Abstract
Polycomb-group (PcG) genes preserve cell identity by gene silencing, and contribute to regulation of lymphopoiesis and malignant transformation. We show that primary nodal large B-cell lymphomas (LBCLs), and secondary cutaneous deposits from such lymphomas, abnormally express the BMI-1, RING1, and HPH1 PcG genes in cycling neoplastic cells. By contrast, tumor cells in primary cutaneous LBCLs lacked BMI-1 expression, whereas RING1 was variably detected. Lack of BMI-1 expression was characteristic for primary cutaneous LBCLs, because other primary extranodal LBCLs originating from brain, testes, and stomach were BMI-1-positive. Expression of HPH1 was rarely detected in primary cutaneous LBCLs of the head or trunk and abundant in primary cutaneous LBCLs of the legs, which fits well with its earlier recognition as a distinct clinical pathological entity with different clinical behavior. We conclude that clinically defined subclasses of primary LBCLs display site-specific abnormal expression patterns of PcG genes of the HPC-HPH/PRC1 PcG complex. Some of these patterns (such as the expression profile of BMI-1) may be diagnostically relevant. We propose that distinct expression profiles of PcG genes results in abnormal formation of HPC-HPH/PRC1 PcG complexes, and that this contributes to lymphomagenesis and different clinical behavior of clinically defined LBCLs.
Polycomb-group (PcG) genes are evolutionary conserved regulators of gene expression that contribute to normal lymphocyte development, and malignant transformation of these cells.1–4 PcG genes are essential for regulation of embryogenesis and preserve cell identity by maintenance of homeobox gene silencing patterns.5 The underlying mechanism is related to formation of chromatin-binding protein complexes composed of multiple PcG proteins and other associated factors.6 Two PcG complexes exist in humans: the HPC-HPH/PRC1 complex contains the BMI-1, MEL-18, RING1, HPC, and HPH PcG proteins, and is also called polycomb repressive complex 1 (PRC1) or maintenance complex. The second complex consists of the EED, EZH, and YY1 PcG proteins and is called the EED-EZH/PRC2 or initiation complex.7–12 These complexes have virtually identical counterparts in Drosophila melanogaster and mice, suggesting evolutionary conservation of interaction between PcG proteins and PcG complex composition.13–20 Individual PcG proteins are expressed in different combinations at different chromosomal sites, tissues, or individual cell populations.21,22 This probably reflects tissue- or cell type-specific formation of PcG complexes with distinct fine composition, and is most likely related to target gene specificity.9 The fine composition of PcG complexes is further affected by non-PcG proteins, including transcription factors that target the complex to DNA, or histone deacetylases and methylases, which probably contribute to gene-silencing by PcG complexes.23–28
Mammalian PcG genes are involved in the regulation of various other processes, including the cell cycle and lymphopoiesis.1–4,29 Interference with expression of individual PcG genes in mutant mice revealed severe defects in hematopoiesis, supportive of a role for PcG genes in lymphoid development.30–35 Involvement of PcG genes in human hematopoiesis has been suggested by several lines of evidence, including PcG transcription patterns in lymphoid progenitors of the bone marrow,36 developing B cells in germinal centers,37 and differentiating T cells in the thymus.38 In addition, altered PcG expression patterns are strongly linked to malignant transformation. The best-known example is induction of lymphomas in BMI-1 transgenic mice, in which BMI-1 overexpression results in down-regulation of p16INK4a/p19ARF, enhanced lymphoproliferation, and eventually malignant transformation of both B and T cells.35,39,40 Other PcG genes that seem to function as oncogenes are RING110 and HPC2,41 whereas EED34 and MEL-1842,43 behave as tumor suppressors. However, despite these observations in experimental models, little is known about a role for PcG genes in development of human lymphoid malignancies. We recently discovered that the human BMI-1 PcG gene is overexpressed in Reed-Sternberg cells of Hodgkin’s disease,44 and in neoplastic cells of various nodal B-cell non-Hodgkin’s lymphomas.45,46 This pattern is in-line with development of lymphomas in BMI-1 transgenic mice, and supports the conclusion that PcG genes contribute to development of hematological cancers in humans.
In the current study we investigated the expression pattern of nine PcG genes, BMI-1, MEL-18, RING1, HPH1, HPC1, and HPC2 (the HPC-HPH/PRC1 complex) and EED, EZH2, and YY1 (the EED-EZH/PRC2 complex) in various subclasses of malignant large B-cell lymphomas (LBCLs). LBCLs can be differentiated according to their site of origin, such as primary nodal and primary cutaneous LBCLs. Within the primary cutaneous LBCLs, primary cutaneous follicle center cell lymphomas originating from head or trunk have a very good prognosis with an overall 5-year survival of 89 to 96%.47–53 By contrast, primary cutaneous LBCLs originating from the leg have an intermediate prognosis with an overall 4-year survival of 58%.47,50,54 We discovered that these lymphomas display altered expression of PcG genes encoding the HPC-HPH/PRC1 complex in a site-specific pattern. Primary nodal LBCLs, and secondary cutaneous deposits from such tumors, overexpressed BMI-1 and RING1, whereas primary cutaneous LBCLs lacked BMI-1 expression and displayed variable expression of RING1. In addition, primary nodal LBCLs and primary cutaneous LBCLs of the legs displayed frequent expression of HPH1, whereas this gene was rarely used by primary cutaneous LBCLs of the trunk. These expression patterns demonstrate that clinically defined subgroups of LBCLs display site-specific expression patterns of PcG genes. We propose that variation of PcG expression results in altered composition of the HPC-HPH/PRC1 PcG complex, and that this may contribute to different mechanisms of lymphomagenesis and different clinical behavior.
Materials and Methods
Patient Material
Lymphomas were selected from the archives of the Department of Pathology of the VU Medical Center, Amsterdam, and the Department of Pathology of the Leiden University Medical Center. Only tissues containing sufficient numbers of tumor cells were used for analysis. We investigated paraffin-embedded and frozen material of 48 large B-cell lymphomas (LBCLs), including 8 primary diffuse nodal LBCLs, 4 secondary cutaneous deposits of primary nodal LBCLs, 24 primary cutaneous LBCLs (comprising 12 primary cutaneous large follicle center cell lymphomas originating from the head or neck, and 12 primary cutaneous LBCLs originating from the leg), and 12 primary extranodal LBCLs including 5 LBCLs from the brain, 5 LBCLs from the testis, and 2 LBCLs from the stomach. Cutaneous lymphomas were classified according to the European Organization for Research and Treatment of Cancer. In primary cutaneous LBCLs diagnoses were based on absence of extracutaneous disease detected by a comprehensive staging procedure at diagnosis and histological and immunophenotypic features showing a majority (ie, >50%) of large cells among neoplastic B cells. The staging procedure included in all cases physical examination, routine laboratory tests, chest radiograph or thoracic computed tomography scan, abdominal computed tomography scan, bone marrow cytology, and bone marrow histology. In addition, histological and immunophenotypical characterization was performed (expression of CD20, CD79a, CD3, CD4, CD8, cyclin D1, CD10, bcl-2, bcl-6, CD5, CD35, Igκ, and Igλ). According to the criteria of the Revised European-American Classification of Lymphoid Neoplasms and the World Health Organization classification all primary cutaneous LBCLs patients were classified as having a diffuse large B-cell lymphoma. Healthy control tissue, such as lymph nodes and extranodal lymphoid tissue, were routinely included in the experiments and obtained from regular surgical pathology archival material.
Immunohistochemistry
Immunohistochemical staining was performed following previously established protocols and antisera specific for human BMI-1,55 RING1,11 HPH1,11 HPC1,41 HPC2,11 EED,12 and EZH2.12 The MEL-18 antiserum was obtained from Santa Cruz (Santa Cruz, CA). Briefly, 4-μm sections of formalin-fixed and paraffin-embedded material were treated with H2O2 to inhibit endogenous peroxidase. After antigen retrieval, slides were rinsed in phosphate-buffered saline (PBS) containing 0.5% Triton X-100 and treated with 0.1 mol/L of glycine. After preincubation with normal swine serum or normal rabbit serum, primary antibodies were applied at optimal dilution. Secondary antibodies were biotinylated swine anti-rabbit or biotinylated rabbit-anti-mouse (DAKO, Glostrup, Denmark), followed by application of horseradish peroxidase-conjugated streptavidin-biotin-complex (sABC-HRP). Immunostaining was performed with 3-amino-9-ethylcarbazole using the streptavidin-biotin complex horseradish peroxidase method (DAKO) and tyramine intensification. Sections were counterstained with hematoxylin, and cells expressing a particular PcG gene were counted at ×20 magnification and higher. If the biopsy was of sufficient size, at least five microscopic fields containing tumor cells were analyzed. Photographs were taken with a Zeiss Axiophot microscope and digitized using an Agfa duoscan.
Double and Triple Immunofluorescence
Immunofluorescence analysis was performed as described previously. In brief, frozen tissue sections were fixed in formaldehyde and endogenous peroxidase was inhibited using H2O2 diluted in PBS. Sections were preincubated with bovine serum albumin, and a combination of two or three primary antibodies were applied at optimal concentration. BMI-1 was detected by incubation with HRP-conjugated goat anti-mouse IgG2b and subsequent rhodamine/tyramine intensification. Detection of HPC1, HPC2, HPH1, RING1, and EZH2 was performed by incubating the slides with ALEXA-conjugated goat anti-rabbit antiserum. The other markers were visualized by incubating the slides with biotinylated goat anti-mouse IgG1 or IgG2a, depending on Ig-subclass of the primary antibody, followed by allophycocyanin coupled to streptavidin. Cross-reactivity of the antisera was excluded by appropriate controls and for each double- or triple-immunofluorescence experiment, single-staining controls were included. In addition, positive and negative controls were routinely included. Sections were analyzed with a Leica DMR confocal laserscan microscope (Leica, Deerfield, IL). Images were stored digitally and processed using Adobe Photoshop 6.
Results
Immunohistochemical Analysis of PcG Expression in Primary Nodal and Cutaneous LBCLs
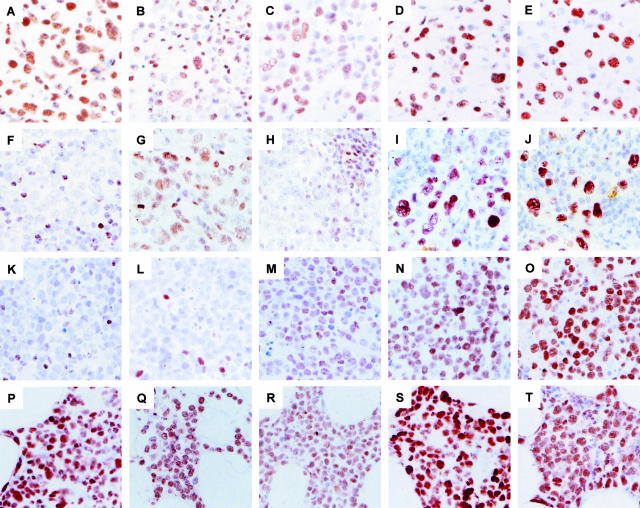
The results of immunohistochemical detection of PcG proteins in primary nodal LBCLs and primary cutaneous LBCLs are shown in Figure 1 and summarized in Table 1. Neoplastic cells in all diffuse primary nodal LBCLs expressed BMI-1 (Figure 1A), confirming our previous findings.45 Human BMI-1 binds several other PcG proteins present in the HPC-HPH/PRC1 complex, such as MEL-18, RING1, HPC1, HPC2, and HPH1. We therefore questioned whether these PcG proteins were also expressed by neoplastic cells in PLBCLs. We detected RING1, MEL-18, HPC2, and HPH1 in all cases of primary nodal LBCLs studied (Table 1; representative examples of RING1 and HPH1 staining are shown in Figure 1, B and C, respectively). These PcG proteins were also frequently detected in the surrounding infiltrate consisting of lymphocytes and monocytes. By contrast, the HPC1 PcG protein was not detectable in neoplastic cells of primary nodal LBCLs or the reactive infiltrating cells (Table 1). Analysis of components of the EED-EZH/PRC2 PcG complex demonstrated that neoplastic cells in primary nodal LBCLs express the EZH2, EED, and YY1 PcG proteins at high frequencies (Table 1; representative examples of EZH2 and EED staining are shown in Figure 1, D and E, respectively). EED and EZH2 were infrequently detected in surrounding reactive lymphocytes, probably because most infiltrating lymphocytes are resting and preferentially express HPC-HPH/PRC1 PcG genes (see immunofluorescence results).37,45 YY1 stained both neoplastic cells and reactive lymphocytes (not shown), which probably reflects the fact that YY1 is a ubiquitous transcription factor. These results collectively demonstrate that most core PcG proteins of the HPC-HPH/PRC1 and EED-EZH/PRC2 complex are co-expressed by neoplastic cells in primary nodal LBCLs.
Figure 1.
Immunohistochemical detection of PcG proteins in primary nodal and primary cutaneous LBCLs. Shown are immunohistochemical staining patterns of HPC-HPH/PRC1 PcG complex proteins (BMI-1, RING1, and HPH1) and EED-EZH/PRC2 PcG complex proteins (EED and EZH2). A–E: Primary nodal LBCLs. Large tumor cells express components of the HPC-HPH/PRC1 complex (BMI-1, RING1, and HPH1) and components of the EED-EZH/PRC2 complex (EED and EZH2). F–J: Primary cutaneous LBCLs originating from the trunk. K–O: Primary nodal LBCLs originating from the leg. Shown are two BMI-1neg examples of primary cutaneous LBCLs; large neoplastic cells in the primary cutaneous LBCLs from the trunk variably express RING1 (G) and are HPH1neg (H). By contrast, neoplastic cells in primary cutaneous LBCLs from the leg are RING1neg (L) and HPH1pos (M). Both primary cutaneous LBCLs use EED (I, N) and EZH2 (J, O). P–T: Secondary cutaneous deposit from a primary nodal LBCL; neoplastic cells display a PcG expression profile that is identical to the profile in primary nodal LBCL (A–E). Note that cells in the reactive infiltrate variably express the HPC-HPH/PRC1 complex proteins BMI-1, RING1, and HPH1, which serves as an internal positive control in cases in which the tumor cells are negative. The EED-EZH/PRC2 PcG complex proteins EED and EZH2 are infrequently detected in reactive lymphocytes. The PcG expression pattern in reactive lymphocytes reflects the mutually exclusive expression pattern of these PcG genes in healthy resting and cycling cells (see Figures 2 and 3), and indicates that the presence of BMI-1, RING1, and HPH1 in tumor cells is abnormal. Original magnifications: ×63.
Table 1.
Immunohistochemical Detection of PcG Proteins in Primary Cutaneous and Nodal LBCL
| Staining | HPC-HPH/PRC1 PcG complex
|
EED-EZH/PRC2 PcG complex
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BMI-1 | MEL-18 | RING1 | HPH1 | HPC1 | HPC2 | EED | EZH2 | YY1 | ||
| Primary nodal LBCL, | Pos | 8 | 8 | 8 | 8 | – | 8 | 8 | 8 | 8 |
| n = 8 | Var | – | – | – | – | – | – | – | – | |
| Neg | – | – | – | – | – | – | – | – | – | |
| Primary cutaneous LBCL | Pos | – | 12 | 7 | 1 | – | 12 | 12 | 12 | 12 |
| (head/trunk), n = 12 | Var | – | – | 2 | 5 | – | – | – | – | – |
| Neg | 12 | – | 3 | 6 | – | – | – | – | – | |
| Primary cutaneous LBCL | Pos | – | 12 | 10 | 7 | – | 12 | 12 | 12 | 12 |
| (legs), n = 12 | Var | – | – | 1 | 4 | – | – | – | – | – |
| Neg | 12 | – | 1 | 1 | – | – | – | – | – | |
| Secondary cutaneous | Pos | 4 | 4 | 4 | 4 | – | 4 | 4 | 4 | 4 |
| deposit, n = 4 | Var | – | – | – | – | – | – | – | – | – |
| Neg | – | – | – | – | – | – | – | – | – | |
| Primary extranodal | Pos | 5 | 5 | 3 | 2 | – | 5 | 5 | 5 | 5 |
| (brain), n = 5 | Var | – | – | 2 | – | – | – | – | – | – |
| Neg | – | – | – | 3 | – | – | – | – | – | |
| Primary extranodal | Pos | 5 | 5 | 3 | 2 | – | 5 | 5 | 5 | 5 |
| (brain), n = 5 | Var | – | – | 2 | – | – | – | – | – | – |
| Neg | – | – | – | 3 | – | – | – | – | – | |
| Primary extranodal | Pos | 5 | 5 | 2 | – | – | 5 | 5 | 5 | 5 |
| (testis), n = 5 | Var | – | – | 2 | – | – | – | – | – | – |
| Neg | – | – | 1 | 5 | – | – | – | – | – | |
| Primary extranodal | Pos | 2 | 2 | 2 | 2 | – | 2 | 2 | 2 | 2 |
| (stomach), n = 2 | Var | – | – | – | – | – | – | – | – | – |
| Neg | – | – | – | – | – | – | – | – | – | |
Staining intensities are represented as positive (pos; >75% of the cells staining), Variable (var; <75% but >5% of the cells staining, often with variable intensity), and negative (neg; <5% of the neoplastic cells staining).
Neoplastic cells in primary cutaneous LBCLs expressed HPC-HPH/PRC1 complex PcG genes in a pattern that was strikingly different from the one observed in primary nodal LBCLs. Whereas neoplastic cells in primary nodal LBCLs strongly expressed the BMI-1 gene, neoplastic cells in primary cutaneous LBCLs were negative for BMI-1 by immunohistochemistry. This difference was statistically significant (two-sided Fisher’s exact test, P < 0,0001) and is exemplified by Figure 1, F and K, of two primary cutaneous LBCLs originating from the trunk and the leg, respectively, which show large BMI-1neg tumor cells surrounded by an infiltrate of reactive BMI-1pos lymphocytes. Primary cutaneous LBCLs originating from head, trunk, or leg all lacked expression of BMI-1 (summarized in Table 1). This suggests that BMI-1 expression in LBCLs is site-specific, because primary nodal LBCLs typically express BMI-1. As found in primary nodal LBCLs, MEL-18 and HPC2 were expressed strongly in all primary cutaneous LBCLs (Table 1), whereas RING1 expression appeared variable (Table 1; exemplified in Figure 1, G and L, for primary cutaneous LBCLs originating from the trunk and the legs, respectively). We were unable to determine whether the variation in RING1 expression was related to location of the tumor, and possible differences did not reach statistical significance because of limited numbers of patients. However, immunohistochemical analysis of HPH1 expression in primary cutaneous LBCLs revealed a location-specific staining pattern: the majority of primary cutaneous LBCLs originating from the legs expressed HPH1 (Table 1, and exemplified by Figure 1M), but primary cutaneous LBCLs originating from head or trunk were negative for HPH1 or expressed this protein at a low level (Table 1, and exemplified by Figure 1M). Statistical analysis of these results demonstrated that the expression of HPH1 was significantly more frequent in primary cutaneous LBCLs originating from the legs than in primary cutaneous LBCLs from the head or trunk (chi-square, P = 0.017). In addition, HPH1 expression in primary cutaneous LBCLs as a group was significantly different from HPH1 expression in primary nodal LBCLs (chi-square, P = 0.005). There were no obvious differences in expression of PcG proteins belonging to the EED-EZH/PRC2 complex: similar to primary nodal LBCLs, EED, EZH2, and YY1 were detected at high frequencies in neoplastic cells of primary cutaneous LBCLs, without any preference for location of the tumor (Figure 1; I, J, N, and O, and summarized in Table 1).
Immunohistochemical Analysis of PcG Expression in Primary LBCLs Originating from Testis, Brain, and Stomach
The lack of BMI-1 expression in primary cutaneous LBCLs prompted us to question whether absence of BMI-1 is characteristic for all primary extranodal LBCLs, or whether it is particular for primary cutaneous LBCLs. We therefore determined the PcG gene expression profile of an additional set of primary extranodal LBCLs, consisting of 12 rare lymphomas originating from the brain (n = 5), the testis (n = 5), and the stomach (n = 2). The immunohistochemical data are summarized in Table 1. Similar to primary nodal LBCLs, all primary extranodal LBCLs in the brain, testes, and the stomach expressed the HPC-HPH/PRC1 PcG complex proteins BMI-1, MEL-18, and HPC2, and lacked HPC1. The two other BMI-1 binding partners, RING1 and HPH1, were expressed at variable levels. Statistical analysis showed that primary extranodal LBCLs expressed significantly more BMI-1 than primary cutaneous LBCLs (Fisher’s exact two-sided test, P < 0.0001). Also, expression of HPH1 in the group of primary extranodal LBCLs was significantly lower compared to the group of primary cutaneous LBCLs (chi-square, P = 0.027) and the group of primary nodal LBCLs (Fisher’s exact two-sided test, P = 0.005).
Detection of PcG Expression in Primary Nodal and Cutaneous LBCLs by Triple Immunofluorescence
We next used immunofluorescence on frozen tissue sections to confirm the PcG expression patterns assessed by immunohistochemistry in primary nodal and cutaneous LBCLs. This allowed us to study PcG proteins simultaneously and in the same nuclei. Immunofluorescent analysis supported the data obtained by immunohistochemistry, and is exemplified for BMI-1, EZH2, and HPH1 in combination with MIB-1, a marker for proliferation (Figures 2 and 3).
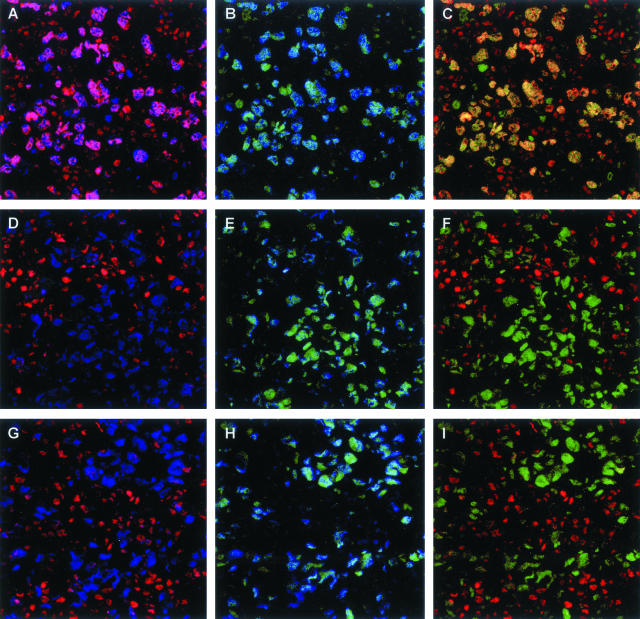
Figure 2.
Triple immunofluorescence for BMI-1, EZH2, and MIB-1 in primary nodal and primary cutaneous LBCLs. BMI-1, EZH2, and the proliferation marker MIB-1 were detected by red, green, and blue immunofluorescence, respectively. A–C: Primary nodal LBCLs. Large cycling (MIB-1pos) neoplastic cells express BMI-1 (A) and EZH2 (B), leading to co-expression of BMI-1 and EZH2 in the same nucleus (C). D–F: Primary cutaneous LBCLs originating from the trunk. G–I: Primary cutaneous LBCLs originating from the legs. In primary cutaneous LBCLs, BMI-1 is undetectable in large cycling (MIB-1pos) neoplastic cells (D and G) whereas EZH2 is present (E and H). F and I: Combination of the BMI-1 and EZH2 signals. Note that the reactive infiltrating cells express BMI-1 in the absence of EZH2. In addition, small EZH2pos cells in the reactive infiltrate are dividing cells because the EZH2 signal overlaps with the signal for MIB-1 (B, E, H). Occasional small EZH2pos cells with undetectable MIB-1 expression may reflect cells that are in transition from being in cycle to being in rest, or they could reflect nonlymphoid cells with a different PcG expression profile.
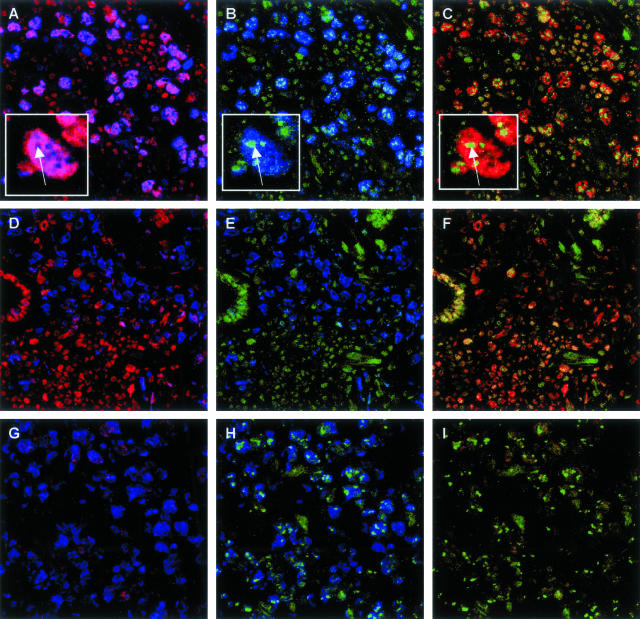
Figure 3.
Triple immunofluorescence for BMI-1, HPH1, and MIB-1 in primary nodal and primary cutaneous LBCLs. BMI-1, HPH1, and the proliferation marker MIB-1 were detected by red, green, and blue immunofluorescence, respectively. A–C: Primary nodal LBCLs. Large cycling (MIB-1pos) neoplastic cells express BMI-1 (A) and HPH1 (B), leading to co-expression of BMI-1 and HPH1 in the same nucleus (C). Co-expression of BMI-1 and HPH1 also occurs in the reactive infiltrate, but is limited to resting (MIB-1neg) cells (see comments in legend of Figure 2, and in the text). D–F: Primary cutaneous LBCLs originating from the trunk. In the example shown, large cycling (MIB-1pos) neoplastic cells are BMI-1neg (D) and HPH1neg (E). By contrast, BMI-1 and HPH1 are co-expressed (F) in resting (MIB-1neg) cells of the reactive infiltrate. G–I: Primary cutaneous LBCLs originating from the legs. In the example shown, large cycling (MIB-1pos) neoplastic cells express HPH1 (H) and are BMI-1neg (G). Note that HPH1pos neoplastic lymphocytes in primary nodal LBCLs (A–C) and primary cutaneous LBCLs originating from the legs contain domains that strongly stain for HPH1. In primary nodal LBCLs, these domains appear to be BMI-1neg (insets in A–C). The faint signal for BMI-1 in tumor cells shown in G is not representative, and most likely reflects weak background staining that was occasionally observed in cutaneous tissue sections.
Figure 2 demonstrates the BMI-1-staining pattern (red fluorescent signal) in combination with EZH2 (green fluorescence) and MIB-1/Ki-67 (blue fluorescence) in primary nodal LBCLs. As found previously, cycling (MIB-1/Ki-67pos) neoplastic cells in primary nodal LBCLs strongly express EZH2 and BMI-1, producing a yellow signal signifying BMI-1/EZH2 co-expression. Note that most reactive lymphocytes are either BMI-1neg/EZH2pos/MIB-1pos (activated cells) or BMI-1pos/EZH2neg/MIB-1neg (resting cells). By contrast to the expression of BMI-1 in primary nodal LBCLs, neoplastic cells in primary cutaneous LBCLs were BMI-1neg. Two representative examples are shown in Figure 2, representing a primary cutaneous lymphoma of the trunk (Figure 2; D to F) and the leg (Figure 2; G to I). Here, cycling (MIB-1pos) tumor cells are strongly positive for EZH2, but lack a signal for BMI-1. These patterns confirm the absence of BMI-1 in neoplastic cells of primary cutaneous LBCLs, as shown earlier by immunohistochemistry.
Figure 3 shows typical results of immunofluorescent staining on high-grade primary nodal LBCLs and primary cutaneous LBCLs for HPH1 (green fluorescence) in combination with BMI-1 (red fluorescence) and MIB-1/Ki-67 (blue fluorescence). Cycling (MIB-1pos) neoplastic cells in primary nodal LBCLs strongly stain for BMI-1 and display a diffuse green signal for HPH1, resulting in a yellow signal signifying BMI-1/HPH1 co-expression (Figure 3; A to C). In addition, tumor cells display distinct nuclear domains that strongly stain for HPH1 but are negative for BMI-1 (Figure 3, insets). Note that reactive infiltrating cells also express HPH1 in combination with BMI-1, but that they lack prominent HPH1pos domains, and are resting (MIB-1neg). Analysis of primary cutaneous LBCLs confirmed the HPH1 staining patterns established by immunohistochemistry. Figure 3, D to F, represents a primary cutaneous LBCL originating from the trunk, and shows neoplastic cells that are negative for BMI-1 and HPH1, while expressing MIB-1. By contrast, neoplastic cells in the primary cutaneous LBCLs originating from the leg, shown in Figure 1, G to I, display a faint diffuse staining pattern for HPH1, with prominent HPH1pos domains, in BMI-1neg/MIB-1pos tumor cells. We were unable to establish a correlation between the occurrence of HPH1pos domains in neoplastic cells of primary nodal LBCLs and primary cutaneous LBCLs, because they were not present in all cases of tumor cells and the number of patients was too limited to correlate the presence of HPH1pos domains with clinical behavior. We currently have no explanation for the presence of BMI-1neg/HPH1pos domains in tumor cells.
PcG Expression Patterns in Cutaneous Secondary Deposits of Primary Nodal LBCLs
Given the observed difference in PcG expression patterns between nodal and cutaneous primary LBCLs, we questioned whether secondary cutaneous deposits originating from a primary nodal LBCL retain the PcG expression profile of the primary lymphoma. Figure 1, P to T, shows a representative example of immunohistochemical staining for PcG proteins on a secondary deposit of a primary nodal LBCLs. Four individual cases were examined and are summarized in Table 1. Neoplastic cells in cutaneous secondary deposits were indistinguishable from neoplastic cells in primary nodal LBCLs: they strongly expressed BMI-1 and other PcG proteins belonging to the HPC-HPH/PRC1 complex, including MEL-18, RING1, HPH1, and HPC2 (exemplified for BMI-1, RING1, and HPH1 in Figure 1, P to R, respectively). Similarly, these tumor cells were positive for the EED-EZH/PRC2 complex PcG proteins EED, EZH2, and YY1 (Table 1, and exemplified for EED and EZH2 in Figure 1, S and T, respectively). These PcG expression patterns collectively demonstrate that secondary cutaneous deposits of primary nodal LBCLs retain their PcG expression profile.
Discussion
Neoplastic Cells in Primary Cutaneous Large B-Cell Lymphomas Express PcG Genes in a Site-Specific Pattern
In this study we determined the expression profile of human PcG proteins in clinically defined subgroups of LBCLs. We discovered that primary nodal LBCLs and primary cutaneous LBCLs display site-specific PcG expression patterns. This is most clearly demonstrated by analysis of the BMI-1 PcG protein. Whereas neoplastic cells of primary nodal LBCLs displayed widespread expression of BMI-1 in cycling neoplastic cells (this study and van Kemenade and colleagues45), BMI-1 was undetectable in cycling tumor cells of primary cutaneous LBCLs. Further analysis of BMI-1 binding partners revealed that PcG expression patterns fit well with the clinically defined subtypes of LBCLs. For instance, primary nodal LBCL and primary cutaneous LBCL are two distinct subtypes of LBCLs that can be distinguished on the basis of BMI-1 expression. In addition, our data suggest that the more aggressive form of primary cutaneous LBCL (originating from the leg) frequently expresses HPH1, whereas primary cutaneous LBCL with a better prognosis (originating from the head or trunk) infrequently express HPH1. Our interpretation of these patterns is that clinically defined subtypes of LBCLs express site-specific HPC-HPH/PRC1 PcG complexes. The current model of PcG-mediated gene silencing predicts that PcG complex composition contributes to target gene specificity, and variation in PcG complex composition theoretically produces different gene-silencing patterns. We propose that formation of site-specific HPC-HPH/PRC1 PcG complexes in clinically defined LBCLs results in different silencing patterns of HPC-HPH/PRC1 target genes and that this contributes to loss of cell identity and different clinical behavior of LBCL subtypes. Whether these distinct HPC-HPH/PRC1 complexes actually exist in LBCL subtypes, is currently under investigation in our laboratory.
Primary LBCLs Express HPC-HPH/PRC1 Complex PcG Proteins that Are Not Normally Present in Cycling Follicular B Cells
Given the observation in experimental models that abnormal expression of PcG genes contributes to development of lymphoid malignancies, it is important to resolve which PcG expression patterns in LBCLs are normal, and which are not. We recently demonstrated that healthy cycling B cells, such as those in the germinal center follicle, express the EZH2, EED, and YY1 PcG genes (Raaphorst and colleagues37 and van Galen and Raaphorst, manuscript submitted). The association of EED-EZH/PRC2 PcG complex proteins with cycling follicular B cells has also been observed in dividing plasma cells,56 and suggests that the presence of EED, EZH2, and YY1 in neoplastic cells of LBCLs is reflective of the normal situation. Previous studies of B-cell non-Hodgkin’s lymphomas showed that cycling neoplastic cells express BMI-1 in the presence of EZH2. Because normal proliferating EZH2pos B cells in the germinal center do not express BMI-1, this pattern suggests overexpression of BMI-1 in tumor cells. The current study on LBCLs supports these findings, and is in-line with development of lymphomas in BMI-1-transgenic mice. However, the distribution pattern of BMI-1 suggests that it is unlikely to function as a common oncogene involved in all human LBCLs, because BMI-1 expression was undetectable in primary cutaneous LBCLs. We further demonstrate that neoplastic cells in primary nodal LBCLs express various known BMI-1 binding partners, some of which have been identified as oncogenes or tumor suppressors in experimental models. Of these, the ubiquitously expressed HPC2 and MEL-18 PcG genes appear to follow a normal expression pattern, because they are also expressed by healthy cycling B cells (van Galen and Raaphorst, manuscript submitted). By contrast, the RING1 and HPH1 PcG proteins are undetectable in healthy dividing B cells (Raaphorst and colleagues37 and van Galen and Raaphorst, in press), and their presence in neoplastic cells of LBCLs suggests that RING1 and HPH1 expression is deregulated in tumor cells. This lends further support for the conclusion that components of the HPC-HPH/PRC1 PcG complex are abnormally expressed in neoplastic cells of LBCLs, leading to abnormal formation of HPC-HPH/PRC1 PcG complexes in cycling cells. Whether these complexes play an active role in cellular transformation and lymphomagenesis in humans remains to be established. Although HPH1 has not been associated with cellular transformation, overexpression of RING1 has been shown to lead to enhanced expression of c-jun and c-fos, induction of anchorage-independent growth, and development of tumors in mice.10 We propose that altered composition of the HPC-HPH/PRC1 complex is one possible result of abnormal expression of individual PcG genes in neoplastic cells. This may lead to changes in gene-silencing pathways of the tumor cells and contribute to altered cellular behavior and loss of cell identity.
Site-Specific PcG Expression in Clinically Defined Subclasses LBCLs: A New Diagnostic Marker?
An important observation in our current study is that abnormal site-specific expression patterns of individual HPC-HPH/PRC1 PcG genes correlate with clinically defined subclasses of primary LBCLs. This is best illustrated by the presence of BMI-1 in primary nodal LBCLs, and its absence from primary cutaneous LBCLs. Importantly, secondary deposits originating from a primary nodal LBCL retain expression of BMI-1, indicating that BMI-1 expression is diagnostically relevant in distinguishing a primary cutaneous LBCL from a secondary deposit. Interestingly, the high frequency of HPH1 expression in primary cutaneous LBCLs originating from the leg, and the low frequency of HPH1 expression in primary cutaneous LBCLs originating from the head or trunk, is in-line with the conclusion that these LBCLs are two distinct subtypes. We expect that other clinically defined tumors will also exhibit characteristic and diagnostically relevant PcG expression profiles, and we are currently pursuing such patterns in other hematological and solid human tumors.
Concluding Remarks
The current study adds to the growing body of evidence linking mammalian PcG genes to regulation of normal lymphopoiesis and development of malignant lymphomas. We showed that clinically defined subclasses of primary LBCLs display site-specific abnormal expression patterns of PcG genes of the HPC-HPH/PRC1 PcG complex, and that some of these patterns (such as the expression profile of BMI-1) may be diagnostically relevant. We propose that distinct expression profiles of PcG genes result in abnormal formation of HPC-HPH/PRC1 PcG complexes, and that this contributes to loss of normal gene regulation and different clinical behavior of clinically defined LBCLs.
Footnotes
Address reprint requests to Frank M. Raaphorst, Ph.D., VU Medical Center, Dept. of Pathology, De Boelelaan 1117, 1081 HV Amsterdam, The Netherlands. E-mail: fm.raaphorst@vumc.nl.
References
- Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- Raaphorst FM, Otte AP, Meijer CJ. Polycomb-group genes as regulators of mammalian lymphopoiesis. Trends Immunol. 2001;22:682–690. doi: 10.1016/s1471-4906(01)02082-8. [DOI] [PubMed] [Google Scholar]
- Smale ST. The establishment and maintenance of lymphocyte identity through gene silencing. Nat Immunol. 2003;4:607–615. doi: 10.1038/ni0703-607. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Polycomb group genes as epigenetic regulators of normal and leukemic hemopoiesis. Exp Hematol. 2003;31:567–585. doi: 10.1016/s0301-472x(03)00081-x. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW. Programming off and on states in chromatin: mechanisms of polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Gunster MJ, Satijn DP, Hamer KM, den Blaauwen JL, de Bruijn D, Alkema MJ, van Lohuizen M, van Driel R, Otte AP. Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol Cell Biol. 1997;17:2326–2335. doi: 10.1128/mcb.17.4.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Otte AP. Polycomb group protein complexes: do different complexes regulate distinct target genes? Biochim Biophys Acta. 1999;1447:1–16. doi: 10.1016/s0167-4781(99)00130-x. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Otte AP. RING1 interacts with multiple polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol. 1999;19:57–68. doi: 10.1128/mcb.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satijn DP, Gunster MJ, van der Vlag J, Hamer KM, Schul W, Alkema MJ, Saurin AJ, Freemont PS, van Driel R, Otte AP. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt RG, van der Vlag J, Gunster MJ, Hamer KM, den Blaauwen JL, Satijn DP, Hendrix T, van Driel R, Otte AP. Characterization of interactions between the mammalian polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S, McCabe D, Pirrotta V. Recruitment of components of polycomb group chromatin complexes in Drosophila. Development. 2001;128:75–85. doi: 10.1242/dev.128.1.75. [DOI] [PubMed] [Google Scholar]
- Ng J, Hart CM, Morgan K, Simon JA. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/mcb.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Harte PJ. The Drosophila polycomb group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development. 1998;125:3483–3496. doi: 10.1242/dev.125.17.3483. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CA, Ng J, Peterson AJ, Morgan K, Simon J, Jones RS. The Drosophila esc and E(z) proteins are direct partners in polycomb group-mediated repression. Mol Cell Biol. 1998;18:2825–2834. doi: 10.1128/mcb.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, DeCamillis M, Zink D, Cheng N, Brock HW, Paro R. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 1992;11:2941–2950. doi: 10.1002/j.1460-2075.1992.tb05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Paro R. The polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol Cell Biol. 1997;17:6773–6783. doi: 10.1128/mcb.17.12.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunster MJ, Raaphorst FM, Hamer KM, den Blaauwen JL, Fieret E, Meijer CJ, Otte AP. Differential expression of human polycomb group proteins in various tissues and cell types. J Cell Biochem. 2001;36:129–143. doi: 10.1002/jcb.1093. [DOI] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Sewalt RG, Lachner M, Vargas M, Hamer KM, den Blaauwen JL, Hendrix T, Melcher M, Schweizer D, Jenuwein T, Otte AP. Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3–K9 methylation contributes to chromosomal targeting of polycomb group proteins. Mol Cell Biol. 2002;22:5539–5553. doi: 10.1128/MCB.22.15.5539-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dahiya A, Wong S, Gonzalo S, Gavin M, Dean DC. Linking the rb and polycomb pathways. Mol Cell. 2001;8:557–569. doi: 10.1016/s1097-2765(01)00346-x. [DOI] [PubMed] [Google Scholar]
- Akasaka T, van Lohuizen M, van der LN, Mizutani-Koseki Y, Kanno M, Taniguchi M, Vidal M, Alkema M, Berns A, Koseki H. Mice doubly deficient for the polycomb group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development. 2001;128:1587–1597. doi: 10.1242/dev.128.9.1587. [DOI] [PubMed] [Google Scholar]
- Akasaka T, Tsuji K, Kawahira H, Kanno M, Harigaya K, Hu L, Ebihara Y, Nakahata T, Tetsu O, Taniguchi M, Koseki H. The role of mel-18, a mammalian polycomb group gene, during IL-7-dependent proliferation of lymphocyte precursors. Immunity. 1997;7:135–146. doi: 10.1016/s1074-7613(00)80516-6. [DOI] [PubMed] [Google Scholar]
- Ohta H, Sawada A, Kim JY, Tokimasa S, Nishiguchi S, Humphries RK, Hara J, Takihara Y. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J Exp Med. 2002;195:759–770. doi: 10.1084/jem.20011911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokimasa S, Ohta H, Sawada A, Matsuda Y, Kim JY, Nishiguchi S, Hara J, Takihara Y. Lack of the polycomb-group gene rae28 causes maturation arrest at the early B-cell developmental stage. Exp Hematol. 2001;29:93–103. doi: 10.1016/s0301-472x(00)00620-2. [DOI] [PubMed] [Google Scholar]
- Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G. Functional antagonism of the polycomb-group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev. 1999;13:2691–2703. doi: 10.1101/gad.13.20.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkema MJ, Jacobs H, van Lohuizen M, Berns A. Pertubation of B and T cell development and predisposition to lymphomagenesis in Emu Bmi1 transgenic mice require the Bmi1 RING finger. Oncogene. 1997;15:899–910. doi: 10.1038/sj.onc.1201262. [DOI] [PubMed] [Google Scholar]
- Lessard J, Baban S, Sauvageau G. Stage-specific expression of polycomb group genes in human bone marrow cells. Blood. 1998;91:1216–1224. [PubMed] [Google Scholar]
- Raaphorst FM, van Kemenade FJ, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Cutting edge: polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J Immunol. 2000;164:1–4. doi: 10.4049/jimmunol.164.1.1. [DOI] [PubMed] [Google Scholar]
- Raaphorst FM, Otte AP, van Kemenade FJ, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Meijer CJ. Distinct bmi-1 and ezh2 expression patterns in thymocytes and mature t cells suggest a role for polycomb genes in human T cell differentiation. J Immunol. 2001;166:5925–5934. doi: 10.4049/jimmunol.166.10.5925. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Olson DJ, van d V, Hamer KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R, Otte AP. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwatari H, Nakanishi K, Kondoh G, Hayasaka N, Li Q, Yamashita A, Inoue H, Hakura A. Suppression of tumor growth by the 3′ untranslated region of mel-18 in 3Y1 cells transformed by the E6 and E7 genes of human papillomavirus type 18. Cancer Lett. 1997;117:57–65. doi: 10.1016/s0304-3835(97)00200-0. [DOI] [PubMed] [Google Scholar]
- Kanno M, Hasegawa M, Ishida A, Isono K, Taniguchi M. mel-18, a polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. EMBO J. 1995;14:5672–5678. doi: 10.1002/j.1460-2075.1995.tb00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM, van Kemenade FJ, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Co-expression of BMI-1 and EZH2 polycomb group genes in Reed-Sternberg cells of Hodgkin’s disease. Am J Pathol. 2000;157:709–715. doi: 10.1016/S0002-9440(10)64583-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kemenade FJ, Raaphorst FM, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood. 2001;97:3896–3901. doi: 10.1182/blood.v97.12.3896. [DOI] [PubMed] [Google Scholar]
- Visser HP, Gunster MJ, Kluin-Nelemans HC, Manders EM, Raaphorst FM, Meijer CJ, Willemze R, Otte AP. The polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol. 2001;112:950–958. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- Grange F, Bekkenk MW, Wechsler J, Meijer CJ, Cerroni L, Bernengo M, Bosq J, Hedelin G, Fink PR, van Vloten WA, Joly P, Bagot M, Willemze R. Prognostic factors in primary cutaneous large B-cell lymphomas: a European multicenter study. J Clin Oncol. 2001;19:3602–3610. doi: 10.1200/JCO.2001.19.16.3602. [DOI] [PubMed] [Google Scholar]
- Willemze R, Meijer CJ. EORTC classification for primary cutaneous lymphomas: a comparison with the R. E. A. L. classification and the proposed WHO classification. Ann Oncol. 2000;11(Suppl 1):S11–S15. [PubMed] [Google Scholar]
- Willemze R, Kerl H, Sterry W, Berti E, Cerroni L, Chimenti S, Diaz-Perez JL, Geerts ML, Goos M, Knobler R, Ralfkiaer E, Santucci M, Smith N, Wechsler J, van Vloten WA, Meijer CJ. EORTC classification for primary cutaneous lymphomas: a proposal from the Cutaneous Lymphoma Study Group of the European Organization for Research and Treatment of Cancer. Blood. 1997;90:354–371. [PubMed] [Google Scholar]
- Rijlaarsdam JU, Toonstra J, Meijer OW, Noordijk EM, Willemze R. Treatment of primary cutaneous B-cell lymphomas of follicle center cell origin: a clinical follow-up study of 55 patients treated with radiotherapy or polychemotherapy. J Clin Oncol. 1996;14:549–555. doi: 10.1200/JCO.1996.14.2.549. [DOI] [PubMed] [Google Scholar]
- Willemze R, Meijer CJ, Sentis HJ, Scheffer E, van Vloten WA, Toonstra J, van der Putte SC. Primary cutaneous large cell lymphomas of follicular center cell origin. A clinical follow-up study of nineteen patients. J Am Acad Dermatol. 1987;16:518–526. doi: 10.1016/s0190-9622(87)70068-1. [DOI] [PubMed] [Google Scholar]
- Wechsler J, Bagot M. Primary cutaneous large B-cell lymphomas. Semin Cutan Med Surg. 2000;19:130–132. doi: 10.1016/s1085-5629(00)80010-3. [DOI] [PubMed] [Google Scholar]
- Pandolfino TL, Siegel RS, Kuzel TM, Rosen ST, Guitart J. Primary cutaneous B-cell lymphoma: review and current concepts. J Clin Oncol. 2000;18:2152–2168. doi: 10.1200/JCO.2000.18.10.2152. [DOI] [PubMed] [Google Scholar]
- Vermeer MH, Geelen FA, van Haselen CW, Voorst Vader PC, Geerts ML, van Vloten WA, Willemze R. Primary cutaneous large B-cell lymphomas of the legs. A distinct type of cutaneous B-cell lymphoma with an intermediate prognosis. Dutch Cutaneous Lymphoma Working Group. Arch Dermatol. 1996;132:1304–1308. [PubMed] [Google Scholar]
- Alkema MJ, Bronk M, Verhoeven E, Otte A, van’t Veer LJ, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J., Jr Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 2003;102:592–600. doi: 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]