Abstract
Recently, there have been a number of studies demonstrating that overexpression of molecules in skeletal muscle can inhibit or ameliorate aspects of muscular dystrophy in the mdx mouse, a model for Duchenne muscular dystrophy. Several such studies involve molecules that increase the expression of dystroglycan, an important component of the dystrophin-glycoprotein complex. To test whether dystroglycan itself inhibits muscular dystrophy in mdx mice, we created dystroglycan transgenic mdx mice (DG/mdx). The α and β chains of dystroglycan were highly overexpressed along the sarcolemmal membrane in most DG/mdx muscles. Increased dystroglycan expression, however, did not correlate with increased expression of utrophin or sarcoglycans, but rather caused their decreased expression. In addition, the percentage of centrally located myofiber nuclei and the level of serum creatine kinase activity were not decreased in DG/mdx mice relative to mdx animals. Therefore, dystroglycan overexpression does not cause the concomitant overexpression of a utrophin-glycoprotein complex in mdx muscles and has no effect on the development of muscle pathology associated with muscular dystrophy.
Duchenne muscular dystrophy (DMD) is an X-linked myopathy that results from the loss of dystrophin protein expression.1 The mdx mouse is a commonly used model for studies on the pathogenesis of DMD and for the testing of potential therapies. In the past few years, studies have suggested that overexpression of a number of muscle molecules can inhibit or ameliorate the development of muscle pathology in the mdx mouse.2 These include dystrophin, utrophin, the cytotoxic T cell (CT) GalNAc transferase, neuronal nitric oxide synthase, ADAM12, calpastatin, and IGF1.3–9 In addition, integrin α7 ameliorates muscular dystrophy in utrn-/-mdx mice, an agrin minigene inhibits muscular dystrophy in dy/dy mice, and antibody blocking of myostatin inhibits muscular dystrophy in mdx mice.10–12 While all of this work is exciting, the plethora of molecules found to have an effect on muscular dystrophy makes one wonder about the specificity of such effects as well as whether they share a common mechanism.
If these models do share a common mechanism, it is likely to involve utrophin. Utrophin is the only known paralog of dystrophin in skeletal muscle, and is very similar to dystrophin at the amino acid level.1 Thus, utrophin is likely to be the only protein that can compensate for the loss of dystrophin by directly replacing its functional properties. Like dystrophin, utrophin can link filamentous actin to the sarcolemmal membrane.13 Unlike dystrophin, however, utrophin is normally confined to the neuromuscular junction in skeletal muscles.14 This is also true in mdx mice, though some increased utrophin expression can occur in regenerating fibers.15,16 Thus, utrophin is unable, at least in part, to compensate for the loss of dystrophin because its subcellular distribution prevents it from doing so. This could be due to local transcription of the utrophin gene from synaptic nuclei, increased stability of utrophin mRNA in synaptic regions, or increased binding of utrophin protein to synaptic membrane proteins such as dystroglycan. Unlike utrophin, dystroglycan is expressed along the entire sarcolemmal membrane, including the neuromuscular junction, in skeletal muscle. Therefore, dystroglycan is likely to contribute to the synaptic localization of utrophin through preferential binding interactions that occur only in the synaptic membrane.
Synaptic dystroglycan-utrophin binding may be defined by synaptic posttranslational modifications of dystroglycan. One such modification is glycosylation with the cytotoxic T cell carbohydrate antigen.17 The CT carbohydrate antigen is made by the CT GalNAc transferase (or Galgt2).18 Expression of both the CT carbohydrate antigen and Galgt2 is confined to the neuromuscular junction in skeletal muscle.19,20 Transgenic overexpression of the Galgt2 in skeletal muscle increases glycosylation of dystroglycan with the CT antigen and concomitantly increases the extrasynaptic expression of utrophin.20 This is the case in both wild-type mice and in mdx mice.5,20 In addition, Galgt2 transgenic mdx mice have a dramatic inhibition in the development of muscle pathology associated with muscular dystrophy,5 and this is similar to Davies and colleagues findings using transgenic overexpression of utrophin.4 Therefore, inhibition of muscular dystrophy by Galgt2 may result from glycosylation of dystroglycan with the CT antigen, which could induce a conformational change or subsequent posttranslational modification in dystroglycan that increases utrophin binding.
An alternative hypothesis would be that overexpression of Galgt2 in mdx animals simply increases the expression of dystroglycan protein along the muscle membrane and that this would inhibit muscular dystrophy irrespective of dystroglycan glycosylation with the CT antigen. Indeed, in vitro studies using recombinant proteins suggest that there is no preference between dystrophin and utrophin for binding to dystroglycan.21 If this is the case, then overexpression of dystroglycan protein in mdx mice may increase utrophin protein expression along myofibers and thereby inhibit muscular dystrophy. Here, we show that this is not the case. In fact, dystroglycan overexpression decreases, rather than increases, utrophin protein expression in mdx mice, and dystroglycan expression does not inhibit muscular dystrophy.
Materials and Methods
Materials
Monoclonal antibodies to α-sarcoglycan (Ad1/20A6), β-sarcoglycan (βSarc1/5B1), β-dystroglycan, (43DAG1/8D5), utrophin (DRP3/20C5), and dystrophin (Dy4/6D3) were obtained from Nova Castra (Newcastle on Tyne, UK). A monoclonal antibody to laminin α2 was purchased from Alexis Biochemicals (San Diego, CA). A monoclonal antibody to α-dystroglycan (IIH6) was a generous gift from Kevin Campbell (HHMI, University of Iowa). An affinity purified anti-peptide antiserum to α-dystroglycan was a generous gift from Stephan Kroger (University of Mainz). A hybridoma producing antibody against the CT antigen (CT2) was a gift from Leo Lefrancois (University of Connecticut). This antibody was produced and purified in our laboratory. Polyclonal antiserum to laminin α4 and laminin α5 was a gift from Joshua Sanes (Washington University). Polyclonal antiserum to actin was obtained from Sigma (St. Louis, MO). All secondary antibodies were purchased from Boehringer Mannheim (Indianapolis, IN) or Zymed (South San Francisco, CA).
Transgenic Mice
Transgenic mice made to overexpress a dystroglycan cDNA (DG) in their skeletal muscles using the human α skeletal actin promoter have been previously described.22 Male DG mice were mated to mdx/mdx females, and transgenic (DG/mdx) and non-transgenic (mdx) male littermates were analyzed. DG and wild-type male littermates were also analyzed in some instances. In all cases, 6- to 8- week old adult animals were used. Genotyping was done by PCR using primers to the transgenic vector and to the dystroglycan cDNA.22 mdx genotypes were confirmed by the absence of dystrophin protein on Western blots. Expression of transgenic dystroglycan protein was also confirmed by Western blot. DG lines 3517 and 3218 were used, with similar results. Both lines of mice expressed transgenic protein in the gastrocnemius, tibialis anterior, diaphragm, quadriceps femoris (vastus intermedius), triceps brachii, trapezius, and gluteus maximus muscles, but did not express in brain, spinal cord, kidney, heart, lung, pancreas, liver, lymph node, bladder, testes, intestine, stomach, thymus, or spleen.22 All results shown were obtained by comparing DG (3218)/mdx mice and mdx littermates.
Western Blotting
For analysis of total muscle proteins by Western blot, protein was extracted from gastrocnemius, quadriceps, diaphragm, gluteus, and trapezius muscle by homogenization in buffer containing SDS, UREA, and DTT as previously described.20 40 μg of protein was loaded per lane (80 μg for utrophin). Antibody binding was visualized by the binding of the appropriate secondary antibody conjugated to horseradish peroxidase, followed by chemiluminescence detection using the ECL detection method. We could not immunoblot mouse protein extracts with the anti-peptide antibody to α-dystroglycan, as it stains but does not blot mouse dystroglycan, as has been previously reported.23 For CT2, antibody binding was visualized by the binding of the goat anti-mouse IgM conjugated to alkaline phosphatase, followed by development in 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium. Horseradish peroxidase-labeled blots were stripped and re-probed with an antiserum to actin to confirm even protein loading and transfer. Blots were scanned and quantitated as previously described.5
Histology
Skeletal muscles from 6- to 8-week-old animals were dissected and snap-frozen in liquid nitrogen-cooled isopentane. Frozen muscles were sectioned on a cryostat at 8 μm thickness. Hematoxylin and eosin staining was done as previously described.20 Quantification of centrally located myofiber nuclei and measurement of myofiber diameters was done using hematoxylin and eosin-stained sections, as previously described.20 In these experiments, significant dystrophy was seen in the diaphragm muscle in mdx and DG/mdx mice, and this is normally not found in adolescent adults.24 Levels of dystrophy for other muscles, including the gastrocnemius, quadriceps, trapezius, gluteus, and triceps, were consistent with previous findings.5 Immunofluorescence-based staining was also done using snap-frozen 8-μm cross-sections, as previously described.20 Each example shown in Figure 2 is indicative of results from three independent experiments. Results similar to those seen for α- and β- sarcoglycan were also seen with antibodies to γ-sarcoglycan, δ-sarcoglycan, and dystrobrevin (data not shown). Mouse monoclonal antibodies were pre-complexed to secondary antibodies before staining to lower background binding of antibodies to sections. Some sections were treated with reagents from the Histomouse kit (Zymed) to block endogenous IgG in mouse sections. Some sections in each experiment were also stained with secondary antibodies alone to verify lack of significant background staining from these reagents. Immunofluorescence was visualized on a Nikon E800 microscope using fluorescein optics. Positive staining for utrophin, α-sarcoglycan, β-sarcoglycan, and CT antigen in mdx mice were confirmed by their presence at the neuromuscular junction.15,19 Neuromuscular junctions were identified by co-labeling sections with 50 nmol/L rhodamine-α-bungarotoxin (Molecular Probes, Eugene, OR), which were then visualized using rhodamine optics. All comparisons of staining for particular antibodies were visualized using identical exposure times.
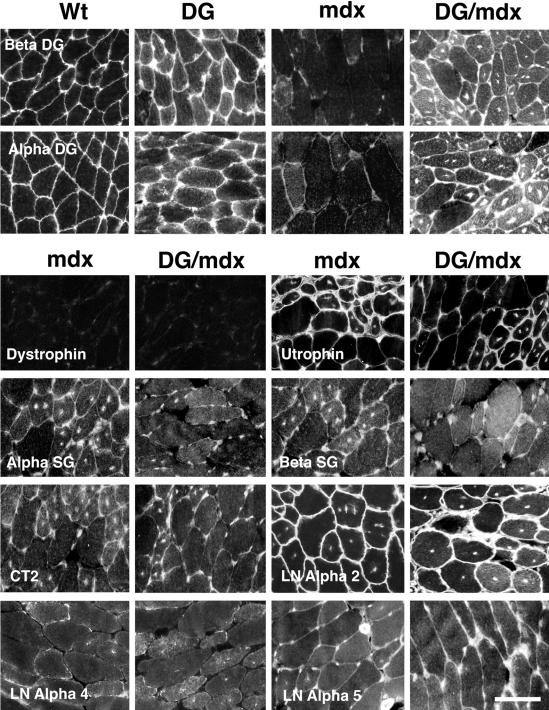
Figure 2.
Immunostaining of dystroglycan and associated proteins in dystroglycan transgenic mdx muscles. α- and β-Dystroglycan were highly overexpressed in most DG/mdx muscles compared to mdx muscle and were also overexpressed in DG muscles compared to wild-type (Wt). Expression was particularly high in some regenerating DG/mdx muscle fibers. No increase in expression was observed for utrophin, α-sarcoglycan, β-sarcoglycan, the CT carbohydrate antigen (CT2), laminin α2, laminin α4, or laminin α5. Scale bar, 40 μm.
Serum Creatine Kinase Activity Assays
Mice were anesthetized with metofane vapor. Tails were nicked with a razor and approximately 200 μl of blood was collected. Blood was allowed to clot for 1 hour at 37°C. Fibrin clot plus cells were centrifuged at 1500 × g for 3 minutes and serum was collected and analyzed without freezing. Creatine kinase activity assays were done using an enzyme-coupled assay reagent from Sigma (CK10) according to the manufacturer’s instructions. Absorbance at 340 nm was measured every 30 seconds for 4 minutes at 25°C to calculate enzyme activity.
Statistics
Determination of significance was done using a paired Student’s t-test in all cases.
Results
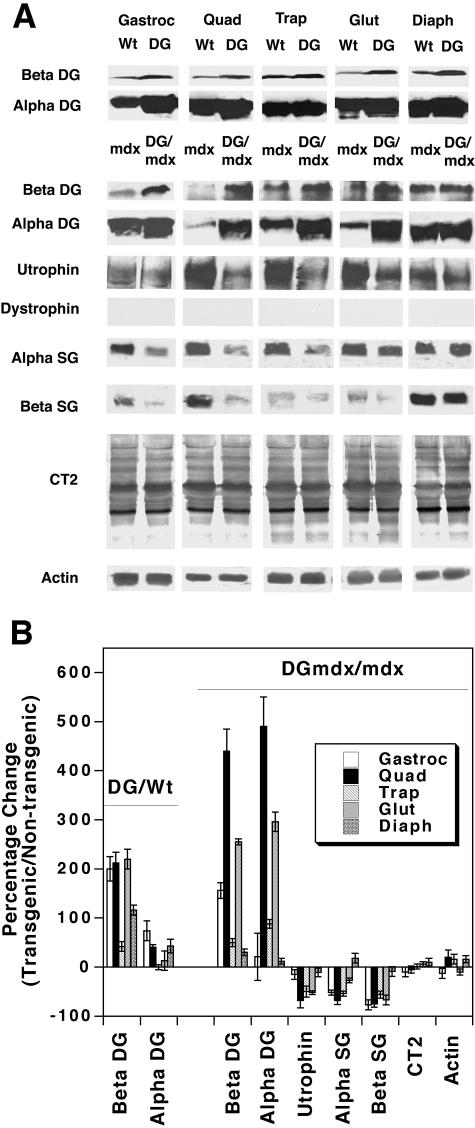
We analyzed protein expression in wild-type (Wt), dystroglycan transgenic (DG), mdx, and dystroglycan transgenic mdx (DG/mdx) muscles to determine the extent of overexpression of dystroglycan and other dystrophin-associated glycoproteins (Figure 1, A and B). β-Dystroglycan was overexpressed in DG muscle (compared to Wt muscle) and in DG/mdx muscle (compared to mdx muscle). In the five muscles we examined, β-dystroglycan was overexpressed by an average of 160 ± 34% (DG versus wild-type) and 180 ± 74% (DG/mdx versus mdx). IIH6, a ligand-blocking carbohydrate-specific monoclonal antibody,25 was used to assess α-dystroglycan expression. α-Dystroglycan expression was increased by 34 ± 12%, on average, in DG muscles (compared with wild-type) and by 181 ± 92% in DG/mdx muscles (compared to mdx). The higher degree of α-dystroglycan overexpression in the mdx background was due to lowered α-dystroglycan expression in mdx muscles compared to wild-type muscles. In a wild-type background, transgenic overexpression of α-dystroglycan, on average, was not nearly as high as it was for β-dystroglycan. This was also true in one mdx muscle, the gastrocnemius. There, α-dystroglycan was overexpressed by 21 ± 48%, while β-dystroglycan was increased by 156 ± 15%. By contrast, the degree of overexpression of α- and β-dystroglycan in other mdx muscles (trapezius, quadriceps, gluteus, and diaphragm) was similar, with a correlation coefficient (R) of 0.94.
Figure 1.
Expression of dystroglycan and associated proteins in dystroglycan transgenic mdx muscles. A: Immunoblots. Transgenic overexpression of dystroglycan in mdx mice (DG/mdx) causes a large increase in the expression of β-dystroglycan (Beta DG, 43 kd) compared to mdx mice, much as occurs in non-mdx transgenic mice (DG) when compared to wild-type mice. α-Dystroglycan (Alpha DG, 160 kd) was also overexpressed in most DG muscles (compared to wild-type) and in most, but not all, DG/mdx muscles (compared to mdx). IIH6, a carbohydrate-specific antibody that recognizes native α-dystroglycan protein was used to determine α-dystroglycan expression. Levels of utrophin (400 kd), α-sarcoglycan (Alpha SG, 50 kd), β-sarcoglycan (Beta SG, 43 kd) were generally reduced in DG/mdx muscles relative to mdx muscles. Levels of the CT carbohydrate antigen (CT2, multiple bands) and actin (50 kd) were unchanged. B: Quantitation of Western blots by densitometric scanning. Errors are SD for n = 3 experiments.
We next determined whether overexpression of dystroglycan would alter the expression of utrophin, α-sarcoglycan, or β-sarcoglycan (Figure 1, A and B). These are proteins that could interact with dystroglycan in a glycoprotein complex. As expected, no expression of dystrophin was evident, as mdx muscles do not make dystrophin protein. In the five muscles examined, utrophin protein levels were reduced on average by 40 ± 11%, α-sarcoglycan by 36 ± 15%, and β-sarcoglycan by 56 ± 12%. The degree of decrease varied significantly between muscles. For example, utrophin protein levels were decreased by 68 + 15% in the quadriceps, while they were decreased by only 11 ± 9% in the diaphragm. Some proteolysis of sarcoglycans was evident in some muscles, but levels of proteolytic products, when present, were also reduced in DG/mdx muscles compared to mdx muscles (data not shown). We observed no increase in the relative level of glycosylation of proteins with the CT antigen in DG/mdx animals relative to mdx animals (0 ± 4%). Levels of actin, a control, were also unchanged (6 ± 7%). Most of the immunoblotting for the CT antigen is due to its presence on proteins in capillaries and small blood vessels within muscle tissue, as it is normally confined to the neuromuscular junction in muscle cells.19 Increased expression of this antigen, however, does occur on muscle cell glycoproteins when levels of the CT GalNAc transferase (Galgt2) are increased.20 Therefore, the fact that these levels did not change suggests that muscle cell glycoproteins do not have increased levels of glycosylation with this antigen in DG/mdx muscles. Thus, transgenic overexpression of dystroglycan did not increase the expression of the CT antigen or utrophin, two molecules that can inhibit muscular dystrophy when increased in mdx muscles.4,5 Rather, increased dystroglycan expression correlated with reduced expression of utrophin protein (R = 0.85 for utrophin loss versus α-dystroglycan increase).
We next performed immunostaining experiments to determine the pattern of protein expression within DG/mdx and mdx muscles (Figure 2). In mdx mice, dystroglycan is expressed at very low levels along the sarcolemmal membrane.15 By contrast, both α- and β-dystroglycan were highly expressed along the membrane in DG/mdx muscles. This was particularly true for some regenerating fibers. Significant intracellular staining also was present in DG/mdx muscles, and this was similar to what we observed in non-dystrophic DG transgenic mice. In the gastrocnemius, where α-dystroglycan was not highly overexpressed, we also observed little increase in staining using an anti-peptide antiserum to the α-dystroglycan protein (data not shown). Therefore, reduced levels of α-dystroglycan, relative to β-dystroglycan, likely result from its preferential degradation in this muscle. No increase in utrophin staining was observed in DG/mdx animals relative to mdx animals. Rather, utrophin staining was reduced in some regenerating myofibers in DG/mdx mice when compared to mdx mice. Staining for α-sarcoglycan, β-sarcoglycan, the CT carbohydrate antigen, and laminin α4 were very low in DG/mdx and mdx myofibers, while staining for laminin α2 and laminin α5 was equally high. Laminin α2 is normally expressed along the entire muscle basal lamina, while laminin α5 and laminin α4 are normally confined to the neuromuscular junction in adult animals.26 Expression of laminin α5, however, is increased along extrasynaptic regions of the muscle basal lamina in mdx mice, while laminin α4 is not.27
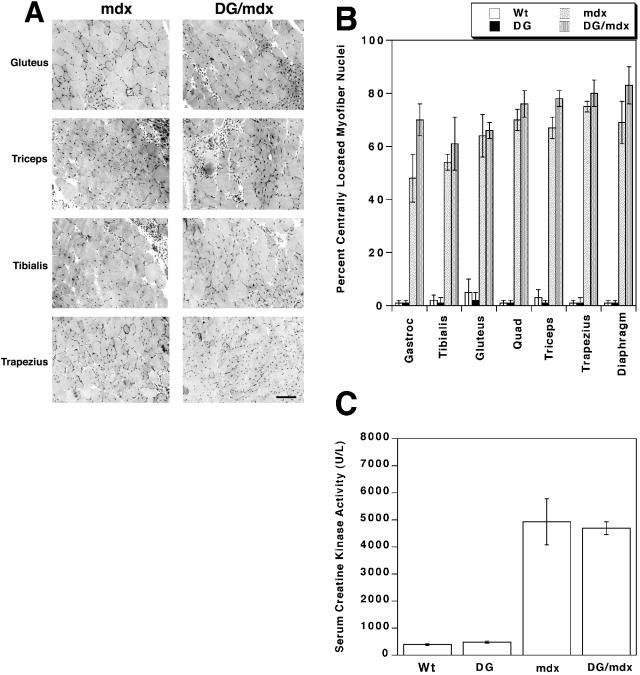
Given the lack of increase in utrophin and the CT carbohydrate antigen, both of which can inhibit muscular dystrophy in mdx mice,4,5 we expected that muscles in DG/mdx mice would have muscular dystrophy. This was indeed the case. We analyzed the extent of muscular dystrophy in DG/mdx and mdx myofibers by quantitating the percentage of centrally located myofiber nuclei (Figure 3, A and B) and by assaying serum creatine kinase activity levels (Figure 3C). The presence of centrally located nuclei in mature myofibers is an indicator of regenerative processes that can result from muscular dystrophy, while creatine kinase is a muscle enzyme that is released into the serum as a result of muscle injury. Transgenic expression of dystroglycan in wild-type mice (DG) did not increase the percentage of myofibers with central nuclei or the level of serum creatine kinase activity relative to non-transgenic (Wt) animals, and transgenic expression of dystroglycan in mdx mice (DG/mdx) did not decrease levels of either of these parameters relative to mdx mice. In fact, levels of central nuclei in DG/mdx mice tended to be slightly higher than levels in mdx littermates, though this was not statistically significant. The increased level may be the result of reduced levels of utrophin or sarcoglycan in DG/mdx muscles, however, central nuclei are very high in both mdx and utrn-/-mdx muscles at this age.28,29 The coefficient of variance in myofiber diameter for the seven DG/mdx muscles examined also was, on average, not significantly different from mdx mice (300 ± 30 for DG/mdx versus 280 ± 20 for mdx). Therefore, expression of the dystroglycan transgene in mdx mice had no effect on the extent of muscular dystrophy as assessed by these measures.
Figure 3.
Muscular dystrophy is not reduced in DG/mdx transgenic mice A: Cross-sections of DG/mdx and mdx muscles were stained with hematoxylin and eosin. Levels of centrally located myofiber nuclei (dark spots) and variations in myofiber diameter were equally high in DG/mdx and mdx muscles. Scale bar, 50 μm. B: The percentage of centrally located myofiber nuclei were quantitated in the gastrocnemius (Gastroc), tibialis anterior, gluteus maximus, quadriceps (Quad) femoris (vastus intermedius), triceps brachii, trapezius, and diaphragm. The percentage of centrally located nuclei was as high in DG/mdx muscles as it was in mdx muscles in all instances. Errors are SEM for n = 6. C: Serum creatine kinase levels in dystroglycan transgenic mice (DG) did not differ from wild-type (Wt) animals. Levels were elevated in mdx and DG/mdx mice, but did not significantly differ from one another. Errors are SEM for n = 10 (Wt or DG) or 6 (mdx or DG/mdx) experiments.
Discussion
The failure of dystroglycan overexpression to inhibit muscular dystrophy in mdx mice stands in contrast to previous results with utrophin or the CT GalNAc transferase (Galgt2), both of which also cause the overexpression of dystroglycan along mdx myofibers.4,5 We can think of three possible explanations for this disparity. First, the inhibition of muscular dystrophy in utrophin- and Galgt2-transgenic mdx mice may not require dystroglycan. Second, these other transgenic models may stimulate utrophin via mechanisms that cannot be duplicated by the dystroglycan transgene. For example, Galgt2 may increase utrophin levels by stimulating utrophin transcription, and dystroglycan overexpression may not do this. Third, the form of dystroglycan that is overexpressed in these transgenic animals is distinct from the form that is overexpressed in DG/mdx animals. We would argue that the latter possibility is the most likely.
Several proteins have synaptic and extrasynaptic forms that define distinct molecular complexes in skeletal muscle.30 Dystrophin- and utrophin-glycoprotein complexes clearly fit into this category.17,31 Despite the fact that dystrophin and utrophin both can bind to dystroglycan in vitro,21 there must be a mechanism that defines their unique subcellular distributions in vivo and in cultured muscle cells. The synaptic alignment of proteins at the neuromuscular junction is so precise that most synaptic proteins use posttranslational mechanisms in addition to local synthesis to ensure their proper synaptic distribution.30 Thus, while some utrophin expression at the neuromuscular junction can be attributed to local mRNA synthesis or stability,1,32 it is likely that local utrophin protein binding also occurs. This must be particularly true in mdx muscles. The fact that utrophin expression can be increased in regenerating mdx myofibers16 and that pharmacological agents can increase utrophin protein in mdx muscles in the absence of increased mRNA33 suggest that utrophin expression is not likely to be solely confined to synaptic nuclei in mdx muscles. Here, we have found that overexpression of dystroglycan actually inhibits utrophin protein expression in mdx muscles. This suggests that the normally extrasynaptic form of dystroglycan does not bind utrophin and may therefore contribute to its synaptic confinement in both normal14 and mdx15 muscle. That utrophin is overexpressed along with dystroglycan in Galgt2 transgenic mdx mice suggests that a synaptic glycoform of dystroglycan, however, does bind utrophin.5 Thus, the presence of the CT antigen on dystroglycan at the neuromuscular junction may dictate the local expression of utrophin through stimulating local binding, while the extrasynaptic form of dystroglycan may inhibit utrophin binding in the extrasynaptic membrane.
The results shown here for dystroglycan are similar to those of Campbell, Chamberlain, and colleagues with transgenic overexpression of Dp71 in mdx mice.34 There, overexpression of a shortened dystrophin transcript, Dp71, in mdx mice caused the overexpression of dystroglycan along the myofiber membrane, yet this did not inhibit the development of muscular dystrophy. It is important to realize, however, that this experiment is fundamentally different from the one we have done here. Dp71 is a 71-kd dystrophin protein that contains a dystroglycan-binding domain. Unlike the native 400-kd muscle forms of dystrophin or utrophin, however, Dp71 does not have an actin-binding domain. Therefore, Dp71 overexpression could compete with utrophin for binding to dystroglycan in mdx muscles. Indeed, the fact that Dp71 overexpression in wild-type muscles can cause muscular dystrophy suggests that it also competes with native dystrophin to inhibit actin binding.35 In the experiment done here, there is no dystrophin, short or long, to interfere with utrophin binding, yet there was still no inhibition of muscular dystrophy. Therefore, these results more clearly demonstrate that dystroglycan expression, in and of itself, is not functionally useful in mdx mice.
Acknowledgments
We thank Joshua Sanes (Washington University), Kevin Campbell (HHMI, University of Iowa), and Stephan Kroger (University of Mainz) for the gift of antibodies used in this study.
Footnotes
Address reprint requests to Paul T. Martin, Department of Neuroscience, Glycobiology Research and Training Center, University of California, San Diego, School of Medicine, La Jolla, CA 92093-0691. E-mail: pmartin@ucsd.edu.
Supported by grants from the Muscular Dystrophy Association and the National Institutes of Health (NS37214) to P.T.M.
References
- Blake DJ, Weir A, Newey SE, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- Martin PT. Role of transcription factors in skeletal muscle and the potential for pharmacological manipulation. Curr Opin Pharmacol. 2003;3:300–308. doi: 10.1016/s1471-4892(03)00050-x. [DOI] [PubMed] [Google Scholar]
- Cox GA, Cole NM, Matsumura K, Phelps S, Hauchka SD, Campbell KP, Faulkner JA, Chamberlain JS. Overexpression of dystrophin in transgenic mdx mice eliminates dystrophic symptoms without toxicity. Nature. 1993;364:725–729. doi: 10.1038/364725a0. [DOI] [PubMed] [Google Scholar]
- Tinsley J, Deconinck N, Fisher R, Kahn D, Sphelps S, Gillis J-M, Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- Nguyen HH, Jayasinha V, Xia B, Hoyte K, Martin PT. Overexpression of the cytotoxic T cell GalNAc transferase in skeletal muscle inhibits muscular dystrophy in mdx mice. Proc Natl Acad Sci USA. 2002;99:5616–5621. doi: 10.1073/pnas.082613599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehling M, Spencer MJ, Tidball JG. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J Cell Biol. 2001;155:123–131. doi: 10.1083/jcb.200105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronqvist P, Kawaguchi N, Albrechtsen R, Xu X, Schroder HD, Moghadaszadeh B, Nielsen FC, Frohlich C, Engvall E, Wewer UM. ADAM12 alleviates the skeletal muscle pathology in mdx dystrophic mice. Am J Pathol. 2002;161:1535–1540. doi: 10.1016/S0002-9440(10)64431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MJ, Mellgren RL. Overexpression of a calpastatin transgene in mdx muscle reduces dystrophic pathology. Hum Mol Genet. 2002;11:2645–2655. doi: 10.1093/hmg/11.21.2645. [DOI] [PubMed] [Google Scholar]
- Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 2002;157:137–148. doi: 10.1083/jcb.200108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkin DJ, Wallace GQ, Nicol KJ, Kaufman DJ, Kaufman SJ. Enhanced expression of the α7β1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J Cell Biol. 2001;152:1207–1218. doi: 10.1083/jcb.152.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll J, Barzaghi P, Lin S, Bezakova G, Lochmuller H, Engvall E, Muller U, Ruegg MA. An agrin minigene rescues dystrophic symptoms in a mouse model for congenital muscular dystrophy. Nature. 2001;413:302–307. doi: 10.1038/35095054. [DOI] [PubMed] [Google Scholar]
- Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LE, Ahima RS, Khurana TS. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420:418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- Rybakova IN, Patel JR, Davies KE, Yurchenco PD, Ervasti JM. Utrophin binds laterally along actin filaments and can couple costameric actin with sarcolemma when overexpressed in dystrophin-deficient muscle. Mol Cell Biol. 2002;13:1512–1521. doi: 10.1091/mbc.01-09-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlendieck K, Ervasti JM, Matsumura K, Kahl SD, Leveille CJ, Campbell KP. Dystrophin-related protein is localized to neuromuscular junctions of adult skeletal muscle. Neuron. 1991;7:499–508. doi: 10.1016/0896-6273(91)90301-f. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360:588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- Galvagni F, Cantini M, Oliviero S. The utrophin gene is transcriptionally upregulated in regenerating muscle. J Biol Chem. 2002;277:19106–19113. doi: 10.1074/jbc.M109642200. [DOI] [PubMed] [Google Scholar]
- Martin PT. Dystroglycan glycosylation and its role in matrix binding in skeletal muscle. Glycobiology. 2003;13:55R–65R. doi: 10.1093/glycob/cwg076. [DOI] [PubMed] [Google Scholar]
- Smith PL, Lowe JB. Molecular cloning of a murine N-acetylgalactosamine transferase cDNA that determines expression of the T lymphocyte-specific CT oligosaccharide differentiation antigen. J Biol Chem. 1994;269:15162–15171. [PubMed] [Google Scholar]
- Martin PT, Scott LJC, Porter BE, Sanes JR. Distinct structures and functions of related pre- and postsynaptic carbohydrates at the mammalian neuromuscular junction. Mol Cell Neurosci. 1999;13:105–118. doi: 10.1006/mcne.1999.0737. [DOI] [PubMed] [Google Scholar]
- Xia B, Hoyte K, Kammesheidt A, Deerinck T, Ellisman M, Martin PT. Overexpression of the CT GalNAc transferase in skeletal muscle alters myofiber growth, neuromuscular structure, and laminin expression. Dev Biol. 2002;242:58–73. doi: 10.1006/dbio.2001.0530. [DOI] [PubMed] [Google Scholar]
- Chung W, Campanelli JT. WW and EF hand domains of dystrophin-family proteins mediate dystroglycan binding. Mol Cell Biol Res Commun. 1999;2:162–171. doi: 10.1006/mcbr.1999.0168. [DOI] [PubMed] [Google Scholar]
- Jayasinha V, Nguyen HH, Xia B, Kammesheidt A, Hoyte K, Martin PT. Inhibition of dystroglycan cleavage causes muscular dystrophy in transgenic mice. Neuromusc Disord. 2003;5:365–375. doi: 10.1016/s0960-8966(03)00040-3. [DOI] [PubMed] [Google Scholar]
- Herrmann R, Straub V, Blank M, Kutzick C, Franke N, Jacob E, Lenard H, Kroger S, Voit T. Dissociation of the dystroglycan complex in caveolin-3-deficient limb girdle muscular dystrophy. Hum Mol Genet. 2000;9:2335–2340. doi: 10.1093/oxfordjournals.hmg.a018926. [DOI] [PubMed] [Google Scholar]
- de La Porte S, Morin S, Koenig J. Characteristics of skeletal muscle in mdx mutant mice. Int Rev Cytol. 1999;191:99–148. doi: 10.1016/s0074-7696(08)60158-8. [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between actin and laminin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton BL, Miner JH, Chiu AY, Sanes JR. Localization, regulation, and function of laminins in the neuromuscular system of developing, adult, and mutant mice. J Cell Biol. 1997;139:1507–1521. doi: 10.1083/jcb.139.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton BL, Connolly AM, Martin PT, Cunningham JM, Mehta S, Pestronk A, Miner JH, Sanes JR. Distribution of ten laminin chains in dystrophic and regenerating muscles. Neuromusc Disord. 1999;9:423–433. doi: 10.1016/s0960-8966(99)00033-4. [DOI] [PubMed] [Google Scholar]
- Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90:729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, Watt DJ, Dickson JG, Tinsley JM, Davies KE. Utrophindystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Development of the neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- Martin PT, Freeze HH. Glycobiology of neuromuscular disorders. Glycobiology. 2003;13:67R–75R. doi: 10.1093/glycob/cwg077. [DOI] [PubMed] [Google Scholar]
- Jasmin BJ, Angus LM, Belanger G, Chakkalakal JV, Gramolini AO, Lunde JA, Stocksley MA, Thompson J. Multiple regulatory events controlling the expression and localization of utrophin in skeletal muscle fibers: insights into a therapeutic strategy for Duchenne muscular dystrophy. J Physiol Paris. 2002;96:31–42. doi: 10.1016/s0928-4257(01)00078-x. [DOI] [PubMed] [Google Scholar]
- Courdier-Fruh I, Barman L, Briguet A, Meier T. Glucocorticoid-mediated regulation of utrophin levels. Neuromusc Disord. 2002;12:S95–S104. doi: 10.1016/s0960-8966(02)00089-5. [DOI] [PubMed] [Google Scholar]
- Greenberg DS, Sunada Y, Cambpell KP, Yaffe D, Nudel U. Exogenous Dp71 restores the levels of dystrophin associated proteins but does not alleviate muscle damage in mdx mice. Nat Genet. 1994;8:340–344. doi: 10.1038/ng1294-340. [DOI] [PubMed] [Google Scholar]
- Leibovitz S, Meshorer A, Fridman Y, Wieneke S, Jockusch H, Yaffe D, Nudel U. Exogenous Dp71 is a dominant negative competitor of dystrophin in skeletal muscle. Neuromusc Disord. 2002;12:836–844. doi: 10.1016/s0960-8966(02)00141-4. [DOI] [PubMed] [Google Scholar]