Abstract
Amyloidoses are a group of disorders characterized by abnormal folding of proteins that impair organ function. We investigated the cellular response of primary cardiac fibroblasts to amyloidogenic light chains and determined the corresponding change in proteoglycan expression and localization. The cellular response to 11 urinary immunoglobulin light chains of κ1, λ6, and λ 3 subtypes was evaluated. The localization of the light chains was monitored by conjugating them to Oregon Green 488 and performing live cell confocal microscopy. Sulfation of the proteoglycans was determined after elution over Q1-columns with a single-step salt gradient (1.5 mol/L NaCl) via dimethylmethylene blue. Light chains were detected inside cells within 4 hours and demonstrated perinuclear localization. Over 80% of the cells showed intracellular localization of the amyloid light chains. The light chains induced sulfation of the secreted glycosaminoglycans, but the cell fraction possessed only minimal sulfation. Furthermore, the light chains caused a translocation of heparan sulfate proteoglycan to the nucleus. The conformation and thermal stability of light chains was altered when they were incubated in the presence of heparan sulfate and destabilization of the amyloid light chains was detected. These studies indicate that internalization of the light chains mediates the expression and localization of heparan sulfate proteoglycans.
The dynamics associated with the observation of proteoglycan deposition and amyloid fibril formation have been of great interest to investigators for a number of years. The amyloidoses are described as a group of disorders characterized by abnormal folding of overexpressed or structurally variant proteins that are deposited extracellularly in tissues and ultimately impair organ function.1 In primary systemic amyloidosis (AL), clonal plasma cells secrete monoclonal immunoglobulin (Ig) light chains (LCs) that aggregate and form fibrillar deposits in the heart, kidney, nerves and other tissues.2 These non-branching fibrils are composed of filaments that are twisted around and form a β-pleated sheet that stains positive with Congo Red and displays apple-green birefringence under polarized light.
Cardiac involvement is of particular concern in AL amyloidosis. Amyloid fibrils are found in the heart in up to 50% of AL patients, often in the interstitium, and are a leading cause of AL-related mortality.3 Cellular toxicity has previously been demonstrated in cardiac myocytes;4 however, the ubiquitous presence of glycosaminoglycans, particularly heparan sulfate, in amyloid deposits suggests that extracellular matrix molecules may play a role in fibril formation.5,6 Since fibroblasts have been show to be major contributors to matrix production,7,8 the goal of our study was to investigate the effect of purified urinary LCs (obtained from patients with AL, multiple myeloma [MM], and MM with AL) on primary cardiac fibroblasts and to assess changes in heparan sulfate proteoglycan (HSPG) expression and localization.
Investigators have hypothesized that glycosaminoglycans (GAGs) and proteoglycans play a critical role in amyloid fibril formation and deposition. In an in vivo model, Snow and Kisilevsky9 showed that there was an increase in GAGs at the time of amyloid deposition. This was supported by studies that demonstrated and quantified GAGs in amyloid fibril preparations and amyloid-rich tissues.10 This study examined the heart tissue of patients with AL and demonstrated that chondroitin sulfate (CS), heparan sulfate (HS), dermatan sulfate (DS), and hyaluronic acid (HA) were all present. More recently, studies have shown that proteoglycans enhance the deposition of β2-microglobulin amyloid fibrils in vivo by acting as a “scaffold” for fibrillogenesis.11
Much of the data implicating HSPGs in the genesis of amyloid in vivo have been obtained in models of AA12 and Aβ amyloid.13 In another model (islet amyloidosis), Castillo et al14 demonstrated that perlecan bound to amylin and that the binding could be inhibited with heparin indicating that the GAG chains and their sulfation were critical components. Furthermore, putative HS-binding sites for GAGs have been demonstrated in a number of amyloid peptides and their precursors (IAPP, SAA, ABPP, and the Aβ protein segment of ABPP).5,15–19
More recently several reports have focused on the cellular responses to amyloidogenic LCs. Investigators showed that human glomerulopathic LCs interact with mesangial cells and they hypothesize that internalization is clathrin-mediated.20 Interestingly, the authors hypothesize that different light chains have a number of intracellular trafficking patterns that determine the resulting pathological changes. Other experiments have demonstrated that polyvinylsulfonates of various lengths can alter the cellular response and formation of amyloid fibrils. 21,22 The cellular response to light chains with the subsequent increase in HSPGs is similar to the injury response exhibited in stromal fibroblasts. In this context, it has been demonstrated that injury results in an increase in HS and CS with a decrease in keratan sulfate (KS), and enhances the sulfation of both HS and CS.23–27 Furthermore, there is an associated change in availability of growth factors such as fibroblast growth factor (FGF) that have been shown to mediate the localization of proteoglycans within the cell.28
Our major goal was to evaluate changes in proteoglycans in response to LCs. Our studies were limited to cardiac fibroblasts as initial studies showed that cardiac myocytes did not internalize light chains unless cellular blebbing was present. As the disease process causes harm to tissues, parameters known to be modified in response to injury such as secretion, sulfation, GAG composition, and translocation were evaluated.24,27,28 Ultimately, as the sulfation of GAGs and deposition of proteoglycans were modified, we performed in vitro experiments to determine whether the GAGs interacted with LCs in a manner that was different from non-AL light chains. We found that in all of the AL light chains studied to date that the glycosaminoglycan, heparan sulfate, altered both conformation and thermal stability. These novel studies allow us to examine the pathophysiology of amyloidosis at a cellular level.
Materials and Methods
Light Chain Purification
Urine was collected from patients with AL, MM, or MM/AL with the approval of the Institutional Review Board of Boston Medical University. Light chains were purified from urine samples extensively dialyzed against water, lyophilized, treated with Affi-Gel Blue (Bio-Rad Laboratories, Hercules, CA) to remove albumin, and chromatographed on Sephacryl S-200 (Amersham Pharmacia Bio-Tech, Buckingham, England).29 The Ig LC proteins were subtyped using a number of cellular and molecular analyses and the sequence examined using both molecular and mass spectrometry. The 11 AL LCs used had the associated clinical disease type and subgroup (κ1, λ6, and λ3) classification seen in Table 1. One non-AL LC κ1 was also purified and the cellular response determined.
Table 1.
Summary of Amyloid Light Chains and Internalization in Cardiac Fibroblasts
| Light chain | Internalization
|
Post-translational modifications | |
|---|---|---|---|
| 4 hours | 24 hours | ||
| AL-96066 (κ1) | + | + | cysteinylated @214 |
| AL-98002 (κ1) | + | + | glycoprot., cysteinylated @214 |
| AL-01102 (κ1)/myeloma | + | + | NK |
| AL-01140 (κ1)/myeloma | ± | + | Mr 23Kd, 12 kd |
| AL-01066 (κ1) | + | + | cysteinylated @214 |
| AL-00131 (κ1) | + | + | NK |
| AL-01090 (κ1) | + | + | NK |
| AL-01029 (λ6) | + | + | dimer Mr 46 kd |
| AL-01148 (λ6) | + | + | cysteinylated |
| AL-00109 (λ3) | − | ± | NK |
| AL-01027 (λ?) | ± | + | NK |
NK, not known
Cell Culture
Primary rat and mouse cardiac fibroblasts were isolated using a collagenase digestion and cells were cultured in Dulbecco’s low glucose medium supplemented with 7% calf serum, 1% non-essential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco BRL/Life Technologies, Grand Island, NY).4 Cells were used in either the first or second passage.
Confocal Laser Scanning Microscopy
Monitoring Localization of Light Chain
Live cell imaging was performed to monitor the localization of each LC. Light chains were conjugated to Oregon Green 488 (Molecular Probes, Eugene, OR) and purified over a sizing column using the suggested methodology from Molecular Probes. The number of Oregon Green molecules/light chain was identified for each conjugation. The LC-conjugated probe was added to cell cultures and evaluated at several time points over a period of 24 hours. For these experiments cells were cultured on 8-well glass coverslips and evaluated at specific time points at an excitation of 496 nm. A single Z-series was taken at each time point/experiment to determine the localization of the LC within the cell compared to the simultaneously acquired differential interference contrast (DIC) images. Negative controls included the addition of unconjugated Oregon Green 488 to cells and acquisition at the same settings as the experimental ones.
Indirect Immunohistochemistry
Indirect immunohistochemistry was performed to localize specific proteins after fixation. The procedure for staining cells was described previously.30–32 Briefly, cells were fixed with freshly prepared 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.2) at room temp for 15 minutes, washed with PBS and permeabilized with 0.1% Triton X-100, and blocked with PBS containing 5% bovine serum albumin (BSA). When the experimental protocol dictated, cells were pretreated with heparinase III (Sigma-Aldrich, St. Louis, MO) before incubation with antibodies. The cells were incubated in PBS/1% BSA containing appropriate monoclonal antibodies (MAbs) for 18 hours at 4°C. After incubation with the primary antibody, cells were rinsed with PBS, washed for 10 minutes in 3% BSA/PBS, and then incubated with a secondary antibody with an appropriate fluorophore conjugated to the IgG (1:100) for 1 hour on a rocker at room temperature. The use of the antibody Δ3G10 (Seikagaku America, E. Falmouth, MA) required a predigestion with heparinase III, since the epitope is the stub created by heparinase III digestion. Negative controls were incubated with either pure goat IgG or pure mouse IgG instead of the primary antibody. In other experiments, cells were incubated with Oregon Green 488 conjugated to LCs, imaged, and then fixed and probed for other proteins such as rhodamine phalloidin (Molecular Probes) and/or mAb to heparan sulfate proteoglycan (Δ3G10) with a secondary antibody conjugated to AlexaFluor 633 (Molecular Probes). Alternatively, To-Pro was used as a nuclear marker (Molecular Probes). Cross-over controls were performed. Cells were imaged using the LSM 510 version 2.8 as described previously.31,32
Biochemical Analysis
Glycosaminoglycan Analysis
Cells were cultured to confluence and treated with 40 μg/ml LC, 50 ng/ml FGF-2, or an equivalent volume of medium. FGF-2 was used as a positive control because it has previously been shown to mediate GAG production.28,33 Cells were incubated for defined times and the medium was removed and treated with 2 volumes of MX buffer (10 mol/L urea, 50 mmol/L Tris-HCl, 10 mmol/L EDTA, pH 7.0). The cell layers were washed with PBS and scraped with ECMX buffer (1 mol/L urea, 50 mmol/L Tris-HCl, 50 mmol/L EDTA, pH 7.0) and then separated into cellular and matrix fractions by centrifugation at 5000 × g for 10 minutes. The supernatant was defined as the matrix fraction and the pellet was resuspended in CX buffer (8 mol/L urea, 50 mmol/L Tris-HCl, 0.1% Triton X-100, pH 7.0). The cell pellet was extracted and the supernatant was saved as the cellular fraction. Each of the fractions were mixed with a 70% Q-Sepharose (Amersham Biosciences, Upsala, Sweden) suspension and rocked for 45 minutes and the slurries were then loaded onto columns, washed with buffer, and the unbound fractions discarded. The columns were washed with 25 column volumes of Q1 buffer (50 mmol/L sodium acetate, 10 mmol/L EDTA, 0.3 mol/L NaCl, 20% propylene glycol, pH 6.0) until the UV monitor reached baseline and then washed further with 5 column volumes of 50 mmol/L sodium acetate buffer containing 0.3 mol/L NaCl. Proteoglycans were eluted with high salt (50 mmol/L sodium acetate buffer containing 1.5 mol/L NaCl). Glycosaminoglycan content was determined for each fraction using 1,9-dimethylmethylene blue (525 nm).34 Selective polysaccharidases were used to identify and quantify GAGs and digestion was carried out using chondroitinase ABC, heparinase I and III, and keratanase as described previously.25 Purified glycosaminoglycans and respective polysaccharidases were run as standards for each experiment.
Further characterization of the proteoglycans present in the cell layer and medium was performed using 12% SDS-PAGE and Western blot analysis. Thirty μg were loaded onto each lane, electrophoresed and transferred to polyscreen PVDF membrane (Perkin Elmer, Boston, MA) using the Semi-Dry Transfer system. Blots were blocked in TBST (10 mmol/L Tris, 100 mmol/L NaCl, 0.1% Tween-20, pH 7.4) containing 0.2% I-block (Applied Biosystems, Foster City, CA) and membranes were incubated with antibodies directed against glypican-1 and syndecan-1, washed and incubated with appropriate secondary antibodies and rinsed again with TBST. Visualization was performed by enhanced chemiluminescence (Perkin Elmer).
In Vitro Association of Glycosaminoglycans with Light Chains
Association between glycosaminoglycans [bovine kidney HS, KS, and CS (Sigma-Aldrich, St. Louis, MO)] and LCs were determined using a solid phase binding assay. Glycosaminoglycans were bound to Maxisorb (Nalge-Nunc, Rochester, NY) for 18 hours at 4°C in a dose-dependent manner, non-specific binding was blocked and κ1 light chains were added to the wells. Excess light chain was removed with extensive washing and an HRP-conjugated secondary antibody was added to the wells. Presence of binding was performed in an ELISA assay at 450 nm.
Circular Dichroism
Circular dichroism (CD) spectra and thermal unfolding studies were performed on an Aviv 62DS Spectropolarimeter (AVIV Association, Inc., Lakewood, NJ), equipped with a thermoelectric temperature controller. Far UV CD spectra were recorded from 250 to 200 nm at 25°C in 0.05 cm cuvettes with 0.40 mg/ml LC with or without HS (LC:HS = 4:1 w/w). Five spectra were recorded, averaged and corrected for buffer, or buffer plus HS, and normalized to molar ellipticity, [θ]. Molar ellipticity values, [θ], were calculated according to the equation: [θ] (deg-cm2/decimol) = θ(MRW)/10(l)(c), where θ = experimental ellipticity in mdeg; MRW = mean residue weight of the amino acids; l = pathlength of the cell in cm; c = concentration of LC in grams per ml.35–37
Thermal unfolding and refolding studies were performed using a Peltier thermoelectric temperature controller in 0.05-cm cuvettes and LC concentration = 0.40 mg/ml at constant wavelength of 217 (β-sheet). Ellipticity was continuously monitored from 5 to 95°C at 0.5°C increments with data collection for 90 seconds per temperature increment. Samples were equilibrated at 5°C for 30 minutes in the sample compartment before unfolding studies. Spectra were recorded at 25°C before and after heating to monitor reversibility. Protein concentration used for CD was measured according to the method of Lowry et al.38
Results
Localization of Exogenous Amyloidogenic and Control Light Chains Added to Cardiac Fibroblasts
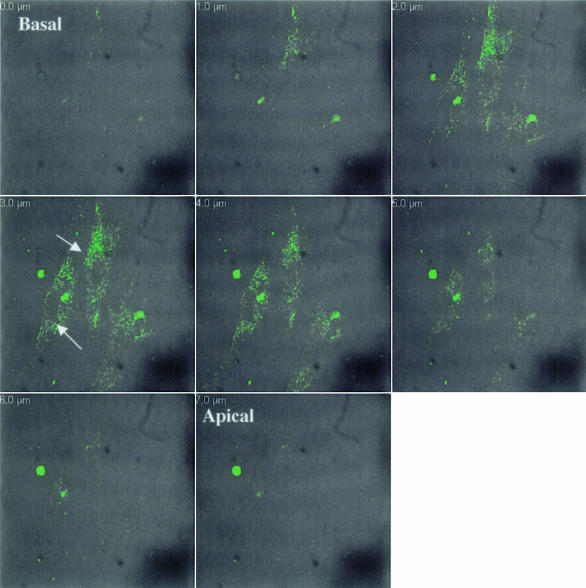
A number of amyloid LCs have been sequenced and post-translational modifications have been characterized.29 To determine the response of cells to amyloidogenic LCs, 11 urinary Ig LCs of κ1, λ6, and λ3 subtypes were individually added to rat and mouse primary cardiac fibroblasts and compared to results obtained with urinary LC from a patient with multiple myeloma, but not AL (non-AL LC κ1 [MM-96100]). Cells were cultured on 8-well coverslips at 2 × 103cells/ml and allowed to achieve confluence. Specific LCs conjugated to Oregon Green 488 were added to independent wells and five live cell confocal images were collected for each LC per experiment at a number of time points. To control for the potential side effects of the dye, unconjugated Oregon Green was added to cell cultures and did not enter the cells. Conversely, unconjugated LCs added to cells and immunostained with antibodies and the Oregon Green did not alter the localization. LCs were repeatedly present in a punctate pattern around the nucleus by 4 hours (Figure 1, Table 1). The punctate pattern was retained and intensified by 24 hours. To determine the localization of each LC within the cell, a Z-series was taken from the apical to basal surface with simultaneous DIC and fluorescent images (Figure 1). Light chain was consistently localized in the central 4 μm out of an 8-μm series. While specific LCs were internalized consistently between experiments, we did detect that each LC displayed a repeatable percentage of cells demonstrating internalization of LCs (ie, three κ1’s showed internalization ranging from 78 to 80%, one λ6 showed internalization of 86%, and one λ3 showed internalization of only 60%). The non-AL LC was also internalized. The differences in internalization may be explained by sequence variability among the LCs. In fact, the λ3 LC has the least sequence homology with the other LCs evaluated to date. In all of the LCs evaluated over time there was a transition in appearance from a punctate pattern to a filamentous one. The extensiveness of the filaments varied with the specific LCs and there was no detectable co-localization with F-actin (Figure 2) or microtubules. Note that the cells that internalized the non-AL LC showed less filamentous morphology. Interestingly, cells that had internalized LC showed a transition to a myofibroblast morphology. This change in cellular architecture has been noted in other fibroblast cells in response to injury.39 Internalization was detected in both primary rat and mouse cardiac fibroblasts and there were no detectable species differences in either the rate of internalization or in the patterns of the LC.
Figure 1.
Light chains conjugated to Oregon Green 488 are localized within cells. Cardiac fibroblasts were cultured for 4 hours in the presence of AL LC κ1 conjugated to Oregon Green 488 and imaged live using confocal microscopy. A Z-series of 1-μm optical sections was taken through cells. Arrows indicate light chain present in middle optical sections. Images are representative of a minimum of five experiments with independent conjugations.
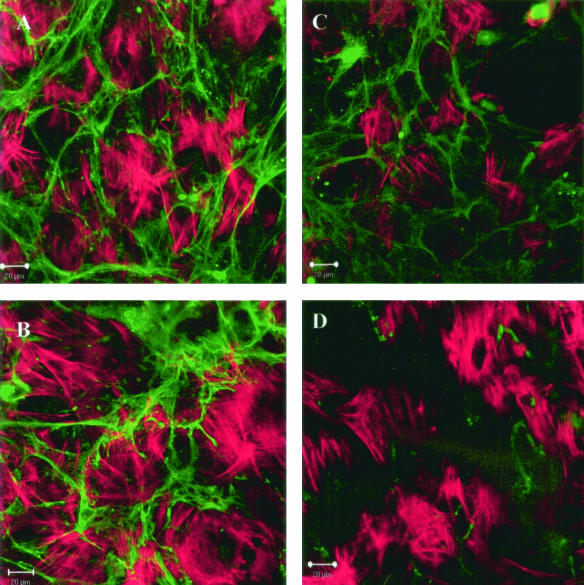
Figure 2.
Long-term incubation (5 days) with AL - κ1 and λ6-LCs induces a filamentous appearance in AL LCs (AL-96066, AL-01066, AL-01029). Cultures were fixed and stained with rhodamine-phalloidin to detect F-actin. A–D: A, κ1; B, κ1; C, λ6; and D, non-AL κ1. Composite images do not show co-localization of F-actin and LC. Images are representative of a minimum of five independent experiments.
The role of cell density on localization was also assessed using three different cell densities (5 × 102, 3 × 103, and 5 × 103 cells/ml). There was no detectable change in the localization of the LC with cell density and the pattern could even be detected through cell layers when cells became multilayered.
Response of Cardiac Fibroblasts to Light Chains
Light Chain Mediates the Translocation of Heparan Sulfate Proteoglycan to the Nucleus
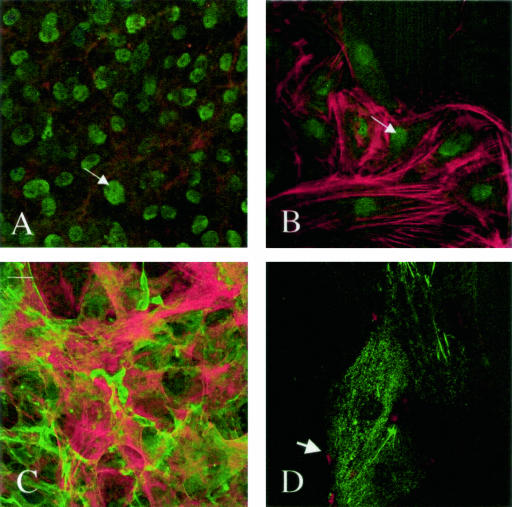
Cells were cultured in the presence or absence of amyloidogenic LCs or a non-AL LC κ1 (MM-96100) for 24 hours. Localization of HSPGs was evaluated in relation to LCs, F-actin, or nuclear markers using confocal microscopy. In short-term experiments, cells were plated and cultured for 18 hours, at which time LCs conjugated to Oregon Green 488 were added and then incubated for 18 additional hours. Heparan sulfate proteoglycans translocated to the nucleus when cells were cultured with AL LC κ1 but not with the non-AL LC (Figure 3). Translocation was detected in both semi-confluent and confluent cultures (Figure 3). A Z-series through the cell verified that the localization of HSPG in response to the AL LC κ1 (AL-96066) was nuclear and that there was overlap with the nuclear marker, To-Pro. HSPG was not detected in the nucleoli. Nuclear translocation was only detected when cells were fixed, incubated with heparinase III for 1 hour, permeabilized with Triton X-100, and then immunostained. When cells were not incubated in heparinase III after fixation, there was no detectable HSPG. In addition, when cells were incubated with heparinase III 1 hour before the end of the incubation and then fixed and stained, HSPGs were only localized at the cell border (Figure 3).
Figure 3.
Nuclear localization of HSPG 24 hours after LC (unconjugated) was added to cell and stained for HSPG with the mAb Δ3G10. Cells were fixed, treated with heparinase III, and stained with Δ3G10 and rhodamine-phalloidin. A and B: The addition of AL LC κ1 (AL-96066) shows HSPG in the nucleus of confluent and in semi-confluent cells (arrows). C: The addition of non-AL LC κ1 (MM-96100) does not induce translocation of HSPG. D: The addition of AL LC κ1 (AL-96066) to cells that were treated with heparinase III 1 hour before fixation and then stained for Δ3G10 shows HSPG on the cell margins (arrowhead). Images are representative of five independent experiments.
Long-Term Incubation with Light Chain Induces Cellular Remodeling
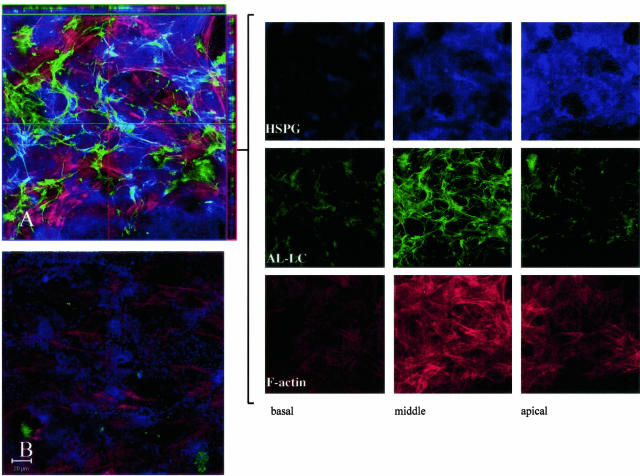
Immunohistochemical and biochemical experiments were performed to evaluate the distribution of the HSPG for a number of the AL LCs compared to control medium. Cells were incubated in the presence of LC-conjugated to Oregon Green 488 for 1, 3, and 7 days and cells were fixed, treated with heparinase III, permeabilized and stained for HSPG using the mAb Δ3G10 and F-actin. The addition of AL LC κ1 (AL-96066) resulted in an increase in the amount of HSPG compared to non-AL LC κ1 (MM-96100) as seen in representative images (Figure 4, A and B). Orthogonal slice analysis on composites of optical serial series throughout the cell showed points of co-localization of HSPG and LCs (Figure 4A). To show the localization of the fluorescent probes within the cell, a series of optical sections 1-μm thick were taken through the cell and three are shown (apical, middle, and basal) in Figure 4. HSPG was detected mainly in the apical regions of the cell while the LC was consistently found at the level of the nucleus (Figure 4). The other AL LCs studied to date induced an enhanced deposition of HSPG throughout the cell. These results indicate that AL LCs enhance the accumulation of HSPG within the cell but in the absence of analytical assays do not provide information as to the degree of sulfation of the GAG chains.
Figure 4.
AL LCs induce remodeling of cellular HSPGs in long-term cultures. Confocal images of cells. Cells were incubated with Oregon Green 488 conjugated to a specific light chain for 7 days, fixed, permeabilized, treated with heparinase III, and stained with mAb Δ3G10 and rhodamine phalloidin. A Z-series of 1-μm optical sections was taken through the cell cultures from the basal to the apical surface of the cell and composites were generated using LSM 5 Image software. A: Cells incubated in the presence of an AL LC κ1 conjugated to Oregon Green 488. Orthogonal slice analysis was performed on the composite using LSM 510 Image software (XZ is depicted on the horizontal and YZ on the vertical axis outside the dimensions of the composite). Co-localization of HSPG and LC is demonstrated. Optical sections of the composite show three regions within the cell and demonstrate localization of HSPG, LC, and F-actin in these regions. HSPG is most prominent in the apical optical section and LC is detected in the central region. B: Cell cultures incubated with non-AL LC κ1 conjugated to Oregon Green 488 for 7 days. Cells were stained and fixed as described above. HSPG is negligible in comparison to A. Images are representative of a minimum of three independent experiments.
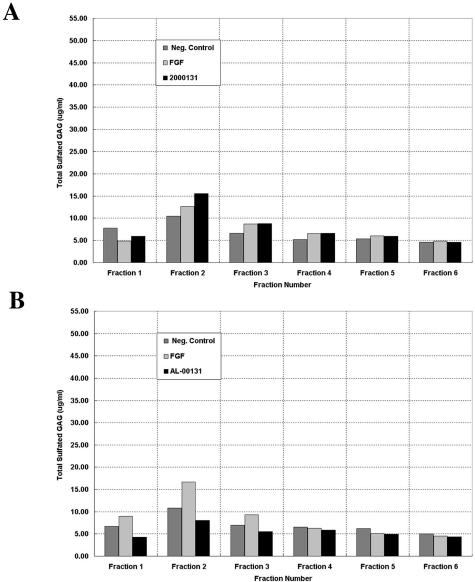
To assess for changes in sulfation, cells were treated with FGF-2, LC or medium for 3 days. FGF-2 was added as a positive control as it is known to mediate HSPG translocation and regulation.28,33 Media and cell lysates were collected and loaded onto Q-Sepharose columns and washed with a sodium acetate buffer with 300 mmol/L NaCl until the A280 nm reached baseline. In low salt fractions there was no detectable sulfated GAG in either the cell lysates or medium. One-ml fractions were collected and after NaCl was increased to 1.5 mol/L sulfation was detected as analyzed by DMB. Sulfated GAGs were consistently found in the medium and cell (Figure 5, A and B) while the matrix extracts were consistently devoid of sulfated GAGs. All of the LCs that we have evaluated to date induced an increase in sulfated GAG. Extractions for one AL LC κ1 (AL-00131) have been repeated five times while other AL LCs (κ1 (AL-96066, AL-01066), λ3 (AL-00109), and λ 6 (AL-01029)) have been repeated at least twice and all reflect the same trend (Figure 5, Table 2). Specifically, one AL LC κ1 (AL-00131) induced an increase in sulfated GAG in the medium that was 53% above control, while FGF-2 induced the secretion of sulfated GAG that was 44% above control (Figure 5A). While the trends do not vary from experiment to experiment or LC to LC, the overall change may vary by 10%. There was substantially less sulfated GAG detected in the cellular fraction (ie, LC: 31% below control; FGF-2: 71% above control) (Figure 5B). These data and the simultaneous readings from the A280 nm, which monitors protein elution, suggest that cellular GAGs are proteoglycan-associated, while the GAGs secreted in the medium are present as free polysaccharides. In summary, the biochemical and immunohistochemical data indicate that cellular GAGs are not highly sulfated, while those that are secreted have a higher sulfation than control.
Figure 5.
FGF-2 and LCs mediate the sulfation of secreted and cellular GAGs. A and B: Representative profiles of media (A) and cell elution profiles (B) from Q1-Sepharose with 1.5 mol/L NaCl single-step gradient. Elution profile is representative of three independent experiments and of nine AL LCs evaluated.
Table 2.
Glycosaminoglycans Present in Medium and Cell after Elution from Column Fractions
| Extract | Fraction | Treatment | Total (μg/ml) | HS (μg/ml) | KS (μg/ml) | CS (μg/ml) |
|---|---|---|---|---|---|---|
| Medium | 2 | Ctrl | 9.32 | 2.1 | 1.99 | 7.3 |
| FGF-2 | 12.5 | 2.48 | 2.48 | 10 | ||
| AL LC κ1 | 13.5 | 1.73 | 1.9 | 10.84 | ||
| 3 | Ctrl | 4.57 | 0.98 | 0.6 | 3.03 | |
| FGF-2 | 7.56 | 1.42 | 1.65 | 4.9 | ||
| AL LC κ1 | 7.79 | 2.05 | 1.26 | 4.91 | ||
| Cell | 2 | Ctrl | 8.95 | 2.02 | 1.1 | 7.41 |
| FGF-2 | 15.93 | 1.87 | 2.68 | 13.21 | ||
| AL LC κ1 | 6.19 | 1.73 | 1.34 | 3.93 | ||
| 3 | Ctrl | 5.13 | 1.38 | 1.11 | 2.73 | |
| FGF-2 | 8.2 | 1.21 | 0.99 | 4.45 | ||
| AL LC κ1 | 3.53 | 0.92 | 0.46 | 1.14 |
Polysaccharidase digestions were performed on medium and cell fractions from two representative κ1 light chains (AL-00131 and AL-96066). Heparan sulfate, CS, and KS were all elevated in the media extracts in response to either FGF-2 or LC. AL LC κ1s caused a change in sulfated GAGs (HS/KS/CS: 23/22/52%) compared to that elicited by FGF-2 (HS/KS/CS: 27/59/44%) (Table 2). In contrast there was a substantial decrease in sulfated GAG in response to LCs in the same respective cellular fractions. All three sulfated GAGs were less than negative control fractions, but the CS was less than 50% of the control. In contrast, FGF-2 induced a large increase in KS and CS (66% and 74% above control, respectively) (Table 2).
To determine the core proteins that were present in response to treatment, cell, matrix and medium fractions were subjected to 12% SDS-PAGE, immunoblotted with antibodies against syndecan-1 and glypican-1 according to manufacturers directions. Perlecan was not investigated in these experiments since only negligible amounts were detected immunohistochemically. Lysates and/or medium samples were loaded onto the gel after treatment with heparinase I and III and chondroitinase ABC. Syndecan-1 and glypican-1 were detected in all three fractions. The addition of AL LC κ1s resulted in cell lysates where the detectable cores were reduced (glypican-1 showed a 37% reduction and syndecan-1 showed a 58% reduction). The differences in the medium and matrix fractions were not as substantial (Figure 6).
Figure 6.
Syndecan-1 and glypican-1 in response to AL LCs. Twelve percent SDS-PAGE and Western blot of media, matrix and cell lysates immunoblotted with MAbs to syndecan-1, and glypican-1. Equivalent amounts of protein were loaded on each lane. Lanes 1–4: Cell; 5–8, Medium; 9–12, ECM. Lanes representing cells treated with AL LC κ1 (AL-00131) 3–4, 7–8, and 11–12. Lanes representing treatment with Heparinase I/III (1.0/2.0U/ml) and Case ABC (0.5U/ml), Cell, 2 and 4; Medium, 6 and 8; Matrix, 10 and 12. Control lanes (1–2, 5–6, 9–10).
To evaluate if the internalization of the LCs was facilitated by GAGs present on the membrane surface of the cells, we enzymatically removed GAGs from the cell surface and asked if LC internalization was decreased using confocal microscopy. Cells were cultured, serum removed and treated with heparinase I and III, chondroitinase, chondroitinase and heparinase together, keratanase or medium alone for 1 hour at 37°C. Cells were washed and LCs conjugated to Oregon Green added and internalization monitored. There was no detectable difference in LC internalization and the punctate pattern was preserved (data not shown). These results indicate that while the LCs alter the production of proteoglycans, their internalization is not mediated by the presence of GAGs on the cell membrane.
Association of Light Chains with Glycosaminoglycans
Solid phase immunobinding assays (ELISA) were performed and absorbance was read at 450 nm. AL-96066 was found to associate with GAGs in the following order (HS>KS>CS≈DS). At 2500 ng of LC, there was a threefold increase in association with HS over that of KS and DS.
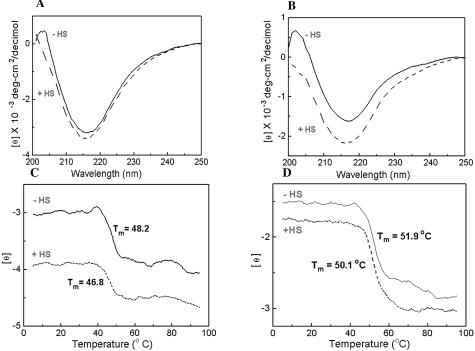
Far UV CD spectroscopy was used to indirectly monitor the binding of HS to selected LCs. This was performed by measuring the induction or alteration of the LC protein secondary structure at 25°C and the thermal stability or folding-unfolding of the LC β-sheets at 217 nm on HS addition in PBS (pH 7.4) containing 50 mmol/L K2HP04 and 0.15 mol/L NaCl. Wavelength spectra of AL LC κ1s (AL-96066 and AL-00131) (Figure 7, A and B) recorded in the presence and absence of HS, showed a change in secondary structure on addition of HS, as indicated by the deeper minimum (more negative molar ellipticity) in the CD spectrum (Figure 7, A and B). Table 3 summarizes Far UV CD results for 2 AL LC κ1s (AL-96066, AL-00131) and the non-AL LC κ1 (MM-96100) reporting molar ellipticity at 217 nm for each LC in the presence and absence of HS and the unfolding temperature of the β-sheets. Thermal stability studies (Figure 7, C and D) demonstrated that addition of HS shifted the midpoint temperatures of unfolding of the β-sheets in AL-96066 and AL-00131, which were reduced by 1.4° and 1.8°C, respectively, suggesting that HS interaction with LCs causes destabilization of LC secondary structure. Studies on the non-AL LC κ1, MM-96100 (Table 3) reveal that while the conformation is altered by the addition of HS, the interaction leads to greater stability in the β-sheets with an increase in unfolding temperature of 1°C.
Figure 7.
Heparan sulfate alters the Far UV spectra and the thermal unfolding of 2 LC κ 1s. Far UV CD Spectra of AL LC κ1 (AL-96066) (A), and AL LC κ1 (AL-00131) (B) with and without HS in PBS at pH 7.4 were recorded at 25°C from 250 to 200 nm. Thermal unfolding of AL-96066 (C) and AL-00131 (D) LCs with and without HS in PBS at pH 7.4 monitored at 217 nm (β-sheet). Ellipticity was continuously monitored from 5 to 95°C every 0.5°C with an averaging time of 90 seconds. Protein concentration, 0.40 mg/ml. LC: HS, 4:1 w/w; pathlength, 0.05 cm.
Table 3.
Light Chain Conformation and Thermal Stability in Presence and Absence of Heparan Sulfate
| LC | [θ] 217 nm, 25°C | TmoC, 217 nm |
|---|---|---|
| AL-96066 (κ1) | ||
| PBS | −3159 | 48.2 |
| +HS | −3367 | 46.8 |
| AL-00131(κ1) | ||
| PBS | −1625 | 51.9 |
| +HS | −2157 | 50.1 |
| MM-96100 (κ1) | ||
| PBS | −3816 | 47.4 |
| +HS | −5091 | 48.4 |
Discussion
In this study we demonstrated that amyloidogenic light chains are internalized by primary cardiac fibroblasts and that the cells respond by altering the regulation of proteoglycan expression and localization. While the disease process of primary amyloidosis has been characterized, it was not known how primary cell systems respond to exogenously added LCs and whether LCs regulate HSPG expression. We hypothesized that LC internalization triggers a cellular response that regulates a number of proteins, including HSPGs. While there is diversity in primary structure and post-translational modification of AL LCs (Table 1)29, there is also a conformational change that occurs, which results in amyloid fibrils. The amyloid fibrils can range from 60 to 120Å in diameter, possess a protein backbone that is predominantly a β-pleated sheet structure and show a green birefringence on polarization microscopy after Congo Red staining.
Light chains were chosen that represent three different LC subtypes (κ1, λ6, and λ 3) and one unidentified lambda subtype. We found that 11 out of 11 AL LCs were internalized by primary cardiac fibroblasts. The LCs present in a perinuclear pattern and optical sections taken through the cell demonstrate that LC was routinely found in the central region of the cell. These indicate that LCs may play a role in mediating synthesis of proteins. Recently, Teng et al20 provided evidence that internalization of LCs into mesangial cells was clathrin-mediated, indicating that entry is a receptor-mediated mechanism. The time course of entry for the LCs in our study does not reflect the one demonstrated by mesangial cells. Time lapse videos with biological markers and inhibitors in our cell model are presently in progress to elucidate the mode of entry and the exact time course of nuclear translocation of HSPGs. We have demonstrated that treatment of cells with GAGases before the addition of LC did not inhibit internalization, however preincubation with heparin or heparan sulfate does inhibit internalization. The processing of the LCs by cardiac fibroblasts differs from that of mesangial cells as the AL LCs that we have evaluated do not have a one-for-one pixel co-localization with lysosomes using the live cell Lysotracker and live cell imaging. In fact some of the AL LCs did not have any demonstrable co-localization. These indicate that while all of the AL LCs are internalized, the processing may differ and this may depend on their tertiary structure. In addition we have data that demonstrate that AL LCs do not enter myocytes unless they demonstrate cellular blebbing (data not shown). Together these data suggest that the mechanism for internalization of LCs may depend on the cell type and on the extracellular environment.
Translocation of HSPGs to the nucleus in response to FGF-2 has been demonstrated in primary fibroblasts.28,33 Translocation seemed to depend on the binding of cells to the heparan-binding domain in fibronectin and the HSPG was hypothesized to play a role as a carrier of heparin-binding proteins. As FGF-2 is a growth factor that requires nuclear localization to induce biological responses40,41, it is reasonable that HSPGs might be a vehicle for heparin-binding growth factors. In addition, matrix molecules have been shown to be altered with cellular injury.42,43 This is interesting as both SAA and ABPP have been demonstrated to have HS binding sequences.5 It is possible that LC may act as transport vehicle for the HSPG to the nucleus where they could bind to nucleosomes as has been described in other systems when cells are stimulated with growth factors.44
Our results indicate that the AL LCs may induce a cellular response that is similar to that of FGF-2.33 All AL LCs evaluated show a similar GAG elution profile and enhance the overall sulfation of GAGs. Furthermore, the medium fractions that were eluted with 1.5 mol/L NaCl had undetectable A280 nm readings and were more highly sulfated than FGF-2. In contrast, the AL-induced HSPGs that are localized within the cell possess a low sulfation and seem to be associated with proteoglycan core. Specific changes that may have occurred in the GAG chains (ie, additional sites of sulfation or length of chain) is not known but is a topic of future studies.
The role of the intact proteoglycan is not well understood. Recently, a transgenic mouse was made where perlecan was overexpressed to study its effect on HSPGs in the development of amyloidosis.45 Surprisingly, while both intracellular accumulation and extracellular deposition were present, there was no increase in amyloidosis.46 These results raise additional questions concerning the underlying mechanisms. The decrease seen in the cellular lysates in our results may reflect an inhibition in the synthesis on the addition of LC. Conversely it may reflect an interaction of the LC and core rendering the epitope inaccessible. Our results demonstrating the shift in thermal stability of two AL LC κ1s (AL-96066 and AL-00131) in the presence of HS indicate that the controlling mechanism may be the GAGs that are secreted into the extracellular milieu. We found that HS seems to increase the thermal stability of non-AL LC κ1s at 217 nm. Our results suggest that the interaction is specific for LCs with an amyloidogenic sequence. Each sequence has a unique post-translational modification.
We propose that the LC causes an injury-like response that alters the regulation of proteoglycans once it is internalized within the cell. It is possible that there is a change in the availability of growth factors that are known to be up-regulated in response to injury and alter expression of proteoglycans and that this may explain the translocation to the nucleus previously shown in response to FGF-2.28,33 The work suggesting that polyvinylsulphonates of various lengths can alter the ultimate fibril formation supports these hypotheses and these and other approaches may be critical in developing treatment strategies to inhibit the role of the LC.21,22 Interestingly, this approach has been used to inhibit the entry of Plasmodium and the circumsporozoite protein has been shown to have a heparan-binding site on its NH2-terminus.46,47 Our model system should allow for a careful study to determine the cooperation between proteoglycans, growth factors, and amyloid light chains. These studies could ultimately provide important insight into the mechanisms that control tissue remodeling, in response to the elevated levels of circulating LCs that occur in AL and the other systemic amyloidoses.
Acknowledgments
We thank the cardiovascular laboratory for supplying the cardiac fibroblasts. We also thank Jeremy Eberhard for assistance with purification, Christopher Schultz for work on confocal microscopy, and Jenny Chiu for work on secondary structure.
Footnotes
Address reprint requests to Dr. V. Trinkaus-Randall, Boston University School of Medicine, L904, 80 E. Concord Street, Boston, MA 02118. E-mail: vickery@bu.edu.
Supported by funds from P01 HL-068705 (to V.T.-R., L.H.C., M.S.), R01 EY4007, P01 HL-26335, the George Burr, Jr. Amyloid Research Fund (to M.T.W.), and the Gerry Foundation.
References
- Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349:583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- Kyle RA, Gertz MA. Primary systemic amyloidosis: clinical and laboratory features in 474 cases. Semin Hematol. 1995;32:45–59. [PubMed] [Google Scholar]
- Pascali E. Diagnosis and treatment of primary amyloidosis. Crit Rev Oncol Hematol. 1995;19:149–181. doi: 10.1016/1040-8428(94)00135-g. [DOI] [PubMed] [Google Scholar]
- Brenner DA, Jain M, Pimentel DR, Wang B, Connors LH, Skinner M, Apstein CS, Liao R. Human amyloidogenic light chains directly impair cardiomyocyte function through an increase in cellular oxidant stress. Circ Res. 2004;94:1008–1010. doi: 10.1161/01.RES.0000126569.75419.74. [DOI] [PubMed] [Google Scholar]
- Stevens FJ, Kisilevsky R. Immunoglobulin light chains, glycosaminoglycans, and amyloid. Cell Mol Life Sci. 2000;57:441–449. doi: 10.1007/PL00000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancsin JB. Amyloidogenesis: historical and modern observations point to heparan sulfate proteoglycans as a major culprit. Amyloid. 2003;10:67–79. doi: 10.3109/13506120309041728. [DOI] [PubMed] [Google Scholar]
- Weber KT, Sun Y, Katwa LC. Local regulation of extracellular matrix structure. Herz. 1995;20:81–88. [PubMed] [Google Scholar]
- Zeydel M, Puglia K, Eghbali M, Fant J, Seifter S, Blumenfeld OO. Properties of heart fibroblasts of adult rats in culture. Cell Tissue Res. 1991;265:353–359. doi: 10.1007/BF00398083. [DOI] [PubMed] [Google Scholar]
- Snow AD, Kisilevsky R. Temporal relationship between glycosaminoglycan accumulation and amyloid deposition during experimental amyloidosis: a histochemical study. Lab Invest. 1985;53:37–44. [PubMed] [Google Scholar]
- Ohishi H, Skinner M, Sato-Araki N, Okuyama T, Gejyo F, Kimura A, Cohen AS, Schmid K. Glycosaminoglycans of the hemodialysis-associated carpal synovial amyloid and of amyloid-rich tissues and fibrils of heart, liver, and spleen. Clin Chem. 1990;36:88–91. [PubMed] [Google Scholar]
- Yamaguchi I, Suda H, Tsuzuike N, Seto K, Seki M, Yamaguchi Y, Hasegawa K, Takahashi N, Yamamoto S, Gejyo F, Naiki H. Glycosaminoglycan and proteoglycan inhibit the depolymerization of beta2-microglobulin amyloid fibrils in vitro. Kidney Int. 2003;64:1080–1088. doi: 10.1046/j.1523-1755.2003.00167.x. [DOI] [PubMed] [Google Scholar]
- Kisilevsky R, Fraser PE. A beta amyloidogenesis: unique, or variation on a systemic theme? Crit Rev Biochem Mol Biol. 1997;32:361–404. doi: 10.3109/10409239709082674. [DOI] [PubMed] [Google Scholar]
- Castillo GM, Lukito W, Wight TN, Snow AD. The sulfate moieties of glycosaminoglycans are critical for the enhancement of beta-amyloid protein fibril formation. J Neurochem. 1999;72:1681–1687. doi: 10.1046/j.1471-4159.1999.721681.x. [DOI] [PubMed] [Google Scholar]
- Castillo GM, Cummings JA, Yang W, Judge ME, Sheardown MJ, Rimvall K, Hansen JB, Snow AD. Sulfate content and specific glycosaminoglycan backbone of perlecan are critical for perlecan’s enhancement of islet amyloid polypeptide (amylin) fibril formation. Diabetes. 1998;47:612–620. doi: 10.2337/diabetes.47.4.612. [DOI] [PubMed] [Google Scholar]
- Ancsin JB, Kisilevsky R. The heparin/heparan sulfate-binding site on apo-serum amyloid A: implications for the therapeutic intervention of amyloidosis. J Biol Chem. 1999;274:7172–7181. doi: 10.1074/jbc.274.11.7172. [DOI] [PubMed] [Google Scholar]
- Watson DJ, Lander AD, Selkoe DJ. Heparin-binding properties of the amyloidogenic peptides Abeta and amylin: dependence on aggregation state and inhibition by Congo red. J Biol Chem. 1997;272:31617–31624. doi: 10.1074/jbc.272.50.31617. [DOI] [PubMed] [Google Scholar]
- Park K, Verchere CB. Identification of a heparin-binding domain in the N-terminal cleavage site of pro-islet amyloid polypeptide: implications for islet amyloid formation. J Biol Chem. 2001;276:16611–16616. doi: 10.1074/jbc.M008423200. [DOI] [PubMed] [Google Scholar]
- Mok SS, Sberna G, Heffernan D, Cappai R, Galatis D, Clarris HJ, Sawyer WH, Beyreuther K, Masters CL, Small DH. Expression and analysis of heparin-binding regions of the amyloid precursor protein of Alzheimer’s disease. FEBS Lett. 1997;415:303–307. doi: 10.1016/s0014-5793(97)01146-0. [DOI] [PubMed] [Google Scholar]
- Clarris HJ, Cappai R, Heffernan D, Beyreuther K, Masters CL, Small DH. Identification of heparin-binding domains in the amyloid precursor protein of Alzheimer’s disease by deletion mutagenesis and peptide mapping. J Neurochem. 1997;68:1164–1172. doi: 10.1046/j.1471-4159.1997.68031164.x. [DOI] [PubMed] [Google Scholar]
- Teng J, Russell WJ, Gu X, Cardelli J, Jones ML, Herrera GA. Different types of glomerulopathic light chains interact with mesangial cells using a common receptor but exhibit different intracellular trafficking patterns. Lab Invest. 2004;84:440–451. doi: 10.1038/labinvest.3700069. [DOI] [PubMed] [Google Scholar]
- Kisilevsky R, Lemieux LJ, Fraser PE, Kong X, Hultin PG, Szarek WA. Arresting amyloidosis in vivo using small-molecule anionic sulphonates or sulphates: implications for Alzheimer’s disease. Nat Med. 1995;1:143–148. doi: 10.1038/nm0295-143. [DOI] [PubMed] [Google Scholar]
- Inoue S, Hultin PG, Szarek WA, Kisilevsky R. Effect of poly(vinylsulfonate) on murine AA amyloid: a high-resolution ultrastructural study. Lab Invest. 1996;74:1081–1090. [PubMed] [Google Scholar]
- Trinkaus-Randall V. Cornea. Lanza RP, Vacanti J, editors. Boston MA: Academic Press; Principles of Tissue Engineering. 1997:pp 471–492. [Google Scholar]
- Brown CT, Applebaum E, Banwatt R, Trinkaus-Randall V. Synthesis of stromal glycosaminoglycans in response to injury. J Cell Biochem. 1995;59:57–68. doi: 10.1002/jcb.240590108. [DOI] [PubMed] [Google Scholar]
- Brown CT, Nugent MA, Lau FW, Trinkaus-Randall V. Characterization of proteoglycans synthesized by cultured corneal fibroblasts in response to transforming growth factor beta and fetal calf serum. J Biol Chem. 1999;274:7111–7119. doi: 10.1074/jbc.274.11.7111. [DOI] [PubMed] [Google Scholar]
- Hassell JR, Cintron C, Kublin C, Newsome DA. Proteoglycan changes during restoration of transparency in corneal scars. Arch Biochem Biophys. 1983;222:362–369. doi: 10.1016/0003-9861(83)90532-5. [DOI] [PubMed] [Google Scholar]
- Funderburgh JL, Cintron C, Covington HI, Conrad GW. Immunoanalysis of keratan sulfate proteoglycan from corneal scars. Invest Ophthalmol Vis Sci. 1988;29:1116–1124. [PubMed] [Google Scholar]
- Richardson TP, Trinkaus-Randall V, Nugent MA. Regulation of heparan sulfate proteoglycan nuclear localization by fibronectin. J Cell Sci. 2001;114:1613–1623. doi: 10.1242/jcs.114.9.1613. [DOI] [PubMed] [Google Scholar]
- Lim A, Wally J, Walsh MT, Skinner M, Costello CE. Identification and location of a cysteinyl post-translational modification in an amyloidogenic kappa1 light chain protein by electrospray ionization and matrix-assisted laser desorption/ionization mass spectrometry. Anal Biochem. 2001;295:45–56. doi: 10.1006/abio.2001.5187. [DOI] [PubMed] [Google Scholar]
- Trinkaus-Randall V, Tong M, Thomas P, Cornell-Bell A. Confocal imaging of the alpha 6 and beta 4 integrin subunits in the human cornea with aging. Invest Ophthalmol Vis Sci. 1993;34:3103–3109. [PubMed] [Google Scholar]
- Trinkaus-Randall V, Kewalramani R, Payne J, Cornell-Bell A. Calcium signaling induced by adhesion mediates protein tyrosine phosphorylation and is independent of pHi. J Cell Physiol. 2000;184:385–399. doi: 10.1002/1097-4652(200009)184:3<385::AID-JCP14>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Song QH, Gong H, Trinkaus-Randall V. Role of epidermal growth factor and epidermal growth factor receptor on hemidesmosome complex formation and integrin subunit beta4. Cell Tissue Res. 2003;312:203–220. doi: 10.1007/s00441-002-0693-x. [DOI] [PubMed] [Google Scholar]
- Hsia E, Richardson TP, Nugent MA. Nuclear localization of basic fibroblast growth factor is mediated by heparan sulfate proteoglycans through protein kinase C signaling. J Cell Biochem. 2003;88:1214–1225. doi: 10.1002/jcb.10470. [DOI] [PubMed] [Google Scholar]
- Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- Greenfield N, Fasman GD. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Walsh MT, Watzlawick H, Putnam FW, Schmid K, Brossmer R. Effect of the carbohydrate moiety on the secondary structure of beta 2-glycoprotein: I. implications for the biosynthesis and folding of glycoproteins. Biochemistry. 1990;29:6250–6257. doi: 10.1021/bi00478a020. [DOI] [PubMed] [Google Scholar]
- Chung CM, Connors LH, Benson MD, Walsh MT. Biophysical analysis of normal transthyretin: implications for fibril formation in senile systemic amyloidosis. Amyloid. 2001;8:75–83. doi: 10.3109/13506120109007348. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Masur SK, Dewal HS, Dinh TT, Erenburg I, Petridou S. Myofibroblasts differentiate from fibroblasts when plated at low density. Proc Natl Acad Sci USA. 1996;93:4219–4223. doi: 10.1073/pnas.93.9.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalric F, Baldin V, Bosc-Bierne I, Bugler B, Couderc B, Guyader M, Patry V, Prats H, Roman AM, Bouche G. Nuclear translocation of basic fibroblast growth factor. Ann NY Acad Sci. 1991;638:127–138. doi: 10.1111/j.1749-6632.1991.tb49023.x. [DOI] [PubMed] [Google Scholar]
- Prudovsky IA, Savion N, LaVallee TM, Maciag T. The nuclear trafficking of extracellular fibroblast growth factor (FGF)-1 correlates with the perinuclear association of the FGF receptor-1alpha isoforms but not the FGF receptor-1beta isoforms. J Biol Chem. 1996;271:14198–14205. doi: 10.1074/jbc.271.24.14198. [DOI] [PubMed] [Google Scholar]
- Rich CB, Nugent MA, Stone P, Foster JA. Elastase release of basic fibroblast growth factor in pulmonary fibroblast cultures results in down-regulation of elastin gene transcription: a role for basic fibroblast growth factor in regulating lung repair. J Biol Chem. 1996;271:23043–23048. doi: 10.1074/jbc.271.38.23043. [DOI] [PubMed] [Google Scholar]
- Buczek-Thomas JA, Nugent MA. Elastase-mediated release of heparan sulfate proteoglycans from pulmonary fibroblast cultures: a mechanism for basic fibroblast growth factor (bFGF) release and attenuation of bfgf binding following elastase-induced injury. J Biol Chem. 1999;274:25167–25172. doi: 10.1074/jbc.274.35.25167. [DOI] [PubMed] [Google Scholar]
- Watson K, Gooderham NJ, Davies DS, Edwards RJ. Nucleosomes bind to cell surface proteoglycans. J Biol Chem. 1999;274:21707–21713. doi: 10.1074/jbc.274.31.21707. [DOI] [PubMed] [Google Scholar]
- Hart M, Li L, Tokunaga T, Lindsey JR, Hassell JR, Snow AD, Fukuchi K. Overproduction of perlecan core protein in cultured cells and transgenic mice. J Pathol. 2001;194:262–269. doi: 10.1002/1096-9896(200106)194:2<262::AID-PATH882>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kisilevsky R, Crandall I, Szarek WA, Bhat S, Tan C, Boudreau L, Kain KC. Short-chain aliphatic polysulfonates inhibit the entry of Plasmodium into red blood cells. Antimicrob Agents Chemother. 2002;46:2619–2626. doi: 10.1128/AAC.46.8.2619-2626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancsin JB, Kisilevsky R. A binding site for highly sulfated heparan sulfate is identified in the N terminus of the circumsporozoite protein: significance for malarial sporozoite attachment to hepatocytes. J Biol Chem. 2004;279:21824–21832. doi: 10.1074/jbc.M401979200. [DOI] [PubMed] [Google Scholar]