Abstract
We previously reported that enhanced expression of the α7β1 integrin ameliorates the development of muscular dystrophy and extends longevity in α7BX2-mdx/utr−/− transgenic mice (Burkin DJ, Wallace GQ, Nicol KJ, Kaufman DJ, Kaufman SJ: Enhanced expression of the α7β1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J Cell Biol 2001, 152:1207–1218). We now report on the mechanism by which these mice were rescued by the integrin. As a result of increased integrin in α7BX2-mdx/utr−/− mice the structural integrity of the myotendinous and neuromuscular junctions are maintained. A twofold increase in satellite cells in α7BX2-mdx/utr−/− skeletal muscle was detected by immunofluorescence using the satellite cell marker c-met. These cells enhanced the regenerative capacity of muscle in the transgenic animals as determined by fusion of BrdUrd-labeled cells into muscle fibers. Increased integrin also leads to hypertrophy. Finally, transgenic expression of α7BX2 integrin chain in skeletal muscle secondarily reduces the development of cardiomyopathy, the ultimate cause of death in these animals. We believe this multiplicity of responses to increased α7β1 integrin collectively inhibits the development of muscle disease and increases longevity in these mice.
The association of the extracellular matrix with the cell cytoskeleton is central to maintaining muscle function and integrity. Defects in protein complexes that mediate this association underlie the pathology in a variety of muscle diseases.1 The α7β1 integrin is a laminin receptor in skeletal and cardiac muscle that links the extracellular matrix to the cell cytoskeleton.2,3 The α7β1 integrin can also mediate both inside-out and outside-in cell signaling. This dual function allows muscle cells to attach to the extracellular matrix and transmit signals into cells that regulate cell attachment, migration, growth, and proliferation.4–7
Multiple isoforms of the α7 chain are produced in skeletal muscle by developmentally regulated RNA splicing. At least two alternative cytoplasmic domains, α7A and α7B, and two alternative extracellular domain isoforms, α7X1 and α7X2, are produced.8–11 The X1 and X2 isoforms bind different laminin isoforms with different affinities.6,12 Two isoforms of the β1 integrin chain are also expressed in skeletal muscle and these add to the structural and functional diversity of the integrin. The β1A integrin chain is present in myoblasts, satellite cells, and immature myofibers, as well as in a wide variety of other cell types. In mature skeletal and cardiac muscle the β1D isoform is expressed and is believed to promote a more tenacious association between the integrin and cytoskeleton.13–16
Several proteins associate with α7β1 integrin and these likely underlie diverse functions. These include galectin-1,17 MIBP,18 ADAM12,19 and acetylcholine receptor.20,21 Mutations in the α7 gene lead to muscular dystrophy in humans22 and mice.23 Moreover, secondary defects in expression of the integrin are relatively common in a variety of human muscle diseases and likely contribute to the observed pathology (Burkin, North, Kaufman, in preparation).24
In patients with Duchenne muscular dystrophy and in mdx mice, mutations in the dystrophin gene result in the absence of dystrophin.25,26 Dystrophin is a key component of a macromolecular complex that also links the cell cytoskeleton to laminin in the extracellular matrix.27–30 In Duchenne muscular dystrophy patients, the lack of dystrophin results in severe muscle disease. Patients are usually confined to a wheelchair in their early teens and death from cardiopulmonary failure often occurs in their early twenties. In contrast, mdx mice show mild muscle pathology and live a normal life span.26,31
Utrophin structurally resembles dystrophin and its localization in mature skeletal muscle is normally confined to neuromuscular junctions.32–35 In mdx mice more utrophin is present and it is distributed around the sarcolemma suggesting that it may partially compensate for the absence of dystrophin.28 In mdx/utr−/− mice, which lack both dystrophin and utrophin, the pathology that develops is severe, more closely resembles that seen in Duchenne patients, and these mice die at 4 to 20 weeks of age.36–38
Both mdx mice and Duchenne patients have elevated levels of α7β1 integrin.39,40 This led us to suggest that the dystrophin and integrin linkage systems may be complementary, and that the increased amount of α7β1 integrin in the absence of dystrophin may impede the development of pathology. Moreover, it led us to question whether further increasing integrin levels in the absence of the dystrophin linkage system could further ameliorate the development of muscle disease. This hypothesis was tested and confirmed with transgenic mdx/utr−/− mice in which expression of the α7BX2 integrin chain was increased in skeletal muscle. An additional twofold increase in integrin did ameliorate the development of muscular dystrophy and longevity was extended threefold in α7BX2-mdx/utr−/− transgenic mice.36
The mechanism by which the mdx/utr−/− mice were rescued by the α7β1 integrin is the focus of these studies. Additional integrin may provide increased mechanical stability to muscle fibers by enhancing attachment to the basal lamina. This could be accomplished by increased integrin-mediated adhesion or regenerative capacity, or by stabilization of components of the dystrophin complex in the plasma membrane that are expressed but markedly reduced in the absence of dystrophin. To examine these possibilities, proteins of the dystrophin glycoprotein complex were analyzed. In addition, muscle regenerative capacity and the structural organization of neuromuscular and myotendinous junctions, which are normally rich in α7β1 integrin,41–44 and which are often damaged in diseased muscle, were also studied.45–48
No changes in dystrophin-associated proteins (DAPs) were detected in skeletal muscle of α7BX2-mdx/utr−/− mice compared to mdx/utr−/− animals. In contrast, mice with enhanced α7β1 integrin levels maintained the normal architecture of myotendinous and neuromuscular junctions. In addition, satellite cell numbers were increased in mice with enhanced α7β1 integrin and these cells were found to participate in muscle repair. Additional integrin also promoted muscle hypertrophy as reflected in increased fiber cross-sectional areas. These results point to a mechanism by which enhanced levels of α7β1 integrin increase the regenerative capacity of muscle as well as maintain the structural organization of junctional sites. As a consequence of maintaining skeletal muscle integrity and function, increased integrin also reduced the development of cardiomyopathy in α7BX2-mdx/utr−/− transgenic mice, a likely key factor in extending the life span of these animals.
Materials and Methods
Transgenic Mice
The generation of transgenic mice expressing the rat α7BX2 protein under control of the MCK promoter and genotyping have been described.36 Transgenic mdx/utr+/− male mice were bred with mdx/utr+/− females to produce transgenic α7BX2-mdx/utr−/− animals. Mice were analyzed at 3, 5, 8, or 10 weeks of age. Transgenic mice used in this study have an approximately fourfold increase in α7BX2 protein in their hindlimb muscle compared to nontransgenic wild-type mice and a twofold increase greater than that detected in mdx/utr−/− mice.36 The original C57BL10 mdx/utr+/− mice were kindly provided by Dr. Joshua Sanes (Harvard University, Cambridge, MA).37
Antibodies
For immunofluorescence analysis the monoclonal antibody O26 was used to detect the rat α7 chain.3 Polyclonal antibodies against α-sarcoglycan, β-sarcoglycan, and sarcospan were gifts from Dr. Kevin Campbell (University of Iowa, Iowa City, IA). Monoclonal antibodies against β-dystroglycan (NCL-β-DG) and dystrobrevin (D63020) were purchased from Novocastra Laboratories (Newcastle upon Tyne, UK) and Transduction Laboratories (Lexington, KY), respectively. Rabbit anti-c-met polyclonal antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Neuromuscular junctions were detected using rhodamine-labeled α-bungarotoxin (Molecular Probes, Eugene, OR) binding to acetylcholine receptors. Horseradish peroxidase- and fluorescein isothiocyanate-labeled donkey anti-mouse and anti-rabbit antibodies were purchased from Jackson ImmunoResearch Laboratories Inc. (West Grove, PA). G3G4 anti-BrdUrd antibody was prepared as described.49
Western Analysis
Muscle tissue from at least six mice of each genotype was extracted in either 200 mmol/L octyl-β-d-glucopyranoside or 10% sodium dodecyl sulfate in 50 mmol/L Tris-HCl, pH 7.4, 2 mmol/L phenylmethyl sulfonyl fluoride, 1:200 dilution of Protease Cocktail Set III (Calbiochem, La Jolla, CA), 1 mmol/L CaCl2, and 1 mmol/L MgCl2 at 4°C for 1 hour. Supernatants were collected and protein concentrations were determined using Bradford assays. Equal amounts of extracted muscle proteins were separated on 8% sodium dodecyl sulfate polyacrylamide gels at 40 mA for 50 minutes. The proteins were transferred to nitrocellulose filters. Blocked filters were incubated with the respective primary antibodies. Horseradish peroxidase linked to anti-rabbit, anti-goat, or anti-mouse secondary antibodies (Jackson Immunoresearch), were used to detect bound primary antibodies. Immunoreactive protein bands were detected using an enhanced chemiluminescence kit (Amersham Biotech, Arlington Heights, IL).
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Mouse heart ventricles were pulverized in liquid nitrogen and RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA). The RNA was treated with RNase-free DNase I (Invitrogen) for 20 minutes at room temperature to remove genomic DNA. RT-PCR reactions were done using the Superscript One-Step RT-PCR kit (Invitrogen). The rat α7 transcript was detected with primers 5′-TTCATGTTGAAATAAGGCAGGTT-3′ and 5′-CACAGGAAAGACTTAGGAGGG-3′. To determine the relative levels of atrial natriuretic factor (ANF) mRNA, multiplex RT-PCR was performed using primers for mouse ANF and GAPDH. Primers were: ANF forward, 5′-CCAGGCCATATTGGAGCAAA-3′; ANF reverse, 5′-GAAGCTGTTGCA GCCTAGTC-3′; GAPDH forward, 5′-CCATGGAGAAGGCCGGGG-3′; GAPDH reverse, 5′-CAAAGTTGTCATGGATGACC-3′. Reactions using 200 ng of ventricular RNA were performed for 20 cycles, which is within the range of exponential accumulation of both ANF and GAPDH products. Control reactions lacking reverse transcriptase were included to ensure that products were not produced from genomic DNA. PCR products were electrophoresed in 1.5% agarose. Band intensities were determined using ImageQuant software (Amersham). ANF transcript intensity was normalized to GAPDH and expressed as fold expression greater than wild type. Three to five animals were used per genotype.
Immunofluorescence
Muscle tissue from wild-type, mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice were embedded in OCT compound (Sakura, Torrance, CA) and frozen in liquid nitrogen-cooled isopentane. Ten-μm sections were cut using a Leica CM1900 series cryostat (Germany), and placed on Surgipath microscope slides (Richmond, IL). Sections were fixed in −20°C acetone for 1 minute, rehydrated in phosphate-buffered saline (PBS) for 10 minutes, and blocked in PBS containing 10% horse serum for 15 minutes. Mouse immunoglobulin was blocked using a MOM kit (Vector Laboratories, Burlingame, CA) when localizations used mouse monoclonal antibodies. Primary antibodies were detected using a 1:100 dilution of fluorescein isothiocyanate-labeled donkey anti-mouse or anti-rabbit antibody in 1% horse serum in PBS. Slides were mounted in Vectashield (Vector Laboratories).
Satellite Cells
Satellite cells were detected by immunofluorescence using anti-c-met antibody diluted 1:500 in PBS. The numbers of c-met-positive cells in 50 fields were scored using the ×40 objective. Sections from the lower hindlimb were taken 50 μm apart to sample the entire musculature. At least six animals per genotype were scored. Localization of the antibody was visualized using a Leica DMRXA2 microscope. Images were acquired using an AxioCam digital camera (Carl Zeiss, Thornwood, NY) and OpenLab software (Improvision, Lexington, MA). Controls lacking primary antibody were routinely negative.
BrdUrd Labeling in Vivo
Mice (3, 5, and 10 weeks old) were injected intraperitoneally with 5-bromo-2-deoxyuridine (50 mg/kg body weight). Lower hindlimbs were excised 72 hours later and flash-frozen in liquid nitrogen-cooled isopentane. Cross-sections (10 μm), cut at least 50 μm apart sampled the entire musculature, were fixed in 95% ice-cold ethanol and the DNA rendered accessible by incubating in 2 N HCl for 20 minutes followed by neutralization in 50 mmol/L NaCl, 100 mmol/L Tris-HCl, pH 7.4. Sections were stained with G3G4 anti-BrdUrd antibody49 and fluorescein isothiocyanate-donkey anti-mouse secondary antibody (1:100), counterstained with a 1:1000 dilution of tetramethyl-rhodamine isothiocyanate-wheat germ agglutinin (Molecular Probes) to delineate fibers, and mounted in Vectashield/DAPI (Vector Laboratories). 4,6-Diamidino-2-phenylindole (DAPI)-stained central nuclei within muscle fibers were identified and those also labeled with anti-BrdUrd antibody were counted. BrdUrd-labeled central nuclei in 50 fields were scored using the ×40 objective for at least six animals of each genotype. Omitting anti-BrdUrd antibody or blocking by addition of soluble BrdUrd yields negative results.49
Transmission Electron Microscopy
The gastrocnemius muscle and associated tendons and the sternomastoid muscle were isolated from three 5-week-old mice. Tissues were immediately fixed in 2.5% glutaraldehyde and 2.5% paraformaldehyde, incubated overnight at 4°C, and rinsed with cold 0.1% cacodylate buffer. The muscle was postfixed in 2% OsO4 for 1 hour at 4°C and incubated in potassium ferricyanide (KCN) for 30 minutes at 4°C. Muscle was stained with saturated uranyl acetate for 90 minutes at room temperature, washed twice with 10% ethanol, and dehydrated in a graded alcohol series. Muscle was infiltrated in 1:1 acetonitrile/Embed 812 (Electron Microscopy Science, Hatfield, PA) for 1 hour, then 1:3 acetonitrile/Embed 812 overnight. The tissue was embedded in Embed 812 and polymerization was allowed to occur at 60°C for 3 to 5 days. Embedded tissue was sectioned at 0.1 μm using a Reichert Ultracut ultramicrotome (Depew, NY) and then stained with uranyl acetate and lead citrate. Sections were viewed with a Hitachi H-600 transmission electron microscope (Japan) at ×15,000 magnification. Folds at the neuromuscular and myotendinous junctions were counted for at least 10 junctions per animal.
Histology, Evans Blue Dye Uptake, and Fiber Cross-Sectional Areas
Ten-μm cryostat sections of cardiac and skeletal muscle were fixed in 100% acetone at −20°C for 10 minutes, rinsed in tap water for 10 minutes, and stained with hematoxylin and eosin using standard histological procedures. Fiber cross-sectional areas were measured for soleus and tibialis anterior muscles from wild-type, mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice using the advanced measurements component of OpenLab software (Improvision). The cross-sectional areas of 200 to 300 fibers from each muscle were measured for each of four animals per genotype and the distributions were plotted using Microsoft Excel. For assessment of cardiomyocyte membrane damage, mice were injected intraperitoneally with 50 μg per 10 g of body weight of a sterile solution of Evans blue dye. Cardiac tissue was harvested 24 to 48 hours after injection and snap-frozen in liquid nitrogen-cooled isopentane. Frozen sections were cut at 10 μm, fixed in 100% acetone at −20°C for 10 minutes, washed with PBS for 10 minutes, and mounted with Vectashield. Images were acquired using an AxioCam digital camera (Zeiss), and OpenLab software. Areas of fibrotic lesions and Evans blue dye-positive areas were measured in five sections from each of five animals per genotype using the advanced measurements component of OpenLab.
Statistical Analysis
All averaged data are presented as the mean ± SEM. Comparisons between groups were performed by analysis of variance or unpaired t-test for BrdUrd, using STATVIEW (SAS Institute Inc., San Francisco, CA). Differences were considered significant at P < 0.05.
Results
Dystrophin-Associated Proteins Remain Unaltered in α7BX2-mdx/utr−/− Transgenic Mice
One possible mechanism by which the development of dystrophic muscle fibers is limited by enhanced levels of integrin may involve an increase and/or stabilization of dystrophin-associated proteins (DAPs) in the sarcolemma of dystrophic muscle. To determine whether enhanced expression of the α7β1 integrin protein altered levels of the DAPs, Western blot analysis and immunofluorescence were used to analyze β-dystroglycan, sarcospan, and β-dystrobrevin in normal and transgenic mice. Western blots showed no significant differences in the levels of these proteins in transgenic and nontransgenic mice at 5, 8, or 10 weeks of age. Compared to wild-type mice, these proteins were reduced or undetected in mdx muscles as reported by Ohlendieck and Campbell.34 The DAPs in mdx/utr−/− and α7BX2-mdx/utr−/− mice were similarly absent or reduced (Figure 1). Likewise, immunofluorescence did not reveal any major changes in the localization of these proteins: considerably less was detected at the sarcolemma of mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice compared to wild-type animals (data not shown). Therefore altered expression or stabilization of the DAPs in α7BX2-mdx/utr−/− skeletal muscle appears not to play a role in preventing the development of pathology in these animals.
Figure 1.
Dystrophin-associated proteins remain unchanged in α7BX2 transgenic mice. Western blot analysis of DAPs isolated from wild-type, mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice. An absence of dystrophin and reduction in β-dystroglycan, dystrobrevin, and sarcospan in 5- and 10-week-old mdx and mdx/utr−/− mice were observed as previously reported.34 Likewise, α7BX2 transgenic mice show a similar absence or reduction in these DAPs. Samples from two mice per genotype are shown and at least six mice per genotype were analyzed.
Junctional Integrity Is Maintained in α7BX2-mdx/utr−/− Mice
The myotendinous junction is the focal point of longitudinal stress during muscle contraction and it is one site where α7β1 integrin is concentrated in normal muscle.42–44,50,51 Mice with targeted disruption of the α7 gene exhibit muscular dystrophy that affects the structure of myotendinous junctions,23,52 thus the integrin appears to have a role in the formation and maintenance of integrity of these sites. We therefore analyzed the ultrastructure of these junctions by electron microscopy of gastrocnemius muscles isolated from wild-type, mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice. Normal myotendinous junctions are characterized by interdigitating finger-like folds that increase contact between muscle and tendon (Figure 2A). In mdx mice there is a slight decrease in the number of folds at these sites compared to wild-type animals, consistent with earlier results.53 In contrast, in the absence of both dystrophin and utrophin there is an approximate sixfold decrease in folding (Figure 2B). The increased integrin in α7BX2-mdx/utr−/− mice maintained near normal junctional folding. Therefore enhanced expression of the α7β1 integrin maintains the structure of the muscle membrane at the myotendinous junction.
Figure 2.
Myotendinous and neuromuscular junctions are maintained in α7BX2 transgenic mice. A: Structure of the myotendinous junctions of gastrocnemius muscle of 5-week-old wild-type, mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice. The sarcolemma of the myotendinous junction in wild-type and mdx mice is highly folded (arrows), increasing the contact between muscle and tendon. In contrast, the sarcolemma of the myotendinous junction in the severely dystrophic mdx/utr−/− mice exhibits little folding. Enhanced expression of the α7BX2 transgene in mdx/utr−/− mice results in maintenance of sarcolemmal folding similar to that seen in wild-type and mdx mice. B: Quantitation of sarcolemmal folds at the myotendinous junction reveals a slight reduction in folding in mdx mice compared to wild-type. In contrast, a sixfold reduction in sarcolemmal folds at the myotendinous junction, was scored in mdx/utr−/− mice. Expression of the α7BX2 transgene maintains near normal folding of the sarcolemma in mdx/utr−/− mice. C: A mild reduction in postsynaptic folding of the neuromuscular junctions of sternomastoid muscles is observed in mdx mice compared to wild-type animals whereas a major reduction in junctional folds occurs in mdx/utr−/− mice.36 Expression of the α7BX2 transgene in mdx/utr−/− mice results in a maintenance of postsynaptic folding comparable to that observed in mdx mice. Folds at the myotendinous and neuromuscular junctions were counted for at least 10 junctions per animal (n = 3 mice per genotype). The standard errors of the mean values for each animal within each group are indicated. P < 0.05 for comparisons of transgenic and nontransgenic mice.
We previously showed that the structure of the neuromuscular junction is also maintained in the α7BX2-mdx/utr−/− transgenic mice.36 The neuromuscular junction is an essential communication point between the motor neuron and muscle fiber. The α7β1 integrin is involved early in the development of the neuromuscular junction, with specific isoforms of the integrin engaging in acetylcholine receptor clustering.20,21 In the adult, α7β1 integrin is concentrated at these junctions, and in the absence of specific isoforms of laminin at these sites there is a commensurate reduction of integrin.41 The postsynaptic membrane of the neuromuscular junction is normally highly folded. This folding increases the surface area, allowing efficient communication and adhesion between the muscle and neuron.54 Because of the role the integrin plays in the development of the neuromuscular junction, the structure of this membrane domain was also analyzed by electron microscopy. Sternomastoid muscles from 5-week-old wild-type, mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice were isolated and the numbers of junctional folds were determined.
Similar to that seen at the myotendinous junction, there is only a minimal reduction in folding at the neuromuscular junction in mdx mice compared to normal animals (Figure 2C). Likewise, in utr−/− mice there is only a mild abnormality at these junctions.55,56 Thus either dystrophin or utrophin appears sufficient to maintain junctional integrity. In contrast, in the absence of both proteins, in mdx/utr−/− mice, the neuromuscular junction is severely compromised.36–38 There is a four to fivefold reduction in junctional folds in mdx/utr−/− mice compared to wild-type animals (Figure 2C). In contrast, in α7BX2-mdx/utr−/− mice, junctional folding is maintained to approximately the same extent as that seen in mdx animals. Therefore enhanced expression of the α7β1 integrin maintains the structure of the neuromuscular junction in the muscles of mice that would otherwise develop severe muscular dystrophy.
Increased Integrin Enhances the Regenerative Capacity of α7BX2-mdx/utr−/− Skeletal Muscle
The α7β1 integrin promotes the organization of laminin in the matrix.7 Moreover, laminin can promote skeletal muscle development in vitro by selectively maintaining the proliferation of myogenic cells.4 This suggested that an increase in the amount of the α7β1 integrin may expand the population of myogenic precursor cells and prompted us to ask whether the transgenic expression of integrin might facilitate repair and regeneration in dystrophic muscle of mdx/utr−/− mice by expanding the population of satellite cells. C-met, a receptor for hepatocyte growth factor, is expressed on myogenic precursor cells57 and was used as an index of satellite cells (Figure 3A). The number of c-met-positive cells detected by immunofluorescence in 5-week-old mice was approximately twofold greater in α7BX2-mdx/utr−/− mice compared to nontransgenic mice (Figure 3B).
Figure 3.
Increased satellite cell number in the muscle of α7BX2 transgenic mice. A: Antibody against the satellite cell marker c-met (hepatocyte growth factor) was used to identify these cells in sections of the lower hindlimb from 5-week-old wild-type, mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice (green, arrow). C-met-positive cells in 50 fields were scored. Mean numbers (±SEM) for at least six animals of each genotype are given. Wheat germ agglutinin staining delineates fibers (red). B: A twofold increase in satellite cells was detected in hindlimb muscle of α7BX2-mdx/utr−/− transgenic mice compared to nontransgenic animals (P < 0.05).
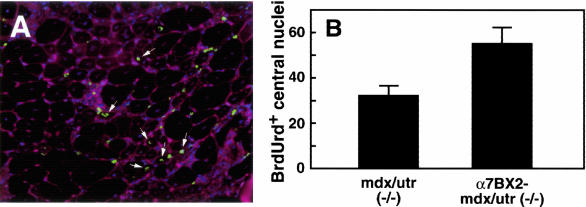
To determine whether the increased number of satellite cells observed in transgenic α7BX2-mdx/utr−/− skeletal muscle contributed to muscle repair, mice were injected with BrdUrd to label proliferating cells. BrdUrd-positive central nuclei in fibers were scored 72 hours later by immunofluorescence (Figure 4A). The number of BrdUrd-labeled central nuclei in 5-week-old α7BX2-mdx/utr−/− transgenic mice was 1.7-fold greater than in nontransgenic mdx/utr−/− mice (Figure 4B). Compared to nontransgenic mice, the increased number of BrdUrd-labeled central nuclei in α7BX2-mdx/utr−/− mice paralleled the increase of c-met-positive cells at 3, 5, and 10 weeks (Table 1). The increase in c-met-positive cells and BrdUrd-labeled central nuclei was maximal at 5 weeks and was sustained until at least 10 weeks of age. Thus, increased integrin expands the reservoir of satellite cells and the regenerative capacity of dystrophic muscle, at least through 10 weeks of age.
Figure 4.
Increased integrin promotes repair. To determine whether the satellite cells were used in repair, mice were injected with BrdUrd to label replicating cells. Seventy-two hours later muscle sections were analyzed by immunofluorescence for BrdUrd incorporation into DNA (green). Wheat germ agglutinin staining delineates fibers (red). Nuclei are stained with DAPI (blue). BrdUrd-labeled central nuclei (arrows) in 50 random fields were scored for each animal. Mean numbers (±SEM) are given for 11 animals for each genotype. For mdx/utr−/− mice, the value is 32.0 ± 4.56 and for α7BX2-mdx/utr−/− 55.0 ± 7.35 (P = 0.0152, unpaired t-test). Thus increased integrin expands the reservoir of satellite cells and the regenerative capacity of dystrophic muscle.
Table 1.
Increased Integrin Expands Regenerative Capacity in α7BX2-mdx/utr−/− Mice
| Age (weeks) | C-met-positive cells | BrdUrd-positive central nuclei |
|---|---|---|
| 3 | 1.2-fold | 1.2-fold |
| 5 | 2.0-fold* | 1.72-fold* |
| 10 | 1.75-fold* | 1.68-fold* |
The numbers of c-met-positive cells and BrdUrd-labeled central nuclei in hindlimb fibers of mdx/utr−/− and α7BX2-mdx/utr−/− mice were determined at 3, 5, and 10 weeks of age. The relative increases in theα 7BX2-mdx/utr−/− mice compared to the mdx/utr−/− animals are indicated. At least six animals per genotype were scored.
Statistical significance by analysis of variance; P < 0.05.
Increased Integrin Promotes a Hypertrophic Response in α7BX2-mdx/utr−/− Mice
Previous studies have shown that an increase in satellite cells can result in increased regenerative capacity of skeletal muscle. This is particularly evident in myostatin-negative mice. In the absence of myostatin, a negative regulator of satellite cell activation and self-renewal, an increase in the number of satellite cells leads to hyperplasia and hypertrophic muscle.58,59 In the case of dystrophic mice lacking myostatin, this can attenuate the severity of muscular dystrophy.60
To determine whether muscle fiber hypertrophy was occurring in transgenic animals, cross-sectional areas of ∼1100 fibers from both the soleus and the tibialis anterior were measured in wild-type, mdx, mdx/utr−/−, and α7BX2-mdx/utr−/− mice. Using the largest fiber cross-sectional areas in wild-type animals as the normal upper limit, α7BX2-mdx/utr−/− mice had four times the number of hypertrophic fibers than mdx/utr−/− mice in both the soleus and the tibialis anterior. Overall, the distribution of fiber sizes in the transgenic mice is similar to the distribution of sizes seen in mdx muscle (Figure 5). Thus, increased expression of integrin promotes a hypertrophic response in α7BX2-mdx/utr−/− mice that may contribute to limiting the development of pathology in these animals.
Figure 5.
Increased expression of α7β1 integrin promotes hypertrophy. Cross-sectional areas of fibers in soleus and tibialis anterior muscles were measured. Using the largest fiber area in wild-type animals as a cutoff point (dashed line), α7BX2-mdx/utr−/− mice were found to have four times more hypertrophic fibers than mdx/utr−/− mice. Thus increasing the level of integrin promotes a hypertrophic response that contributes to the rescue of these animals. The cross-sectional areas of 200 to 300 fibers from each muscle were measured for each of four animals per genotype.
Transgenic Expression of α7BX2 Integrin Chain in Skeletal Muscle Reduces Cardiomyopathy in mdx/utr−/− Mice
Unlike mdx mice, which develop little if any cardiomyopathy, the mdx/utr−/− mouse develops severe cardiomyopathy characterized by cardiomyocyte degeneration and fibrotic lesions.37,38 This cardiomyopathy is abolished on targeted re-expression of utrophin in skeletal muscle, suggesting that the pathology is a secondary consequence of skeletal muscle abnormalities.61 It was therefore of interest to determine whether enhanced levels of the α7β1 integrin in skeletal muscle also reduced cardiomyopathy in α7BX2-mdx/utr−/− mice. The muscle creatine kinase promoter used in these experiments supports 100-fold less transcriptional activity in cardiac compared to skeletal muscle62 and no expression of the rat α7 transgene was detected in cardiac tissue of α7BX2-mdx/utr−/− mice by Western blots, immunofluorescence, or RT-PCR (data not shown).
Histological analyses showed that the extent of fibrotic lesions in mdx/utr−/− mice is largely reduced in α7BX2-mdx/utr−/− mice (Figure 6A). Measurement of fibrotic lesions as a percentage of total cardiac section area demonstrated a significant decrease in lesion area in α7BX2-mdx/utr−/− mice compared to mdx/utr−/− animals (5.14 ± 2.22% versus 1.18 ± 1.16%, respectively; P < 0.02). Fibrotic lesions were not observed in wild-type and mdx hearts. Similarly, significant uptake of Evans blue dye (an index of myocyte damage) was detected in cardiomyocytes of mdx/utr−/− mice, whereas α7BX2-mdx/utr−/− mice showed little or no dye uptake (Figure 6B). Measurement of areas of dye-positive lesions demonstrated 10-fold less Evans blue-positive areas in α7BX2-mdx/utr−/− mice compared to mdx/utr−/− animals (3.58 ± 1.81% versus 0.31 ± 0.10%; P < 0.02). Both wild-type and mdx hearts showed no uptake of Evans blue dye.
Figure 6.
α7BX2-mdx/utr−/− mice exhibit reduced cardiomyopathy. A: α7BX2-mdx/utr−/− mice have a fourfold reduction in the area of fibrotic lesions compared to mdx/utr−/− mice as evidenced by H&E staining of 8-week cardiac muscle (dashed outline). No lesions were observed in wild-type or mdx mice. B: Similarly, transgenic α7BX2-mdx/utr−/− mice showed a 10-fold reduction in the area of Evans blue dye uptake compared to mdx/utr−/−mice (arrows) and no dye uptake was observed in wild-type or mdx mice. C: Ventricular expression of ANF is an indicator of cardiomyopathy. RT-PCR analysis of ANF RNA shows that it is highly elevated in 8-week ventricular muscle of mdx/utr−/− mice and comparatively it is markedly reduced in α7BX2-mdx/utr−/− mice (P < 0.0002). All values are normalized to GAPDH.
RT-PCR analyses of ventricular expression of atrial natriuretic factor (ANF) (an index of cardiomyopathy) demonstrated that mdx mice, which exhibit mild or no cardiomyopathy, have 2.5-fold more ventricular ANF transcripts than wild-type animals (Figure 6C). In contrast, mdx/utr−/− mice have severe cardiomyopathy and exhibit a 7.2-fold increase in ANF transcripts compared to wild-type mice. This increase in ventricular ANF gene expression in dystrophic mdx/utr−/− mice is significantly (P < 0.0002) reduced in α7BX2-mdx/utr−/− animals to 3.8-fold that in controls. Thus increasing the amount of α7β1 integrin in skeletal muscle significantly ameliorates the development of pathology in skeletal muscle and thereby secondarily prevents cardiac tissue damage.
Discussion
Duchenne muscular dystrophy is an X-linked disease affecting 1 in 3500 newborn boys. Mutations in the dystrophin gene result in a lack of the dystrophin protein in both Duchenne patients and mdx mice. The loss of dystrophin leads to a breakdown of a transmembrane system linking laminin in the extracellular matrix to the actin cytoskeleton resulting in muscle disease. Attempts to treat muscular dystrophy have involved both direct gene replacement and gene repair as well as myoblast and precursor cell therapy.63–72 Recently, several additional approaches have emerged based on the study of proteins with complementary functions. In addition to the α7β1 integrin,36 increasing levels of utrophin,73,74 an agrin minigene,75 nitric oxide synthase,76 the disintegrin ADAM12,77 GalNAc transferase,78 calpastatin,79 and IGF-1,80 circumvent to various degrees the development of muscle disease in the absence of the dystrophin glycoprotein complex. The α7β1 integrin is a laminin receptor on the surface of muscle cells that interacts with the actin cytoskeleton and has complementary functions to the dystrophin glycoprotein complex. Enhanced expression of the α7β1 integrin in mice lacking dystrophin and utrophin results in a threefold increase in life span and a significant decrease in the development of muscle pathology in mice that would otherwise develop severe muscle disease.36 The mechanism by which enhanced levels of the α7β1 integrin rescued these mice is unclear and has been the focus of this investigation.
In the skeletal muscle of mdx mice the absence of dystrophin results in the loss of membrane-associated dystroglycans, sarcoglycans, and syntrophins.34 These complexes have vital roles in muscle function. The importance of DAPs has been demonstrated in transgenic mice in which elements of the complex have been partially restored. For example, transgenic expression of nNOS was shown to reduce levels of pathology in mdx mice.76 Therefore, a mechanism by which α7β1 integrin may rescue severely dystrophic mice may involve stabilization of the DAPs at the sarcolemma. To investigate this possibility, several members of the dystrophin-associated complex were analyzed in transgenic α7BX2-mdx/utr−/− mice and compared to nontransgenic animals. These studies revealed the expected reduction of DAPs in mdx/utr−/− mice compared to wild type. In α7BX2-mdx/utr−/− mice, the amounts of the sarcoglycans, dystrobrevins, and dystroglycans analyzed were similar to those seen in nontransgenic mdx/utr−/− mice. In addition, immunofluorescence revealed no changes in the membrane localization of any DAPs analyzed in transgenic mice. Therefore enhanced transgenic expression of the integrin does not appear to increase the expression or stabilization of DAPs in mdx/utr−/− mice. In contrast, ADAM12 interacts with the integrin19 and overexpression of ADAM12 in mdx mice increases expression of α7 integrin, utrophin, and associated proteins.81 This difference may reflect the more severe state of pathology that develops in the absence of both utrophin and dystrophin and/or a requirement for either utrophin or dystrophin to stabilize their associated proteins.
The β1A integrin chain is expressed in myoblasts and in a wide variety of other cell types. On myoblast differentiation into multinucleate fibers, RNA splicing results in an accompanying switch in integrin β1 chain synthesis from the β1A to the β1D isoform. It is believed that this change in the structure of the β1 chain cytoplasmic domain underlies a switch from α7β1A’s roles in replication and migration to the α7β1D integrin mediating a more tenacious attachment of the extracellular matrix and muscle fiber cytoskeleton.13,16 We previously reported that transgenic expression of the α7BX2 integrin chain in skeletal muscle results in an increase in the β1D chain.36 In contrast, in the absence of increased integrin in the mdx/utr−/− mice, there is a reversion to β1A expression. Thus the α7β1 integrin present in the dystrophic muscle of mdx/utr−/− mice is less adapted to maintaining muscle integrity. In contrast, the β1D integrin chain predominates in the α7BX2-mdx/utr−/− mice and thereby contributes to maintaining the normal association between muscle and the extracellular matrix. This is clearly reflected at the neuromuscular and myotendinous junctions, sites of integrin concentration.
The structural integrity of neuromuscular and myotendinous junctions is central to skeletal muscle function. The α7β1 integrin is normally enriched at these sites and functions in the development and maintenance of these domains.21,40,41 The importance of the α7β1 integrin to junctional integrity is demonstrated in mice lacking the α7 chain; these mice exhibit a muscular dystrophy caused by defects in myotendinous junctions.23 Our results demonstrate that the sarcolemma at both myotendinous and neuromuscular junctions in transgenic mice retain a normal highly folded structure that is absent in nontransgenic dystrophic animals. These results suggest that significant pathology may be due to compromised junctional sites in mdx/utr−/− mice that are not maintained by the limited increase (twofold) in endogenous α7β1 integrin. The further twofold increase in levels of α7β1 in the transgenic α7BX2-mdx/utr−/− mice, that is a fourfold increase greater than wild-type, appears to be required to maintain the normal organization of myotendinous and neuromuscular sarcolemma. It is possible that additional integrin would promote even better protection.
An increase in the capacity of muscle to regenerate may also underlie the mechanism by which the α7β1 integrin reduces the development of severely dystrophic mice. In this study a twofold increase in satellite cells, which can repair damaged muscle, was observed in mice with enhanced α7 chain expression. Although the MCK promoter used to drive expression of the rat α7 transgene is not active in myoblasts or satellite cells,82,83 the increase in satellite cell number may still be attributed to the increase in α7BX2 integrin chain expressed in transgenic skeletal muscle. Satellite cells are located under the basal lamina in close proximity to the myofiber sarcolemma. The α7β1 integrin has been shown to promote the organization of laminin in the extracellular matrix,7 and laminin has been shown to maintain the proliferation of myogenic precursor cells.4 Therefore increased transgenic α7β1 integrin in myofibers may enhance laminin organization in the matrix surrounding myofibers and stimulate satellite cell proliferation. BrdUrd labeling of satellite cells that subsequently fuse with muscle fibers and give rise to centrally localized nuclei indicate that this population is functional. The increase in satellite cell number enhances the capacity for muscle regeneration and may underlie the increase in fiber size in the transgenic mice. Integrin-mediated signaling in muscle fibers may also contribute to hypertrophy (Burkin, Bopport, Wallace, Kaufman, manuscript in preparation), perhaps by promoting more efficient presentation of autocrine factors such as IGF-1, that sustain repair and regeneration.80,84
Lastly, analysis of cardiomyopathy in mdx/utr−/− mice revealed α7BX2 transgenic mice have a reduced cardiac pathology compared to nontransgenic animals, reflected in reduced Evans blue dye uptake and fewer ventricular ANF transcripts. Expression of transgenic α7 integrin chain was not detected in the hearts of these mice, consistent with reduced expression in cardiac tissue from the MCK promoter.62 Therefore preventing development of the cardiomyopathy can only be attributed to expression of the integrin in skeletal muscle. These results suggest the cardiomyopathy observed in mdx/utr−/− mice is a secondary manifestation of the skeletal muscle pathology and is consistent with previous reports that show skeletal muscle expression of utrophin also rescues cardiomyopathy in these mice.61
In summary, the mechanism by which increased α7β1 integrin ameliorates the development of muscular dystrophy is complex and involves mechanical reinforcement or maintenance, especially at junctional sites, increased muscle regenerative capacity and hypertrophy. As a collective consequence, increased α7β1 integrin also reduces cardiomyopathy, which is probably a key factor in the extended life span of these mice. This study also suggests that enhanced expression of the α7β1 integrin in other neuromuscular diseases in which some or all of these functions are compromised may provide a novel avenue for treatment of these diseases. Whereas integrins are adhesive proteins that provide mechanical integration, they also engage signaling pathways that promote cell survival and hypertrophy. In addition to the increased mechanical stability provided by the integrin, signaling through the α7β1 complex may further contribute to the inhibition of development of pathology.
Acknowledgments
We thank Dr. Joshua Sanes (Harvard University) for providing the mdx/utr+/− mice and Dr. Kevin Campbell (University of Iowa) for providing antibodies.
Footnotes
Address reprint requests to Stephen Kaufman, Ph.D., Department of Cell and Structural Biology, University of Illinois, B107 Chemical and Life Sciences Laboratory, 601 South Goodwin Ave., Urbana, IL 61801. E-mail: stephenk@life.uiuc.edu.
Supported by the National Institutes of Health and the Muscular Dystrophy Association.
D.J.B and G.Q.W. contributed equally to this study.
Current address of D.J.B.: Department of Pharmacology, University of Nevada School of Medicine, Manville Medical Science Building-318, Reno, NV 89557.
References
- Straub V, Campbell KP. Muscular dystrophies and the dystrophin-glycoprotein complex. Curr Opin Neurol. 1997;10:168–175. doi: 10.1097/00019052-199704000-00016. [DOI] [PubMed] [Google Scholar]
- von der Mark H, Durr J, Sonnenberg A, von der Mark K, Deutzmann R, Goodman SL. Skeletal myoblasts utilize a novel beta 1-series integrin and not alpha 6 beta 1 for binding to the E8 and T8 fragments of laminin. J Biol Chem. 1991;266:23593–23601. [PubMed] [Google Scholar]
- Song WK, Wang W, Foster RF, Bielser DA, Kaufman SJ. H36-alpha 7 is a novel integrin alpha chain that is developmentally regulated during skeletal myogenesis. J Cell Biol. 1992;117:643–657. doi: 10.1083/jcb.117.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster RF, Thompson JM, Kaufman SJ. A laminin substrate promotes myogenesis in rat skeletal muscle cultures: analysis of replication and development using anti-desmin and anti-BrdUrd monoclonal antibodies. Dev Biol. 1987;122:11–20. doi: 10.1016/0012-1606(87)90327-7. [DOI] [PubMed] [Google Scholar]
- Yao CC, Ziober BL, Sutherland AE, Mendrick DL, Kramer RH. Laminins promote the locomotion of skeletal myoblasts via the alpha 7 integrin receptor. J Cell Sci. 1996;109:3139–3150. doi: 10.1242/jcs.109.13.3139. [DOI] [PubMed] [Google Scholar]
- Crawley S, Farrell EM, Wang W, Gu M, Huang HY, Huynh V, Hodges BL, Cooper DN, Kaufman SJ. The α7β1 integrin mediates adhesion and migration of skeletal myoblasts on laminin. Exp Cell Res. 1997;235:274–286. doi: 10.1006/excr.1997.3671. [DOI] [PubMed] [Google Scholar]
- Colognato H, Winkelmann DA, Yurchenco PD. Laminin polymerization induces a receptor-cytoskeleton network. J Cell Biol. 1999;145:619–631. doi: 10.1083/jcb.145.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collo G, Starr L, Quaranta V. A new isoform of the laminin receptor integrin α7β1 is developmentally regulated in skeletal muscle. J Biol Chem. 1993;268:19019–19024. [PubMed] [Google Scholar]
- Song WK, Wang W, Sato H, Bielser DA, Kaufman SJ. Expression of alpha 7 integrin cytoplasmic domains during skeletal muscle development: alternate forms, conformational change, and homologies with serine/threonine kinases and tyrosine phosphatases. J Cell Sci. 1993;106:1139–1152. doi: 10.1242/jcs.106.4.1139. [DOI] [PubMed] [Google Scholar]
- Ziober BL, Kramer RH. Identification and characterization of the cell type-specific and developmentally regulated alpha7 integrin gene promoter. J Biol Chem. 1996;271:22915–22922. doi: 10.1074/jbc.271.37.22915. [DOI] [PubMed] [Google Scholar]
- Hodges BL, Kaufman SJ. Developmental regulation and functional significance of alternative splicing of NCAM and α7β1 integrin in skeletal muscle. Basic Appl Myol. 1996;6:437–446. [Google Scholar]
- von der Mark H, Williams I, Wendler O, Sorokin L, von der Mark K, Poschl E. Alternative splice variants of alpha 7 beta 1 integrin selectively recognize different laminin isoforms. J Biol Chem. 2002;277:6012–6016. doi: 10.1074/jbc.M102188200. [DOI] [PubMed] [Google Scholar]
- van der Flier A, Kuikman I, Baudoin C, vanderNeut R, Sonnenberg A. A novel beta 1 integrin isoform produced by alternative splicing: unique expression in cardiac and skeletal muscle. FEBS Lett. 1995;369:340–344. doi: 10.1016/0014-5793(95)00814-p. [DOI] [PubMed] [Google Scholar]
- Zhidkova NI, Belkin AM, Mayne R. Novel isoform of beta 1 integrin expressed in skeletal and cardiac muscle. Biochem Biophys Res Commun. 1995;214:279–285. doi: 10.1006/bbrc.1995.2285. [DOI] [PubMed] [Google Scholar]
- Belkin AM, Zhidkova NI, Balzac F, Altruda F, Tomatis D, Maier A, Tarone G, Koteliansky VE, Burridge K. Beta 1D integrin displaces the beta 1A isoform in striated muscles: localization at junctional structures and signaling potential in nonmuscle cells. J Cell Biol. 1996;132:211–216. doi: 10.1083/jcb.132.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkin AM, Retta SF, Pletjushkina OY, Balzac F, Silengo L, Fassler R, Koteliansky VE, Burridge K, Tarone G. Muscle beta1D integrin reinforces the cytoskeleton-matrix link: modulation of integrin adhesive function by alternative splicing. J Cell Biol. 1997;139:1583–1595. doi: 10.1083/jcb.139.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Wang W, Song WK, Cooper DNW, Kaufman SJ. Selective modulation of the interaction of α7β1 integrin with fibronectin and laminin during skeletal muscle differentiation. J Cell Sci. 1994;107:175–181. doi: 10.1242/jcs.107.1.175. [DOI] [PubMed] [Google Scholar]
- Li J, Burkin DJ, Kaufman SJ, Wu C. The muscle integrin binding protein (MIBP) interacts with α7β1 integrin and regulates cell adhesion and laminin matrix deposition. Dev Biol. 2003;261:209–219. doi: 10.1016/s0012-1606(03)00304-x. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Gruszczynska-Biegala J, Cheuvront T, von der Mark H, von der Mark K, Kaufman SJ, Zolkiewska A. Interaction of the disintegrin and cysteine-rich domain of ADAM12 with integrin α7β1. Exp Cell Res. 2004;298:28–37. doi: 10.1016/j.yexcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Burkin DJ, Gu M, Hodges BL, Campanelli JT, Kaufman SJ. A functional role for specific spliced variants of the α7β1 integrin in acetylcholine receptor clustering. J Cell Biol. 1998;143:1067–1075. doi: 10.1083/jcb.143.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkin DJ, Kim JE, Gu M, Kaufman SJ. Laminin and α7β1 integrin regulate agrin-induced clustering of acetylcholine receptors. J Cell Sci. 2000;113:2877–2886. doi: 10.1242/jcs.113.16.2877. [DOI] [PubMed] [Google Scholar]
- Hayashi YK, Chou F-L, Engvall E, Ogawa M, Matsuda C, Hirabayashi S, Yokochi K, Ziober BL, Kramer RH, Kaufman SJ, Ozawa E, Goto Y, Nonaka I, Tsukahara T, Wang J, Hoffman EP, Arahata K. Mutations in the integrin α7 gene cause congenital myopathy. Nat Genet. 1998;19:94–97. doi: 10.1038/ng0598-94. [DOI] [PubMed] [Google Scholar]
- Mayer U, Saher G, Fassler R, Bornemann A, Echtermeyer F, von der Mark H, Miosge N, Poschl E, von der Mark K. Absence of integrin alpha 7 causes a novel form of muscular dystrophy. Nat Genet. 1997;17:318–323. doi: 10.1038/ng1197-318. [DOI] [PubMed] [Google Scholar]
- Pegoraro E, Cepollaro F, Prandini P, Marin A, Fanin M, Trevisan CP, El-Messlemani AH, Tarone G, Engvall E, Hoffman EP, Angelini C. Integrin α7β1 in muscular dystrophy/myopathy of unknown etiology. Am J Pathol. 2002;160:2135–2143. doi: 10.1016/s0002-9440(10)61162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Campbell KP. Dystrophin-glycoprotein complex: its role in the molecular pathogenesis of muscular dystrophies. Muscle Nerve. 1994;17:2–15. doi: 10.1002/mus.880170103. [DOI] [PubMed] [Google Scholar]
- Matsumura K, Ervasti JM, Ohlendieck K, Kahl SD, Campbell KP. Association of dystrophin-related protein with dystrophin-associated proteins in mdx mouse muscle. Nature. 1992;360:588–591. doi: 10.1038/360588a0. [DOI] [PubMed] [Google Scholar]
- Campbell KP. Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- Rybakova IN, Amann KJ, Ervasti JM. A new model for the interaction of dystrophin with F-actin. J Cell Biol. 1996;135:661–672. doi: 10.1083/jcb.135.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DR, Hill DF, Dickson G, Spurr NK, Byth C, Marsden RF, Walsh FS, Edwards YH, Davies KE. An autosomal transcript in skeletal muscle with homology to dystrophin. Nature. 1989;339:55–58. doi: 10.1038/339055a0. [DOI] [PubMed] [Google Scholar]
- Khurana TS, Hoffman EP, Kunkel LM. Identification of a chromosome 6-encoded dystrophin-related protein. J Biol Chem. 1990;265:16717–16720. [PubMed] [Google Scholar]
- Ohlendieck K, Campbell KP. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J Cell Biol. 1991;115:1685–1694. doi: 10.1083/jcb.115.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana TS, Watkins SC, Chafey P, Chelly J, Tome FM, Fardeau M, Kaplan JC, Kunkel LM. Immunolocalization and developmental expression of dystrophin related protein in skeletal muscle. Neuromuscul Disord. 1991;1:185–194. doi: 10.1016/0960-8966(91)90023-l. [DOI] [PubMed] [Google Scholar]
- Burkin DJ, Wallace GQ, Nicol KJ, Kaufman DJ, Kaufman SJ. Enhanced expression of the α7β1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J Cell Biol. 2001;152:1207–1218. doi: 10.1083/jcb.152.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deconinck AE, Rafael JA, Skinner JA, Brown SC, Potter AC, Metzinger L, Watt DJ, Dickson JG, Tinsley JM, Davies KE. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell. 1997;90:717–727. doi: 10.1016/s0092-8674(00)80532-2. [DOI] [PubMed] [Google Scholar]
- Grady RM, Teng H, Nichol MC, Cunningham JC, Wilkinson RS, Sanes JR. Skeletal and cardiac myopathies in mice lacking utrophin and dystrophin: a model for Duchenne muscular dystrophy. Cell. 1997;90:729–738. doi: 10.1016/s0092-8674(00)80533-4. [DOI] [PubMed] [Google Scholar]
- Hodges BL, Hayashi YK, Nonaka I, Wang W, Arahata K, Kaufman SJ. Altered expression of the α7β1 integrin in human and murine muscular dystrophies. J Cell Sci. 1997;110:2873–2881. doi: 10.1242/jcs.110.22.2873. [DOI] [PubMed] [Google Scholar]
- Burkin DJ, Kaufman SJ. The α7β1 integrin in muscle development and disease. Cell Tissue Res. 1999;296:183–190. doi: 10.1007/s004410051279. [DOI] [PubMed] [Google Scholar]
- Martin PT, Kaufman SJ, Kramer RH, Sanes JR. Synaptic integrins in developing, adult, and mutant muscle: selective association of α1, α7A, and α7B integrins with the neuromuscular junction. Dev Biol. 1996;174:125–139. doi: 10.1006/dbio.1996.0057. [DOI] [PubMed] [Google Scholar]
- Miosge N, Klenczar C, Herken R, Willem M, Mayer U. Organization of the myotendinous junction is dependent on the presence of α7β1 integrin. Lab Invest. 1999;79:1591–1599. [PubMed] [Google Scholar]
- Kaariainen M, Liljamo T, Pelto-Huikko M, Heino J, Jarvinen M, Kalimo H. Regulation of alpha7 integrin by mechanical stress during skeletal muscle regeneration. Neuromuscul Disord. 2001;11:360–369. doi: 10.1016/s0960-8966(00)00193-0. [DOI] [PubMed] [Google Scholar]
- Kaariainen M, Nissinen L, Kaufman SJ, Sonnenberg A, Jarvinen M, Heino J, Kalimo H. Expression of α7β1 integrin splicing variants during skeletal muscle regeneration. Am J Pathol. 2002;161:1023–1031. doi: 10.1016/s0002-9440(10)64263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel A, Lehmann-Horn F, Engel AG. Neuromuscular transmission in the mdx mouse. Muscle Nerve. 1990;13:742–749. doi: 10.1002/mus.880130813. [DOI] [PubMed] [Google Scholar]
- Lyons PR, Slater CR. Structure and function of the neuromuscular junction in young adult mdx mice. J Neurocytol. 1991;20:969–981. doi: 10.1007/BF01187915. [DOI] [PubMed] [Google Scholar]
- Andersson PB, Rando TA. Neuromuscular disorders of childhood. Curr Opin Pediatr. 1999;11:497–503. doi: 10.1097/00008480-199912000-00004. [DOI] [PubMed] [Google Scholar]
- Rafael JA, Townsend ER, Squire SE, Potter AC, Chamberlain JS, Davies KE. Dystrophin and utrophin influence fiber type composition and post-synaptic membrane structure. Hum Mol Genet. 2000;9:1357–1367. doi: 10.1093/hmg/9.9.1357. [DOI] [PubMed] [Google Scholar]
- George-Weinstein M, Foster RF, Gerhart JG, Kaufman SJ. In vitro and in vivo expression of α7 integrin and desmin define the primary and secondary myogenic lineages. Dev Biol. 1993;156:209–229. doi: 10.1006/dbio.1993.1071. [DOI] [PubMed] [Google Scholar]
- Bao ZZ, Lakonishok M, Kaufman S, Horwitz AF. Alpha 7 beta 1 integrin is a component of the myotendinous junction on skeletal muscle. J Cell Sci. 1993;106:579–589. doi: 10.1242/jcs.106.2.579. [DOI] [PubMed] [Google Scholar]
- Paul AC, Sheard PW, Kaufman SJ, Duxson MJ. Localization of alpha7 integrins and dystrophin suggests potential for both lateral and longitudinal transmission of tension in large mammalian muscles. Cell Tissue Res. 2002;308:255–265. doi: 10.1007/s00441-002-0526-y. [DOI] [PubMed] [Google Scholar]
- Nawrotzki R, Willem M, Miosge N, Brinkmeyer H, Mayer U. Defective integrin switch and matrix composition at alpha 7-deficient myotendinous junctions precede the onset of muscular dystrophy in mice. Hum Mol Genet. 2003;12:483–495. doi: 10.1093/hmg/ddg047. [DOI] [PubMed] [Google Scholar]
- Law DJ, Tidball JG. Dystrophin deficiency is associated with myotendinous junction defects in prenecrotic and fully regenerated skeletal muscle. Am J Pathol. 1993;142:1513–1523. [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- Grady RM, Merlie JP, Sanes JR. Subtle neuromuscular defects in utrophin-deficient mice. J Cell Biol. 1997;136:871–882. doi: 10.1083/jcb.136.4.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deconinck AE, Potter AC, Tinsley JM, Wood SJ, Vater R, Young C, Metzinger L, Vincent A, Slater CR, Davies KE. Postsynaptic abnormalities at the neuromuscular junctions of utrophin-deficient mice. J Cell Biol. 1997;136:883–894. doi: 10.1083/jcb.136.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- McCroskery S, Maxwell TM, Sharma M, Kambadur R. Myostatin regulates satellite cell activation and self-renewal. J Cell Biol. 2003;162:1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- Wagner KR, McPherron AC, Winik N, Lee SJ. Loss of myostatin attenuates severity of muscular dystrophy in mdx mice. Ann Neurol. 2002;52:832–836. doi: 10.1002/ana.10385. [DOI] [PubMed] [Google Scholar]
- Rafael JA, Tinsley JM, Potter AC, Deconinck AE, Davies KE. Skeletal muscle-specific expression of a utrophin transgene rescues utrophin-dystrophin deficient mice. Nat Genet. 1998;19:79–82. doi: 10.1038/ng0598-79. [DOI] [PubMed] [Google Scholar]
- Donoviel DB, Shield MA, Buskin JN, Haugen HS, Clegg CH, Hauschka SD. Analysis of muscle creatine kinase gene regulatory elements in skeletal and cardiac muscles of transgenic mice. Mol Cell Biol. 1996;16:1649–1658. doi: 10.1128/mcb.16.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DJ, Wells KE, Walsh FS, Davies KE, Goldspink G, Love DR, Chan-Thomas P, Dunckley MG, Piper T, Dickson G. Human dystrophin expression corrects the myopathic phenotype in transgenic mdx mice. Hum Mol Genet. 1992;1:35–40. doi: 10.1093/hmg/1.1.35. [DOI] [PubMed] [Google Scholar]
- Lee CC, Pons F, Jones PG, Bies RD, Schlang AM, Leger JJ, Caskey CT. Mdx transgenic mouse: restoration of recombinant dystrophin to the dystrophic muscle. Hum Gene Ther. 1993;4:273–281. doi: 10.1089/hum.1993.4.3-273. [DOI] [PubMed] [Google Scholar]
- Phelps SF, Hauser MA, Cole MN, Rafael JA, Hinkle RT, Faulkner JA, Chamberlain JS. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum Mol Genet. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- Decrouy A, Renaud JM, Davis HL, Lunde JA, Dickson G, Jasmin BJ. Mini-dystrophin gene transfer in mdx4cv diaphragm muscle fibers increases sarcolemmal stability. Gene Ther. 1997;4:401–408. doi: 10.1038/sj.gt.3300407. [DOI] [PubMed] [Google Scholar]
- Cox GA, Cole NM, Matsumura K, Phelps SF, Hauschka SD, Campbell KP, Faulkner JA, Chamberlain JS. Overexpression of dystrophin in transgenic mdx mice eliminates dystrophic symptoms without toxicity. Nature. 1993;364:725–729. doi: 10.1038/364725a0. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Hauser MA, DelloRusso DC, Duan D, Crawford RW, Phelps SF, Harper HA, Robinson AS, Engelhardt JF, Brooks SV, Chamberlain JS. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Yuasa K, Yohimura M, Yokota T, Ikemoto T, Suzuki M, Dickson G, Miyagoe-Suzuki Y, Takeda S. Micro-dystrophin cDNA ameliorates dystrophic phenotypes when introduced into mdx mice as a transgene. Biochim Biophys Res Commun. 2002;293:1265–1272. doi: 10.1016/S0006-291X(02)00362-5. [DOI] [PubMed] [Google Scholar]
- Bertoni C, Rando TA. Dystrophin gene repair in mdx muscle precursor cells in vitro and in vivo mediated by RNA-DNA chimeric oligonucleotides. Hum Gene Ther. 2002;13:707–718. doi: 10.1089/104303402317322276. [DOI] [PubMed] [Google Scholar]
- Gussoni E, Soneoka Y, Strickland SD, Buzney EA, Kahn MK, Flint AF, Kunkel LM, Mulligan RC. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:309–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D’Antona D, Pellegrino MA, Barresi R, Bresolin N, De Angelis MG, Campbell KP, Bottinelli R, Cossu G. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- Squire S, Raymackers JM, Vandebrouck C, Potter A, Tinsley J, Fisher R, Gillis JM, Davies KE. Prevention of pathology in mdx mice by expression of utrophin: analysis using an inducible transgenic expression system. Hum Mol Genet. 2002;11:3333–3344. doi: 10.1093/hmg/11.26.3333. [DOI] [PubMed] [Google Scholar]
- Moll J, Barzaghi P, Lin S, Bezakova G, Lochmuller H, Engvall E, Muller U, Ruegg MA. An agrin minigene rescues dystrophic symptoms in a mouse model for congenital muscular dystrophy. Nature. 2001;413:302–307. doi: 10.1038/35095054. [DOI] [PubMed] [Google Scholar]
- Wehling M, Spencer MJ, Tidball JG. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J Cell Biol. 2001;155:123–131. doi: 10.1083/jcb.200105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronqvist P, Kawaguchi N, Albrechtsen R, Xu X, Schroder HD, Moghadaszadeh B, Nielsen FC, Frohlich C, Engvall E, Wewer UM. ADAM12 alleviates the skeletal muscle pathology in mdx dystrophic mice. Am J Pathol. 2002;161:1535–1540. doi: 10.1016/S0002-9440(10)64431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HH, Jayasinha V, Xia B, Hoyte K, Martin PT. Overexpression of the cytotoxic T cell GalNAc transferase in skeletal muscle inhibits muscular dystrophy in mdx mice. Proc Natl Acad Sci USA. 2002;99:5616–5621. doi: 10.1073/pnas.082613599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MJ, Mellgren RL. Overexpression of a calpastatin transgene in mdx muscle reduces dystrophic pathology. Hum Mol Genet. 2002;11:2645–2655. doi: 10.1093/hmg/11.21.2645. [DOI] [PubMed] [Google Scholar]
- Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 2002;157:137–148. doi: 10.1083/jcb.200108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadaszadeh B, Albrechtsen R, Guo LT, Zaik M, Kawaguchi N, Borup RH, Kronqvist P, Schroder HD, Davies KE, Voit T, Nielsen FC, Engvall E, Wewer UM. Compensation for dystrophin-deficiency: ADAM12 overexpression in skeletal muscle results in increased alpha 7 integrin, utrophin and associated glycoproteins. Hum Mol Genet. 2003;12:2467–2479. doi: 10.1093/hmg/ddg264. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Wold BJ, Hauschka SD. Muscle creatine kinase sequence elements regulating skeletal and cardiac muscle expression in transgenic mice. Mol Cell Biol. 1989;9:3393–3399. doi: 10.1128/mcb.9.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes JB, Chamberlain JS, Buskin JN, Johnson JE, Hauschka SD. Transcriptional regulation of the muscle creatine kinase gene and regulated expression in transfected mouse myoblasts. Mol Cell Biol. 1986;6:2855–2864. doi: 10.1128/mcb.6.8.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ME, DeMayo F, Yin FKC, Lee HM, Geske R, Montgomery C, Schwartz RJ. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J Biol Chem. 1995;270:12109–12116. doi: 10.1074/jbc.270.20.12109. [DOI] [PubMed] [Google Scholar]