Abstract
Survivin is a member of the inhibitor of apoptosis protein family that is expressed in G2/M phase. Survivin is overexpressed and associated with parameters of poor prognosis in different human tumors. The role of survivin in the pathogenesis of mantle cell lymphoma (MCL) was examined in a series of typical and blastoid tumors. Survivin was detected as a nuclear pattern in a variable number of tumor cells. Mitotic figures were always positive with a strong delineation of the chromosomes. Western blot analysis confirmed the presence of survivin only in nuclear fractions. Protein expression detected by immunohistochemistry correlated with mRNA levels analyzed by quantitative real-time reverse transcription-polymerase chain reaction (P < 0.0001). Survivin expression levels were higher in blastoid MCL variants (P < 0.0001) and were associated with the proliferative activity (P = 0.001), but not with the ploidy status of the tumors. The number of apoptotic cells was independent of survivin or Ki-67 expression. Overall survival was significantly shorter in patients with high survivin expression. However, in a multivariate analysis, proliferative index was a better predictor of survival than survivin score. These findings indicate that survivin is commonly expressed in MCL with a nuclear and mitotic pattern. The expression levels are strongly associated with the proliferative activity of the tumors and the survival of the patients, suggesting a potential role in cell cycle regulation and tumor progression.
Mantle cell lymphoma (MCL) is a malignant lymphoproliferative disorder characterized by the proliferation of CD5+, CD23- monoclonal B cells containing the chromosomal translocation t(11;14)(q13;q32) which places the cyclin D1 gene under the transcriptional control of the immunoglobulin heavy chain enhancer elements.1,2 Additional alterations in the tumor suppressor genes p16INK4a and p53 have been described in aggressive variants of MCL, suggesting that these genes may cooperate with cyclin D1 activation in the progression of these lymphomas.3–6 Classical cytogenetic and comparative genomic hybridization (CGH) studies have also demonstrated complex karyotypes and high number of additional recurrent chromosomal imbalances.7–9 Furthermore, a high incidence of tetraploidy has been described in blastoid variants of MCL.10 This high number of secondary chromosomal aberrations are associated with inactivation of DNA damage response genes such as ATM and CHK2.11,12 MCL has an aggressive clinical course, with a median survival of less than 5 years, and a poor response to conventional treatments. The mechanisms accounting for the resistance of these tumor cells to chemotherapeutic drugs are poorly understood. It has been postulated that most chemotherapeutic agents induce cell death by apoptosis triggering,13 but the molecular basis for chemoresistance in this pathology is not well known.
Inhibitors of apoptosis proteins (IAP) are a family of negative regulators of apoptosis.14 Survivin is a unique mammalian IAP family protein that is expressed during mitosis in a cell-cycle-dependent manner and localized to different components of the mitotic apparatus.15,16 Survivin is virtually undetectable in most normal adult tissues, but it is highly expressed during embryogenesis and in most human cancers. This overexpression in malignant tumors has been associated with an aggressive behavior and poor prognosis of the patients.17–20 The anti-apoptotic function and topographic distribution during cell cycle progression have suggested that survivin may play a role in the mitotic checkpoint control, regulating chromosome segregation and cell division.21,22 Its overexpression in cancer cells may facilitate tumor progression by providing a mechanism to tolerate increasing chromosomal abnormalities and promoting resistance to treatment with anticancer drugs and irradiation.19,20
The aim of this study was to determine the potential role of survivin in the pathogenesis of MCL, analyzing the relationship of its expression with the proliferative activity, genomic alterations, apoptotic levels of the tumors, as well as response to therapy and survival of the patients.
Materials and Methods
Patients
Tumor specimens from 80 patients diagnosed with MCL between 1988 and 2000 were obtained from the Department of Pathology of the Hospital Clinic, University of Barcelona (Spain) and the Institute of Pathology, University of Würzburg (Germany). Tumors were classified as classical MCL (n = 57; 71%), and blastoid variant, including pleomorphic (n = 9; 11%) and blastic (n = 14; 18%) subtypes.1,23 In addition, paired samples from two patients that had progressed from classical to blastoid MCL variants (5 months to 45 months for progression) and 10 paired samples from 5 classical MCL at different times of disease (15 months to 68 months of follow-up) were also examined. The immunophenotype of the tumors was analyzed by immunohistochemistry on tissue sections and/or by flow cytometry on cell suspensions. Cyclin D1 overexpression was demonstrated in all cases by Northern blot analysis and/or immunohistochemistry.
For each patient the following initial data were recorded and evaluated for analysis: 1) clinical data: age, sex, performance status (PS) according to the Eastern Cooperative Oncology Group (ECOG) scale, and presence of B-symptoms; 2) hematological and biochemical parameters: Hb, WBC count, presence of atypical lymphocytes in peripheral blood, platelet count, serum β2-microglobulin, and serum LDH level; 3) tumor extension data: number of nodal and extranodal involved sites, spleen palpable below the left costal margin, hepatomegaly, Ann Arbor stage, and bone marrow infiltration; and 4) the International Prognostic Index (IPI), as defined by the International Non-Hodgkin’s Lymphoma Prognostic Factors Project.24 Moreover, response to treatment and survival, measured from the time the sample was obtained, was also recorded and analyzed.
Cell Lines
The following human cell lines were used: Raji and Daudi, derived from a Burkitt lymphoma and Granta 519, derived from a MCL. Cell lines were purchased from the American Type Culture Collection (ATCC) and were cultured in RPMI 1640 medium (Gibco BRL, Paisley, Scotland) containing 10% heat-inactivated fetal calf serum (Gibco BRL), 2 mmol/L glutamine and 50 μg/ml penicillin-streptomycin, at 37°C in a humidified atmosphere containing 5% carbon dioxide.
Immunohistochemical Studies
MCL tumors were analyzed immunohistochemically using a rabbit polyclonal antibody anti-survivin directed against the full-length recombinant survivin (Novus Biologicals, Littleton, CO), a mouse monoclonal anti-Ki-67 (Immunotech, Marseille, France) and a rabbit polyclonal antibody against cleaved caspase-3, which only recognizes the large fragment of caspase-3 that results after cleavage of procaspase-3 at Asp175 position (Cell Signaling, Beverly, MA). Antigen retrieval conditions and antibody dilutions are shown in Table 1. Briefly, paraffin sections on silane-coated slides were dewaxed and then subjected to antigen retrieval using a hot start method with the adequate buffer solution in a pressure cooker for 2 to 5 minutes at highest pressure, depending on the antibody used (Table 1). Slides were stained using an automated immunostainer TechMate 500 Plus (DAKO, Carpinteria, CA) and the Envision System (DAKO), counterstained in Gill’s hematoxylin and mounted in Pertex (Histolab GmbH, Göteborg, Germany).
Table 1.
Immunohistochemical Study: Antibodies and Conditions of Usage
| Antibody | Source | Dilution | Antigen retrieval*
|
|||
|---|---|---|---|---|---|---|
| Buffer | pH | Time | Temperature | |||
| Survivin | Novus | 1:1000 | Cytrate | 6 | 5 min | 100°C |
| Cleaved caspase-3 | Cell Signaling | 1:10 | EDTA | 8 | 2 min | 100°C |
| Ki-67 (MIB-1) | Immunotech | 1:400 | Cytrate | 6 | 5 min | 100°C |
Simultaneous demonstration of Ki-67 or cleaved caspase-3 and survivin expression was determined in 20 tumor samples using an Universal DAKO Envision Doublestain System (DAKO) following the supplier instructions. Briefly, a unique common retrieval treatment was made for double staining of Ki-67 and survivin, but for double staining of cleaved caspase-3 and survivin, samples were subjected to pressure cooker in adequate buffer before each primary antibody incubation. Survivin immunostaining was performed using the DAB-peroxidase Envision method. After washing, slides were re-incubated with either anti-Ki-67 or anti-cleaved caspase-3 and then an APAAP-Envision method was used.
Immunohistochemical results were expressed as percentage of positive cells. A minimum of 1000 cells per case was counted in representative areas of the tumor. Entrapped and involved follicular germinal centers were excluded for quantification.
RT-PCR Analysis
Total RNA was isolated from each frozen tumor sample and from cell lines using guanidinium thiocyanate method (Ultraspec; Biotecx Laboratories, Houston, TX). RNA was treated with DNase (Ambion, Austin, TX) to eliminate contaminating DNA. For cDNA synthesis, 1 μg of RNA and Taqman Reverse Transcriptions reagents (including Multiscribe reverse transcriptase and random hexamers) were used, as described by the manufacturer (Applied Biosystems, Foster City, CA). The primers and probe used to amplify and quantify survivin cDNA by real-time were designed using Primer Express software (Applied Biosystems) and were as follows: survivin F, 5′-ATT TGA ATC GCG GGA CCC-3′; survivin R, 5′-GAG AAA GGG CTG CCA GGC-3′ and survivin probe, FAM-5′-CAT GGG TGC CCC GAC GTT GC-3′-TAMRA. The primers and the probe were located in a region with no homology to the EPR-1 gene. Real-time monitoring of PCR amplification of cDNAs was done with Taqman Universal master mix (Applied Biosystems) using 200 nmol/L of probe and 100 nmol/L of each survivin primer, in the ABI Prism 7700 sequence Detection System (Applied Biosystems). Relative quantification of gene expression was performed as described in the Taqman user’s manual using human β-glucoronidase (GUSB)(Applied Biosystems) as an internal control.
To examine separately the three splice variants of survivin mRNA, the same forward primer but different reverse primers were used, as described previously.25 Amplification was performed in a total volume of 25 μl in the presence of 3 mmol/L MgCl2, 0.5 μmol/L of each primer and 2.5 μl of each cDNA. RT-PCR amplification was performed with an initial denaturation step of 10 minutes at 95°C, followed by 40 cycles of 30 seconds at 95°C, 30 seconds annealing at 60°C, and 30 seconds elongation at 72°C.
DNA Ploidy
DNA ploidy of the tumors was studied by flow cytometry in 27 cases in which suitable material was available. The analysis was performed on 50-μm-thick sections obtained from formalin-fixed, paraffin-embedded tissues as previously described26 and analyzed with an Epics Profile II flow cytometer (Coulter Company, Hialeah, FL). Non-neoplastic cells in the section under study were used as the internal standard of the diploid channel.
Western Blot Analysis
Western blot analysis was performed on whole, cytoplasmic, and nuclear protein lysates. Cryostat frozen sections were lysed in ice-cold buffer containing 10 mmol/L N-2-hydroxyethylpiperazine-N-2-ethanesulphoric acid (HEPES)(pH 7.9), 10 mmol/L KCl, 1.5 mmol/L MgCl2, 0.1% Nonidet P-40, 0.5 mmol/L dithiothreitol, 2 μg/ml leupeptin, 5 μg/ml aprotinin, and 0.5 mmol/L phenylmethylsulfonyl fluoride for 15 minutes. After centrifugation at 3500 rpm, the supernatant was collected (cytosolic fraction) and the nuclear fraction was obtained by lysis of the precipitated nuclei in a buffer containing 80 mmol/L Tris-HCl (pH 6.8), 2% sodium dodecyl sulfate, 10% glycerol and 0.1 mol/L DTT. Total extracts were obtained by lysis of cryostat sections in the latter buffer. Fifty μg of protein were separated by electrophoresis on 12% polyacrylamide gel and transferred to Immobilon-P (Millipore, Bedford, MA) membranes. The membranes were incubated with a polyclonal antibody against survivin. This antibody was the same used in the immunohistochemical studies. Antibody binding was detected using a secondary antibody conjugated to horseradish peroxidase and an enhanced chemiluminescence (ECL) detection kit (Amersham, Buckinghamshire, UK). Equal protein loading was confirmed with α-tubulin antibody (Oncogene Science, Inc, Cambridge, MA).
Statistical Analysis
The Fisher’s exact, χ2, t-test and analysis of variance tests were used to analyze the association between the clinico-biological parameters and the following categorized variables: immunohistochemical survivin protein expression (cutpoint 20%), survivin mRNA expression by real-time RT-PCR (cutpoint, 2), Ki-67 (cutpoint 50%), and cleaved caspase 3 (cutpoint 0.8%). Testing and estimation of possible cutoff values for survivin and Ki-67 expression was done by maximally selected log-rank statistics. Correlation between survivin, Ki-67 and cleaved caspase-3 was ascertained by means of linear regression. Survival time was measured from the time the sample was obtained. Probability of survival was calculated by the method of Kaplan and Meier, and curves were compared by means of the log-rank test. All statistical tests were two-sided and the significance level was established at 0.05. All significant prognostic variables in the univariate study, as well as survivin expression, proliferative index and Ki-67 expression, were considered for multivariate analysis performed by the stepwise proportional hazard regression method of Cox (survival). All statistical tests were performed with the SPSS v10.06 statistical package (SPSS Inc., Chicago, IL).
Results
Analysis of Survivin Expression by Western Blot
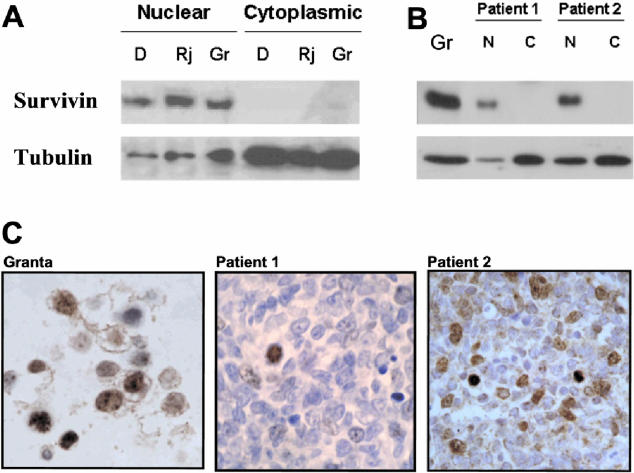
Survivin expression was first studied in nuclear and cytoplasmic protein lysates from Granta 519, Raji, and Daudi human lymphoma cell lines. Western blot analysis using a polyclonal antibody directed against full-length recombinant survivin showed a dominant band of approximately 16.5 kd that was only observed in nuclear extracts from all of the cell lines tested (Figure 1A). In MCL tumors, survivin was also only detected in nuclear protein lysates (Figure 1B).
Figure 1.
Western Blot analysis of survivin protein (Rj) in nuclear and cytoplasmic cell lysates from B human lymphoma cells and MCL patients. Nuclear and cytoplasmic protein lysates from Daudi (D), Raji (R) and Granta 519 (Gr) human cell lines (A) and from two representative MCL patients (B) were obtained and survivin expression was analyzed by Western blot. Expression of α-tubulin was used as a loading control. C: Immunostaining for survivin in cells from Granta 519 cell line and two representative MCL patients confirm the nuclear localization of survivin.
Analysis of Survivin Expression by Immunohistochemistry
The nuclear expression of survivin observed by Western blot was confirmed by immunohistochemistry in all of the cell lines tested and MCL tumors (Figure 1C). Some cells showed weak cytoplasmic staining that tended to disappear with progressive antibody dilutions (data not shown). In reactive lymph nodes and adult tonsils, survivin-positive cells were found mainly in the nucleus of proliferating germinal center cells, and only scattered cells in the mantle zone were positive (Figure 2A). To further confirm this nuclear localization in other lymphoid tumors, additional immunohistochemical studies were performed in a short series of diffuse large B-cell lymphomas (DLBCL) and chronic lymphocytic leukemia (CLL). In all cases, survivin staining was exclusively nuclear as in MCL cases.
Figure 2.
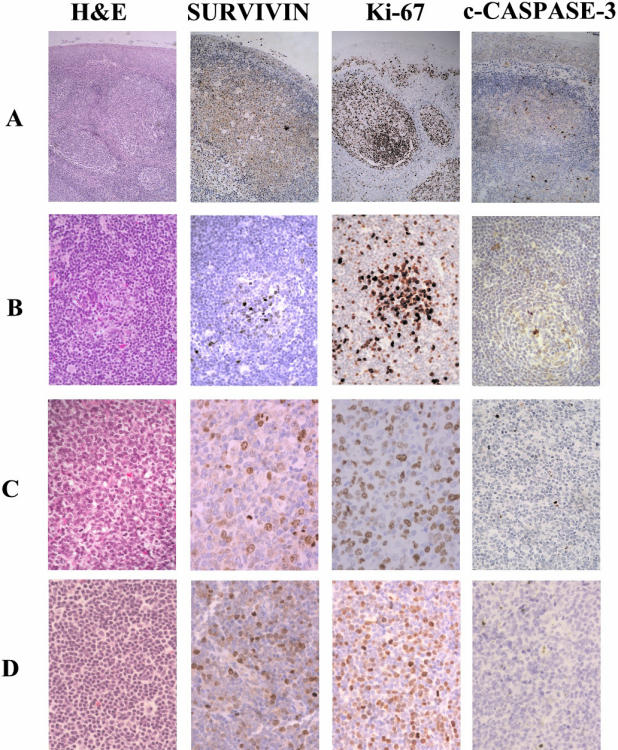
Survivin, Ki-67, and cleaved caspase-3 expression in reactive tonsil and MCL. A: Adult tonsil. Reactive tonsil showing a number of hyperplastic follicles (hematoxylin and eosin, H&E). Cells expressing survivin, Ki-67, and cleaved caspase-3 were mainly localized in the germinal center of secondary follicles. B: MCL, classical variant. An entrapped reactive germinal center was colonized by tumor cells (H&E). Cyclin D1 staining confirmed the presence of tumor cells inside the reactive germinal center (not shown). Survivin-positive tumor and reactive cells were clustered in a partially involved germinal center. Only very few scattered cells were positive outside the follicle. The proliferative activity of the tumor was low. A very low number of positive cleaved caspase-3 cells were detected in the tumor. Apoptotic activity was mostly concentrated in the entrapped follicles. C: MCL, pleomorphic variant. A high mitotic activity was observed in this tumor (H&E). Survivin-positive cells were widely distributed through the tumor. A high number of Ki-67 positive cells were also detected. A few cells were positive for cleaved caspase-3. D: MCL, blastic variant. Monotonous infiltration by medium-sized lymphocytes with rounded nuclei and dispersed chromatin (H&E). Survivin was widely expressed in tumor cells. Increased proliferative activity was noticed with Ki-67 antigen. A few number of cells were positive for cleaved caspase-3. Original magnification, ×200
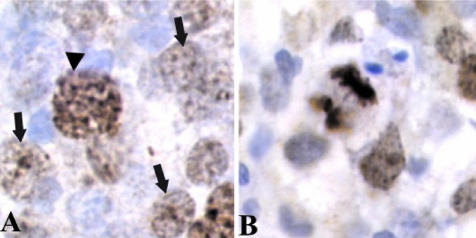
Survivin expression was examined immunohistochemically in 80 MCL. A variable number of survivin nuclear-positive cells ranging from 5% to 95% was detected (Figure 2, B to D). This nuclear distribution of survivin was homogeneous with nucleolar enhancement in non-mitotic nuclei and more intense and granular in prophase (Figure 3A). In metaphase, anaphase, and telophase chromosomes were strongly stained, whereas mitotic spindles were also occasionally observed with a weaker labeling (Figure 3B). Survivin-positive cells were diffusely distributed throughout the tumor. However, in some cases, reactive and colonized germinal centers were easily recognized as nodular clusters of cells with strong survivin nuclear positivity (Figure 2B). Colonization of germinal centers by tumor cells was confirmed by detection of numerous cyclin D1-positive cells within the germinal centers.
Figure 3.
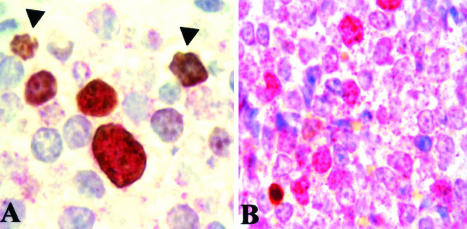
Nuclear and mitotic pattern of survivin expression in primary MCL cells. A: Survivin expression showed a homogeneous pattern with nucleolar reinforcement in non-mitotic nuclei (arrows) and a strong multi-granular pattern in large-sized prophasic appearing nuclei (arrowheads). B: In late mitotic nuclei, chromosomes were strongly stained. Original magnification, ×1000
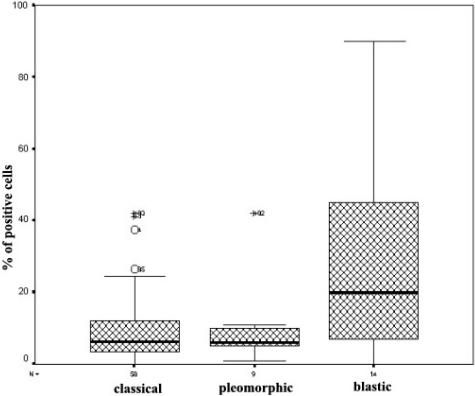
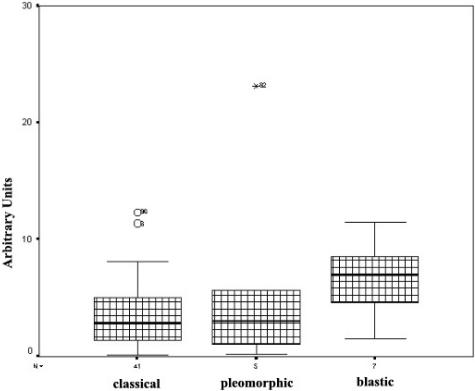
The number of survivin-positive cells correlated with the histological subtype of MCL. Thus, the highest expression of survivin was observed in the blastic variant (30 ± 29% of positive cells, n = 14), compared to classical MCL (10 ± 10%, n = 57) (analysis of variance, P < 0.0001). The expression of survivin in pleomorphic MCL was similar to that observed in classical tumors (10 ± 12%, n = 9; P = 0.89) (Figure 4).
Figure 4.
Immunohistochemical analysis of survivin expression in MCL. Blastic MCL variant showed higher survivin expression than classical and pleomorphic MCL variants. Symbols above the bars represent outlier cases.
No changes in survivin levels were observed in 10 paired samples of five classical MCL at different times of disease, without evidence of tumor progression (11 ± 9% vs. 13 ± 8%). In two additional patients who had progressed from classical to blastoid MCL, an increase in the number of survivin-positive cells was observed in the blastoid variant (1.4- and three-fold, respectively).
Analysis of Survivin Expression by Real-Time RT-PCR
Quantification of survivin expression was assessed by real-time RT-PCR. The comparative CT (cycle threshold) method was used for relative quantification of survivin mRNA expression, after confirming that survivin cDNA and GUS cDNA were amplified with the same efficiency. Survivin expression in normal lymphocytes was used as a reference control, and the survivin expression levels of these cells adopted the arbitrary value of 1. Granta 519, a MCL-derived cell line, showed 100-fold increase in survivin levels compared to normal lymphocytes. The expression of survivin by real-time RT-PCR was analyzed in 51 MCL tumors. Survivin mRNA was detected in all MCL samples, with a mean relative value of 4 ± 4. Higher levels of survivin mRNA were detected in the blastic MCL variants (10 ± 7, n = 7), compared to those obtained in classical (4 ± 3, n = 41) and pleomorphic (3 ± 2, n = 3) tumors (Figure 5). Moreover, a significant linear correlation was found between mRNA survivin levels and the number of survivin-positive cells analyzed by immunohistochemistry (Pearson Regression Test, P < 0.0001).
Figure 5.
Real-time RT-PCR quantitative analysis of survivin mRNA expression in MCL. Blastic MCL variant showed higher survivin mRNA expression than classical and pleomorphic MCL variants. Survivin mRNA expression levels are given in arbitrary units using normal lymphocytes as reference control. PCR arbitrary units were defined as described in Material and Methods. Symbols above the bars represent outlier cases.
Analysis of Survivin Variants by RT-PCR
Using specific primers, the different survivin variants were detected in seven MCL tumors. The specificity of amplification products was confirmed by DNA sequencing (data not shown). In all MCL cases, survivin, survivin-2B and survivin-ΔEx3 mRNA were detected. A series of six DLBCL and five CLL cases were also analyzed. The three survivin variants were detected in all DLBCL, with an expression pattern similar to MCL samples. The expression pattern of these splicing variants was heterogeneous in the CLL patients since survivin was detected in four patients, survivin-2B in two and survivin-ΔEx3 only in one of the five patients.
Correlation of Survivin Expression with Cell Proliferation, Apoptosis, and Ploidy Status in MCL
Survivin expression was compared to the proliferative activity and apoptotic index of the tumors. The proliferative activity was analyzed in 82 MCL tumors and it was calculated as the percentage of Ki-67 positive cells in the most proliferative areas, excluding colonized follicles. The apoptotic index was calculated in 55 cases as the percentage of positive cells stained with anti-cleaved caspase-3, the active form of caspase-3.
Blastic MCL cases showed a higher proliferative index (63 ± 29%, n = 14) than classical (30 ± 23%, n = 57) (analysis of variance, P < 0001) or pleomorphic (40 ± 29%, n = 9) MCL variants (Figure 2, B to D). The proliferative activity of the tumors showed a statistically significant correlation with protein and mRNA survivin expression levels measured by immunohistochemistry (Pearson Regression Test, P = 0.001) or real-time quantitative RT-PCR (Pearson Regression Test, P = 0.0001), respectively.
Double immunolabeling for Ki-67 and survivin was performed in 20 selected tumors. In all cases, a nuclear co-localization of both proteins was observed, although the number of Ki-67 positive cells was higher than survivin stained cells. Of note, virtually all cells positive for survivin also expressed the Ki-67 antigen (Figure 6A).
Figure 6.
Double immunohistochemical staining of survivin and Ki-67 or cleaved caspase-3. A: Survivin/Ki-67 staining. All survivin-positive cells (red) were also Ki-67-positive cells (brown) (×60). Some cells were only positive for Ki-67 (arrows). B: Survivin/cleaved caspase-3 staining. A wide number of tumor cells were positive for survivin expression (red) but only few cells were stained with the antibody against cleaved caspase-3 (brown). No double-positive cells were found. Original magnification, ×1000.
In all MCL cases, the number of cells labeled with anti-cleaved caspase-3 was low, ranging from 0.1% to 5.6% (Figure 2). In all of the samples analyzed, the areas showing the highest positivity for cleaved caspase-3 were also the most proliferative areas (data not shown). Although a higher apoptotic activity was observed in blastic and pleomorphic variants, no significant differences in cleaved caspase-3 expression among the histological subtypes were found. Moreover, no correlation between survivin expression and the apoptosis index was observed. Using a double-immunohistochemical labeling for cleaved caspase-3 and survivin, no cells simultaneously positive for both antigens were identified (Figure 6B).
In 27 cases, the ploidy status was assessed by flow cytometry. Tetraploidy was found in 5 of 6 pleomorphic MCL (83%), in 2 of 6 blastic variant (33%) and only in 2 of 15 classical tumors (13%) (χ2, P = 0.03). By immunohistochemistry, no differences in survivin levels were observed between diploid (17 ± 16%) and tetraploid (16 ± 16%) MCL cases.
Prognostic Significance of Survivin Expression
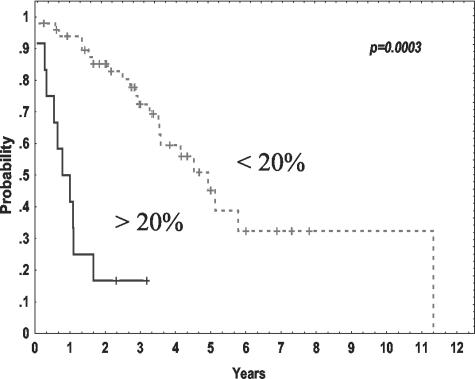
The main clinical characteristics and follow-up were available in 62 patients. Median follow-up of the series was 36 months. Among the clinical features at diagnosis, mRNA survivin expression correlated only with the performance status of the patients (ECOG ≥2, χ2, P = 0.05). No relationship between survivin levels and other clinico-biological parameters was found. Moreover, no correlation of survivin levels with the degree of therapeutical response was detected. Interestingly, increased survivin expression assessed by both real-time RT-PCR and immunohistochemistry correlated with a shorter survival of the patients. Thus, patients with survivin expression in more than 20% of tumor cells had a median survival of 8 months, whereas it was of 60 months for those patients with survivin staining in ≤20% (Kaplan-Meier, log survival P = 0.0003) (Figure 7). Furthermore, the median survival of the patients with high and low survivin mRNA expression was 24 and 80 months, respectively (Kaplan-Meier, log survival P = 0.00023).
Figure 7.
Overall survival of 62 MCL patients according to survivin protein expression analyzed by immunohistochemistry.
Ki-67 expression was also able to predict survival in MCL patients. Patients with Ki-67 expression in more than 50% of tumor cells had a median survival of 9 months, whereas it was 62 months for patients with less than 50% of Ki-67-positive cells (Kaplan-Meier, log survival P = 0,0006). Furthermore, Ki-67 expression was able to predict survival in typical histological variants (P = 0.022), but did not predict survival in either the blastic or pleomorphic variants. This was probably due to the fact that virtually all blastic variants (8 of 10) exhibited high Ki-67 expression. When Ki-67 expression (considering 50% cut-off) was compared to the histological variants (typical versus blastoid) in a multivariate analysis for survival, only the Ki-67 expression remained into the model (Cox Multivariate Test, risk ratio (RR) = 4.4; 0.95 CI of RR = 1.9 to 9.9). In addition, in a Cox multivariate analysis comparing survivin versus the proliferative index for survival, Ki-67 expression, but not the survivin levels, retained its prognostic value (RR = 10.88; P = 0.0001).
Discussion
In this study, we have examined survivin expression in a large series of MCL and the results have been compared to different pathological characteristics of the tumors and clinical parameters of the patients. Our findings indicate that survivin is commonly expressed in these tumors with a nuclear and mitotic pattern. The expression levels were significantly associated with the histological variants and proliferative activity of the tumors as well as with the survival of the patients. However, survivin levels were not related to the ploidy or caspase-3 activation in these tumors.
Survivin was only detected in the nuclear fraction of tumor cells. In mitotic cells, after nuclear membrane disruption, survivin strongly labeled chromosomes and the mitotic spindle. These immunohistochemical observations were also confirmed by Western blot analysis showing survivin expression mainly in the nuclear but not in the cytoplasmic fraction of cell lysates. A similar nuclear staining was also observed in normal lymphocytes of reactive germinal centers. These staining patterns in a human lymphoid neoplasm are concordant with previous observations in human cancer cell lines in which survivin has been associated with centromeres, mitotic spindle microtubules, centrosomes, and cytokinesis.16,27–30 Previous studies analyzing survivin expression by immunohistochemistry have shown a predominant cytoplasmic distribution.15,31 However, nuclear accumulation of survivin has been recently described in other human tumors.32,33 The reason for these differences in cell distribution between tumors is not clear. It has been proposed that the subcellular distribution of survivin is regulated by an active import into the nucleus and a CRM1-mediated export to the cytoplasm, suggesting that survivin may be considered a nuclear shuttling protein. Therefore, the almost exclusively cytoplasmic localization in a high number of tumors may be the result of a higher rate of nuclear export.34
Survivin expression was detected in all tumors with a variable number of positive cells ranging from 5% to 95%. The number of positive cells detected by immunohistochemistry correlated significantly with the mRNA levels detected by real-time quantitative RT-PCR (P < 0.0001), suggesting that survivin expression in these tumors is predominantly regulated at the transcriptional level. The survivin gene gives three alternatively spliced transcripts. In addition to wild-type survivin, which exhibits a three-intron-four exon structure, two survivin isoforms were generated by insertion of an alternative exon 2 (survivin-2B) or removal of exon 3 (survivin-ΔEx3).25 In MCL cases the three survivin variants were detected.
Higher levels of survivin were detected in blastic MCL than in classic or pleomorphic variants. In addition, survivin expression levels were significantly associated with the proliferative activity of the tumors. Blastic variant of MCL has a higher proliferative activity and a more aggressive biological behavior than classical forms of this tumor.35 Concordantly, MCL patients with high survivin expression had a shorter overall survival. In this regard, it is of note that survivin overexpression in other human cancers has been associated with unfavorable prognostic features, poor response to therapy, high relapse rate and shortened survival.17–20 Furthermore, survivin has been considered an independent prognostic factor in DLBL.36 However, the multivariate analysis performed in our study indicates that survival in MCL was better predicted by the proliferative activity of the tumors than by survivin levels. This finding is concordant with previous studies indicating that proliferation is one of the best prognosis parameters in MCL.35,37 The relationship between survivin expression, proliferation, and survival in other human tumors has not been explored. Recently, it has been described that in CLL cells, survivin is the only IAP whose expression is induced by CD40L, suggesting a role of survivin in the control of CLL proliferative pool interfacing apoptosis.38 The protective anti-apoptotic effect of survivin in proliferating malignant cells may be a mechanism to stabilize tumor cells with chromosomal abnormalities favoring the survival of these cells and the progression of the tumors.29
Survivin seems to be required for the targeting of members of the Aurora family of kinases to metaphase chromosomes, thereby controlling chromosome segregation and possibly cytokenesis.28,29,39 Recently, it has been described that the preponderant survivin pool is associated with microtubules and participates in the assembly of a bipolar mitotic spindle.40 This function is consistent with its localization to centrosomes, spindle pores, and spindle microtubules.16,40 Furthermore, suppression of human survivin expression induces a catastrophic defect of microtubule assembly, with abnormal mitotic spindles and formation of multinucleated cells.27,29,41,42 MCL is characterized by an increased number of chromosomal imbalances and ploidy alterations.7–9 In addition, tetraploidy is a common numerical chromosomal aberration in blastic and pleomorphic MCL variants.10 In this sense, we found tetraploidy in 83% of pleomorphic MCL and in 33% of blastic tumors but only in 13% of classical variants. However, no differences in survivin levels were detected in MCL in correlation with the ploidy status, suggesting that survivin expression may not be involved in the generation of this type of chromosomal aberrations.
Survivin has been implicated in a dual role connecting suppression of apoptosis to regulation of chromosomal segregation and cell division.16,19 Targeting experiments using antisense survivin or dominant-negative mutants resulted in spontaneous apoptosis, increased caspase activity, and inhibition of cell proliferation.26,40,42 A role for survivin in blocking apoptosis has also been demonstrated in vivo.43 However, the mechanisms of apoptotic suppression by survivin are not completely clear. Initial observations suggested that survivin could directly suppress activation of caspase-344,45; however, more recent studies have demonstrated that survivin lacks the ability to directly inhibit caspase-3.46,47 Other groups have proposed that survivin acts like other IAPs by binding to caspase-9 and/or by neutralizing the new IAP-inhibiting protein, Smac/Diablo,48,49 raising the possibility that it might suppress caspases indirectly by freeing other IAP family members from the constraints of this protein. In this study we have investigated the apoptotic activity in MCL by examining the expression of the cleaved form of caspase-3. This immunohistochemical assay has been recently used in other B cell lymphomas, showing that the number of apoptotic cells was variable in different types of tumors.50 However, in this study, MCL tumors were not examined. Our findings demonstrate that expression of cleaved caspase-3 in MCL was virtually absent or very low in most tumors. Furthermore, survivin and cleaved caspase-3 were never expressed in the same MCL cell. This low percentage of apoptotic cells and the detection of survivin in MCL are in agreement with a potential role of survivin in inhibiting apoptosis.
In conclusion, our results indicate that survivin is commonly expressed in MCL with a nuclear and mitotic pattern and that its expression levels are strongly associated with the proliferative activity of the tumors and the survival of the patients, suggesting a potential role in cell cycle regulation and tumor progression. The dual role of survivin in apoptosis inhibition and regulation of cell cycle may facilitate evasion from checkpoint mechanisms of growth arrest and promote resistance to chemotherapeutic regimens targeting the mitotic spindle. Since MCL cells showed alterations in cell cycle regulator genes as well as a poor response to conventional treatments, manipulation of survivin expression might be of interest in the treatment of MCL.
Acknowledgments
We thank Montse Sanchez for the excellent technical assistance. Montse Sanchez was supported by Dako.
Footnotes
Address reprint requests to Elias Campo, Hematopathology Unit, Department of Pathology, Hospital Clinic, Villarroel 170, 08036 Barcelona, Spain. E-mail: campo@medicina.ub.es.
Supported in part by grants FIS 00/946, and 03-398 European Union European Framework 5 grant QLG1-CT-2000–0687, CICYT SAF 02–3261, Asociación Española contra el Cancer (AECC) and FIJC-02/P-EM and P-CR. A.M. had a fellowship from Hospital Clínic and Fondo de Investigacion Sanitaria (FIS) and B.B. was the recipient of a research fellowship from the Instituto de Salud Carlos III.
A.M. and B.B. contributed equally to this study.
References
- Campo E, Raffeld M, Jaffe ES. Mantle-cell lymphoma. Semin Hematol. 1999;36:115–127. [PubMed] [Google Scholar]
- Bosch F, Jares P, Campo E, Lopez-Guillermo A, Piris MA, Villamor N, Tassies D, Jaffe ES, Montserrat E, Rozman C, Cardesa A. PRAD-1/cyclin D1 gene overexpression in chronic lymphoproliferative disorders: a highly specific marker of mantle cell lymphoma. Blood. 1994;84:2726–2732. [PubMed] [Google Scholar]
- Hernandez L, Fest T, Cazorla M, Teruya-Feldstein J, Bosch F, Peinado MA, Piris MA, Montserrat E, Cardesa A, Jaffe ES, Campo E, Raffeld M. p53 gene mutations and protein overexpression are associated with aggressive variants of mantle cell lymphomas. Blood. 1996;87:3351–3359. [PubMed] [Google Scholar]
- Greiner TC, Moynihan MJ, Chan WC, Lytle DM, Pedersen A, Anderson JR, Weisenburger DD. p53 mutations in mantle cell lymphoma are associated with variant cytology and predict a poor prognosis. Blood. 1996;87:4302–4310. [PubMed] [Google Scholar]
- Pinyol M, Hernandez L, Cazorla M, Balbín M, Jares P, Fernández PL, Montserrat E, Cardesa A, Lopez-Otin C, Campo E. Deletions and loss of expression of p16INK4a and p21Waf1 genes are associated with aggressive variants of mantle cell lymphomas. Blood. 1997;89:272–280. [PubMed] [Google Scholar]
- Dreyling MH, Bullinger L, Ott G, Stilgenbauer S, Muller-Hermelink HK, Bentz M, Hiddemann W, Dohner H. Alterations of the cyclin D1/p16-pRB pathway in mantle cell lymphoma. Cancer Res. 1997;57:4608–4614. [PubMed] [Google Scholar]
- Bea S, Ribas M, Hernandez JM, Bosch F, Pinyol M, Hernandez L, Garcia JL, Flores T, Gonzalez M, Lopez-Guillermo A, Piris MA, Cardesa A, Montserrat E, Miro R, Campo E. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood. 1999;93:4365–4374. [PubMed] [Google Scholar]
- Bentz M, Plesch A, Bullinger L, Stilgenbauer S, Ott G, Muller-Hermelink HK, Baudis M, Barth TF, Moller P, Lichter P, Dohner H. t(11;14)-positive mantle cell lymphomas exhibit complex karyotypes and share similarities with B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2000;27:285–294. [PubMed] [Google Scholar]
- Allen JE, Hough RE, Goepel JR, Bottomley S, Wilson GA, Alcock HE, Baird M, Lorigan PC, Vandenberghe EA, Hancock BW, Hammond DW. Identification of novel regions of amplification and deletion within mantle cell lymphoma DNA by comparative genomic hybridization. Br J Haematol. 2002;116:291–298. doi: 10.1046/j.1365-2141.2002.03260.x. [DOI] [PubMed] [Google Scholar]
- Ott G, Kalla J, Ott MM, Schryen B, Katzenberger T, Muller JG, Muller-Hermelink HK. Blastoid variants of mantle cell lymphoma: frequent bcl-1 rearrangements at the major translocation cluster region and tetraploid chromosome clones. Blood. 1997;89:1421–1429. [PubMed] [Google Scholar]
- Camacho E, Hernandez L, Hernandez S, Tort F, Bellosillo B, Bea S, Bosch F, Montserrat E, Cardesa A, Fernández PL, Campo E. ATM gene inactivation in mantle cell lymphoma mainly occurs by truncating mutations and missense mutations involving the phosphatidylinositol-3 kinase domain and is associated with increasing numbers of chromosomal imbalances. Blood. 2002;99:238–244. doi: 10.1182/blood.v99.1.238. [DOI] [PubMed] [Google Scholar]
- Tort F, Hernandez S, Bea S, Martinez A, Esteller M, Herman JG, Puig X, Camacho E, Hernández L, Sanchez M, Nayach I, Lopez-Guillermo A, Fernández PL, Colomer D, Campo E. CHK2-decreased protein expression and infrequent genetic alterations mainly occur in aggressive types of non-Hodgkin’s lymphomas. Blood. 2002;100:4602–4608. doi: 10.1182/blood-2002-04-1078. [DOI] [PubMed] [Google Scholar]
- Reed JC. Apoptosis-based therapies. Nat Rev Drug Discov. 2002;1:111–121. doi: 10.1038/nrd726. [DOI] [PubMed] [Google Scholar]
- Deveraux QL, Reed JC. IAP family proteins-suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58:5071–5074. [PubMed] [Google Scholar]
- Islam A, Kageyama H, Takada N, Kawamoto T, Takayasu H, Isogai E, Ohira M, Hashizume K, Kobayashi H, Kaneko Y, Nakagawara A. High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617–623. doi: 10.1038/sj.onc.1203358. [DOI] [PubMed] [Google Scholar]
- Altieri DC. The molecular basis and potential role of survivin in cancer diagnosis and therapy. Trends Mol Med. 2001;7:542–547. doi: 10.1016/s1471-4914(01)02243-2. [DOI] [PubMed] [Google Scholar]
- Ikeguchi M, Ueda T, Sakatani T, Hirooka Y, Kaibara N. Expression of survivin messenger RNA correlates with poor prognosis in patients with hepatocellular carcinoma. Diagn Mol Pathol. 2002;11:33–40. doi: 10.1097/00019606-200203000-00007. [DOI] [PubMed] [Google Scholar]
- Kallio MJ, Nieminen M, Eriksson JE. Human inhibitor of apoptosis protein (IAP) survivin participates in regulation of chromosome segregation and mitotic exit. EMBO J. 2001;15:2721–2723. doi: 10.1096/fj.01-0280fje. [DOI] [PubMed] [Google Scholar]
- Bolton MA, Lan W, Powers SE, McCleland ML, Kuang J, Stukenberg PT. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol Biol Cell. 2002;13:3064–3077. doi: 10.1091/mbc.E02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe ES, Harris NL, Stein H, Vardiman JWE. World Health Organization classification of tumors. Jaffe ES, Harris NL, Stein H, Vardiman JWE, editors. Lyon, France: IARC Press; Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. 2001:pp 168–170. [Google Scholar]
- A predictive model for aggressive non-Hodgkin’s lymphoma. The International Non-Hodgkin’s Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- Mahotka C, Krieg T, Krieg A, Wenzel M, Suschek CV, Heydthausen M, Gabbert HE, Gerharz CD. Distinct in vivo expression patterns of survivin splice variants in renal cell carcinomas. Int J Cancer. 2002;100:30–36. doi: 10.1002/ijc.10450. [DOI] [PubMed] [Google Scholar]
- Hedley DW, Friedlander ML, Taylor IW, Rugg CA, Musgrove EA. Method for analysis of cellular DNA content of paraffin-embedded pathological material using flow cytometry. J Histochem Cytochem. 1983;31:1333–1335. doi: 10.1177/31.11.6619538. [DOI] [PubMed] [Google Scholar]
- Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, Tognin S, Villa A, Marchisio PC, Altieri DC. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
- Skoufias DA, Mollinari C, Lacroix FB, Margolis RL. Human survivin is a kinetochore-associated passenger protein. J Cell Biol. 2000;151:1575–1582. doi: 10.1083/jcb.151.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- Giodini A, Kallio MJ, Wall NR, Gorbsky GJ, Tognin S, Marchisio PC, Symons M, Altieri DC. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 2002;62:2462–2467. [PubMed] [Google Scholar]
- Adida C, Recher C, Raffoux E, Daniel MT, Taksin AL, Rousselot P, Sigaux F, Degos L, Altieri DC, Dombret H. Expression and prognostic significance of survivin in de novo acute myeloid leukaemia. Br J Haematol. 2000;111:196–203. doi: 10.1046/j.1365-2141.2000.02328.x. [DOI] [PubMed] [Google Scholar]
- Okada E, Murai Y, Matsui K, Isizawa S, Cheng C, Masuda M, Tacaño Y. Survivin expression in tumor cell nuclei is predictive of a favorable prognosis in gastric cancer patients. Cancer Lett. 2001;163:109–116. doi: 10.1016/s0304-3835(00)00677-7. [DOI] [PubMed] [Google Scholar]
- Frost M, Jarboe EA, Orlicky D, Gianani R, Thompson LC, Enomoto T, Shroyer KR. Immunohistochemical localization of survivin in benign cervical mucosa, cervical dysplasia, and invasive squamous cell carcinoma. Am J Clin Pathol. 2002;117:738–744. doi: 10.1309/6V09-38K3-JQ40-UR50. [DOI] [PubMed] [Google Scholar]
- Rodriguez JA, Span SW, Ferreira CG, Kruyt FA, Giaccone G. CRM1-mediated nuclear export determines the cytoplasmic localization of the antiapoptotic protein survivin. Exp Cell Res. 2002;275:44–53. doi: 10.1006/excr.2002.5492. [DOI] [PubMed] [Google Scholar]
- Bosch F, Lopez-Guillermo A, Campo E, Ribera JM, Conde E, Piris MA, Vallespi T, Woessner S, Montserrat E. Mantle cell lymphoma: presenting features, response to therapy, and prognostic factors. Cancer. 1998;82:567–575. doi: 10.1002/(sici)1097-0142(19980201)82:3<567::aid-cncr20>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Adida C, Haioun C, Gaulard P, Lepage E, Morel P, Briere J, Dombret H, Reyes F, Diebold J, Gisselbrecht C, Salles G, Altieri DC, Molina TJ. Prognostic significance of survivin expression in diffuse large B-cell lymphomas. Blood. 2000;96:1921–1925. [PubMed] [Google Scholar]
- Velders GA, Kluin-Nelemans JC, De Boer CJ, Hermans J, Noordijk EM, Schuuring E, Kramer MH, Van Deijk WA, Rahder JB, Kluin PM, Van Krieken JH. Mantle-cell lymphoma: a population-based clinical study. J Clin Oncol. 1996;14:1269–1274. doi: 10.1200/JCO.1996.14.4.1269. [DOI] [PubMed] [Google Scholar]
- Granziero L, Ghia P, Circosta P, Gottardi D, Strola G, Geuna M, Montagna L, Piccoli P, Chilosi M, Caligaris-Cappio F. Survivin is expressed on CD40 stimulation and interfaces proliferation and apoptosis in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2777–2783. doi: 10.1182/blood.v97.9.2777. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- Fortugno P, Wall NR, Giodini A, O’Connor DS, Plescia J, Padgett KM, Tognin S, Marchisio PC, Altieri DC. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci. 2002;115:575–585. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- Chen J, Wu W, Tahir SK, Kroeger PE, Rosenberg SH, Cowsert LM, Bennett F, Krajewski S, Krajewska M, Welsh K, Reed JC, Ng SC. Down-regulation of survivin by antisense oligonucleotides increases apoptosis, inhibits cytokinesis and anchorage-independent growth. Neoplasia. 2000;2:235–241. doi: 10.1038/sj.neo.7900091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olie RA, Simoes-Wust AP, Baumann B, Leech SH, Fabbro D, Stahel RA, Zangemeister-Wittke U. A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res. 2000;60:2805–2809. [PubMed] [Google Scholar]
- Grossman D, Kim PJ, Schechner JS, Altieri DC. Inhibition of melanoma tumor growth in vivo by survivin targeting. Proc Natl Acad Sci USA. 2001;98:635–640. doi: 10.1073/pnas.230450097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, Reed JC. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K, Nakano T, Miura M, Shiraki K. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346–1353. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol. 2000;7:602–608. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- Banks DP, Plescia J, Altieri DC, Chen J, Rosenberg SH, Zhang H, Ng SC. Survivin does not inhibit caspase-3 activity. Blood. 2000;96:4002–4003. [PubMed] [Google Scholar]
- O’Connor DS, Grossman D, Plescia J, Li F, Zhang H, Villa A, Tognin S, Marchisio PC, Altieri DC. Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc Natl Acad Sci USA. 2000;97:13103–1310. doi: 10.1073/pnas.240390697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- Dukers DF, Oudejans JJ, Vos W, ten Berge RL, Meijer CJ. Apoptosis in B-cell lymphomas and reactive lymphoid tissues always involves activation of caspase 3 as determined by a new in situ detection method. J Pathol. 2002;196:307–315. doi: 10.1002/path.1046. [DOI] [PubMed] [Google Scholar]