Abstract
Germline mutations in the fumarate hydratase (FH) gene at 1q43 predispose to dominantly inherited cutaneous and uterine leiomyomas, uterine leiomyosarcoma, and papillary renal cell cancer (HLRCC syndrome). To evaluate the role of FH inactivation in sporadic tumorigenesis, we analyzed a series of 299 malignant tumors representing 10 different malignant tumor types for FH mutations. Additionally, 153 uterine leiomyomas from 46 unselected individuals were subjected to and informative in loss of heterozygosity analysis at the FH locus, and the five (3.3%) tumors displaying loss of heterozygosity were subjected to FH mutation analysis. Although mutation search in the 299 malignant tumors was negative, somatic FH mutations were found in two nonsyndromic leiomyomas; a splice site change IVS4 + 3A>G, leading to deletion of exon four, and a missense mutation Ala196Thr. The occurrence of somatic mutations strongly suggests that FH is a true target of the 1q43 deletions. Although uterine leiomyomas are the most common tumors of women, specific inactivating somatic mutations contributing to the formation of nonsyndromic leiomyomas have not been reported previously. Taking into account the apparent risk of uterine leiomyosarcoma associated with FH germline mutations, the finding raises the possibility that also some nonsyndromic leiomyomas may have a genetic profile that is more prone to malignant degeneration. Our data also indicate that somatic FH mutations appear to be limited to tumor types observed in hereditary leiomyomatosis and renal cell cancer.
Germline mutations in a nuclear gene encoding a tricarboxylic acid cycle (Krebs cycle) enzyme fumarate hydratase (fumarase, FH, NM_000143.2), have recently been associated with tumor predisposition. Germline FH defects were found in ∼60% (24 of 42) of families segregating hereditary leiomyomatosis and renal cell cancer (HLRCC; OMIM 605839) and multiple cutaneous and uterine leiomyomata 1 (MCUL1, OMIM 150800), which thus appear to be allelic conditions.1 In these families, affected individuals typically develop leiomyomas of the skin and/or uterus.1,2 In addition to leiomyomas, some families are also predisposed to papillary renal cell carcinoma and uterine leiomyosarcoma.3 Mutations in FH have also been observed in nonfamilial lesions, especially in tumor types associated with the HLRCC phenotype.4 To date, only two pilot studies have been performed examining the nonsyndromic counterpart tumors of HLRCC.4,5 Between these two studies, only one case of somatic bi-allelic loss-of-function FH mutation was found, in a soft-tissue sarcoma of the lower limb. Unfortunately a more specific classification of this high-grade lesion was not possible.4 Interestingly in two unselected cases of uterine leiomyosarcoma and cutaneous leiomyoma, a germline FH mutation was unexpectedly identified.4 Thus, to date only one tumor, a soft-tissue sarcoma,4 has been found to have a purely somatic FH defect.
The first observation that a Krebs cycle component could be involved in tumorigenesis was made by Baysal and colleagues6 Germline mutations in subunits of succinate-ubiquinone oxidoreductase (mitochondrial complex II), succinate dehydrogenase subunit D (SDHD), C (SDHC), and B (SDHB), were observed in patients with hereditary head and neck paraganglioma (PGL) and pheochromocytoma, and somatic mutations were detected in respective nonsyndromic lesions.6–10 Pheochromocytomas are also associated with von Hippel-Lindau syndrome (VHL; OMIM 193300), which is the most common condition predisposing to hereditary renal cell carcinoma.11
Benign fibroids or leiomyomas of the uterus are the most common tumors in women of reproductive age. Uterine leiomyomas are clinically apparent in at least 25% of women, but the prevalence could be as high as 77%.12 Although most leiomyomas are symptomless, many women suffer from heavy menstrual bleeding, pelvic pain, or reproductive dysfunction.12 Little is known about the molecular background of these lesions. Other than activating K-RAS mutations reported in one study,13 no specific point mutations underlying nonsyndromic leiomyomas have been identified. However, contribution of chromosomal rearrangements in development of these lesions has long been evident.14 Chromosomal abnormalities have been observed in 40% of uterine leiomyomas, the most common aberration being deletion of chromosome 7q.15 In a genome-wide allelotyping effort, loss of heterozygosity (LOH) was observed most commonly (9%) on 7q22-23, and less frequently (2%) on 1q42-42 and 16q12-22.16 Other rearrangements typically include the translocation t(12;14)(q15;q23-24), involving a chromosomal segment of a gene encoding a member of high-mobility group proteins, HMGA2.17,18 Altered gene expression, aberrant splicing, or cytogenetic translocation leading to chimeric fusion of HMGA2 and a target gene is a frequent event in uterine leiomyomas.18,19 HMGA2 and the DNA repair gene RAD51L1 have been hypothesized to create pathologically significant fusion transcripts.20 Although HMGA2-RAD51L1 fusion transcripts or their misspliced products have been detected, the pattern of rearrangements suggests that dysregulated expression of HMGA2, not the formation of fusion transcripts, is the principal mechanism in the development of uterine leiomyomata.21 Elevated expression of the high-mobility group AT-hook 1 (HMGA1) gene has also been observed in uterine leiomyomas. The dysregulating mechanism may be similar to HMGA2, including posttranslational modifications or cytogenetic rearrangements at HMGA1 locus on 6p21.22,23 Estrogen- and progesterone-induced growth factors such as insulin-like growth factor 1, fibroblast growth factor, and transforming growth factor-β, and their receptors, are other candidates contributing to the pathogenesis of uterine leiomyomas.24
Although mitochondrial defects have been suspected to play an important role in tumorigenesis, the detailed mechanisms and signaling pathways involving mitochondrial proteins in tumor development and progression are not fully understood.25 Somatic mutations in the mitochondrial genome have been identified in a substantial proportion of papillary thyroid carcinomas, and subsequently in various other tumors, such as ovarian, breast, and prostate carcinomas.26–28 It has been speculated that oxidative damage-associated DNA mutations could promote tumorigenesis. Other explanations include that the tricarboxylic cycle dysfunction could activate hypoxic pathways or promote anti-apoptotic effects.29 In the great majority of studied FH defective tumors, the event of the second hit has been loss of a wild-type allele.1,30 FH has been suggested to act as a tumor suppressor according to Knudson’s31 two-hit hypothesis.
Our previous FH mutation analysis study in nonsyndromic tumors focused on tumor types observed in Finnish HLRCC families; uterine and cutaneous leiomyomas, renal cell carcinomas, prostate cancers, sarcomas, and lobular breast cancers.4 In the current study, to evaluate further the role of FH in human tumor development, we analyzed 299 malignant tumors for FH mutations. We wished to include as large a selection as was feasible. The material included 10 different malignant tumor types: colorectal, breast (nonlobular), lung, ovarian, testicular, thyroid, and head and neck cancers, as well as pheochromocytomas, glioblastomas, and melanomas (Table 1). A large series of unselected uterine leiomyomas was analyzed for 1q43 LOH, and positive cases were subjected to FH mutation analysis to scrutinize further the possible contribution of FH to the nonsyndromic form of this disease.
Table 1.
Malignant Tumor Types Included in FH Mutation Analysis by DHPLC
| Malignant tumors | Number of samples | |
|---|---|---|
| Papillary thyroid carcinomas | 52 | |
| Glioblastomas | 25 | |
| Head and neck tumors | 14 | |
| Hypopharynx carcinoma | 5 | |
| Oral or lingual carcinoma | 5 | |
| Other | 4 | |
| Adrenal pheochromocytomas | 18 | |
| Ovarian tumors | 60 | |
| Carcinoma adenomatosum | 18 | |
| Cystadenocarcinoma | 27 | |
| Other | 15 | |
| Nonlobular mammary gland cancers | 44 | |
| Lung tumors | 34 | |
| Colorectal cancers | 23 | |
| MSI− | 12 | |
| MSI+ | 11 | |
| Testicular cancers of germ cell origin | 14 | |
| Melanoma | 15 | |
| Total | 299 | |
Materials and Methods
Tumor Material
Malignant tumors from 299 Finnish individuals were subjected to FH mutation screening. Samples included 52 papillary thyroid carcinomas, 60 ovarian tumors, 44 nonlobular breast carcinomas, 14 testicular carcinomas of germ cell origin, 34 lung carcinomas, 23 colorectal cancers, 15 cutaneous melanomas, 18 adrenal pheochromocytomas, 25 glioblastomas, and 14 head and neck squamous cell carcinomas. RNA (for the 18 pheochromocytomas, only RNA was available) and DNA (for analysis of other tumor types) extractions from tumor samples were performed by standard procedures. In addition, 166 fresh-frozen uterine leiomyomas from 51 anonymous and unselected Finnish patients were collected and subjected to LOH analysis of markers on chromosome band 1q43 closely flanking the FH locus. Leiomyomas that displayed allelic loss were subsequently screened for FH mutations (Table 2). Although this series mostly represents nonsyndromic lesions, inclusion of some syndromic cases cannot be excluded.
Table 2.
Results of LOH and Mutation Analysis of Unselected Uterine Leiomyomas
| LOH and mutation analysis | Number of samples |
|---|---|
| LOH analysis flanking FH locus | 166 |
| Informative with at least one marker | 153 |
| LOH detected flanking FH locus | 5 |
| > FH mutation analysis | 5 |
| FH mutation detected | 2 |
Analysis of LOH in Leiomyomas
We identified two microsatellite repeats adjoining the FH locus. Polymerase chain reaction (PCR) primers (Sigma-Genosys, Cambridgeshire, UK) for the repeats were as follows: FH-T (telomeric) forward primer, AGCAATGATGGTTTCTCTCTCA; FH-T reverse primer, CAGCACTAGCAGAA TATGTGTAA; FH-C (centromeric) forward primer, CCTTACCATTGCTCCCAAGA; FH-C reverse primer, ACCTTCATCCCTGTCCTGTG. Fluorescence-labeled PCR products were detected by ABI377 sequencer and analyzed by Genotyper 2.5.2 software (Applied Biosystems, Foster City, CA).
Denaturing High-Performance Liquid Chromatography (DHPLC) Analysis
Mutation screening of tumor samples and leiomyomas showing allelic imbalance at the FH locus were performed by DHPLC, except pheochromocytomas and testicular tumors. The amplification of the exons and the mutation analysis were performed as described by Lehtonen and colleagues32 Samples from two individuals were pooled to enable mutation detection in cases of a missing wild-type allele.
Genomic Sequencing
Any pool of samples showing heteroduplex, ambiguous, or missing DHPLC peaks were reanalyzed by genomic sequencing. The PCR reactions, conditions, and oligonucleotide primers used were identical to the study of Kiuru and colleagues.4 PCR products were purified using a NucleoSpin PCR purification kit (Matcherey-Nagel, Duren, Germany). Direct sequencing of PCR products was performed using the BigDye3 termination chemistry (Applied Biosystems) with ABI 3100 Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions. The mutation analysis of the testicular tumors was also performed by genomic sequencing.
cDNA Amplification and Sequencing
Pheochromocytomas were screened by sequencing of cDNA, because only corresponding RNA was available. Primers for the 5′ fragment of FH were CTCCCTCAGCACCATGTACC (forward) and CCACTTTTGCAGCAACCTTT (reverse); for 3′ fragment CTTGGGCAGGAATTTAGTGG (forward) and GCAGTTTCCTTTCAAACTTATCC (reverse). PCR reactions were performed in a 50-μl reaction volume containing 200 ng cDNA, 1× PCR buffer (Applied Biosystems), 300 μmol/L each dNTP (Finnzymes, Espoo, Finland), 1.25 of μmol/L forward and reverse primer, and 2.5 U of AmpliTaqGOLD polymerase (Applied Biosystems). MgCl2 concentrations were 2.8 mmol/L for the 5′ fragment and 1.4 mmol/L for the 3′ fragment. The following cycling conditions were used for the 5′ fragment: 10 minutes at 95°C, followed by 40 cycles of denaturation at 95°C for 45 seconds, annealing at 56°C for 1 minute, and elongation at 72°C for 1 minute, and final extension at 72°C for 10 minutes. Equal conditions for the 3′ fragment were used despite the annealing temperature, which was decreased from 60°C to 57°C, 1 degree per two cycles, and from 57°C to 56°C after five cycles. The corresponding cDNA of leiomyoma P4M3 was analyzed to detect a splicing defect on the mRNA level. Primers for cDNA amplification were as follows: forward primer, TGGTATGGCAGACTGGATCA; reverse primer, CACCACGCAGTTTTCTGTA. PCR reactions and conditions were slightly different from the pheochromocytoma 5′ fragment amplification in MgCl2 concentration (4.2 mmol/L), annealing temperature (57°C), and number of denaturing/annealing/elongation cycles.33 The sequencing protocol for cDNA was equal to genomic sequencing.
Results
FH mutation analysis of this large series of non-HLRCC-component neoplasias, including 10 malignant tumor types, revealed that 94% (2675) of 2846 fragments were successfully examined, but no coding region mutations or splice site changes were identified (Table 1). Probable polymorphisms detected were as follows: silent change 798G>A (10 tumors), and three intronic alterations IVS2-21A>T (6 tumors), IVS3 + 32A>G (1 tumor), and IVS2 + 61T>A (1 tumor). The heterozygous substitution from C to T at position −11 upstream from the translation initiation codon was observed in one papillary thyroid carcinoma. The corresponding germline change has been reported in a familial prostate cancer patient,32 and therefore seems to be an infrequent polymorphism. The numbering of the exons, amino acid residues, and nucleotide positions are based on the mature cytosolic isoform of FH (NM_000143.2).
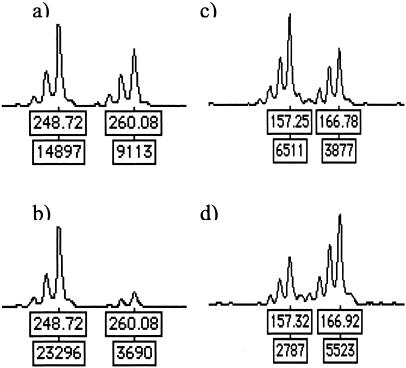
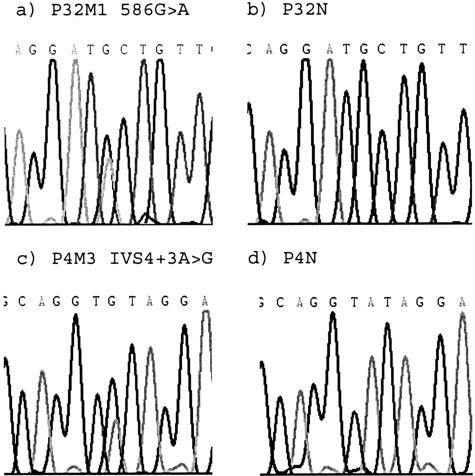
Allelic loss at the FH locus was analyzed by polymorphic microsatellite markers in the set of 166 unselected uterine leiomyomas from 51 individuals. Forty-six individuals (153 leiomyomas) were informative with at least one of the markers. Allelic loss was observed in five myomas (5 of 153, 3.3% of myomas), derived from five different patients (5 of 46, 11% of patients). One of these lesions was informative with both markers and showed LOH at both marker loci (P21M1; patient 21, myoma 1), whereas the other four were informative with one marker (Figure 1). These five leiomyomas were subjected to FH mutation screening to detect the putative second alteration in the remaining allele. Three lesions were mutation-negative, and two displayed a FH mutation. Myoma P32M1 harbored a missense change Ala196Thr in exon 4 (Figure 2a). The altered amino acid is completely conserved among species from human to Escherichia coli. This change was not found in six other leiomyomas from patient P32. Allelic imbalance was not present at the Ala196Thr mutation site, suggesting that the deletion observed in the marker analysis was small. Myoma P4M3 harbored a splice site change IVS4 + 3A>G (Figure 2c), which is predicted to result in deletion of exon 4. The remaining wild-type allele opposite the splice site mutations was under-represented in sequence chromatographs, reflecting loss of the wild-type FH allele and the presence of some normal tissue contamination. Both mutations affect exon 4, one of the two FH mutation hot spots.29 Genomic normal tissue DNA samples from these individuals were sequenced for all FH exons, but no alterations were detected confirming the somatic origin of the mutations (Figure 2, b and d) and nonsyndromic nature of the cases. The predicted consequence of the splice site change, deletion of exon 4, was confirmed by cDNA amplification and sequencing. Histopathological evaluation of the FH mutation-positive lesions confirmed that they were typical leiomyomas without any distinct features such as atypia.
Figure 1.
LOH flanking FH locus was analyzed by two microsatellite markers. Allelic loss was observed in myoma P4M3 (b) and in P32M1 (d) by markers FH-T and FH-C, respectively. Alleles of normal tissue samples are presented above the tumor samples (a and c). Myomas from three additional individuals harbored allelic imbalance in at least one of these markers at the FH locus (data not shown).
Figure 2.
Sequences of FH mutations detected in uterine leiomyomas Ala196Thr (a) and IVS4 + 3A>G (c) and the chromatographs of the corresponding normal tissues (b and d, respectively). LOH at the splice site change (c) is noticeable by the lower wild-type allele signal in the presence of normal tissue contamination.
Discussion
The spectrum of tumor types in HLRCC families varies from rare and aggressive renal cell carcinoma to common and benign leiomyomas of the uterus. Our previous effort to evaluate FH in nonsyndromic tumorigenesis focused on tumor types associated with HLRCC. Only one tumor with a purely somatic FH defect was identified in that work; biallelic FH defect was present in a soft-tissue sarcoma of the lower limb.4
In this effort, we analyzed 299 cancers representing a wide range of common malignancies for FH mutations. No pathogenic mutations were found in this series of malignant tumors. Cancer types that might be associated with defects in Krebs cycle or mitochondrial DNA were included in the screening set of 299 tumors. Frequent allelic loss at, and examples of somatic mutations in, SDHB and SDHD have been reported in nonsyndromic pheochromocytomas.7,9 The absence of FH mutations in pheochromocytomas may speak for differing pathways of tumorigenesis in SDH and FH deficiencies, although the number of analyzed cases is limited. This would be in line with the fact that the tumor spectra associated with hereditary FH and SDH defects show little similarities. Somatic FH mutations occur at a low frequency and seem to be limited to counterpart tumor types segregating in HLRCC families.
The sensitivity of DHPLC in detecting substitutions and small deletions has been shown to vary from 93 to 100% in many data sets.34 Like all PCR-based mutation-detection technologies,35 our mutation analysis methods were not capable of detecting large genomic deletions or defects in the regulatory or untranslated regions of FH. The polymorphisms detected by DHPLC were identical to those found in previous sequencing efforts (and unpublished data). Although no mutation analysis method has complete sensitivity, it is unlikely that the absence of pathogenic mutations in this set of malignant tumors was because of technical reasons.
Recent studies have suggested that biallelic inactivation of FH, in the majority of cases by LOH, is essential for FH deficiency related tumor growth.1,30 We hypothesized that if FH is associated with the development of nonsyndromic uterine leiomyomas, some mutations should be found in lesions displaying allelic loss at 1q43. Biallelic inactivation could occur also by a second mutation in the coding sequence, by nondetected small deletions, or by hypermethylation of the promoter region. Multiple reports of cytogenetic rearrangements, alterations in gene expression, and analysis of candidate genes in uterine leiomyomas have been published in the last few decades.24 Demonstration of FH mutations in nonsyndromic leiomyomas would be of particular value because other than activating K-RAS alterations,13 no specific point mutations have been associated with these lesions previously.
In this study, allelic loss at the FH locus was detected in 5 of 153 (3.3%) informative unselected leiomyomas by microsatellite markers. Although low, this percentage is significant considering the benign nature of the tumors in question, and indeed based on this result the FH locus appears to be one of the repeatedly deleted regions in uterine leiomyomas though 7q deletions are by far the most frequently observed change.16 That FH is a true target of the 1q43 deletions is demonstrated by the positive findings in sequence analysis. In our leiomyoma series, somatic FH mutations were detected in two of five cases displaying LOH at the FH locus. A negative germline FH mutation analysis of all exons, performed from normal tissue DNA, confirmed the nonsyndromic nature of the two cases. The FH mutation data obtained is quite convincing. The missense mutation is located in a conserved amino acid, and changed the amino acid subclass from nonpolar to uncharged side chain. The splice site change results in deletion of the entire exon. Both changes are located in the FH mutation hot spot detected in HLRCC families. Mutations were not detected in germline excluding the possibility of polymorphism. Three lesions displayed LOH at the FH locus but no mutations were detected in DHPLC. The role of FH inactivation in these lesions remains unknown.
Previously we in collaboration with others have demonstrated that FH mutations predispose to myomas of the uterus and skin with high penetrance in HLRCC families.1 This is the first time, to our knowledge, when specific inactivating point mutations in nonsyndromic uterine leiomyomas have been detected in any gene. Although mutational FH inactivation in nonsyndromic uterine leiomyomas is not common on the tumor level, the frequent co-existence of multiple myomas in patients suggests that the occurrence of FH-deficient leiomyomas is not a rarity. Overall, a minimum of 4.3% (2 of 46) of our uterine leiomyoma patients harbored FH somatic mutations in one of their leiomyomas. The proportion of patients with a leiomyoma displaying a deletion at the FH locus was 5 of 46 (11%). Our current study demonstrates that allelic loss at 1q43 is not rare in nonsyndromic uterine leiomyomas, and that FH is a target of these deletions. This is of interest considering the suggested link between FH germline mutations and risk of uterine leiomyosarcoma. Although the degree of the risk needs to be determined in more detail, and may well be population-specific because of modifying factors, the phenotypes of the Finnish FH mutation carriers provide strong evidence that the association exists and can be quite strong in some populations. We have thus far identified 48 individuals with FH germline mutations (unpublished data).1,3,4,30 Of these, 31 are women. Among these 31 individuals there are 4 (13%) cases of histologically verified uterine leiomyosarcoma, as well as 2 cases with leiomyomas with atypia (unpublished data).3,4,30 The ages at diagnosis of uterine leiomyosarcoma have been 30, 32, 35, and 39 years, and atypia 27 and 36 years. In the general population uterine leiomyosarcomas are rare and typically occur in old age. The proportion of affected women and the remarkably young age at onset of the lesions, are highly unlikely incidental. A persuasive further piece of evidence demonstrating the association is that in the one HLRCC uterine leiomyosarcoma that has been available for molecular studies a somatic FH nonsense mutation had inactivated the wild-type FH allele.4 Although previous studies have been unable to provide a link between nonsyndromic leiomyomas and leiomyosarcoma,33 this study together with what is known on fumarase defects in the hereditary setting suggest that some sporadic leiomyomas, characterized by a FH defect, may have malignant potential. Thus far no purely somatic inactivation of FH in uterine leiomyosarcomas have been reported, only one FH germline mutation associated with a somatic nonsense mutation being the finding in two separate studies examining altogether 44 lesions.4,5 Similarly, the two first studies on FH and nonsyndromic uterine leiomyomas were negative. Our present study identifies FH as the first gene known to undergo inactivation in nonsyndromic uterine leiomyomas by specific point mutations, and calls for additional work to examine the hypothesis that FH defects form a link between uterine leiomyomas and leiomyosarcoma.
Acknowledgments
We thank Mikko Aho, Tomi Kempas, Annika Korvenpää, Kirsi Laukkanen, Inga-Lill Svedberg, Sini Marttinen, Laura Sonninen, and Tuula Lehtinen for technical assistance.
Footnotes
Address reprint requests to Lauri A. Aaltonen, Department of Medical Genetics, University of Helsinki, P. O. Box 63, Helsinki, FIN-00014, Finland. E-mail: lauri.aaltonen@helsinki.fi.
Supported by grants from the Finnish Cancer Society, the Helsinki University Central Hospital, the Sigrid Juselius Foundation, and the Academy of Finland (grant 44870, Finnish Center of Excellence Programme 2000 to 2005).
References
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Alam NA, Bevan S, Churchman M, Barclay E, Barker K, Jaeger EE, Nelson HM, Healy E, Pembroke AC, Friedmann PS, Dalziel K, Calonje E, Anderson J, August PJ, Davies MG, Felix R, Munro CS, Murdoch M, Rendall J, Kennedy S, Leigh IM, Kelsell DP, Tomlinson IP, Houlston RS. Localization of a gene (MCUL1) for multiple cutaneous leiomyomata and uterine fibroids to chromosome 1q42.3-q43. Am J Hum Genet. 2001;68:1264–1269. doi: 10.1086/320124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launonen V, Vierimaa O, Kiuru M, Isola J, Roth S, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci USA. 2001;98:3387–3392. doi: 10.1073/pnas.051633798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuru M, Lehtonen R, Arola J, Salovaara R, Jarvinen H, Aittomaki K, Sjoberg J, Visakorpi T, Knuutila S, Isola J, Delahunt B, Herva R, Launonen V, Karhu A, Aaltonen LA. Few FH mutations in sporadic counterparts of tumor types observed in hereditary leiomyomatosis and renal cell cancer families. Cancer Res. 2002;62:4554–4557. [PubMed] [Google Scholar]
- Barker KT, Bevan S, Wang R, Lu YJ, Flanagan AM, Bridge JA, Fisher C, Finlayson CJ, Shipley J, Houlston RS. Low frequency of somatic mutations in the FH/multiple cutaneous leiomyomatosis gene in sporadic leiomyosarcomas and uterine leiomyomas. Br J Cancer. 2002;87:446–448. doi: 10.1038/sj.bjc.6600502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, van der Mey A, Taschner PE, Rubinstein WS, Myers EN, Richard CW, III, Cornelisse CJ, Devilee P, Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- Gimm O, Armanios M, Dziema H, Neumann HP, Eng C. Somatic and occult germ-line mutations in SDHD, a mitochondrial complex II gene, in nonfamilial pheochromocytoma. Cancer Res. 2000;60:6822–6825. [PubMed] [Google Scholar]
- Baysal BE, Willett-Brozick JE, Lawrence EC, Drovdlic CM, Savul SA, McLeod DR, Yee HA, Brackmann DE, Slattery WH, III, Myers EN, Ferrell RE, Rubinstein WS. Prevalence of SDHB, SDHC, and SDHD germline mutations in clinic patients with head and neck paragangliomas. J Med Genet. 2002;39:178–183. doi: 10.1136/jmg.39.3.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Skoldberg F, Husebye ES, Eng C, Maher ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti D, Douglas F, Lennard TW, Aligianis IA, Woodward ER, Evans DG, Eng C, Latif F, Maher ER. Germline SDHD mutation in familial phaeochromocytoma. Lancet. 2001;357:1181–1182. doi: 10.1016/S0140-6736(00)04378-6. [DOI] [PubMed] [Google Scholar]
- Maddock IR, Moran A, Maher ER, Teare MD, Norman A, Payne SJ, Whitehouse R, Dodd C, Lavin M, Hartley N, Super M, Evans DG. A genetic register for von Hippel-Lindau disease. J Med Genet. 1996;33:120–127. doi: 10.1136/jmg.33.2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EA. Uterine fibroids. Lancet. 2001;357:293–298. doi: 10.1016/S0140-6736(00)03622-9. [DOI] [PubMed] [Google Scholar]
- Hall TM, Taylor RR, Lemon S, Ebina M, Linnoila RI, Norris JH, Park RC, Birrer MJ. Analysis of Ki-ras, p53, and MDM2 genes in uterine leiomyomas and leiomyosarcomas. Gynecol Oncol. 1997;65:330–335. doi: 10.1006/gyno.1997.4653. [DOI] [PubMed] [Google Scholar]
- Pandis N, Heim S, Bardi G, Floderus UM, Willen H, Mandahl N, Mitelman F. Chromosome analysis of 96 uterine leiomyomas. Cancer Genet Cytogenet. 1991;55:11–18. doi: 10.1016/0165-4608(91)90229-n. [DOI] [PubMed] [Google Scholar]
- Ishwad CS, Ferrell RE, Davare J, Meloni AM, Sandberg AA, Surti U. Molecular and cytogenetic analysis of chromosome 7 in uterine leiomyomas. Genes Chromosom Cancer. 1995;14:51–55. doi: 10.1002/gcc.2870140109. [DOI] [PubMed] [Google Scholar]
- Mao X, Barfoot R, Hamoudi RA, Easton DF, Flanagan AM, Stratton MR. Allelotype of uterine leiomyomas. Cancer Genet Cytogenet. 1999;114:89–95. doi: 10.1016/s0165-4608(99)00053-9. [DOI] [PubMed] [Google Scholar]
- Pandis N, Bardi G, Sfikas K, Panayotopoulos N, Tserkezoglou A, Fotiou S. Complex chromosome rearrangements involving 12q14 in two uterine leiomyomas. Cancer Genet Cytogenet. 1990;49:51–56. doi: 10.1016/0165-4608(90)90163-5. [DOI] [PubMed] [Google Scholar]
- Mine N, Kurose K, Nagai H, Doi D, Ota Y, Yoneyama K, Konishi H, Araki T, Emi M. Gene fusion involving HMGIC is a frequent aberration in uterine leiomyomas. J Hum Genet. 2001;46:408–412. doi: 10.1007/s100380170059. [DOI] [PubMed] [Google Scholar]
- Tallini G, Dal Cin P. HMGI(Y) and HMGI-C dysregulation: a common occurrence in human tumors. Adv Anat Pathol. 1999;6:237–246. [PubMed] [Google Scholar]
- Takahashi T, Nagai N, Oda H, Ohama K, Kamada N, Miyagawa K. Evidence for RAD51L1/HMGIC fusion in the pathogenesis of uterine leiomyoma. Genes Chromosom Cancer. 2001;30:196–201. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1078>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Quade BJ, Weremowicz S, Neskey DM, Vanni R, Ladd C, Dal Cin P, Morton CC. Fusion transcripts involving HMGA2 are not a common molecular mechanism in uterine leiomyomata with rearrangements in 12q15. Cancer Res. 2003;63:1351–1358. [PubMed] [Google Scholar]
- Williams AJ, Powell WL, Collins T, Morton CC. HMGI(Y) expression in human uterine leiomyomata. Involvement of another high-mobility group architectural factor in a benign neoplasm. Am J Pathol. 1997;150:911–918. [PMC free article] [PubMed] [Google Scholar]
- Sornberger KS, Weremowicz S, Williams AJ, Quade BJ, Ligon AH, Pedeutour F, Vanni R, Morton CC. Expression of HMGIY in three uterine leiomyomata with complex rearrangements of chromosome 6. Cancer Genet Cytogenet. 1999;114:9–16. doi: 10.1016/s0165-4608(99)00054-0. [DOI] [PubMed] [Google Scholar]
- Newbold RR, DiAugustine RP, Risinger JI, Everitt JI, Walmer DK, Parrott EC, Dixon D. Advances in uterine leiomyoma research: conference overview, summary, and future research recommendations. Environ Health Perspect. 2000;108(Suppl 5):S769–S773. doi: 10.1289/ehp.00108s5769. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, Nagley P, Wong LC, Ngan HY. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res. 2001;61:5998–6001. [PubMed] [Google Scholar]
- Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E, Cuomo C, Cohen D, Pandit S, Spencer M, Rabitti C, Fazio VM, Sidransky D. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001;61:7623–7626. [PubMed] [Google Scholar]
- Yeh JJ, Lunetta KL, van Orsouw NJ, Moore FD, Jr, Mutter GL, Vijg J, Dahia PL, Eng C. Somatic mitochondrial DNA (mtDNA) mutations in papillary thyroid carcinomas and differential mtDNA sequence variants in cases with thyroid tumours. Oncogene. 2000;19:2060–2066. doi: 10.1038/sj.onc.1203537. [DOI] [PubMed] [Google Scholar]
- Eng C, Kiuru M, Fernandez MJ, Aaltonen LA. A role for mitochondrial enzymes in inherited neoplasia and beyond. Nat Rev Cancer. 2003;3:193–202. doi: 10.1038/nrc1013. [DOI] [PubMed] [Google Scholar]
- Kiuru M, Launonen V, Hietala M, Aittomaki K, Vierimaa O, Salovaara R, Arola J, Pukkala E, Sistonen P, Herva R, Aaltonen LA. Familial cutaneous leiomyomatosis is a two-hit condition associated with renal cell cancer of characteristic histopathology. Am J Pathol. 2001;159:825–829. doi: 10.1016/S0002-9440(10)61757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- Lehtonen R, Kiuru M, Rokman A, Ikonen T, Cunningham JM, Schaid DJ, Matikainen M, Nupponen NN, Karhu A, Kallioniemi OP, Thibodeau SN, Schleutker J, Aaltonen LA. No fumarate hydratase (FH) mutations in hereditary prostate cancer. J Med Genet. 2003;40:e19:1–e19:5. doi: 10.1136/jmg.40.3.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy B, Mukherjee T, Hirschhorn K. Molecular cytogenetic analysis of uterine leiomyoma and leiomyosarcoma by comparative genomic hybridization. Cancer Genet Cytogenet. 2000;121:1–8. doi: 10.1016/s0165-4608(00)00225-9. [DOI] [PubMed] [Google Scholar]
- Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: a review. Hum Mutat. 2001;17:439–474. doi: 10.1002/humu.1130. [DOI] [PubMed] [Google Scholar]
- Eng C, Brody LC, Wagner TM, Devilee P, Vijg J, Szabo C, Tavtigian SV, Nathanson KL, Ostrander E, Frank TS. Interpreting epidemiological research: blinded comparison of methods used to estimate the prevalence of inherited mutations in BRCA1. J Med Genet. 2001;38:824–833. doi: 10.1136/jmg.38.12.824. [DOI] [PMC free article] [PubMed] [Google Scholar]