each tumor sample, the measured expression level for each gene on the array is scaled so that its average measured expression is equal to the average measured for our reference sample. These normalized expression measures were then used as input to our classifier. We chose to use rescaled expression values rather than ratios because neural networks perform best when the input data have as wide a range as possible.
For cross-platform comparisons (cDNA and oligonucleotide), an additional normalization step was performed. Common genes were identified across platforms using RESOURCERER16 (http://www.tigr.org/tdb/tgi.shtml). For each gene in common, expression levels for the reference RNA sample on the spotted arrays was averaged and compared to expression measured for the reference RNA applied to the appropriate Affymetrix GeneChip to calculate a gene-specific scaling factor. This scaling factor was used to adjust the remaining data (GeneChip) to make it comparable to the spotted arrays. Whenever multiple representatives of a single gene were represented on array, their values were averaged. The final scaled values were used as input for the classifier.
Statistical Analysis
The Kruskal-Wallis H-test is a nonparametric statistical test that was used to rank the importance of each gene. The null hypothesis for this test is that the distribution of gene expressions is identical across tumor types relative to the alternative hypothesis that expression distribution differs between types. The Kruskal-Wallis test was used as a guide in selecting the subsets of genes used to train each of the neural networks (see below). Specifically, this test is applied only to the set of training tumors to identify genes that best distinguished tumor types. The genes were sorted in ascending order by the P value assigned in the Kruskal-Wallis test. A subset of genes was then selected from this list for use in construction of the ANNs. The entire list of calculated P values, including the average P value used in gene selection for each random data split, can be found in the supplemental data (all supplemental data are accessible on website http://cancer.tigr.org/data/classifier.html).
The Artificial Neural Network (ANN)
An ANN is a versatile algebraic construct that can arbitrarily closely approximate any nonlinear function. It is an ideal tool for classification problems associated with complex microarray data sets because it requires no a priori assumptions about the relative importance of any particular gene in the classification. The neural network package used was a locally modified version of Scott Fahlman’s QuickProp17 neural network software. We modified the C implementation to provide a cleaner input/output interface and to operate within a Beowulf cluster environment. For these experiments we disabled the quickprop modifications, making the classifier essentially a standard feed-forward back-propagation neural network. We used a learning rate of 1.0, a momentum term of 0.2, and trained in incremental mode.
Before the ANN can be used for classification, however, it must first be trained to perform this function. Training data consists of input gene expression vectors that are paired with target vectors representing tumors with defined histological classifications. The ANN uses the weighted combination of these input genes for generating a prediction of a particular tumor type. A single hidden layer feed-forward, back-propagation neural network was chosen for this study. The standard sigmoid transfer function was used and the learning rate was set to 1.0. Classification accuracy was estimated by running 10 random training and test set splits. For each microarray platform (cDNA, oligonucleotide, and mixed), 10 different stratified splits of the data were created. A neural network was constructed from a training set and validated on the corresponding test set. This technique more accurately estimates the true generalization error rate for classifier and allows the ability to assess the variance in predicting correct classifications. All data are reported as mean accuracies of the 10 training and test splits with an associated 95% confidence interval unless otherwise specified. An electronic table of the accuracies of the individual cDNA, oligonucleotide, and mixed platform classifiers are included in the supplemental data. See the supplemental data also for a list of those genes selected in common across all 10 training and test splits.
Results
Unweighted Approaches Were Unsuccessful in Accurate Tumor Classification
To assess the feasibility of constructing a multi-class tumor classifier, we first analyzed data derived from 78 primary adenocarcinomas representing eight different organ sites (n = 10 samples/organ site) of origin, using a single platform based on 32K cDNA microarrays. Tumors were selected based on their similar histomorphological appearance (see supplemental data). Two of 80 tumors did not produce informative hybridizations. All 78 tumors were subjected to two flip-dye hybridizations. The results, shown in Figure 1, illustrate that using a simple, unweighted clustering approach, we were initially able to separate tumor types into distinct groups. Closer inspection of the cluster dendrogram, however, illustrates that this unweighted, unsupervised approach failed to fully distinguish all tumor types. Subsequent attempts at classifying independent, blinded data sets using informative genes identified by clustering approaches were primarily unsuccessful, with 30% or more tumors being misclassified (data not shown). Even when new samples were added to this data set and the clustering rerun, our ability to classify samples accurately was limited. Similar results were obtained with principal components analyses (data not shown).
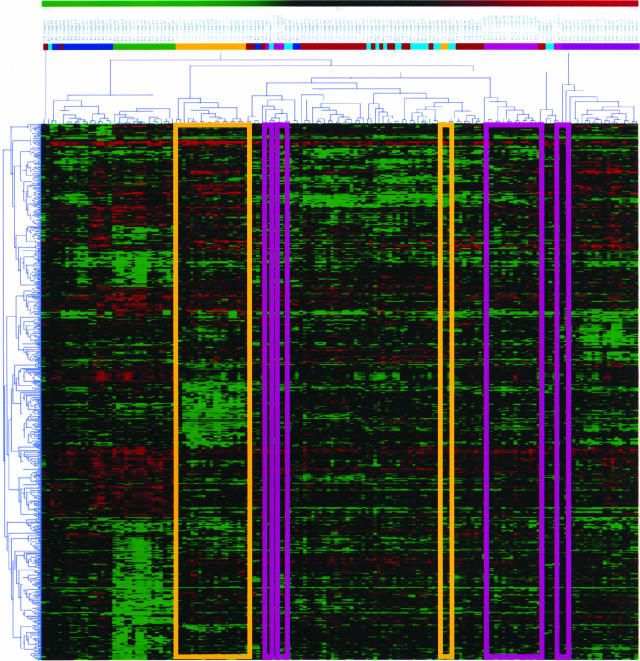
Figure 1.
Hierarchical clustering of eight different types of adenocarcinoma. The Kruskal-Wallis H-test was used to identify those genes most correlated with each tumor type, selecting ∼700 genes from 30,849 distinct transcripts on the cDNA chip. Average linkage hierarchical clustering of spotted cDNA array expression data using a Pearson correlation coefficient distance matrix illustrates the problems with this approach to classification, which typically weights each gene equally. Even for ovarian cancer samples (yellow boxes), which are generally well classified, there are two outlying samples that are grouped within a set of diverse tumors. For other tissues of origin such as lung (pink boxes), the situation is worse. Similar results are obtained for samples assayed using Affymetrix GeneChips. Although hierarchical clustering can be used with weights for each gene, we have no a priori means of determining the appropriate weights. This is the rationale that underlies the use of the ANN in tumor classification.
Construction of cDNA-Based Tumor Classifier Using a Gene-Weighted Approach
Examination of the cluster heat map suggested that some genes were more informative than others in classifying samples and should be weighted appropriately. However, we had no rational means of knowing the appropriate weights to assign to each gene in the classification. To address this problem, we combined a nonparametric statistical screen to select discriminating genes, with an ANN18–25 to assign weights to individual genes, that could then be used for classification. A nonparametric Kruskal-Wallis H-test was used to identify a set of 700 genes (see supplemental data) most correlated with tumor histological classification from a randomly selected training set of ∼75% of informative primary tumor hybridizations. These genes and their expression vectors were then used to train an ANN to identify specific tumor types. In constructing our first ANN classifier, we used 700 input nodes (the number of genes in the training set), 200 hidden nodes, and 8 output nodes (equal to the number of distinct tumor types). A learning rate of 1.0 was used and the training and testing tolerances were both 0.1. To validate our classification technique, we randomly partitioned the entire available data set into truly independent training and test sets. Independence is crucial, therefore the test set of data were not exposed to either the Kruskal Wallis H-test set or the ANN learning algorithm.
After training a series of 10 classifiers using this set of 78 tumors, we achieved a mean accuracy of 83% with a 95% confidence interval of (76.4%, 88.6%) as calculated from those tumors held out in each of the splits (see supplemental data).
Construction of Oligonucleotide-Based Tumor Classifier
Based on our initial success in classification of tumors arrayed on the cDNA platform, we next sought to extend this approach to oligonucleotide-based microarray data, to develop a more general, clinically applicable, and robust classifier. The approach we used is summarized in Figure 2. We combined new data (kindly provided by R. Jove and G. Bepler, H. Lee Moffitt Cancer Center) with published data to produce a collection of 466 tumors, which were profiled on both Affymetrix HFL6800 and U95A GeneChips (Table 1) at six different performance sites, representing 21 tumor types, and accounting for more than 95% of all human tumors. We chose only those data sets that included at least 10 independent measurements for each tumor type, as fewer in any single group greatly reduced the ability to develop an accurate classifier. By incorporating the 5296 genes in common on these two chip platforms, we again applied our classification strategy. The Kruskal-Wallis H-test was used to select a subset of 2000 discriminate genes from a training set of tumors (n = 346) representing ∼75% of the entire tumor collection. These genes were used to train a series of ANNs using intraplatform (Affymetrix HFL6800 and U95A GeneChips) data, appropriately scaled and normalized. By applying these trained ANNs to the remaining 25% (n = 120) of the entire collection of tumor samples—not included in gene selection via the Kruskal-Wallis test and not exposed to the training algorithm—we were able to correctly predict the known pathology of 88% of the test samples with a 95% confidence interval of (84.3%, 89.5%) (see supplemental data). This high level of accuracy in predicting the blinded, independent test sets suggests that these data were not subject to over-fitting, a potential pitfall of ANNs. Further, by replicating the experiment using 10 independent, randomly-selected, training tumor sets, we have ensured that the reported accuracy of predicting the independent test sets is not dependent on any individual training/test split of the data. Finally, the error rates achieved are clinically acceptable when compared with the probable rate of error in routine pathological diagnosis.1,2 Importantly, these errors were distributed relatively evenly across multiple tissue classes (Table 2).
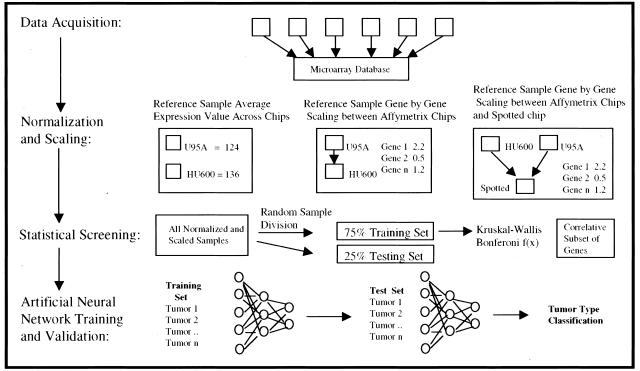
Figure 2.
Graphical depiction of classifier development separated into the four major stages. Data acquisition involves a literature search for suitable published microarray data and the collection of this and newly generated data into a microarray database. Normalization and scaling shows the three major steps in data preparation. Namely, calculation of an average Gene expression value across the reference sample for the two Affymetrix chip types, gene by gene scaling between Affymetrix chip types and the gene by gene scaling between Affymetrix chip types and the spotted microarray. A nonparametric statistical screening was then used to find a subset of genes correlative with tumor type. This set of genes was used to train and validate an ANN.
Table 2.
Performance of the Oligonucleotide Classifier across 21 Tumor Types Shown as an Average of the 10 Different Accuracies from the Train/Test Splits
| Tumor type | Average classification success rate as % |
|---|---|
| Bladder | 77 |
| Breast | 67 |
| Central-nervous AT/RT | 98 |
| Central-nervous glioma | 95 |
| Central-nervous meduloblastoma | 97 |
| Colon | 99 |
| Stomach/EG junction | 57 |
| Kidney | 70 |
| Leukemia acute lymphocytic B cell | 88 |
| Leukemia acute lymphocytic T cell | 83 |
| Leukemia, acute myelogenous | 91 |
| Lung adenocarcinoma | 94 |
| Lung squamous cell | 87 |
| Lymphoma, follicular | 97 |
| Lymphoma large B cell | 96 |
| Melanoma | 96 |
| Mesothelioma | 89 |
| Ovary | 85 |
| Pancreas | 80 |
| Prostate | 94 |
| Uterus | 74 |
Construction of a Mixed-Platform Multi-Tissue Tumor Classifier
Because gene expression profiling is currently being produced on at least two different microarray platforms, oligonucleotide and cDNA, we sought to develop a means to exploit both types of data, for the ultimate purpose of compiling many sources of data to build a multi-tissue, extensible, tumor classifier. For this purpose, oligonucleotide-based microarray data were combined with the cDNA expression data we produced from spotted arrays to develop a mixed-platform classifier based on seven performance sites. We first selected 2252 genes common to all microarrays under consideration using RESOURCERER16 (http://www.tigr.org/tdb/tgi.shtml) and used the approach described previously to select those genes most highly correlated with particular histological classifications. To provide ratiometric measures of gene expression, we used the same GeneChips to profile the same RNA sample used as a reference in our spotted array assays (see Normalization and Scaling in Materials and Methods). We selected 400 tumors representing a combination of all available array platforms and tumor types to train a series of 10 mixed-platform ANNs. The resulting tumor classifiers were applied to the remaining 140 tumor samples in their respective test sets. This approach was able to correctly classify 85% of the 140 tumors from the blinded test sets with a 95% confidence interval of (82.2%, 87.6%) (see supplemental data). This high level of accuracy agrees with our previous results for the independent cDNA- or oligonucleotide-based classifiers, suggesting that it is feasible to produce a multi-tissue classifier capable of incorporating more than one data format derived from more than one performance site.
Classification of Metastases to Liver, Lung, and Brain from Known Primary Sites
It has been previously reported that metastatic lesions may be difficult to classify because these lesions have lost some of the expression of their differentiating genes.9 Moreover, these metastatic tumors represent the majority of tumors that are difficult to diagnose by standard pathological methods, particularly when a primary tumor has not been identified. We sought to validate our classification technique and address this difficult clinical problem by evaluating a large set of 50 tumors metastatic to brain, lung, and liver, and derived from multiple known primary sites. Data were produced on both our cDNA arrays as well as oligonucleotide arrays. When assessed by our cDNA- and oligonucleotide tumor classifiers, a classification accuracy of 84% was achieved using the entire 50 tumors as a blinded, independent test set (Table 3). These data support the concept that classification of tumors, without a known primary site of origin, may now be feasible.
Estimation of Minimal Gene Set for Adequate Tumor Classification
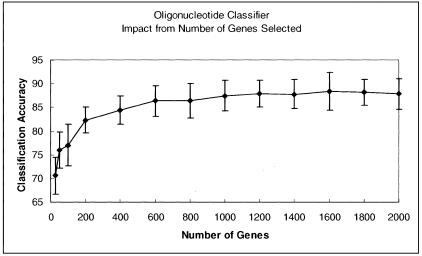
One potential limitation of our classifiers is their apparent dependence on a large number of genes to function effectively, reducing the potential to derive meaningful biological information from a diverse set of genes. At the same time, a large number of genes may make the classifier more robust in a clinical setting, and more forgiving of data derived from multiple performance sites, while at the same time permitting separation of a large number of different tumor types. To investigate the number of genes required for adequate classification, we performed a feature reduction experiment using the oligonucleotide classifier as an example. The Kruskal-Wallis H-test was used to identify the 2000 genes with the lowest P values, from a total of 5296 available (common to both Affymetrix platforms), for use in the classifier algorithm. Ten random train and test splits were used and the mean accuracy for the 10 classifiers was calculated. This process was repeated for sequentially smaller numbers of genes until the ANN was no longer able to train. The summary of this experiment is reported in the supplemental data. As can be seen from Figure 3 the accuracy of the classifier is relatively unaffected by the number of genes used in training the ANN until the number drops to ∼400. This demonstrates both that the neural network is very robust with respect to the number of input genes, and that the number of genes can be significantly limited, still obtaining useful classification accuracies. When fewer than 25 genes were used, the classifier performed very poorly. When we tested the classifier using the smallest number of effective genes (n = 25), we confirmed the importance of gene selection based on P values. Accuracy using the 25 genes from the Kruskal-Wallis selected set of 2000 with the most significant P values was 76%, but was reduced to 63% using the genes with the least significant P values, and further reduced to 55% with a random selection of 50 genes from the genes excluded by the initial Kruskal-Wallis test.
Figure 3.
Analysis of the effect of removing genes from the oligonucleotide classifier on classifier accuracy. Genes were sequentially removed from the 2000 genes selected by the Kruskal-Wallis test, starting with the least significant to the most significant P values.
Discussion
We have produced the first multi-tissue classifier with both sufficiently broad scope and classification accuracy to demonstrate potential for clinical application. Whereas our first attempt at producing a cDNA classifier produced accuracy levels less than optimal, we were encouraged that these levels exceeded those reported by other published studies and might be improved by larger sample sizes. This prediction was validated with the production of an oligonucleotide classifier and a mixed-platform classifier, both providing high levels of accuracy. At present, our classifiers are capable of interrogating 21 different tumor types, representing 95% of all cancers, with up to a 88% accuracy rate using as few as 400 genes. Moreover, we have demonstrated the ability to classify metastatic tumors, often representing the most difficult of diagnostic challenges. Here we demonstrate an 84% accuracy rate classifying metastatic tumors to liver, lung, and brain when derived from multiple sites of origin. Our results are, at present, limited by the number of available data sets and the relatively small number of genes assayed on the early Affymetrix HU6800 GeneChips, as well as by the number of genes shared by multiple platforms. By continuing to add data to our classifier, which we have shown is possible using data from a variety of sources and performance sites, we believe that both the accuracy and scope of these ANN-based classifiers can be further improved. In addition, it is likely that this approach can be extended to subtype classification, making it possible to classify even difficult to diagnose tumors such as those of unknown primary origin.
Classifiers can be constructed using a variety of learning algorithms, including support vector machines.9 We sought to compare our approach directly with support vector machines to ensure an equivalent result. When we applied our approach to the same published training data set using a leave one out cross validation, our classification technique resulted in an accuracy of 84% versus the reported accuracy rate of 78% using a support vector machine (see supplemental data).
To classify a large number of distinct tumor types, we used a relatively large number of informative genes with a relatively forgiving analysis tool (ANN) to permit accurate classification. As expected, using gene ontology analysis, there was no underlying discriminating biological function that would permit separation of 21 different tumor types; however, highly accurate classification was achievable with hundreds of genes when using our classification technique. Although these classifiers are incomplete for actual clinical application, we believe these models and collected data sets will provide a cornerstone for the construction of expandable, versatile tumor classifiers. Although neural networks are only one of many tools that can be used for classification, the approach we developed demonstrates that one can accurately partition samples into large numbers of different classes even though the data are collected at multiple sites and on multiple platforms. These data demonstrate that microarray-based tumor classifiers hold promise to objectively compliment existing histomorphological techniques in the accurate diagnosis of cancer origin, with significant implications for cancer therapy.
Acknowledgments
We thank the Research Oriented Computing Center, University of South Florida, for the services provided.
Footnotes
Address reprint requests to Timothy J. Yeatman, Department of Interdisciplinary Oncology, H. Lee Moffitt Cancer Center, University of South Florida, 12902 Magnolia Dr., Tampa, Florida 33612-9497. E-mail: yeatman@moffitt.usf.edu or to johnq@tigr.org.
Supported in part by the National Institutes of Health (National Cancer Institute grants CA85052-01A1 and CA85429-01 to T. J. Y. and 6120-119-LO-A to J. Q.) and the Department of Health and Human Services (to J. Q.).
G. B. and I. V. Y. contributed equally to this work.
All supplemental data are accessible on website (http://cancer.tigr.org/data/classifier.html).
References
- Nakhleh RE, Zarbo RJ. Amended reports in surgical pathology and implications for diagnostic error detection and avoidance: a College of American Pathologists Q-probes study of 1,667,547 accessioned cases in 359 laboratories. Arch Pathol Lab Med. 1998;122:303–309. [PubMed] [Google Scholar]
- Zarbo RJ. Monitoring anatomic pathology practice through quality assurance measures. Clin Lab Med. 1999;19:713–742. [PubMed] [Google Scholar]
- van de Wouw AJ, Janssen-Heijnen ML, Coebergh JW, Hillen HF. Epidemiology of unknown primary tumours; incidence and population-based survival of 1285 patients in Southeast Netherlands, 1984–1992. Eur J Cancer. 2002;38:409–413. doi: 10.1016/s0959-8049(01)00378-1. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Golub TR. DNA microarrays in clinical oncology. J Clin Oncol. 2002;20:1932–1941. doi: 10.1200/JCO.2002.20.7.1932. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, Hampton GM. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Yang IV, Chen E, Hasseman JP, Liang W, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, Quackenbush J. Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol. 2002;3(11):Research 0062. doi: 10.1186/gb-2002-3-11-research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–550. doi: 10.2144/00293bi01. [DOI] [PubMed] [Google Scholar]
- Tsai J, Sultana R, Lee Y, Pertea G, Karamycheva K, Antonescu V, Cho J, Parvizi B, Cheung F, Quackenbush J. RESOURCERER: a database for annotating and linking microarray resources within and across species. Genome Biol. 2001;2:0002.0001–0002.0004. doi: 10.1186/gb-2001-2-11-software0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlman SE. Los Altos, CA: Morgan-Kaufmann; Faster-Learning Variations on Back-PropagationAn Empirical Study. 1988 “Proceedings, 1988 Connectionist Models Summer School”. [Google Scholar]
- Alkon DL, Blackwell KT, Barbour GS, Rigler AK, Vogl TP. Pattern-recognition by an artificial network derived from biologic neuronal systems. Biol Cybern. 1990;62:363–376. doi: 10.1007/BF00197642. [DOI] [PubMed] [Google Scholar]
- Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoncini G, Sacchi G, Viazzi F, Ravera M, Parodi D, Ratto E, Vettoretti S, Tomolillo C, Deferrari G, Pontremoli R. Microalbuminuria identifies overall cardiovascular risk in essential hypertension: an artificial neural network-based approach. J Hypertens. 2002;20:1315–1321. doi: 10.1097/00004872-200207000-00018. [DOI] [PubMed] [Google Scholar]
- Mulsant BH. A neural network as an approach to clinical diagnosis. MD Comput. 1990;7:25–36. [PubMed] [Google Scholar]
- Okon K, Tomaszewska R, Nowak K, Stachura J. Application of neural networks to the classification of pancreatic intraductal proliferative lesions. Anal Cell Pathol. 2001;23:129–136. doi: 10.1155/2001/657268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibnegger G, Weiss G, Werner-Felmayer G, Judmaier G, Wachter H. Neural networks as a tool for utilizing laboratory information: comparison with linear discriminant analysis and with classification and regression trees. Proc Natl Acad Sci USA. 1991;88:11426–11430. doi: 10.1073/pnas.88.24.11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly DL, Cooper LN, Elbaum C. A neural model for category learning. Biol Cybern. 1982;45:35–41. doi: 10.1007/BF00387211. [DOI] [PubMed] [Google Scholar]
- Wu FY, Slater JD, Honig LS, Ramsay RE. A neural network design for event-related potential diagnosis. Comput Biol Med. 1993;23:251–264. doi: 10.1016/0010-4825(93)90024-u. [DOI] [PubMed] [Google Scholar]