Abstract
Diffuse large B-cell lymphoma (DLBCL) patients are treated using relatively homogeneous protocols, irrespective of their biological and clinical variability. Here we have developed a protein-expression-based outcome predictor for DLBCL. Using tissue microarrays (TMAs), we have analyzed the expression of 52 selected molecules in a series of 152 DLBCLs. The study yielded relevant information concerning key biological aspects of this tumor, such as cell-cycle control and apoptosis. A biological predictor was built with a training group of 103 patients, and was validated with a blind set of 49 patients. The predictive model with 8 markers can identify the probability of failure for a given patient with 78% accuracy. After stratifying patients according to the predicted response under the logistic model, 92.3% patients below the 25 percentile were accurately predicted by this biological score as “failure-free” while 96.2% of those above the 75 percentile were correctly predicted as belonging to the “fatal or refractory disease” group. Combining this biological score and the International Prognostic Index (IPI) improves the capacity for predicting failure and survival. This predictor was then validated in the independent group. The protein-expression-based score complements the information obtained from the use of the IPI, allowing patients to be assigned to different risk categories.
Diffuse large B-cell lymphoma (DLBCL) is the most frequent type of lymphoma, with a 5-year survival probability of around 50%. Although a significant proportion of DLBCL patients can be cured with current combination chemotherapy regimes, at present there is no clinical or biological score available that can accurately distinguish patients who can be cured with standard therapy and those who require new treatment approaches.1
Outcome with DLBCL, as in other types of cancer, is the result of interactions between the genetic abnormalities in the tumor and the clinical status of the patients. Information concerning the molecular abnormalities present in DLBCL, derived from genome-wide expression analysis, allows us to identify multiple markers that suggest the existence of a vast number of underlying genetic events in all of the major cell pathways involved in control of proliferation, apoptosis, signal transduction, DNA repair, and other processes.2–4 Nevertheless, until recently, outcome-predictor systems have been based on single genetic abnormalities, or the integration of clinical data into predictive models, such as the International Prognostic Index (IPI).5
Tissue microarrays (TMAs) are a powerful and reproducible technique for demonstrating the biological variability inherent in cancer and, when applied to lymphoma samples, are capable of identifying multiple alterations in the regulation of critical genes and pathways.6,7
In the present study we have investigated the expression of a large number (52) of markers in a DLBCL series using TMAs. The results yield information concerning the variety of molecular markers that predict clinical response. These can be integrated into a single predictive model that identifies the probability of failure with 78% accuracy. This biological score can be used to complement the information obtained by the use of the IPI, allowing patients to be stratified into different risk categories.
Materials and Methods
DLBCL Samples
235 cases of DLBCL were collected. These were diagnosed between 1990 and 1999, the stages being evaluated according to standard protocols. Patients were treated with regimes including polychemotherapy (mainly adriamycin-based) with or without adjuvant radiotherapy and/or surgery. Diagnostic paraffin blocks were selected on the basis of the availability of suitable formalin-fixed paraffin-embedded tissue, containing enough remaining tissue as for a minimum of 60 sections. Histological confirmation of DLBCL was achieved in all cases by central review using standard tissue sections. Histological criteria used for diagnoses and classification of cases were those described in the World Health Organization classification.8 Paraffin-embedded blocks from reactive lymphoid tissue, cell lines and different B- and T-cell lymphoma samples, used for control purposes, were obtained from the tissue archives of the CNIO Tumor Bank.
Tissue Microarray Design
We used a Tissue Arrayer device (Beecher Instruments, Sun Prairie, WI) to construct three different TMA blocks, containing 502 cylinders in total, according to conventional protocols.7 All cases were histologically reviewed and the most tumor-rich areas were marked in the paraffin blocks. Two selected 0.6-mm-diameter cylinders from two different areas were included in each case, along with 16 separate controls to ensure the quality, reproducibility and homogenous staining of the slides. Selected controls include reactive lymph nodes and tonsils, and paraffin-embedded cell lines.
Immunohistochemical staining was performed and evaluated for the 50 different antibodies using standard procedures.7 The selected markers correspond to sets of key proteins involved in cell cycle, apoptosis (extrinsic and intrinsic pathways), and B-cell differentiation, additionally including a large majority of the markers previously identified as survival predictors in DLBCL.
Staining of TMA sections was evaluated by three different pathologists (A.S., J.F.G., F.C.), using uniform criteria. To guarantee the reproducibility of this method, we decided to employ straightforward and clear-cut criteria. After initial analysis, the pattern of staining for each Ab was recorded as positive versus negative, or high versus low level of expression, taking into account the expression in reactive and tumoral cells and specific cut-offs for each marker. Specific details of the threshold used in each case are given in Table 1. As a general criterion, these thresholds were preferentially selected on the basis of their reproducibility and, when possible, their ability to correlate with previous findings using these markers and/or specific biological events.
Table 1.
Antibodies Used in the Analyses, Indicating Source, Dilution, Threshold and Pattern of Reactivity Used and Positive Controls
| Protein | Clone | Source | Dilution | Reactivity | Threshold | Internal control |
|---|---|---|---|---|---|---|
| Bcl-2 | 124 | DAKO | 1:25 | High/low | >50% positive cells | Small lymphocytes |
| Bax | POLYCLONAL | Santa Cruz | 1:1000 | Positive/negative | >10% positive cells | Benign B lymphocytes |
| Bcl-XL | 2H12 | ZYMED | 1:10 | High/low | >10% positive cells | TMA controls |
| Mcl1 | POLYCLONAL | DAKO | 1:100 | High/low | >50% positive cells | Proliferating cells |
| Survivin | POLYCLONAL | RD Systems | 1:1500 | High/low | >10% positive cells | TMA controls |
| p65/RelA | F-6 (p65) | Santa Cruz | 1:2000 | Positive/negative | Nuclear expression | TMA controls |
| Caspase 3 | C92-605 | PharMingen | 1:25 | Positive/negative | >10% positive cells | TMA controls |
| Bcl-10 | 331.3 | Santa Cruz | 1:1000 | Positive/negative | >10% positive cells | Reactive lymphocytes |
| CD95 | GM30 | Novocastra | 1:50 | Positive/negative | >10% positive cells | Reactive lymphocytes |
| Oct-1 | 12F11 | Santa Cruz | 1:10 | Positive/negative | >10% positive cells | Reactive lymphocytes |
| Oct-2 | POLYCLONAL | Santa Cruz | 1:500 | Positive/negative | >10% positive cells | Reactive lymphocytes |
| Bob-1 | POLYCLONAL | Santa Cruz | 1:3000 | Positive/negative | >10% positive cells | Reactive lymphocytes |
| PU1 | G148-74 | PharMingen | 1:50 | Positive/negative | >10% positive cells | Benign B-lymphocytes |
| Pax-5 | POLYCLONAL | Santa Cruz | 1:200 | Positive/negative | >10% positive cells | CG (germinal center) B cells |
| MUM-1 | POLYCLONAL | Santa Cruz | 1:200 | High/low | >80% positive cells | Plasma cells |
| STAT3 | F-2 | Santa Cruz | 1:500 | Positive/negative | Nuclear expression | Reactive lymphocytes, macrophages |
| Bcl-6 | PG-B6p | DAKO | 1:10 | Positive/negative | >10% positive cells | CG (germinal center) B cells and B-cell lymphomas |
| CD38 | VS38 | DAKO | 1:25 | High/low | >80% positive cells | Plasma cells |
| CD138 | MI15 | DAKO | 1:50 | High/low | >80% positive cells | Plasma cells |
| CD5 | 4C7 | Novocastra | 1:50 | Positive/negative | >10% positive cells | Reactive lymphocytes |
| CD10 | 56C6 | Novocastra | 1:10 | Positive/negative | >10% positive cells | CG (germinal center) B cells |
| CD20 | L-26 | DAKO | 1:100 | Positive/negative | Any positive tumoral cell | Reactive lymphocytes |
| CD30 | 15B3 | Novocastra | 1:100 | Positive/negative | >10% positive cells | TMA controls |
| EMA | E29 | DAKO | 1:50 | Positive/negative | >10% positive cells | TMA controls |
| CD27 | 137B4 | Novocastra | 1:150 | Positive/negative | >10% positive cells | Reactive lymphocytes |
| Cyclin A | 6E6 | Novocastra | 1:100 | Positive/negative | >10% positive cells | Proliferating cells (G2/M) |
| Cyclin B1 | 7A9 | Novocastra | 1:25 | Positive/negative | >50% positive cells | Proliferating cells (G2/M) |
| Cyclin D1 | DCS-6 | DAKO | 1:100 | Positive/negative | Any positive tumoral cell | Macrophages and endothelial cells |
| Cyclin D3 | DCS-22 | Novocastra | 1:10 | Positive/negative | >50% positive cells | Proliferating cells |
| Cyclin E | 13A3 | Novocastra | 1:10 | High/low | >80% positive cells | TMA controls, proliferating cells |
| CDK1 | 1 | Transduction | 1:1500 | Positive/negative | >80% positive cells | TMA controls, proliferating cells |
| CDK2 | 8D4 | NeoMarkers | 1:500 | Positive/negative | >50% positive cells | TMA controls, proliferating cells |
| CDK6 | K6.83 | Chemicon | 1:10 | Positive/negative | >80% positive cells | TMA controls |
| P21 | EA10 | Oncogene | 1:50 | Positive/negative | >10% positive cells | Scattered GC cells |
| P16 | POLYCLONAL | Santa Cruz | 1:50 | High/low | >10% positive cells | Normal cells |
| P27 | 57 | Transduction | 1:1000 | High/low | >10% positive cells | Resting lymphoid cells |
| Ki67 | MIB1 | DAKO | 1:100 | High/low | >50% positive cells | Proliferating cells |
| SKP2 | 1G12E9 | ZYMED | 1:10 | Positive/negative | >80% positive cells | Proliferating cells |
| P53 | DO-7 | Novocastra | 1:50 | High/low | >80% positive cells | Scattered GC cells |
| Hdm2 | IF2 (Mdm2) | Oncogene | 1:10 | High/low | >10% positive cells | Macrophages, endothelial cells |
| Rb | G3-245 | BD PharMingen | 1:250 | High/low | >80% positive cells | Proliferating cells |
| Rb-P (Phospho-Rb) | sc-7986-R | Santa Cruz | 1:250 | High/low | >80% positive cells | Proliferating cells |
| PTEN | 28H6 | Novocastra | 1:50 | Positive/negative | Any positive tumoral cell | Normal cells |
| DP-1 | 1DP06 | NeoMarkers | 1:50 | Positive/negative | >80% positive cells | Proliferating cells |
| PKCβ | 28 | Serotec | 1:500 | Positive/negative | >10% positive cells | Plasma cells, endothelial cells |
| TOPO | Ki-S1 | DAKO | 1:400 | High/low | >50% positive cells | Proliferating cells |
| GST | 353-10 | DAKO | 1:150 | High/low | >50% positive cells | Proliferating cells |
| c-kit | POLYCLONAL | DAKO | 1:25 | High/low | >10% positive cells | Stromal cells |
| ALK | ALK1 | DAKO | 1:50 | High/low | >10% positive cells | TMA internal controls |
| CD3 | F7.2.38 | DAKO | 1:25 | Positive/negative | Any positive tumoral cell | Reactive lymphocytes |
DAKO, Glostrup, Denmark; Santa Cruz Biotechnology, Santa Cruz, CA; BD PharMingen, San Diego, CA; Novocastra, Newcastle, UK; Transduction Laboratories, Lexington, KY; Neomarkers, Fremont, CA; Chemicon, Temecula, CA; Oncogene Research Products, Darmstadt, Germany; Serotec, Oxford, UK.
As cytoplasmic STAT1, STAT3, and NFκB expression can generally be found in normal lymphoid cells and lymphomas, we have considered as positive cases only those showing distinct nuclear expression in the tumoral cells, thereby indicating the activated form of these proteins.9
Discrepancies between the two cylinders included for each case were resolved through a reviewed joint analysis of both cores. The same procedure was applied to discrepancies among pathologists.
The reactivity of most of the antibodies used here has been validated in previous studies.7
In situ detection of apoptosis and EBER in situ hybridization (ISH) were performed using standard procedures,7 using the appropriate controls. Apoptosis was detected using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Intergen Co., Oxford, UK). Epstein-Barr virus (EBV) was detected by ISH with fluorescein-conjugated Epstein-Barr Virus (EBER) PNA probe (DAKO, Glostrup, Denmark). EBV-positive cases were considered to be those showing EBER nuclear expression in a majority of the tumoral cells.
Validation of the Technique
The reproducibility of the results obtained was confirmed by comparing them with those from whole sections from 42 randomly selected cases that had been stained using the same procedures for a selection of markers including CD20, bcl-2, and bcl-6.
Statistical Study
The Pearson χ2 statistic and the Spearman correlation coefficient were used as appropriate to analyze relationships between the 52 markers studied.
Survival analyses were performed on all patients for whom follow-up information was available for a minimum of 24 months (approximately 70% of the overall series) and who had complete expression analysis data. HIV-positive patients9 were excluded from the outcome analysis. The final number of patients included in the survival analysis was 152, all of them treated with curative intention.
Failure was defined as the absence of complete remission, progression, or death attributable to the tumor. The series was divided into a training group of 103 cases for the purpose of building the predictor, and a second, smaller group of 49 cases, to validate the model.
Overall Survival (OS) and Failure-Free Survival (FFS) curves were plotted using the Kaplan-Meier method. Statistical significance of associations between individual variables and OS or FFS was determined using the log-rank test.
Cox’s univariate proportional hazard analysis was also performed independently for each variable. Results were validated by multiple testing and the random permutation test.
For multivariate analysis, the series was divided into a training group of 103 cases for the purpose of building the predictor, and a second, smaller group of 49 cases, to validate the model.
A logistic regression model was used to predict failure. Only variables identified in the univariate analysis associated with FFS with values of P < 0.2 and in which at least 5 cases were considered positive or negative were included. Highly variable components in the model were excluded, since they could have introduced uncertainty in predictions. For comparative purposes, multivariate models using step-up (forward) variable selection and other heuristic procedures were also fitted. The final model estimates values of the odds ratio (OR), 95% confidence interval (CI) and P for each variable. General applicability of the model was tested by leave-one-out cross-validation. The stability of the model was evaluated by influence statistics (DfBeta). Different predictor models were found, when using the leave-one-out cross-validation, but these showed only small variations in the weight of each marker, or selection of markers. Accuracy was also tested by the Receiver Operating Characteristic (ROC) curve, which allows the discriminating ability of the model to be estimated.
To demonstrate the predictive capacity of the model, patients were ranked according to this score and then divided into four equal groups, or quartiles. To validate the model overall, the specific weight or coefficient assigned to each gene, as determined in the preliminary group, was applied to calculate the outcome-predictor score in the validation group. Once the model had been validated, a final logistic regression model was fitted to the entire data, allowing adjustment of the coefficients. Statistical analyses were performed using the SPSS program and the tools at http://bioinfo.cnio.es/ for random permutation tests.
Results
The percentage of informative individual cores was 90.4%. As each TMA included 2 different core cylinders from each marker, the final percentage of missing expression data values was 12% (Table 2).
Table 2.
Expression of 51 of the 52 Markers Analyzed in the Entire DLBCL Series, Indicating Number of Positive/Total Analyzed Cases
| Protein | Positive cases | Percentage |
|---|---|---|
| Apoptosis | ||
| MBcl-2 | 122/224 | 54.46 |
| Bax | 194/215 | 90.23 |
| Bcl-XL | 73/188 | 38.83 |
| Mcl1 | 107/186 | 57.53 |
| Survivin | 70/217 | 32.26 |
| p65/RelA | 116/225 | 51.56 |
| Caspase 3 active | 17/194 | 8.77 |
| Bcl-10 | 75/188 | 39.89 |
| CD95 | 69/169 | 40.83 |
| TUNEL | 141/191 | 73.82 |
| Transcription factors | ||
| Oct-1 | 186/187 | 99.46 |
| Oct-2 | 189/192 | 98.44 |
| Bob-1 | 176/180 | 97.78 |
| PU1 | 11/194 | 5.67 |
| Pax-5 | 209/215 | 97.21 |
| MUM-1 | 113/206 | 54.85 |
| STAT3 | 23/224 | 10.27 |
| B-cell differentiation | ||
| Bcl-6 | 168/207 | 81.16 |
| CD38 | 73/204 | 35.78 |
| CD138 | 15/219 | 6.85 |
| CD5 | 53/188 | 28.19 |
| CD10 | 51/182 | 28.02 |
| CD20 | 224/231 | 96.97 |
| CD30 | 41/206 | 19.90 |
| EMA | 8/214 | 3.74 |
| CD27 | 30/209 | 14.35 |
| Cell cycle | ||
| Cyclin A | 107/202 | 52.97 |
| Cyclin B1 | 37/221 | 16.74 |
| Cyclin D1 | 0/235 | 0 |
| Cyclin D3 | 50/229 | 21.83 |
| Cyclin E | 25/214 | 11.68 |
| CDK1 | 40/179 | 22.35 |
| CDK2 | 151/198 | 76.26 |
| CDK6 | 111/174 | 63.79 |
| P21 | 20/226 | 8.85 |
| P16 | 166/212 | 78.30 |
| P27 | 78/216 | 36.11 |
| MIB1 | 131/210 | 63.38 |
| SKP2 | 26/214 | 12.15 |
| P53 | 37/222 | 16.66 |
| Hdm2 | 120/221 | 54.30 |
| Rb | 112/215 | 52.09 |
| Rb-P | 57/189 | 30.16 |
| Other | ||
| PTEN | 211/211 | 100 |
| DP-1 | 114/155 | 73.55 |
| PKCβ | 55/186 | 29.57 |
| TOPO | 186/207 | 89.85 |
| GST | 150/209 | 71.77 |
| c-kit | 60/213 | 28.17 |
| ALK | 1/213 | 0.47 |
| EBER | 20/221 | 9.05 |
CD3 was negative in all cases.
To check the reliability and accuracy of TMA for this measure of protein expression, TMA and quantitative whole tissue stainings were compared in a subset of 42 cases. Concordances of 100%, 91.1%, and 90% were obtained for CD20, bcl-2 and bcl-6, respectively, thus coinciding with the results of other NHL analysis studies.10,11
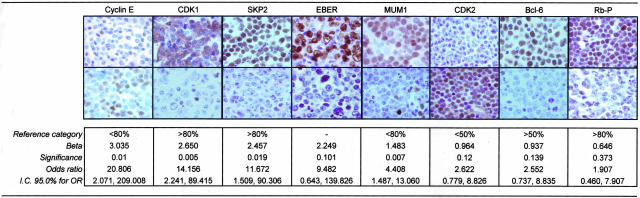
Results of the overall DLBCL series are summarized in Table 2. Figure 1 shows the expression of the markers found to predict failure after the multivariate analysis.
Figure 1.
Characteristics and variables included in the biological model for failure prediction in DLBCL. Representative immunohisto chemistry and in situ hybridzation results for the eight markers selected in the multivariate analysis. A positive and a negative tumor are shown for each marker. β represents the weight of each variable estimated from the multivariate analysis.
Correlation between Markers
The Pearson test revealed a large number of significant associations between the different markers analyzed. Full details of the correlation between markers are given in Supplementary Appendix 2 at http://bioinfo.cnio.es/data/DLBCL_TMA.
The most striking findings were as follows:
• Higher levels of expression of specific cyclins and CDKs were observed in varying numbers of this series: 11.7% (25 of 214) in the case of cyclin E, 52.9% (197 of 202) for cyclin A, 22.3% (40 of 179) in the case of CDK1 and 76.3% (151 of 198) for CDK2. There was a close relationship between proliferation and apoptosis, including the positive association observed between proliferation and apoptotic indices, and between the apoptotic index and different CDKs and cyclins.
• EBV presence was accompanied by changes in the expression of numerous proteins, including an increase in CDK1, cyclin B1, SKP2, p21, CD30, and a loss of BOB1, pax-5, and bcl-6.
• SKP2 expression, observed in 12.1% (26 of 214) of cases, was significantly associated with changes in numerous apoptosis and cell-cycle regulators, including a strongly positive correlation with CDK1, Rb, cyclin A, B1, D3, survivin, and a negative association with Bax and bcl-2. A significant relationship was also observed between SKP2 expression and the increased expression of Rb-P, CDK6, MDM2, p53, bcl-6, CD10, c-kit, EBER, NF-kB, caspase 3 active, MCL1, MIB1, and TUNEL.
• An unexpected finding was the association of c-kit expression with various cell cycle markers (increased p27, SKP2, CDK1, cyclin E, and Rb-P), apoptosis (loss of Bax and increase in bcl-XL, bcl-10, survivin), high PKCβ, and B-cell differentiation (elevated CD27, CD38, CD5, CD10).
• Finally, bcl-6 expression, detected in 81% (169 of 207) of cases, was associated with profound changes in molecules regulating cell cycle (high SKP2, CDK6, MDM2, Rb, Rb-P, and loss of p21), apoptosis (increase in bcl-xL and NF-kB), and B-cell differentiation (increase in Pax5, CD10, and Bob1 expression). Notably, it was found to be inversely correlated with EBV and epithelial membrane antigen (EMA). Another interesting finding was the existence of a group (47% of cases) that simultaneously expressed bcl-6 and MUM1, two proteins that normal lymphoid B cells do not express at the same time.
Correlation between Protein-RNA Expression and Outcome in DLBCL
To detect any possible selection bias, the 152 included patients (Table 3) were compared with those who had been excluded due to insufficient follow-up. Comparison of age, gender, clinical stage and IPI revealed no significant differences.
Table 3.
Clinical Characteristics of the 152 DLBCL Patients Included in the Outcome Analysis
| Age (mean, range) | 58.4 (5–96) | |
| Gender | Female | 47.6% |
| Male | 52.4% | |
| IPI | 0–1 | 41.8% |
| 2 | 27.1% | |
| 3 | 17.1% | |
| 4–5 | 14.1% | |
| Follow-up (median) | 60.9 | |
| Overall survival | 5-year cumulative survival | 59.8% |
| Failure | Cured versus fatal/refractory disease | 50.6%/49.4% |
| Failure-free survival | 5-year cumulative survival | 50.4% |
All 52 individual variables were analyzed using Kaplan-Meier plots and Cox proportional hazard models to determine whether the expression was significantly associated with changes in FFS (Table 4). Ten variables (cyclin E, CDK1, SKP2, bcl-6, p21, Oct-2, BOB1, EMA, Bax, bcl-2) were significantly correlated with FFS (P < 0.05) and nine showed a non-significant trend (P < 0.2). All of the significantly FFS-correlated variables, except Rb-P, were also associated with OS probability (P < 0.05) (data not shown). Furthermore, EBER, which showed a non-significant trend in the FFS analysis, was found to be associated with OS (P < 0.05) (data not shown). The result of the Cox proportional hazard analysis was then validated using multiple testing and random permutation tests (n = 1000).
Table 4.
Univariate Analysis for OS and FFS in the Current Series (n: 152 Patients) and Logistic Regression Model for Failure Prediction in the Training Set of Patients (n: 103)
| Protein | Reference category | Univariate analysis for FFS (Cox regression)
|
Multivariate analysis for failure protein, RNA-expression-based model and model integrating IPI; (logistic regression)
|
|||||
|---|---|---|---|---|---|---|---|---|
| P | RR | 95% CI
|
Beta in PEB model | Beta in PEB + IPI model | Difference between models | |||
| Lower | Higher | |||||||
| IPI | IPI (0–2) | 0.000 | 3.257 | 2.121 | 5.001 | 3.260 | ||
| cyclin E | <80% | 0.000 | 3.293 | 1.839 | 5.894 | 3.035 | 2.477 | 0.184 |
| CDK1 | >80% | 0.029 | 2.281 | 1.090 | 4.771 | 2.650 | 2.975 | −0.123 |
| SKP2 | >80% | 0.019 | 3.999 | 1.261 | 12.683 | 2.457 | 2.329 | 0.052 |
| EBER | − | 0.086 | 1.898 | 0.913 | 3.945 | 2.249 | 2.569 | −0.142 |
| MUM1 | <80% | 0.071 | 0.065 | 0.409 | 1.037 | 1.483 | 1.758 | −0.185 |
| CDK2 | <50% | 0.114 | 0.623 | 0.347 | 1.120 | 0.964 | 0.739 | 0.233 |
| Bcl-6 | >50% | 0.040 | 1.747 | 1.027 | 2.972 | 0.937 | 0.655 | 0.301 |
| Rb-P | >80% | 0.117 | 1.648 | 0.882 | 3.078 | 0.646 | 1.037 | −0.391 |
| p21 | − | 0.001 | 3.042 | 1.601 | 5.780 | |||
| cyclin B1 | >50% | 0.094 | 1.869 | 0.899 | 3.883 | |||
| cyclin A | >50% | >0.2 | ||||||
| MDM2 | − | 0.097 | 1.446 | 0.936 | 2.234 | |||
| Rb | >80% | 0.192 | 1.342 | 0.863 | 2.088 | |||
| CD38 | <80% | 0.188 | 1.369 | 0.858 | 2.184 | |||
| CD138 | <80% | 0.090 | 1.882 | 0.906 | 3.907 | |||
| Oct_2* | + | 0.015 | 4.270 | 1.325 | 13.757 | |||
| BOB1* | + | 0.003 | 8.836 | 2.074 | 37.657 | |||
| EMA* | − | 0.040 | 2.891 | 1.052 | 7.948 | |||
| CD95 | − | >0.2 | ||||||
| Bax | − | 0.037 | 3.428 | 1.080 | 10.879 | |||
| Bcl-2 | <50% | 0.015 | 1.740 | 1.114 | 2.719 | |||
, <5 values in one category; −, no data available.
PEB, protein-expression-based; RR, relative risk.
Specific weight (beta) of each variable for predicting failure in the protein and RNA-expression-based model, compared with values for model integrating the IPI. Differences were calculated as (beta1-beta2)/beta1.
Predicting Failure in DLBCL
Logistic regression analysis was used to find a DLBCL outcome predictor, making it possible to recognize which patients could be cured by the application of chemotherapeutic regimes. The group of 103 cases was used to build the predictor. Only variables identified in the univariate analysis associated with FFS with values of P < 0.2, and in which at least five cases were considered positive or negative, were included (19 variables, excluding EMA, Oct-2, BOB1). The final logistic regression model included the following markers: cyclin E, CDK1, SKP2, EBER, MUM1, CDK2, bcl-6, and Rb-P (Figure 1).
The predictor is a biological score, the probability of “failure” for one patient, which is calculated as
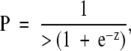 |
where
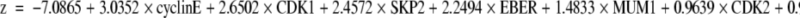 |
and where coefficients from the logistic model are used as weights for the corresponding markers.
The percentage of correct classification for this model, using the training series, was 78.64% (81.13% for predicting FFS and 76% for patients with treatment failure).
In a second step, patients were ranked according to their protein-expression-based score (0 to 1) and divided into four different quartiles, according to their specific risk. Stratifying patients according to these quartiles, 92.3% of patients beneath the 25 percentile were accurately predicted as “failure-free” by the score, and 96.2% of the patients above the 75 percentile were correctly predicted as belonging to the group of “fatal or refractory disease”. Between the 25 and 75 percentiles the accuracy of prediction fell below 90% for both categories (64% in the second quartile and 53.8% in the third quartile). Thus, when assigning each patient a specific risk, the capacity for predicting the upper and lower quartile is much higher than for patients with intermediate quartiles.
Validating the Biological Score for Failure in DLBCL
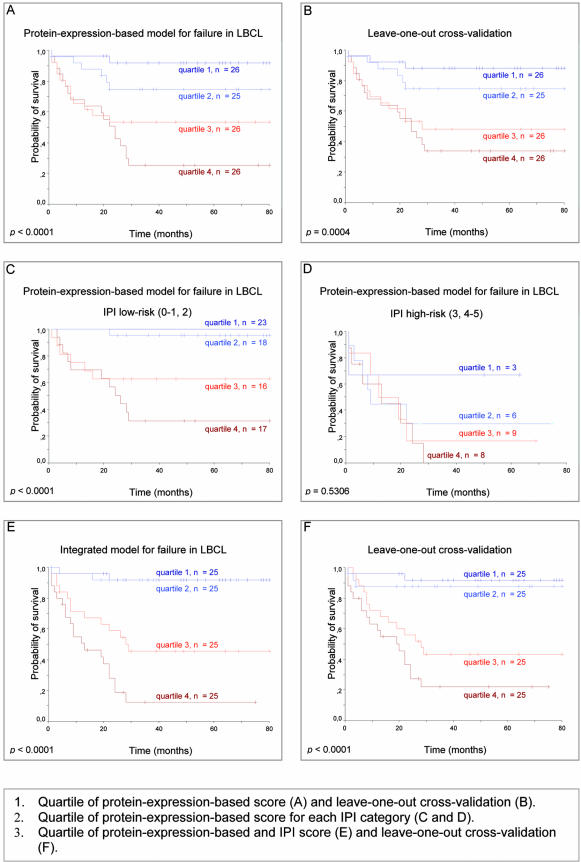
A Kaplan-Meier survival analysis, classifying patients according to the quartile of assigned probability, confirmed that the patients predicted to be cured had significantly improved long-term survival compared with those predicted to have fatal/refractory disease (5-year OS: 91.97% below the 25 percentile, vs. 25.45% above the 75 percentile; P < 0.0001) (Figure 2A).
Figure 2.
Protein-expression-based model for failure prediction in DLBCL. Kaplan-Meier estimation of OS according to the assigned probability for each model in the training set of patients. Quartiles of protein-expression-based score (A) and leave-one-out cross-validation (B). Quartiles of protein-expression-based score for each IPI category (C) and leave-one-out cross-validation (D). Quartiles of protein-expression-based and IPI score (E) and leave-one-out cross-validation (F).
The prediction accuracy of the score was then assessed using a leave-one-out cross-validation testing method, withholding one case and using the remaining set of tumors to train the model, predicting the “failure” probability of the withheld case. The process was repeated until all 103 samples had been predicted in turn. The results confirmed, with minor differences, the FFS and OS predictive capacity of the biological score (Figure 2B). Different predictor models were found, when using the leave-one-out cross-validation, showing only small variations in the weight of each marker, or selection of markers.
Although the majority of the patients of this series received anthracycline-based chemotherapy, 12 of 103 (11.6%) patients were treated with different drugs. To examine whether the biological model was independent of the treatment regimes used, treatment was included as a new variable. The specific weight of each variable in the model remained similar (3.064 × cyclin E + 2.499 × CDK1 + 2.364 × SKP2 + 2.264 × EBER + 1.391 × MUM1 + 1.088 × CDK2 + 0.898 × bcl-6 + 0.828 × Rb-P). Moreover, the correct classification percentage in this new model with the variable “treatment” decreased imperceptibly (77.2% for the overall prediction). Correct prediction percentage in the different quartiles was 92% (quartile 1 for failure-free) vs. 96.2% (quartile 4 for failure). These percentages are very similar to those obtained previously.
Integration of Protein-Expression-Based Score and IPI
This biological score yielded a 13.616-fold odds ratio (OR) [95% CI (5.288, 35.063), P < 0.0001] for failure of treatment (percentile 50). IPI (low risk versus high risk), the standard clinical score for predicting the outcome in DLBCL,5 in this series yielded a 10.151-fold OR [95% CI (3.159, 32.616), P < 0.0001] for failure. A multivariate analysis including both the IPI and the protein-expression-based score showed that the significance of the biological score for failure [percentile 50; OR = 18.983; 95% CI (5.988, 60.180); P < 0.0001] seemed to be superior to and independent of the IPI [OR = 15.359; 95% CI (3.672, 64.244); P < 0.0001].
To determine whether the information contained in the protein and RNA-expression-based model was the same as or additional to the variables included in the IPI, patients were classified into low-risk (IPI: 0–2) and high-risk groups (IPI: 3–5), and then the protein-expression-based score quartiles were used in both groups. Low-risk IPI patients were accurately stratified by the protein-expression-based score into groups with a failure probability of 95.24% (quartile 4), 81.89% (quartiles 3 and 2) and 31.59% (quartile 1), P < 0.00001. High-risk IPI patients were also discriminated into two main groups using the protein-expression-based score, although the difference was not significant. These results suggest that an integrated use of the IPI and the protein-expression-based score could improve the predictive capacity of the model (Figure 2, C and D).
The joint predictive capacity of the protein-expression-based score and IPI was analyzed in a multivariate model. The specific weight of each component of the biological score in this new model remained quite similar (Table 4), confirming that the biological and clinical scores contain at least partially independent information. The predictive capacity of the model incorporating the IPI and the variables integrated in this biological score was slightly higher than that based purely on the protein and RNA- expression-based model, with 83% overall correct classification of failure (92% for quartile 1 and 96% for quartile 4).
This was correlated with a better discrimination of patients with different outcomes. Thus, patients allocated above the 50 percentile of the integrated score had 91.73% 5-year OS versus 29.71% for patients predicted for “failure” (Figure 2E).
Blind Test for Validation of the Predictor
The leave-one-out cross-validation confirmed the high predictive capacity of this integrated model, with a probability of failure in each respective quartile of 12%, 24%, 68%, and 88%, reflected in the overall survival probability (Figure 2F). The discriminating ability of this model was better than that of the protein and RNA-expressed-based model [ROC curve area: 0.901; P < 0.0001, 95% CI (0.840, 0.961)].
As this evaluation was based on the same training set of patients from which the predictive model was derived, we decided to estimate the accuracy of the classifier with an additional cohort of 49 patients who had not previously been included. In this independent series, the failure prediction and the outcome were evaluated by the model integrating the 8 markers and IPI, using the threshold from the training set of patients. The immunostaining and evaluation of these tumors were performed independently of the previous cases. The predictive capacities of the validation and preliminary group were comparable with respect to the assigned score for each patient by the model (76.9% and 83.3% of correct classification into quartiles 1 and 4, P < 0.001). Furthermore, values for 5-year OS were closely related with the assigned failure probability for each patient (5-year OS: 100%, 81.48%, 75%, and 25% for each quartile of the score; P < 0.0001).
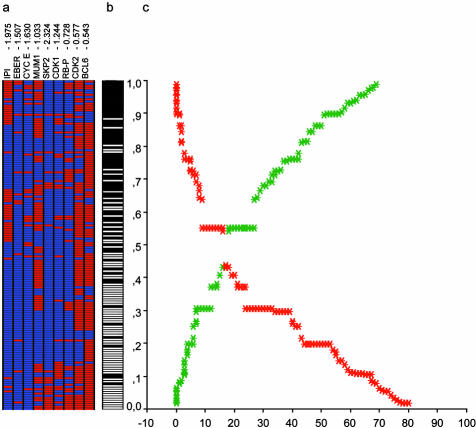
Once the model had been validated, a final model with the 8 biological markers and IPI was fitted to the entire data (training + validation series). Finally, the biological-IPI score allowed assignment of a case-specific probability of failure, as can be observed in Figure 3.
Figure 3.
Final biological and clinical predictor model. a: Tree-view representation of the eight markers and IPI. Each column represents a marker, while each row corresponds to a patient, ordered according to the assigned failure probability. Specific weight of each marker is included at the top of each column. b: Real status of each patient (failure, black vs. maintained complete response, white). c: Graphic representation of the relation between the assigned probability and the real status. The graphic represents the accuracy of the predictor model. If the probability assigned to each patient (y axis) is less than 0.5, the model classifies the case into the group of maintained response. If the probability is greater than 0.5, the system classifies the case as a failure. The curves represent the number of patients erroneously classified as failure (in red), and those cases erroneously predicted to maintain a complete response (in green). Eventually, a threshold for each curve of cumulative error could be chosen to select a group of patients with a high probability of failure or of maintained complete remission.
Discussion
DLBCL seems to be the result of deregulation of multiple genes involved in the control of cell cycle, apoptosis, cell growth, DNA repair, ubiquitin degradation, and other processes. Particularly striking is the existence of multiple concurrent abnormalities in the genes and pathways in the control of cell cycle and apoptosis. Subtle alterations in this exquisitely regulated balance between cell proliferation and apoptosis seem to contribute critically to DLBCL pathogenesis.
Some of the observed changes affect the large majority of cases analyzed here, such as the expression of bcl-6. The hypothetical relevance of bcl-6 in DLBCL pathogenesis is underlined by the increasing number of bcl-6 targets that are being described in B cells, and for its capacity to contribute to oncogenesis by rendering cells unresponsive to antiproliferative signals from the p19(ARF)-p53 pathway, as demonstrated by Shvarts et al.12 In this respect, it is noteworthy that in this series bcl-6 expression appears to be associated with down-regulation of p21 and overexpression of MDM2. The potential role of bcl-6 as a promoter of cell-cycle progression beyond the G1/S restriction point is suggested by the existence of an additional significant relationship with increased phosphorylated Rb. Our data also confirm the prognostic significance of bcl-6 expression in DLBCL, as previously pointed out, when taking into account bcl-6 mRNA expression levels.13
According to the results of this study, Skp2 expression, which increased in one-fifth of the cases analyzed, is associated with many changes in apoptosis and cell-cycle regulators. Protein degradation throughout the ubiquitin pathway thus seems to be indicated as a potential contributory factor in the deregulation of proliferation and apoptosis in DLBCL.14,15 In addition to the confirmed role of Skp2 for inducing the degradation of p27 and Cdk2-unbound cyclin E, an accelerated degradation of unknown additional substrates is likely to play a role in oncogenic events mediated by Skp2.15
Cyclin E overexpression is highlighted by the uni- and multivariate analyses as a clinically highly relevant adverse prognostic marker, thus confirming previous observations in specific lymphoma types16,17 and other tumors.18 A possible explanation for these findings is provided by the recent demonstration that overexpression of cyclin E leads to increased chromosome instability and impaired S-phase progression.19
In general, the results of the univariate analysis confirm those previously published concerning single markers, such as the case for bcl-2 or others.20,21 Nevertheless, some of the significant markers in the univariate analysis, can prove not significant in the multivariate analysis.
Results of this study, not based on previous hypotheses of DLBCL subclassification, are difficult to match with the three DLBCL subgroups defined by Rosenwald et al4: germinal-center B-cell-like, activated B-cell-like, and type 3 diffuse large B-cell lymphoma. Instead, it seems that the tumors accumulate alterations in critical pathways stochastically, leading to the increased proliferation and loss of apoptosis observed here. The existence of a large group of double bcl-6+ MUM1+ cases demonstrates that the mutual exclusion of these markers, as observed in reactive germinal centers, is not preserved in DLBCLs.22 Tumoral cells probably take advantage of the simultaneous expression of both proteins.
The technique used here is based on large-scale analysis of protein expression, detected by immunohistochemistry. The use of tissue microarrays is limited by the relatively small number of markers chosen (52 in this case), although it has the advantage of using protein profiling, which probably reflects more closely the characteristics of the tumoral cells than does RNA detection.
The integration of these markers into a single model allows the assignment of a specific probability of failure to each patient, according to the biological and clinical characteristics of each case. This information could eventually be used for individualized treatments, in which patients are stratified into therapeutic groups. A clinical application of this and other studies should, nevertheless, first fulfill the necessity of demonstrating the reproducibility of immunohistochemistry techniques among different groups, which would be facilitated by the application of automated systems for scoring immunohistochemical expression.
Acknowledgments
We thank Teresa Flores, M.D., from the Hospital Clínico, Salamanca, Carlos Perez-Seoane, M.D., from Hospital Reina Sofía, Córdoba, and Manuel Medina, M.D., from Hospital de la Merced, Osuna, for their kind help. We also extend our appreciation to the staff of the CNIO Tumor Bank for their efficient provision of tumor samples.
Footnotes
Address reprint requests to Dr. Miguel A. Piris, Programa de Patología Molecular, Centro Nacional de Investigaciones Oncológicas, c/Sinesio Delgado, 4–12. 28029 Madrid, Spain. E-mail: mapiris@cnio.es.
Supported by grants from the Fondo de Investigaciones Sanitarias (FIS 98/993, 01/0035–01, 02/0201), Ministerio de Sanidad y Consumo; from the Ministerio de Ciencia y Tecnología (SAF2001–0060); and from Xunta de Galicia (XUGA20810B96), Spain. A.I. Sáez was supported by a grant from the Ministerio de Sanidad y Consumo, Spain. F. Camacho was supported by a grant from the Madrid City Council and the CNIO.
References
- The Non-Hodgkin’s Lymphoma Classification Project: A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin’s lymphoma. Blood. 1997;89:3909–3918. [PubMed] [Google Scholar]
- Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- Sanchez-Beato M, Saez AI, Navas IC, Algara P, Sol Mateo M, Villuendas R, Camacho F, Sanchez-Aguilera A, Sanchez E, Piris MA. Overall survival in aggressive B-cell lymphomas is dependent on the accumulation of alterations in p53, p16, and p27. Am J Pathol. 2001;159:205–213. doi: 10.1016/S0002-9440(10)61686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A, Staudt LM. Clinical translation of gene expression profiling in lymphomas and leukemias. Semin Oncol. 2002;29:258–263. doi: 10.1053/sonc.2002.32901. [DOI] [PubMed] [Google Scholar]
- The International Non-Hodgkin’s Lymphoma Prognostic Factors Project: A predictive model for aggressive non-Hodgkin’s lymphoma. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- Torhorst JBC, Kononen J, Haas P, Zuber M, Kochli OR, Mross F, Dieterich H, Moch H, Mihatsch M, Kallioniemi OP, Sauter G. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol. 2001;159:2249–2256. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF, Camacho FI, Morente M, Fraga M, Montalban C, Alavaro T, Bellas C, Castano A, Diez A, Flores T, Martin C, Martinez MA, Mazorra F, Menarguez J, Mestre MJ, Mollejo M, Saez AI, Sanchez L, Piris MA, Spanish Hodgkin Lymphoma Study Group Hodgkin’s and Reed-Sternberg cells harbor alterations in the major tumor suppressor pathways and cell-cycle checkpoints: analyses using tissue-microarrays. Blood. 2003;101:681–689. doi: 10.1182/blood-2002-04-1128. [DOI] [PubMed] [Google Scholar]
- Jaffe ES, Harris NL, Stein H, Vardiman JW. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. Lyon: IARC Press; World Health Organization Classification of Tumours. 2001 [Google Scholar]
- Hinz MLP, Mathas S, Krappmann D, Dorken B, Scheidereit C. Constitutive NF-κB maintains high expression of a characteristic gene network, including CD40, CD86, and a set of antiapoptotic genes in Hodgkin/Reed-Sternberg cells. Blood. 2001;97:2798–2807. doi: 10.1182/blood.v97.9.2798. [DOI] [PubMed] [Google Scholar]
- Sáez AI AM, Romero C, Rodríguez S, Cigudosa JC, Pérez-Rosado A, Fernández I, Sánchez-Beato M, Sánchez E, Mollejo M, Piris MA. Development of a real-time RT-PCR assay for C-MYC expression that allows the identification of a subset of C-MYC+ diffuse large B-cell lymphoma. Lab Invest. 2003;83:143–152. doi: 10.1097/01.lab.0000057000.41585.fd. [DOI] [PubMed] [Google Scholar]
- Hedvat CV HA, Chaganti RS, Chen B, Qin J, Filippa DA, Nimer SD, Teruya-Feldstein J. Application of tissue microarray technology to the study of non-Hodgkin’s and Hodgkin’s lymphoma. Hum Pathol. 2002;33:368–374. doi: 10.1053/hupa.2002.127438. [DOI] [PubMed] [Google Scholar]
- Shvarts A, Brummelkamp TR, Scheeren F, Koh E, Daley GQ, Spits H, Bernards R. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 2002;16:681–686. doi: 10.1101/gad.929302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Chiarle R, Fan Y, Piva R, Boggino H, Skolnik J, Novero D, Palestro G, De Wolf-Peeters C, Chilosi M, Pagano M, Inghirami G. S-phase kinase-associated protein 2 expression in non-Hodgkin’s lymphoma inversely correlates with p27 expression and defines cells in S phase. Am J Pathol. 2002;160:1457–1466. doi: 10.1016/S0002-9440(10)62571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, Pagano M. Role of the F-box protein Skp2 in lymphomagenesis. Proc Natl Acad Sci USA. 2001;98:2515–2520. doi: 10.1073/pnas.041475098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreri AJ, Ponzoni M, Pruneri G, Freschi M, Rossi R, Dell’Oro S, Baldini L, Buffa R, Carboni N, Villa E, Viale G. Immunoreactivity for p27(KIP1) and cyclin E is an independent predictor of survival in primary gastric non-Hodgkin’s lymphoma. Int J Cancer. 2001;94:599–604. doi: 10.1002/ijc.1509. [DOI] [PubMed] [Google Scholar]
- Erlanson M, Portin C, Linderholm B, Lindh J, Roos G, Landberg G. Expression of cyclin E and the cyclin-dependent kinase inhibitor p27 in malignant lymphomas-prognostic implications. Blood. 1998;92:770–777. [PubMed] [Google Scholar]
- Muller-Tidow C, Metzger R, Kugler K, Diederichs S, Idos G, Thomas M, Dockhorn-Dworniczak B, Schneider PM, Koeffler HP, Berdel WE, Serve H. Cyclin E is the only cyclin-dependent kinase 2-associated cyclin that predicts metastasis and survival in early stage non-small cell lung cancer. Cancer Res. 2001;61:647–653. [PubMed] [Google Scholar]
- Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- Sanchez E, Chacon I, Plaza MM, Munoz E, Cruz MA, Martinez B, Lopez L, Martinez-Montero JC, Orradre JL, Saez AI, Garcia JF, Piris MA. Clinical outcome in diffuse large B-cell lymphoma is dependent on the relationship between different cell-cycle regulator proteins. J Clin Oncol. 1998;16:1931–1939. doi: 10.1200/JCO.1998.16.5.1931. [DOI] [PubMed] [Google Scholar]
- Gascoyne RD, Adomat SA, Krajewski S, Krajewska M, Horsman DE, Tolcher AW, O’Reilly SE, Hoskins P, Coldman AJ, Reed JC, Connors JM. Prognostic significance of Bcl-2 protein expression and Bcl-2 gene rearrangement in diffuse aggressive non-Hodgkin’s lymphoma. Blood. 1997;90:244–251. [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Larocca LM, Capello D, Pierconti F, Canzonieri V, Tirelli U, Dalla-Favera R, Gaidano G. Expression profile of MUM1/IRF4, BCL-6, and CD138/syndecan-1 defines novel histogenetic subsets of human immunodeficiency virus-related lymphomas. Blood. 2001;97:744–751. doi: 10.1182/blood.v97.3.744. [DOI] [PubMed] [Google Scholar]