Abstract
Tobacco is a known cause of oral disease but the mechanism remains elusive. Nicotine (Nic) is a likely culprit of pathobiological effects because it displaces the local cytotransmitter acetylcholine from the nicotinic receptors (nAChRs) expressed by oral keratinocytes (KCs). To gain a mechanistic insight into tobacco-induced morbidity in the oral cavity, we studied effects of exposures to environmental tobacco smoke (ETS) versus equivalent concentration of pure Nic on human and murine KCs. Both ETS and Nic up-regulated expression of cell cycle and apoptosis regulators, differentiation marker filaggrin, and signal transduction factors at both the mRNA and protein levels. These changes could be abolished in cultured human oral KCs transfected with anti-α3 small interfering RNA or treated with the α3β2-preferring antagonist α-conotoxin MII. Functional inactivation of α3-mediated signaling in α3−/− mutant KCs prevented most of the ETS/Nic-dependent changes in gene expression. To determine relevance of the in vitro findings to the in vivo situation, we studied gene expression in oral mucosa of neonatal α3+/+ and α3−/− littermates delivered by heterozygous mice soon after their exposures to ETS or equivalent concentration of pure Nic in drinking water. In addition to reverse transcriptase-polymerase chain reaction and Western blot, the ETS/Nic-dependent alterations in gene expression were also detected by semiquantitative immunofluorescence assay directly in KCs comprising murine oral mucosa. Only wild-type mice consistently developed significant (P < 0.05) changes in the gene expression. These results identified α3β2 nAChR as a major receptor mediating effects of tobacco products on KC gene expression. Real-time polymerase chain reaction demonstrated that in all three model systems the common genes targeted by α3β2-mediated ETS/Nic toxicity were p21, Bcl-2, NF-κB, and STAT-1. The expression of the nAChR subunits α5 and β2 and the muscarinic receptor subtypes M2 and M3 was also altered. This novel mechanism offers innovative solutions to ameliorate the tobacco-related cell damage and intercede in disease pathways, and may shed light on general mechanisms regulating and driving tobacco-related morbidity in human cells.
Tobacco is the single most important cause of avoidable human deaths. The mechanisms contributing to the illnesses that cause these deaths are under extensive investigation. Several studies have demonstrated a strong positive correlation between the use of tobacco products and increased incidence and severity of periodontal disease.1,2 Both nonkeratinizing and keratinizing oral epithelia may become targeted by tobacco-related morbidity.3 Nevertheless, the nature of the relationship between smoking and periodontal disease is not clear.4
Approximately 430,000 persons die in the United States annually of causes related to smoking cigarettes and ∼30% of those deaths are because of some form of cancer.5,6 Oral squamous cell carcinomas comprise 2 to 3% of all new malignancies diagnosed in the United States, making it the 10th most common malignancy.7 The complex process of epithelial carcinogenesis is composed of discrete biological events including the early activation events of initiation and promotion.8 Despite the identification of possible target genes and their mutations, the initiation events for oral cancer remain poorly understood. The intracellular signals these factors provide are ultimately translated into cellular growth via steps involving nuclear transcription factors. Cigarette smoke condensate activates nuclear transcription factor-κB (NF-κB), which is not cell type-specific and can be seen in T cells (Jurkat), lung cells (H1299), and head and neck squamous cell lines.9 Exposure of Swiss 3T3 cells to mainstream cigarette smoke results in the dose-dependent expression of mRNA and protein c-Fos,10 an early response gene encoding nuclear transcription factors that are expressed in cancer cells.8
Although tobacco smoke contains at least 4000 chemicals, it is generally believed that nicotine (Nic), a major pharmacologically active component of tobacco smoke, is one of the factors responsible for the deleterious consequences of cigarette smoking.11–16 Cumulative results revealed negative effects of Nic on cell function in various nonneuronal locations, some of which may provide a mechanism for the development of tobacco-related illnesses. Furthermore, the safety of local applications of Nic replacement products to mucocutaneous tissues remains to be determined in long-term studies.17,18
The neuronal nicotinic acetylcholine receptors (nAChRs) subserve a range of Nic effects in both neurons and nonneuronal cells. These nAChRs are pentamers variously composed of α (α2 to α10) and β subunits (β2 to β4). The nAChRs are classic representatives of a large superfamily of ligand-gated ion channel proteins, or ionotropic receptors, which mediate the influx of Na+ and Ca2+ and efflux of K+.19 Pharmacological and ligand-binding studies have shown that the different subunits vary in their distribution and channel properties. Studies of the mechanisms of Nic action on various types of cells inhabiting mucocutaneous tissues have identified a number of biological events mediated by a complex of genomic and nongenomic effects resulting from downstream signaling of nAChRs ligated by Nic on the cell membrane.20,21
Regulatory genes can be used to study the nicotinic signals for differential expression of the gene products.5 Nic is known to affect the expression of a number of genes implicated in the mechanisms of tobacco-related human diseases. For instance, Nic has been shown to enhance the AP-1 and ENKCRE-2 DNA binding activities,22,23 modulate activation of NF-κB,24 and up-regulate the mRNA levels of the immediate early genes c-Fos and c-Jun that function as transcription factors.25,26 Thus, although it has been shown that both cigarette smoke and its major constituent, Nic, can alter the expression of a number of genes in neuronal and nonneuronal cells, the regulatory pathways involved primarily remain to be defined.
Because Nic rapidly crosses the plasma membrane, it could affect gene expression either through a nAChR-dependent signaling pathway or through nAChR-independent pathway. To distinguish between these possibilities, Dunckley and Lukas27 studied the ability of nAChR antagonist to abolish Nic-dependent modulation of gene expression. The results revealed a critical role for nAChR activation. In a separate study, it has been demonstrated that the nAChR agonist (+/−)-epibatidine increases fibroblast growth factor-2 mRNA and protein levels in the rat brain.28 Taken together, these observations point out that activation of nAChRs with Nic has a broad role in affecting cellular physiology through modulating gene expression.
The epithelial nAChRs in human gingival and esophageal epithelia may contribute to the development of tobacco-associated morbidity, because pathobiological mechanisms of aberrant cell cycle progression leading to mucosal lesions in chronic tobacco users appear to involve Nic-induced alterations of the keratinocyte (KC) acetylcholine (ACh) axis. The epithelial cells lining the upper digestive tract, KCs, have a functional nonneuronal cholinergic system for signal transduction with ACh as a single cytotransmitter that may exert a plethora of biological effects. The system includes the synthesizing enzyme choline acetyltransferase, two molecular forms of the degrading enzyme acetylcholinesterase, and two types of cholinergic receptors, the nicotinic and the muscarinic receptors. The KC nAChRs can be comprised of the α3, α5, α7, α9, β2, and β4 subunits,29–31 and KC muscarinic ACh receptors (mAChRs) are represented by the M2, M3, M4, and M5 subtypes.32 Hence, the physiological control of KCs by ACh can be mediated by two distinct types of biochemical events: the ionic events, generated by opening of ACh-gated ion channels represented by KC nAChRs; and the metabolic events, elicited by ACh binding to the G protein-coupled single-subunit transmembrane glycoproteins, or mAChRs. Simultaneous stimulation of KC nAChRs and mAChRs by endogenously secreted ACh may be required to synchronize and balance the development of ionic and metabolic events in a single KC. In this model, binding of ACh to the cell membrane simultaneously elicits several diverse biochemical events the biological sum of which, taken together with cumulative effects of hormonal and environmental stimuli, determines a distinct change in the cell cycle. Thus, by simultaneously activating both cholinergic receptor classes, ACh can play a role of pace maker for KC development.
Some pathobiological effects of tobacco products in oral tissues may stem from Nic-induced alterations of the structure and function of KC nAChRs.30 We have demonstrated that chronic stimulation of KCs with Nic alters the genetically determined program of the cell differentiation-dependent expression of nAChR subunits. Exposure of KCs to Nic also changed the mRNA and protein levels of the cell cycle and cell differentiation markers Ki-67, proliferating cell nuclear antigen (PCNA), p21, cyclin D1, p53, filaggrin, loricrin, and cytokeratin 10. The nicotinic antagonist mecamylamine prevented these changes, indicating that the Nic-induced alterations in the expression of both the nAChR and the cell cycle and cell differentiation genes resulted from pharmacological stimulation of nAChRs.30 To establish the relevance of these findings to the pathobiological effects of tobacco products in vivo, we studied the above parameters in the oral tissue of rats and mice after their exposures for 3 weeks to environmental tobacco smoke (ETS) or drinking water containing equivalent concentrations of Nic that are pathophysiologically relevant.30 The changes of the nAChRs, and the cell cycle and cell differentiation genes were similar to those found in vitro. Thus, by interfering with ACh signaling in KCs, Nic can alter the normal balance of cell growth and differentiation, which accelerates squamatization, and increases the risk for malignant transformation. The type of nAChR(s) mediating pathobiological effects on KCs, however, remains to be identified.
The net biological effect produced by Nic depends on the type of nAChRs predominantly binding this ligand at the cell membrane because the signal transduction pathway downstream from different nAChR subtypes are different, which may reflect differences in the ionic permeability of the ACh-gated channels formed.33 Identification of a nAChR subtype-selective control of the gene expression responsible for acquisition of a particular cell phenotype will provide a mechanistic insight into a general regulatory mechanism subserving the deleterious effects of Nic in the oral epithelium. Immature basal KCs provide the main target for Nic-dependent pathology in the upper digestive tract.34 These cells predominantly express α3 and β2 nAChR subunits29 that can form a functional channel.35 The crucial role of ion channels, such as nAChRs, in maintenance of cell viability has been previously demonstrated.36 Under pathological conditions, such as chronic tobacco usage, aberrant signaling through α3-containing nAChRs was shown to play a role in atherosclerotic plaque development and tumor growth.37 In a previous study, we investigated functional coupling of fibroblast α3 nAChR to regulation of cell development and function.38 A 24-hour exposure to Nic up-regulated expression of p21, cyclin D1, PCNA, Ki-67, Bcl-2, and caspase 3 in human dermal fibroblasts. Transfection with α3 anti-sense oligonucleotide practically eliminated the responsiveness of cells to Nic. The α3 deletion in knockout (KO) mice was associated with a decrease of fibroblast p21, cyclin D1, PCNA, Ki-67, and Bcl-2.38 Thus, α3 nAChR is a likely candidate for mediating effects of Nic on mucocutaneous cells.
To gain a mechanistic insight into the mode of pathophysiological action of Nic in the epithelium lining the upper portion of the digestive tract, we in this study investigated effects of exposures to ETS versus equivalent concentration of pure Nic on human and murine oral KCs. Both ETS and Nic up-regulated expression of several cell cycles, signal transduction, apoptosis, and differentiation genes that could be abolished because of pharmacological or functional inactivation of α3 nAChR-mediated signaling. The similarity of the effects observed with Nic and ETS exposures argued that Nic was the major component of smoke responsible for the cellular effects observed. Results of pharmacological experiments identified α3β2 nAChR as the major receptor mediating tobacco effects on KC gene expression, and suggested that a switch in nAChR subtypes expressed by KCs can, at least in part, mediate pathobiological effects of tobacco products in oral mucosa.
Materials and Methods
Human KC Cultures
Normal human KCs were obtained from attached gingiva (this study has been approved by the University of California Davis Human Subjects Review Committee), as detailed elsewhere.29 Briefly, attached gingival samples were freed of clotted blood and rinsed in Ca2+- and Mg2+-free phosphate-buffered saline (Life Technologies, Inc., Gaithersburg, MD). Samples were then cut into 3- to 4-mm pieces, placed epithelium up into a sterile cell culture dish containing 2.5 ml of 0.125% trypsin (Sigma-Aldrich, Inc., St. Louis, MO) and 2.5 ml of minimum essential medium (Life Technologies, Inc.) supplemented with 50 μg/ml gentamicin, 50 μg/ml kanamycin sulfate, 10 U/ml penicillin G, 10 μg/ml streptomycin, and 5 μg/ml amphotericin (all from Life Technologies, Inc.). Tissue was incubated overnight at 37°C in a humidified atmosphere with 5% CO2. The epithelial sheets were then separated from the lamina propria in minimum essential medium containing 20% heat-inactivated newborn calf serum (Life Technologies, Inc.), and individual KCs were isolated by gentle pipetting followed by centrifugation. The KCs were grown at 37°C and 5% CO2 in 25-cm2 or 75-cm2 Falcon culture flasks (Corning Glass Works, Corning, NY) in serum-free keratinocyte growth medium (KGM; Life Technologies, Inc.) containing 0.09 mmol/L Ca2+. Cell culture medium was changed every 3 days, and cultures were passaged at ∼80% confluence. Cell number was computed using a hemocytometer, and the percentage of dead cells was determined based on trypan blue dye positivity. The KCs undergoing apoptosis were visualized using the DeadEnd Fluorometric TUNEL system purchased from Promega (Madison, WI). The enzymatic activities of proapoptotic caspase 3 and caspase 8 were determined using the respective fluorometric assay kits purchased from Calbiochem (San Diego, CA), following the protocols provided by the manufacturer.
α3 nAChR KO Mice and Murine KCs
The generation of α3 nAChR-deficient mice (α3−/−) by deletion of exon 5 in the α3 gene was described previously.39 All experiments were conducted by an experimenter who was blind to the genotype of the mice. The genotyping of the α3 mutant mice was performed by PCR analysis of mouse tail DNA, as detailed elsewhere.38 The α3+/+ littermates of α3−/− mice were used as controls. Genomic DNA extracted from neonatal mice was used in reverse transcriptase (RT)-PCR assay using the following primers: 5′-GTGGATCCCTCCGGCCATCTTTAAGAG-3′ (wild-type forward), 5′-GACTGTGATGACAATGGACAAGGTGAC-3′ (wild-type reverse), and 5′-GAGACTAGTGAGACGTGCTACTTCCATTTG-3′ (mutant reverse). After 30 cycles, the PCR analysis identified the α3 homozygous null (−/−), heterozygous (+/−), and wild-type (+/+) phenotypes. Murine KCs were obtained from oral mucosa of 2- to 4-day-old mice as reported elsewhere,38,40 and grown at 37°C and 5% CO2 in 25-cm2 Falcon culture flasks using the cell culture techniques optimized for mouse KCs.41,42
ETS/Nic Exposure Experiments
The heterozygous α3−/+ mice in their last week of pregnancy were exposed to ETS (see below) or to an equivalent concentration of Nic in drinking water. This study was approved by University of California Davis Committee on the Use of Animals in Research. We used a sidestream smoke exposure system that provides for whole body exposure.43 Briefly, a TE-10 smoking machine (Teague Enterprises, Davis, CA) burned 2RF4 reference cigarettes (Tobacco and Health Research Institute, University of Kentucky, Louisville, KY) that had been temperature- and humidity-conditioned to generate aged and diluted sidestream cigarette smoke as a surrogate to ETS. Each 2RF4 cigarette delivers ∼0.8 mg of Nic.43 Each cigarette was smoked under rigid conditions of one puff (35 ml volume for 2 seconds duration)/minute throughout a period of 8 minutes. A portion of the air and smoke from the conditioning chamber was further mixed with fresh filtered air before introducing the cigarette smoke into the 0.44-m3 animal exposure chambers. Daily measurements of total suspended particulates of Nic and carbon monoxide were performed. Mean concentrations throughout the course of the study were 1 ± 0.07 mg/m3 of total suspended particulates, 344 ± 85 μg/m3 of Nic, and 4.9 ± 0.7 parts/million of carbon monoxide. Past experiments in our laboratory using identical conditions of exposure to ETS have found plasma Nic levels in rats to range from 2 to 17 ng/ml with an average concentration of 12 ng/ml (ie, in the μm range).44,45 This concentration of sidestream cigarette smoke was selected because it represents a high ambient level that individuals could encounter at home or in other settings where smoking occurs.46 In another series of exposure experiments, pregnant α3+/−mice drank water containing 10 μmol/L of pure Nic. The nonexposed control mice were housed in a chamber and received pure water and filtered clean air only. The pups delivered by exposed and nonexposed mice were euthanized, and oral tissue samples were collected. The fresh oral tissue was immediately used for RNA and protein extractions or embedded in the O.C.T. Tissue Tek compound (Sakura, Tokyo, Japan) for use in indirect immunofluorescence (IF) experiments.
In in vitro ETS/Nic exposure experiments, the monolayers of murine α3−/− and α3+/+ KCs from the littermates of the same progeny were exposed for 6 hours per day for 5 consecutive days to ETS, as described above, or to 10 μmol/L of pure Nic dissolved in KGM. To block KC nAChR, some cells were exposed to ETS/Nic in the absence or presence of 100 nmol/L of the α3β2 selective antagonist α-conotoxin MII (αCtxMII Advanced ChemTech, Louisville, KY) targeting α3β2 nAChRs with the IC50 of 0.5 nmol/L, and other nAChR subunit combinations with 2 to 4 orders of magnitude less potent.47 The experiments were done in triplicates for both exposed and control, nonexposed cultures, and the cells from each culture were harvested and used in experiments separately. In each individual culture, 2.5 × 106 viable KCs were used to extract total RNA and proteins.
Small Interfering RNA (siRNA) Preparation and KC Transfection
The siRNA experiments were performed in accordance to the standard procedure detailed elsewhere.48 To design target-specific siRNA duplexes, we selected sequences of the type AA (N19) UU (N, any nucleotide) from the open reading frame of the target mRNAs, to obtain a 21-nucleotide sense and 21-nucleotide anti-sense strand with symmetric 2-nucleotide 3′ overhangs of identical sequence.49–51 We used 2′-deoxythymidines instead of uridine residues in the 3′ overhangs to enhance nuclease resistance. The 2′-protection ensures that the RNA is not degraded before use. The siRNA-α3 was custom synthesized by Dharmacon (Lafayette, CO) and used for transfection according to the protocol provided by the manufacturer. The target sequence for the human CHRNA3 gene (NM_000743) was: 5′-AACUGCCAGUGGCCAGGGCCU-3′ (mRNA target region, 262 to 280). Before use in experiments, the pair of RNA oligonucleotides was simultaneously 2′-deprotected and annealed in the same reaction as a further precaution against degradation. The siRNA duplex was then desalted via ethanol precipitation directly from the aqueous 2′-deprotection/annealing reaction.52 After deprotection and annealing, the RNA pellet was dissolved in 400 μl of 2′-deprotection buffer, and 40 μl of 10 mol/L ammonium acetate was added to adjust the ammonium concentration to 1.0 mol/L. After addition of 1.5 ml of 100% ethanol, the solution was vortexed, incubated for 5 hours at 70°C, and RNA was pelleted by centrifugation and dissolved in 1.0 ml of 1× buffer (20 mmol/L KCl, 6 mmol/L HEPES-KOH, pH 7.5, 0.2 mmol/L MgCl2). To prepare the transfection solution, the TransIT-TKO transfection reagent (Mirus, Madison, WI) was mixed with Opti-MEM medium (Life Technologies, Inc.) at the ratio of 1:12.5, incubated for 20 minutes at room temperature, after which time the siRNA duplex was added and incubated for additional 20 minutes. For transfection, human KCs were seeded at a density of 5 × 104 cells per well of a 24-well plate, and incubated for 16 to 24 hours to achieve ∼70% confluence. To each well, 60 pmol of siRNA duplex (3 μl of 20 μmol/L annealed duplex) in the transfection solution was added, and the transfection was continued for 16 hours at 37°C in a humid, 5% CO2 incubator. On the next day, the transfection medium was replaced by KGM, and the cells were incubated for 72 hours to achieve maximum inhibition of α3 protein expression, as was experimentally determined by Western blot (WB) and IF assays with anti-α3 antibody at different time points after transfection. After 72 hours of incubation, the cells were harvested, and the proteins were extracted and used in WB assays of KC gene expression (see Results). The siRNA transfection efficiency in KCs was also assayed using fluorescein isothiocyanate (FITC)-labeled Luciferase GL2 duplex (Dharmacon) with the target sequence 5′-CGTACGCGGAATACTTCGA-3′ (data not shown). The fluorescein-labeled Luciferase duplex was used as a negative control for all RNA inhibition experiments.
RT-PCR Assay
Total RNA was extracted from cultured KCs and oral tissue samples using the guanidinium-thiocyanate-phenol-chloroform extraction procedure as described elsewhere.53 One μg of dried, DNase-treated, RNA was reverse transcribed in 20 μl of RT-PCR mix [50 mmol/L Tris (pH 8.3), 6 mmol/L MgCl2, 40 mmol/L KCl, 25 mmol/L dNTPs, 1 μg Oligo-dt (Life Technologies, Inc.), 1 mmol/L dithiothreitol, 1 U RNase inhibitor (Boehringer Mannheim, Mannheim, Germany) and 10 U SuperScript II (Life Technologies, Inc.)] at 42°C for 2 hours. The PCR was performed in a final volume of 50 μl containing 1 μl of the single-strand cDNA product, 10 mmol/L Tris-HCl (pH 9.0), 5 mmol/L KCl, 5 mmol/L MgCl2, 0.2 mmol/L dATP, 0.2 mmol/L dCTP, 0.2 mmol/L dGTP, 0.2 mmol/L dTTP, and 2.5 U TaqDNA polymerase (Perkin Elmer, San Jose, CA) and 20 pmol for each forward (5′) and reverse (3′) primers. To allow a quantitative determination of relative gene expression levels,30 the cDNA content of the samples was normalized, and the linear range of amplification was determined for each primer set. For each experiment the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified with 20 to 30 cycles to normalize the cDNA content of the samples. The amplification was performed at 94°C (1 minute), 60°C (2 minutes), and 72°C (3 minutes) for 24 to 30 cycles. The studied human and murine genes are listed in Tables 1 and 2, respectively. For quantitative determination of the relative gene expression levels, the 20-μl samples were collected during PCR after the completion of three different cycle numbers in a linear range. The amplicons were analyzed on a 2% Sea Kem LE agarose gel (FMC, Riceland, ME) stained with SYBRGreen I (Molecular Probes, Eugene, OR). Pictures of the bands were taken using a digital imaging system (α Imager 2000, San Leandro, CA). Band intensities were determined by area integration. The experimental samples were always run in parallel with control samples. In each experiment, the relative gene expression level was determined after at least two different cycle numbers in a linear range, and then averaged. To standardize the analysis, the results were expressed as ratio of the gene expression level in experimental sample compared to that in control sample. To obtain this ratio, the intensity of the band in experimental sample was divided by the value obtained in the control sample, and the results from three independent experiments were averaged. Therefore, the gene expression ratio in the control samples was always set equal to 1. The images shown represent typical results from three independent experiments (n = 3).
Table 1.
Human Genes Studied by RT-PCR
| Name | Abbreviation | Gene name | Accession no. | cDNA size (bp) |
|---|---|---|---|---|
| Cell cycle regulators | ||||
| p53, transition from G0 to G1 | p53 | p53 | NM_019845 | 1496 |
| Cdk, inhibitor p21 binding protein 1A | p21 | CDK1A | BC000312 | 2265 |
| Apoptosis markers | ||||
| Bcl-2, apoptosis inhibitor | Bcl2 | BCL2 | NM_000633 | 6030 |
| Caspase-3, cysteine protease | CASP3 | CASP3 | BC016926 | 2640 |
| Keratinocyte differentiation marker | ||||
| Filaggrin | Flgn | Flg | M24355 | 1248 |
| Signal transduction factors | ||||
| GATA-3, promoter enhancer | GATA3 | GATA3 | NM_002051 | 3067 |
| NF-κB, transcription enhancer | NF-κB | NF-κB | NM_003998 | 4104 |
| STAT-1, transcription activator | STAT1 | STAT-1 | NM_139266 | 2762 |
| JAK-1, tyrosine kinase | JAK1 | JAK1 | M64174 | 3541 |
| nAChR subunits | ||||
| α5 | α5 | CHRNA5 | M83712 | 1679 |
| β2 | β2 | CHRNB2 | U62437 | 2448 |
| mAChR subtypes | ||||
| M2 | M2 | CHRM2 | XM_004724 | 1401 |
| M3 | M3 | CHRM3 | NM_000740 | 2757 |
| Glyceraldehyde-3 phosphate dehydrogenase | GAPDH | GAPD | NM_002046 | 1283 |
Table 2.
Murine Genes Studied by RT-PCR
| Name | Abbreviation | Gene name | Accession no. | cDNA size (bp) |
|---|---|---|---|---|
| Cell cycle regulators | ||||
| p53, transition from G0 to G1 | p53 | p53 | AB043586 | 1305 |
| Cdk, inhibitor p21 binding protein 1A | p21 | Cdk1a | NM_007669 | 1909 |
| Apoptosis markers | ||||
| Bcl-2, apoptosis Inhibitor | Bcl2 | Bcl2 | NM_177410 | 1897 |
| Caspase 3, cysteine protease | Casp3 | Casp3 | NM_009810 | 1466 |
| Keratinocyte differentiation marker | ||||
| Filaggrin | Flgn | Flg | J03458 | 1488 |
| Signal transduction factors | ||||
| GATA-3, promoter enhancer | GATA3 | Gata3 | NM_008091 | 2853 |
| NF-κB, transcription enhancer | NF-κB | Nf-κB | NM_008689 | 3892 |
| STAT-1, transcription activator | STAT1 | Stat-1 | NM_009283 | 4248 |
| JAK-1 tyrosine kinase | JAK1 | Jak1 | NM_146145 | 4191 |
| nAChR subunits | ||||
| α5 | α5 | Chrna5 | NM_176844 | 1317 |
| β2 | β2 | Chrnb2 | AF299083 | 1548 |
| mAChR subtypes | ||||
| M2 | M2 | Chrm2 | XM_145245 | 1872 |
| M3 | M3 | Chrm3 | NM_033269 | 3171 |
| Glyceraldehyde-3 phosphate dehydrogenase | Gapdh | Gapd | XM_132897 | 1231 |
Real-Time PCR
Primers for the genes encoding p53, p21, Bcl-2, caspase-3, NF-κB, JAK-1, STAT-1, and GATA-3 are shown in Table 3. The nucleotide databases were searched to confirm gene specificity. To avoid amplification of contaminating genomic DNA, when possible one of the two primers was placed at the junction between two exons. Primer pairs were chosen to minimize primer dimerization and to generate an amplicon between 75 and 150 bp. For each primer pair, we used no-template control and no-reverse transcriptase controls, which produced no signals, suggesting that primer-dimer formation and genomic DNA contamination effects were negligible.54,55 All PCR reactions were performed using an ABI Prism 7500 sequence detection system (Applied Biosystems) and the SYBR Green PCR Core Reagents kit (Applied Biosystems) in accordance to the manufacturer’s protocol. Briefly, 22.5 μl of diluted cDNA sample produced from 1 μg of total RNA was added to 25 μl of the PCR master-mix. The amplification included a 2-minute 50°C step required for optimal AmpErase UNG activity, an initial denaturation step for 10 minutes at 95°C, followed by 40 cycles consisting of 15 seconds at 95°C and 1 minute at 60°C. Obtained gene expression values were normalized using the housekeeping gene GAPDH to correct minor variations in mRNA extraction and reverse transcription. The threshold cycle (Ct value), ie, the point at which target-derived fluorescence can be distinguished against background fluorescence, was determined. By constructing a standard curve, the Ct value was translated into a quantitative result.56 The data from triplicate samples were analyzed with sequence detector software (Applied Biosystems) and expressed as mean ± SD of mRNA in question relative to that of control. Relative expression was determined using the Ct method57 that calculates relative expression through the equation: -fold induction = 2[ΔΔCt], where ΔΔCt = Ct gene of interest − Ct GAPDH.
Table 3.
Primers Used in Real-Time PCR Assays
| Name | Human primers (forward/reverse) | Product size (bp) | Murine primers (forward/reverse) | Product size (bp) |
|---|---|---|---|---|
| p53 | 5′-GGAGGGGCGATAAATACC-3′ | 5′-TCTCGCTCACTGTGGTTTTTGG-3′ | ||
| 5′-AACTGTAACTCCTCAGGCAGGC-3′ | (131) | 5′-TCCACCTCTTTAGACGGC-3′ | (111) | |
| p21 | 5′-GATTTCTACCACTCCAAACGCC-3′ | 5′-AATCCTGGTGATGTCCGACC-3′ | ||
| 5-AGAAGATGTAGAGCGGGC-3′ | (115) | 5-TCAAAGTTCCACCGTTCTCGGG-3′ | (145) | |
| Bcl-2 | 5′-TTTTTCTCCTTCGGCGGG-3′ | 5′-GGAAGGTAGTGTGTGTGG-3′ | ||
| 5-GGTGGTCATTCAGGTAAGTGGC-3′ | (105) | 5-ACTCCACTCTCTGGGTTCTTGG-3′ | (103) | |
| Cs-3 | 5′-GTGGCATTGAGACAGACAGTGG-3′ | 5′-GTGGCATTGAGACAGACAGTGG-3′ | ||
| 5-GCCAAGAATAATAACCAGGTGC-3′ | (110) | 5-TCCAGGAATAGTAACCAGGTGC-3′ | (110) | |
| NF-κB | 5′-TCCGTTATGTATGTGAAGGC-3′ | 5′-ATTTGAAACACTGGAAGCACGG-3′ | ||
| 5-TTTGCTGGTCCCACATAGTTGC-3′ | (111) | 5-CCGCCTTCTGCTTGTAGATAGG-3′ | (101) | |
| JAK-1 | 5′-TACCTCTATGGCGTCTGTGTCC-3′ | 5′-TGGCACGGAACCAATGACAACG-3′ | ||
| 5-TGGTAAGGACATCACTTTTCCG-3′ | (108) | 5-ATCAAGGAGTGGGGTTGC-3′ | (107) | |
| STAT-1 | 5′-GAACCCAGGAATCTGTCC-3′ | 5′-ATAGAGCAGGAAATCAAGACCC-3′ | ||
| 5-TCCACATTGAGACCTCTTTTGG-3′ | (112) | 5-CTGTTTTTGGTCGCTCTTCGCC-3′ | (116) | |
| GATA-3 | 5′-GGGCTCTATCACAAAATGAACG-3′ | 5′-ATGGGTTAGAGAGGCAGAGC-3′ | ||
| 5-TTGTGGTGGTCTGACAGTTCGC-3′ | (111) | 5-ATACTGCTCCTGCGAAAAACGC-3′ | (103) |
WB Assay
Proteins were isolated from the phenol-ethanol supernatant of homogenized human or murine KCs, or murine oral tissues, by adding 1.5 ml of isopropyl alcohol per 1 ml of TRIzol reagent (Life Technologies, Inc.) and analyzed via quantitative immunoblotting as described.30 The specificities and working concentrations of the primary antibodies used are listed in Table 4. The membranes were developed using the ECL + Plus chemiluminescent detection system (Amersham Pharmacia Biotech, Inc.). To visualize antibody binding, the membranes were scanned with Storm/FluorImager (Molecular Dynamics, Mountain View, CA), and band intensities were determined by area integration using ImageQuant software (Molecular Dynamics). To normalize data for protein content, the housekeeping protein actin was visualized in each sample with anti-β-actin antibody (Table 4). The results were standardized by expressing the density of each protein band under investigation in the experimental sample relative to the value determined in the control sample. The ratios obtained in three independent experiments were averaged to obtain the mean value. The protein content ratio in each control sample was always set equal to 1. The images shown represent typical appearances of protein band in the gels.
Table 4.
Primary Antibodies Used in Western Blot Assays
| Antibody | Isotype | MW | Dilution (μg/ml) | Epitope | Reactivity |
|---|---|---|---|---|---|
| α3 nAChR* | IgG | 60 | 1 | PLMAREDA | Human and rodents |
| α5 nAChR* | IgG | 60 | 1 | PVHIGNANK | Human and rodents |
| β2 nAChR* | IgG | 60 | 1 | HSDDHSAPSSK | Human and rodents |
| M2 mAChR* | IgG | 65 | 1 | PVHIGNANK | Human and rodents |
| M3 mAChR* | IgG | 70 | 1 | FVEAVSKDFA | Human and rodents |
| p21† | IgG | 21 | 1 | TSMTDFYHSKRR | Human and rodents |
| p53† | IgG | 53 | 5 | RHSVV | Human and rodents |
| Bcl-2† | IgG | 24 | 0.5 | 20–30 a.a. | Human and rodents |
| Caspase-3† | IgG | 32 | 5 | Whole protein | Human and rodents |
| Filaggrin‡ | IgG | 38 | 0.5 | DSQVHSGVQVEGRRGH | Human and rodents |
| GATA-3§ | IgG | 50 | 1 | 167–214 a.a | Human and rodents |
| NF-κB§ | IgG | 50 | 1 | 120–239 a.a | Human and rodents |
| STAT-1§ | IgG | 91 | 1 | Carboxy terminus | Human and rodents |
| JAK-1§ | IgG | 130 | 1 | 270–375 a.a | Human and rodents |
| β-Actin¶ | IgG1 | 42 | 0.2 | PPIAALVIPSGSGL | Human and rodents |
Research & Diagnostic Antibodies, Benicia, CA.
Oncogene Research Products, Boston, MA.
BabCO, Richmond, CA.
Santa Cruz Biotechnology, Santa Cruz, CA.
Sigma Chemical Co., St. Louis, MO.
IF Assay
The IF experiments with oral tissues from experimental and control mice were performed as detailed previously,58,59 using a computer-assisted image analysis with a software package purchased from Scanalytics (Fairfax, VA). The intensity of fluorescence was calculated pixel by pixel by dividing the summation of the fluorescence intensity of all pixels by the area occupied by the pixels (ie, segment), and then subtracting the mean intensity of fluorescence of a tissue-free segment (ie, background). For each tissue specimen, a minimum of three different segments in at least three different microscopic fields was analyzed, and the results compared. To visualize membrane-associated molecules, such as nAChR subunits, the specimens were fixed for 3 minutes with 3% fresh depolymerized paraformaldehyde that contained 7% sucrose, so as to avoid cell permeabilization. To visualize molecules located intracellularly, such as signal transduction factors, oral tissue sections were permeabilized with 100% acetone for 10 minutes. The fixed specimens were washed and incubated overnight at 4°C with a primary antibody (Table 4). Binding of primary antibody was visualized by incubating the specimens for 1 hour at room temperature with the appropriate secondary, FITC-conjugated antibody purchased from Pierce (Rockford, IL). The specimens were examined with an Axiovert 135 fluorescence microscope (Carl Zeiss Inc., Thornwood, NY). The specificity of antibody binding in IF experiments was demonstrated by omitting the primary antibody or by replacing primary antibody with an irrelevant antibody of the same isotype and species as the primary antibody. In the cell state assays, we used FITC-labeled polyclonal antibody to the marker of undifferentiated KCs cytokeratin 5, purchased from BAbCO (Richmond, CA), and monoclonal antibody to the marker of differentiated KCs cytokeratin 10 (Novocastra Laboratories Ltd., Newcastle on Tyne, UK), and FITC-labeled secondary antibody purchased from Sigma-Aldrich, Inc.
Statistical Analysis
All experiments were performed in triplicates, or quadruplicates, and the results were expressed as mean ± SD. Statistical significance was determined using Student’s t-test. Differences were deemed significant if the calculated P value was <0.05.
Results
The Role of α3 nAChR in Mediating ETS and Nic Effects on Gene Expression in Human KCs
Previous studies have demonstrated profound effects of ETS and pure Nic on KCs and pointed out that KC nAChRs mediate most of these effects.30 Because α3-containing ACh-gated ion channels are the most abundant subtype of nAChRs expressed by basal KCs,29 they might play a role in mediating deleterious effects of ETS/Nic in oral mucosa. To test this hypothesis, we sought to determine the role for α3-containing nAChRs in mediating changes produced by both ETS and Nic in oral KCs in in vitro and in vivo experiments. We therefore studied the effects of pharmacological or functional (ie, gene silencing) inactivation of α3 nAChR on KC response to ETS and Nic. The candidate genes were identified based on the published results of cDNA microarray studies with Nic-stimulated cells,27,60,61 and our own previously reported30 and unpublished observations of ETS/Nic effects on oral KCs. The genes that responded solely to ETS, eg, GATA-1, JAK-2, and STAT-4, were excluded from this study. In a series of in vitro experiments using monolayers of normal human KCs and RT-PCR and WB, we measured effects of ETS and equivalent concentration of pure Nic on transcription and translation of the genes encoding the cell cycle and apoptosis regulators p53, p21, Bcl-2, and caspase 3, the differentiation marker filaggrin, transcription transduction factors NF-κB, JAK-1, STAT-1, and GATA-3, as well as the nAChR subunits α5 and β2, and the mAChR subtypes M2 and M3 mediating effects of the local cytotransmitter ACh on KCs. We used RT-PCR primers and antibodies listed in Tables 1 and 2, respectively.
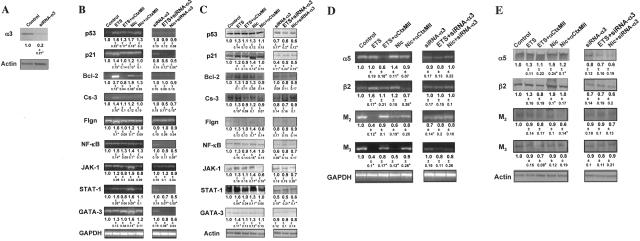
To dissect the role of α3β2 nAChR in mediating ETS/Nic effects on KCs, the cells were exposed to ETS/Nic in the presence of the α3β2-selective antagonist αCtxMII62 at the concentration of 100 nmol/L, which is ordinarily used in cell culture experiments.63,64 High-affinity binding of αCtxMII to α6-containing nAChRs65 was not concerning to us, because the RT-PCR experiments using a set of forward (5′-atgatggcattgagactcttcg-3′) and reverse (5′-ttcaccacagtccgaaggaagg-3′) primers and human genomic DNA as a positive control revealed no expression of this subunit in human oral KCs used for ETS/Nic exposures (data not shown). To functionally inactivate all α3-containing AChRs, some cells were transfected with siRNA-α3 before exposures. The uptake of siRNA by KCs was determined in IF experiments using FITC-conjugated siRNA (data not shown). The efficacy of α3 gene knock-down was measured in WB assays wherein the relative amount of α3 nAChR subunit protein was estimated using α3-specific antibody characterized in the past.29 Transfection with anti-α3 siRNA decreased the relative amount of subunit protein by ∼80% (Figure 1A).
Figure 1.
Gene expression changes in human KCs exposed to ETS or pure Nic. Total RNA and proteins were isolated from intact or siRNA-α3-transfected human KCs exposed for 6 hours per day for 5 consecutive days to ETS or 10 μmol/L Nic. The relative amounts of mRNA transcripts and protein levels of the regulatory molecules p53, p21, Bcl-2, caspase-3 (Cs-3), NF-κB, JAK-1, STAT-1, GATA-3, the differentiation markers filaggrin (Flgn), the α5 and β2 nAChR subunits, and the M2 and M3 mAChR subtypes were measured, and the results expressed as described in the Materials and Methods section. Asterisks indicate significant (P < 0.05) differences from control. A: Representative results of WB analysis of the effect of siRNA-α3 on α3 nAChR subunit expression in human KCs. The numbers underneath the bands are ratios of the densitometry value of α3 subunit to that of β-actin, compared to the values obtained in control KCs (taken as 1). B and D: RT-PCR analysis of ETS and Nic effects on gene expression in human KCs. Gene-specific RT-PCR primers were designed to amplify the human p53, p21, Bcl-2, Cs-3, Flgn, NF-κB, JAK-1, STAT-1, and GATA-3 (B), or α3 and β2 nAChR subunit, or M2 and M3 mAChR subtype (D) genes (Table 1). To standardize the analysis, the gene expression ratios in the control cells, ie, intact KCs in experiments with αCtxMII (shown on the gels) and KCs transfected with a nonspecific siRNA in experiments with siRNA-α3 (not shown), were taken as 1. The ratio data underneath the bands are the means ± SD of the values obtained in at least three independent experiments. The images show representative bands in gels. C and E: WB analysis of ETS and Nic effects on gene expression in human KCs. After the exposure experiments described in the B and D, the protein levels of KC p53, p21, Bcl-2, Cs-3, Flgn, NF-κB, JAK-1, STAT-1, and GATA-3 (C), or α3 and β2 nAChR subunits, or M2 and M3 mAChR subtypes (E) were analyzed by WB. The gene expression ratio of 1 was assigned to control KCs, as explained above. The ratio data are the means ± SD of the values obtained in at least three independent experiments. The images show typical bands appearing at the expected molecular weights (Table 4). Specific staining was absent in the negative control experiments in which the membranes were treated without primary antibody or with irrelevant primary antibody of the same isotype and host (not shown).
The RT-PCR assays yielded products of expected sizes (Figure 1B). Overall, the KCs exposed to ETS or pure Nic produced similar alterations of gene expression. ETS and Nic elevated the message of p53, 1.6- and 1.7-fold; p21, 1.4- and 1.2-fold; Bcl-2, 3.7- and 1.9-fold; caspase-3, 1.4- and 1.2-fold; filaggrin, 1.6- and 1.2-fold; and signal transduction factors (NK-κB, JAK-1, STAT-1, and GATA-3) in the range of 1.2- to 1.5-fold and 1.4- to 1.6-fold, respectively. αCtxMII abolished or attenuated these effects (Figure 1B), indicating that ETS/Nic-induced alterations resulted from the intracellular events downstream from activation of the α3β2 nAChR type. The α3-containing nAChR comprising other subunits, ie, β4 and/or α5, could have mediated effects of ETS and Nic on the transcription of those genes that remained up-regulated in the presence of αCtxMII, such as p53, Bcl-2, and NF-κB. Functional inactivation of all α3-containing nAChRs with siRNA-α3 totally blocked the ETS/Nic effects, compared to control cells transfected with a nonspecific siRNA (Figure 1B). The GAPDH gene amplification remained constant in each experiment. The negative control experiments failed to produce any amplified product.
The ETS/Nic-dependent changes in the expression of the KC genes under consideration detectable by WB were consistent with those determined by RT-PCR (Figure 1C). Each protein band was visualized at the expected molecular weight (Table 4). In addition, we also found that α3 knock-down resulted in significant (P < 0.05) decrease of the relative amounts of the regulatory proteins p53 and p21, and the signal transduction factors NF-κB and STAT, indicating that α3 nAChR are coupled to regulation of these molecules in human KCs.
To determine the biological endpoint of the gene expression changes induced by ETS and pure Nic in KCs, we performed a series of functional assays measuring cell number and viability, expression of the cell state, apoptosis and differentiation markers, and activities of caspase 3 and 8 using the preferential blocker of KC α3β2 receptor αCtxMII (Table 5). We identified functional alterations of the proapoptotic enzymes, and the cell state and viability that are consistent with the gene expression changes found through the RT-PCR and WB approaches.
Table 5.
Functional Analysis of the Endpoint Effects of ETS and Nic on Human Oral KCs
| Studied parameters | Experimental conditions
|
||||
|---|---|---|---|---|---|
| Control (no treatment) | ETS | ETS+αCtxMII | Nic | Nic+αCtxMII | |
| Cell number (1 × 104/well) | 20.4 ± 2.2 | 24.6 ± 2.4 | 18.9 ± 2.1 | 22 ± 2.2 | 20.8 ± 2.4 |
| Number of TBD+ cells (% of total cells) | 3.7 ± 2.1 | 24.2 ± 4.5* | 5.4 ± 2.1 | 15.3 ± 4.2* | 3.1 ± 1.3 |
| Number of TUNEL+ cells (% of total cells) | 3.3 ± 1.8 | 29.1 ± 4.7* | 6.9 ± 2.3 | 21.1 ± 3.7* | 4.7 ± 1.2 |
| Number of CK 5+ cells (% of total cells) | 79.1 ± 5.7 | 49.8 ± 4.7* | 80.7 ± 5 | 60.2 ± 4.7* | 90.4 ± 6 |
| Number of CK 10+ cells (% of total cells) | 11.1 ± 3.7 | 45.8 ± 4.7* | 10.9 ± 4.3 | 35.3 ± 5* | 7.6 ± 3 |
| Caspase 3 activity (RFU) | 304 ± 50 | 1220 ± 78* | 634 ± 55 | 962 ± 36* | 512 ± 58 |
| Caspase 8 activity (RFU) | 372 ± 63 | 1413 ± 125* | 556 ± 55 | 1084 ± 87* | 484 ± 70 |
Data shown are means ± SD.
αCtxMII, α-conotoxin MII; CK, cytokeratin; ETS, environmental tobacco smoke; KCs, keratinocytes; Nic, nicotine; RFU, relative signal; TBD, Trypan Blue dye.
P < 0.05, the rest are P > 0.05, compared to control.
As a result of exposures to ETS and Nic, the relative amounts of mRNA (Figure 1D) and protein (Figure 1E) of KC nAChR subunits and mAChR subtypes also changed. Both ETS and Nic up-regulated expression of the β2 gene, detectable by both RT-PCR and WB. Nic also up-regulated α5 (P < 0.05). In marked contrast, the expression of the mAChR subtypes M2 and M3 was significantly (P < 0.05) down-regulated at the mRNA level. However, changes in the protein levels of these receptors did not reach significance (P < 0.05). The ETS/Nic-dependent alterations were attenuated or completely blocked by αCtxMII, and siRNA-α3. Taken together, these experimental results suggested strongly that the nAChR channels comprised with contribution of α3 subunit, α3β2 in particular, play a central role in mediating effects of tobacco smoke on the expression of the genes that regulate vital function of KCs as well as major types of ACh receptors operating in these cells.
In Vivo Studies of the Effects of Functional Inactivation of Murine KC α3 nAChR on ETS/Nic-Dependent Changes in the Regulatory, and the ACh Receptor Gene Expression
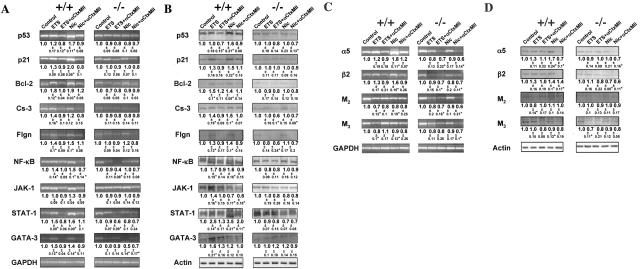
To determine the role of α3-containing nAChRs in mediating in vivo effects of tobacco exposures on oral mucosa, we studied alterations in the expression of the aforementioned KC genes in neonates delivered by the exposed α3+/− mice, followed by genotyping. This approach was justified by the fact that homozygous α3−/− mutant mice have impaired growth and increased mortality before and after weaning.39 In addition to RT-PCR and WB analysis of the oral tissue, we also used semiquantitative IF assay to measure the relative amounts of protein molecules under consideration directly in KCs residing in the oral mucosa of α3+/+ versus α3−/− mice.
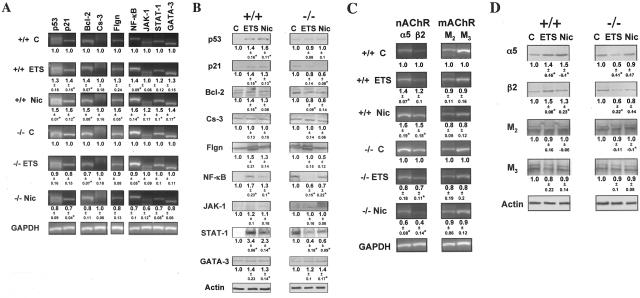
Gene-specific primers for murine p53, p21, Bcl-2, caspase-3, filaggrin, NK-κB, JAK-1, STAT-1, and GATA-3 (Table 2) amplified products of the expected sizes, and the GAPDH gene amplification remained constant in each experiment (Figure 2A). As was expected from the results with human KCs (Figure 1), both ETS exposure and drinking water containing equivalent concentration of pure Nic up-regulated expression of most of studied genes in wild-type mice. The changes in gene expression were detectable by both RT-PCR (Figure 2A) and WB (Figure 2B). In marked contrast, no up-regulation could be seen in the exposed α3−/− KO mice, except for an increased protein level of GATA-3 (Figure 2B). Also in agreement with the results obtained with human KCs were the changes in the relative amounts of KC nAChR subunits and mAChR subtypes induced by exposures to ETS or Nic. Both by RT-PCR (Figure 2C) and WB (Figure 2D), the levels of α5 and β2 mRNA transcripts and proteins, respectively, were increased in wild type, but not in α3−/− KO mice, whereas those of M2 and M3 were somewhat decreased.
Figure 2.
Null mutation of α3 nAChR subunit abolishes ETS- and Nic-dependent changes in KC gene expression in in vivo experiments. To determine the role of α3 nAChRs in mediating effects of ETS and pure Nic on KCs, the gene expression at the mRNA and protein levels was analyzed using RT-PCR and WB assays, respectively. Asterisks indicate significant (P < 0.05) differences from control. A and C: RT-PCR analysis of ETS and Nic effects on gene expression in oral mucosa of exposed α3−/− mice. Gene-specific RT-PCR primers were designed to amplify the murine p53, p21, Bcl-2, Cs-3, Flgn, NF-κB, JAK-1, STAT-1, and GATA-3 (A), or α3 and β2 nAChR subunit, or M2 and M3 mAChR subtype (C) genes (Table 2). The ratio data underneath the bands are the means ± SD of the values obtained in at least three independent experiments. The images show representative bands in gels. B and D: WB analysis of ETS and Nic effects on gene expression in oral mucosa of exposed α3−/− mice. The proteins levels of p53, p21, Bcl-2, Cs-3, Flgn, NF-κB, JAK-1, STAT-1, and GATA-3 (B), or α3 and β2 nAChR subunits, or M2 and M3 mAChR subtypes (D) were analyzed by WB of total protein isolated from the same α3+/+ and α3−/− mice described in B and D, and analyzed by WB as detailed in the legend to Figure 1, C and E, using primary antibodies listed in Table 4. The gene expression ratio of 1 was assigned to oral tissue samples from α3+/+ mice. The ratio data underneath the bands are the means ± SD of the values obtained in at least three independent experiments. The images show typical bands in gels.
To assure that the changes in the gene expression detected by the RT-PCR and WB analysis of the oral epithelium from α3+/+ and α3−/− mice adequately reflected alterations induced by ETS and Nic in KCs, we used IF assay to measure relative amounts of the protein molecules in question directly in the cells comprising oral mucosa of the exposed animals. As seen in Table 6, functional deletion of α3 nAChR dramatically modified cell response to ETS and Nic exposures. Most of the changes in the expression of the study genes were similar to those detected by RT-PCR and WB. We observed from ∼150 to 300% increase of the protein levels of p53, p21, Bcl-2, filaggrin, NF-κB, STAT-1, GATA-3, α5, and β2, and ∼50% decrease of M2 and M3. In α3−/− mice, most of these changes could not be detected. The exposed α3−/− mice showed an increased level of GATA-3, indicating that in vivo this signal transduction molecule is regulated through an additional or an alternative ETS/Nic-sensitive pathway. The relative amounts of caspase-3 and JAK-1 remained within the normal range in exposed α3+/+ and α3−/− mice (P > 0.05). Thus, our observation that the ETS/Nic-dependent changes with p53, p21, Bcl-2, filaggrin, NF-κB, and STAT-1 were either completely or partially blocked in α3−/− mice suggested that KC α3 nAChRs mediated, at least in part, the effects of ETS and Nic on the expression of these genes.
Table 6.
Semiquantitative IF Analysis of KC Gene Expression in the Oral Mucosa of Wild-Type and α3−/− Mutant Mice Exposed to ETS or Pure Nic
| α3+/+ mice
|
α3−/− mice
|
|||||
|---|---|---|---|---|---|---|
| Control | ETS | Nic | Control | ETS | Nic | |
| p53 | 1.0 | 1.63 ± 0.22 | 1.48 ± 0.24 | 1.0 | 1.1 ± 0.11 | 1.12 ± 0.12 |
| P | 0.01 | 0.03 | 0.18 | 0.15 | ||
| p21 | 1.0 | 1.62 ± 0.28 | 1.54 ± 0.25 | 1.0 | 0.97 ± 0.09 | 1.13 ± 0.19 |
| P | 0.02 | 0.02 | 0.62 | 0.31 | ||
| Bcl-2 | 1.0 | 1.78 ± 0.22 | 1.74 ± 0.24 | 1.0 | 1.15 ± 0.17 | 0.99 ± 0.16 |
| P | <0.01 | 0.01 | 0.2 | 0.89 | ||
| Caspase-3 | 1.0 | 1.17 ± 0.12 | 1.19 ± 0.28 | 1.0 | 1.06 ± 0.32 | 0.87 ± 0.18 |
| P | 0.08 | 0.31 | 0.75 | 0.28 | ||
| Filaggrin | 1.0 | 2.05 ± 0.28 | 2.1 ± 0.29 | 1.0 | 1.07 ± 0.15 | 1.15 ± 0.21 |
| P | <0.01 | <0.01 | 0.45 | 0.27 | ||
| NF-κB | 1.0 | 2.6 ± 0.31 | 2.67 ± 0.11 | 1.0 | 1.16 ± 0.27 | 1.22 ± 0.32 |
| P | <0.01 | <0.01 | 0.35 | 0.31 | ||
| JAK-1 | 1.0 | 1.12 ± 0.22 | 1.06 ± 0.26 | 1.0 | 1.03 ± 0.22 | 1.23 ± 0.23 |
| P | 0.4 | 0.73 | 0.81 | 0.17 | ||
| STAT-1 | 1.0 | 3.06 ± 0.54 | 2.98 ± 0.38 | 1.0 | 1.15 ± 0.33 | 1.08 ± 0.21 |
| P | <0.01 | <0.01 | 0.48 | 0.58 | ||
| GATA-3 | 1.0 | 1.79 ± 0.35 | 2.11 ± 0.25 | 1.0 | 2.12 ± 0.4 | 1.62 ± 0.2 |
| P | 0.02 | <0.01 | 0.01 | 0.01 | ||
| α5 | 1.0 | 2.04 ± 0.46 | 2.25 ± 0.16 | 1.0 | 1.06 ± 0.34 | 0.99 ± 0.41 |
| P | 0.02 | <0.01 | 0.78 | 0.98 | ||
| β2 | 1.0 | 2.14 ± 0.25 | 2.07 ± 0.45 | 1.0 | 1.1 ± 0.24 | 1.23 ± 0.49 |
| P | <0.01 | 0.02 | 0.54 | 0.46 | ||
| M2 | 1.0 | 0.39 ± 0.16 | 0.42 ± 0.22 | 1.0 | 1.05 ± 0.19 | 1.09 ± 0.53 |
| P | <0.01 | 0.01 | 0.69 | 0.79 | ||
| M3 | 1.0 | 0.36 ± 0.13 | 0.48 ± 0.04 | 1.0 | 1.11 ± 0.2 | 0.87 ± 0.39 |
| P | <0.01 | <0.01 | 0.4 | 0.6 | ||
Cryostat sections of oral mucosa were stained with the indicated antibodies (Table 4). The intensities of fluorescence produced by antibody binding to KCs in the samples from experimental mice are expressed relative to levels determined in KCs residing on the oral mucosa of intact wild-type mice (taken as 1). Data are means ± SD of at least two independent experiments. No specific staining could be seen in cryostat sections of murine oral mucosa in negative control experiments in which the primary antibody was either omitted or replaced with an irrelevant, isotype-matching antibody (not shown).
In Vitro Studies of the Effects of Functional Inactivation of Murine KC α3 nAChR on ETS/Nic-Dependent Changes in the Regulatory, and ACh Receptor Gene Expression
To ultimately establish the role of α3 deletion in the prevention of ETS/Nic-dependent alterations in KC gene expression, we performed a series of exposure experiments using single cell-type cultures of murine KCs grown from α3−/− versus α3+/+ mice. The α3+/+ KCs exposed to either ETS or Nic in both cases featured increased amounts of p21, Bcl-2, caspase-3, filaggrin, NF-κB, STAT-1, and GATA-3 at both mRNA (Figure 3A) and protein (Figure 3B) levels, ranging from 1.3- to 3.5-fold. ETS/Nic-dependent alterations of p53 and JAK-1 at the mRNA and protein levels were inconsistent. In the exposed α3−/− KCs, no up-regulation of the studied genes could be detected, except for slightly increased protein level of GATA-3.
Figure 3.
Null mutation of α3 nAChR subunit abolishes ETS- and Nic-dependent changes in KC gene expression in in vitro experiments. Monolayers of murine KCs were established from the oral mucosa of neonatal α3+/+ and α3−/− mice and grown to ∼70% confluence in KGM, after which the monolayers were exposed to ETS or Nic as detailed in Materials and Methods. Asterisks indicate significant (P < 0.05) differences from control. A and C: RT-PCR analysis of ETS and Nic effects on gene expression in exposed α3−/− murine KCs. Gene-specific RT-PCR primers were designed to amplify the murine p53, p21, Bcl-2, Cs-3, Flgn, NF-κB, JAK-1, STAT-1, and GATA-3 (A), or α3 and β2 nAChR subunit, or M2 and M3 mAChR subtype (C) genes (Table 2). To standardize analysis, the gene expression ratio in α3+/+ KCs was taken as 1. The ratio data underneath the bands are the means ± SD of the values obtained in at least three independent experiments. The images show representative bands in gels. B and D: WB analysis of ETS and Nic effects on gene expression in exposed α3−/− murine KCs. The KC proteins under consideration were visualized by primary antibodies (Table 4) using the WB procedure described in detail in the legend to Figure 1, C and E. The gene expression ratio of 1 was assigned to α3+/+ KCs. The ratio data underneath the bands are the means ± SD of the values obtained in at least three independent experiments.
Alterations in the expression of α5 and β2 nAChR subunits, and M2 and M3 mAChR subtypes in ETS/Nic-exposed wild-type KCs were found to be very similar to those observed in exposed wild-type mice. The relative amounts of mRNA transcripts (Figure 3C) and protein molecules (Figure 3D) of both nAChR subunits were increased, whereas those of mAChR subtypes were decreased, except for the normal level of M3 in ETS-exposed KCs, as judged from the WB results. To further elucidate the role of KC α3 nAChR in mediating ETS/Nic-induced shift in mucosal ACh receptors, we asked if functional inactivation of α3 nAChR in receptor knockout KCs prevents ETS/Nic-dependent alterations in the expression of ACh receptors under consideration. The α3−/− KCs failed to respond to ETS/Nic with up-regulation of α5 and β2 and down-regulation of M2 and M3 mRNA transcripts (Figure 3C) and proteins (Figure 3D). These results provided additional evidence that the α3 nAChR-coupled pathway of ACh signaling in KCs could mediate nicotinic effects on the expression of the cell cycle, apoptosis, differentiation, signal transduction factor, and ACh receptor genes detected in in vivo experiments. As one can see from the schematic presentation of summarizing results obtained in human and murine KCs via different experimental approaches (Table 7), both ETS and pure Nic significantly up-regulated the expression of predominantly those genes that were down-regulated because of functional inactivation of the α3-containing nAChRs. This correlation links KC α3 nAChR activity with specific gene expression changes.
Table 7.
Summary of Changes of the KC Regulatory Gene Expression
| Experimental conditions | Regulatory genes
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p53
|
p21
|
Bcl-2
|
Caspase 3
|
Filaggrin
|
NF-κB
|
JAK-1
|
STAT-1
|
GATA-3
|
||||||||||
| PCR | WB | PCR | WB | PCR | WB | PCR | WB | PCR | WB | PCR | WB | PCR | WB | PCR | WB | PCR | WB | |
| ETS + | ||||||||||||||||||
| HKC | ↑* | ↑ | ↑* | ↑ | ↑* | ↑ | ↑* | ↑ | ↑* | ↑ | ↑* | ↑* | ↑ | ↑ | ↑* | ↑* | ↑ | ↑* |
| −/−mice | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑ | ↑ | ↑* | ↑* | ↑ | ↑ | ↑* | ↑ | ↑ | |||
| −/−MKC | ↑ | ↑ | ↑ | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ||
| Nic + | ||||||||||||||||||
| HKC | ↑* | ↑* | ↑* | ↑* | ↑* | ↑ | ↑ | ↑* | ↑* | ↑* | ↑* | ↑* | ↑ | ↑ | ↑* | ↑* | ↑ | ↑ |
| −/−mice | ↑* | ↑* | ↑* | ↑* | ↑* | ↑ | ↑* | ↑ | ↑* | ↑* | ↑ | ↑ | ↑* | ↑* | ↑* | ↑* | ||
| −/−MKC | ↑* | ↑* | ↑* | ↑* | ↑* | ↑* | ↑ | ↑* | ↑* | ↑* | ↑* | ↑* | ↑ | ↓* | ↑* | ↑* | ↑* | ↑ |
| siRNA-α3 | ↓* | ↓ | ↓* | ↓ | ↓ | ↓ | ↓* | ↓* | ↓ | |||||||||
ETS, environmental tobacco smoke; HKC, human keratinocytes; MKC, murine keratinocytes; Nic, nicotine; WB, Western blot.
P < 0.05, the rest are P > 0.05, compared to appropriate controls.
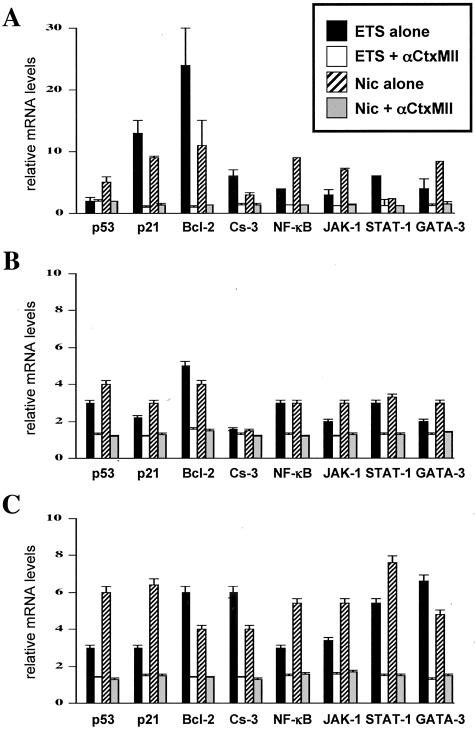
Evaluation of the Effects of ETS/Nic on the KC Gene Expression by Real-Time PCR
Because the changes in the mRNA and protein levels as measured by RT-PCR and WB were, in most cases, marginal, a more sensitive quantitative approach such as real-time PCR was used to raise confidence in the obtained results. Among various regulatory gene studies, the statistically significant (P < 0.05) up-regulations because of exposures to ETS and Nic that could be abolished by the α3β2 selective antagonist αCtxMII (Figure 4A), or because of α3 gene KO in vivo (Figure 4B) and in vitro (Figure 4C) was observed with p53, p21, Bcl-2, caspase-3, NF-κB, JAK-1, STAT-1, and GATA-3. In all three model systems used in this study, similar degrees of transcription elevation because of exposure to either ETS or Nic was found only for the p21, Bcl-2, NF-κB, and STAT-1 genes. The level of up-regulation of the p21 gene ranged from 2.2- to 12.7-fold, that of Bcl-2 from 3.9- to 23.7-fold, NF-κB from 2.2- to 8.7-fold and that of STAT-1 from 2.4- to 7.6-fold. The obtained results indicated that these cell regulators represent the most sensitive targets for the α3β2-mediated toxicity of tobacco products in gingival KCs.
Figure 4.
Real-time PCR analysis of the effects of ETS and Nic on KC gene expression revealing the most sensitive targets of the tobacco toxicity. The real-time PCR analysis of the gene expression was performed using RNA isolated from ETS- or Nic-exposed human gingival KCs in the absence of presence of αCtxMII (A), oral mucosa of α3−/−, and α3+/+ littermates (B), and α3−/− and α3+/+ murine oral KCs (C) in experiments described above in the legend to Figures 1, 2, and 3, respectively. The real-time PCR was performed exactly as described in the Materials and Methods section using the primers listed in Table 3. The alterations in the gene expression levels are presented relative to the rates of expression of corresponding genes in control samples, taken as the baseline.
Discussion
The use of tobacco products is associated with increased incidence of periodontal disease and poor response to periodontal therapy.66 Previous studies have shown the detrimental effects of tobacco usage on periodontal tissue.67–70 The purpose of this study was to gain a mechanistic insight into nAChR-mediated morbidity of tobacco products in the oral cavity. We studied effects of exposures to ETS versus equivalent concentration of pure Nic on human and murine oral KCs. We demonstrated for the first time that both ETS and Nic up-regulate expression of KC genes encoding several cell cycle, signal transduction, apoptosis, and differentiation markers at both the mRNA and protein levels, and that these alterations can be abolished because of pharmacological or functional inactivation of α3 nAChR. The obtained results identified KC α3β2 nAChR as a major candidate for mediating deleterious effects of tobacco products in the epithelium lining the upper digestive tract, and suggested that a switch in the subtype of nAChR expressed by KC may play a mechanistic role.
Both cigarette smoke and pure Nic have been shown to affect proliferation and differentiation of various types of nonneuronal cells.71,72 The classical studies showed direct effects of Nic on oral epithelial cells,73,74 but the types of cellular alterations and the nAChR type(s) mediating Nic effects remained unclear. More recently, after the nonneuronal cholinergic system had been characterized in oral epithelium,29 it became clear that Nic can exhibit its pathobiological effects by displacing the local cytotransmitter ACh from the nAChR expressed on the cell-surface of KCs.30 The cytotransmitter ACh is released locally by nonneuronal cells and can regulate tissue homeostasis in an autocrine and paracrine manner by exhibiting a plethora of biological effects on different cell types inhabiting mucocutaneous tissues.20 ACh, Nic, and cholinergic compounds elicit biological effects on KCs through binding to two different classes of cholinergic receptors, nAChRs and mAChRs. Acting via its nAChRs, ACh has been shown to affect KC apoptosis, differentiation, and cell cycle progression.30,75 The downstream coupling of oral KC mAChRs to regulation of cell cycle progression was demonstrated by the ability of the muscarinic agonist muscarine to increase relative amounts of Ki-67, PCNA, and p53 mRNAs and PCNA, cyclin D1, p21, and p53 proteins in KCs. Thus, the downstream signaling from mAChRs expressed in oral mucosa proceeds via a pathway that up-regulates the expression of cell-cycle progression regulators at both transcriptional and translational levels. Nic, too, may use these signaling pathways to produce its pathobiological effect on oral mucosa.
In this study, we demonstrated profound nicotinic effects of the gene expression in KCs that may provide a mechanism for smoking-related alterations in mucosal cell growth and function. We compared effects of ETS and pure Nic to determine the role for chronic nicotinic stimulation of KCs in tobacco-related oral morbidity. Both in vitro and in vivo ETS exposure experiments used an established system that distributes smoke to each animal and tissue culture plate.43 Previous studies showed that tobacco smokers have saliva concentrations of Nic as high as 1.3 μg/ml, which is more than 100,000 times higher than the level in the blood.76–78 In keeping with the concentration of Nic encountered by tobacco users, we30,75 and others37,44,45,79,80 have reported that in in vitro and in vivo experiments the maximal effects of Nic on nonneuronal cells occur at the dose ranging from 10−8 to 10−6 mol/L. Therefore, the dosing of Nic in this study was chosen to correlate well with the levels found in mucous secretions of smokers and snuffers.
As revealed by real-time PCR assays, among the studied genes the most consistent changes were observed with p21, Bcl-2, NF-κB, and STAT-1, which effectively identified them as the most sensitive targets for Nic toxicity in KCs. The multifold up-regulations of these genes because of ETS/Nic exposures were abolished in the absence of α3 in receptor KO KCs, and completely blocked by the α3β2-selective antagonist αCtxMII, indicating that the downstream signaling from this receptor played a pivotal role. Some inconsistent findings of relative changes of the mRNA and protein levels of the same molecule between human and murine cells may be attributed to posttranslational modifications and/or protein degradation as well as known differences in mRNA half lives, stability, and so forth.81
In the past, using the neuronal SH-SY5Y cell line, it has been demonstrated that binding of Nic to the cell membrane modulates the expression of a diverse set of genes that may be broadly categorized into four groups: transcription factors, protein processing factors, RNA-binding proteins, and plasma membrane-associated proteins.27 Cumulative results of experiments with nonneuronal cells showed that Nic-induced effects on cell growth and differentiation can be mediated through growth factors, such as β-fibroblast growth factor, insulin-like growth factor-1, vascular endothelial growth factor, and transforming growth factor-β1, and their receptors;28,82,83 signal transduction effectors, such as transcription factor NF-κB, CREB, and various AP-1 proteins;22–24,84 oncogenes, such as c-Fos, c-Jun, c-Myc, and H-Ras;5,10,25,26,85,86 cell cycle regulator and tumor suppressor genes, such as p53, TOP2A, CCNB1, CCNA, and CDKN3;87,88 local hormones, growth factors or cytokines, and/or their synthesizing enzymes and receptors, such as the catecholamine biosynthetic enzymes tyrosine hydroxylase, dopamine β-hydroxylase, and phenylethanolamine N-methyltransferase, prolactin, nerve growth factor, neuropeptide Y, nitric oxide synthase, endothelin-1, 5-HT(1A) receptor;89–95 various inflammatory mediators, such as COX-2, macrophage inflammatory protein-1α, tissue-type plasminogen activator, plasminogen activator inhibitor-1, and vascular cell adhesion molecule-1,93,96,97 and a variety of oxidative stress-responsive and xenobiotic enzymes.88,98
Elevation of the early KC differentiation marker filaggrin99 simultaneously with stimulation of the cyclin-dependent kinase inhibitor p21, anti-apoptotic regulator Bcl-2, and the signal transduction effectors NF-κB and STAT-1 observed by us in this study suggest that KCs exposed to cigarette smoke go through dramatic alterations in their growth and function, which may decrease a threshold for malignant transformation. Previously, it was opined that in tobacco users such changes could disrupt the critical balance between cell death and proliferation, resulting in the unregulated growth of cells.100
Pronounced effects of ETS/Nic on KC Bcl-2 levels are in keeping with previous reports that signaling pathways mediating nicotinic regulation of apoptosis predominately involve changes in the expression/activity of the Bcl-2 protein,101 occurring through Bcl-2 phosphorylation by the mitogen-activated protein kinases ERK1 and ERK2.102 In other cell types, treatment with Nic also has been reported to attenuate apoptosis caused by etoposide, ultraviolet irradiation, or hydrogen peroxide.103,104 The central role of nAChRs in mediating tobacco effects on apoptosis is illustrated by the ability of Nic and nicotinic agonists to suppress apoptosis in human neutrophils,105 rat cardiac myocytes,106 murine immune cells,107 chick ciliary ganglion neurons,108 and cultured spinal cord neurons,89 as well as human lung cancer cells.109 Nic inhibition of apoptosis suggested a role in tumor promotion.110 Additionally, Nic has been shown to rapidly activate the serine/threonine kinase Akt,104 a well-known inhibitor of apoptosis.111
Although it has been shown that cigarette smoke prevents apoptosis through inhibition of caspase-3 activation and induces necrosis,112 in this study both ETS and Nic elevated caspase 3. This is in keeping with reports that cigarette smoke can activate caspase-3 to induce apoptosis of human umbilical venous endothelial cells.113 Cigarette smoke also increases apoptosis of alveolar macrophages,114 and in the gastric mucosa115 and rat testis.116 Peripheral blood lymphocytes of chronic cigarette smokers exhibit enhanced expression of FasL protein without in vitro mitogen stimulation.117 The presumable controversy of the results on ETS/Nic effects on caspase-3 can be easily explained by differences in the duration of the exposures, concentrations of Nic used, as well as the presence of other biologically active substances in tobacco smoke. For instance, exposure to cigarette smoke increased apoptosis in the rat gastric mucosa through a reactive oxygen species-mediated and p53-independent pathway.118 Therefore, the nAChR-mediated effects of ETS/Nic that involve regulation of p53 and other apoptosis-related genes may be predominantly anti-apoptotic, whereas the nonreceptor effects of ETS/Nic that induce oxidative stress, may be mainly proapoptotic.117,119
The ETS/Nic-induced morbidity in the upper digestive tract may stem from genomic and nongenomic events resulting from overstimulation of the nicotinic pathways of physiological control of KCs by ACh. Results of this study indicate that this novel pathophysiological mechanism includes Nic-induced changes in the repertoire of α3-made nAChRs, favoring overexpression of α5-containing α3β2 nAChR channels, as well as down-regulation of M2 and M3 mAChRs in KCs. For instance, a switch in the nAChR subunit composition from predominantly α3β2 channels to α3β2α5 channels can alter Ca2+ permeability of the cell membrane,120 leading to altered cell regulation and function. It is well known that overstimulation of nAChRs with agonists can modify ACh metabolism, secretion, and signaling through both the nicotinic and the muscarinic pathways with toxicological implications.121–124 Nic-induced alterations in the structure and function of the cellular cholinergic system, caused by its chronic pharmacological stimulation, occur in the neural system125–127 as well as various types of nonneuronal cells, including both epithelial cells, such as KCs30,128 and bronchial epithelial cells,58 and nonepithelial cells, such as blood polymorphonuclear cells,129 human leukemic T-cell line,130 and dermal fibroblasts.38 Thus, binding of Nic to α3-nAChRs can cause an imbalance in the entire cholinergic network. Alterations in the structure and function of the KC cholinergic system, resulting from chronic stimulation of the nicotinic pathway, may provide a novel mechanism of tobacco-related morbidity.
In conclusion, this study was designed to identify the role of the α3 nAChR type in mediating the effects of both ETS and pure Nic on oral KCs. This study demonstrates for the first time that abnormalities in the KC gene expression caused by tobacco exposure result from receptor-mediated action of Nic, and identify the type of KC nAChR involved. The results convincingly showed an involvement of α3-containing nAChRs in mediating ETS/Nic effects. In future studies, we plan to address the role for α3 nAChR subtypes, ie, α3β2(β4)±α5. The mechanism of pathobiological effects encompasses alterations of the expression of genes encoding regulatory molecules affecting apoptosis, cell cycle progression, and differentiation. The α3β2 nAChR is apparently a major mediator of nicotinic effects on oral KCs. The expression of the nAChR subunits α5 and β2 and the muscarinic receptor subtypes M2 and M3 was also altered, suggesting that a switch in KC cholinergic receptors mediates, at least in part, pathobiological effects of tobacco products in oral mucosa. This novel mechanism offers innovative solutions to ameliorate the tobacco-related cell damage and intercede in disease pathways, and may shed light on general mechanisms regulating and driving tobacco-related morbidity in human cells.
Acknowledgments
We thank Mr. Michael Goldsmith for help with the animal ETS exposure experiments, Mansi R. Chovatia for excellent technical assistance, and Arlene D. Gonzales (Biology and Biotechnology Research Program, Lawrence Livermore National Laboratory, Livermore, California) for help with experiments with siRNA.
Footnotes
Address reprint requests to Sergei A. Grando, M.D., Ph.D., D.Sc., Department of Dermatology, University of California Davis Medical Center, 4860 Y St., Suite #3400, Sacramento, California 95817. E-mail: sagrando@ucdavis.edu.
Supported by the National Institutes of Health (grants PO1 DA12661 to A.L.B. and L.M.M. and R01 DE14173 to S.A.G.) and the Flight Attendant Medical Research Institute (research grant to S.A.G.).
References
- Calsina G, Ramon JM, Echeverria JJ. Effects of smoking on periodontal tissues. J Clin Periodontol. 2002;29:771–776. doi: 10.1034/j.1600-051x.2002.290815.x. [DOI] [PubMed] [Google Scholar]
- Palmer RM, Scott DA, Meekin TN, Poston RN, Odell EW, Wilson RF. Potential mechanisms of susceptibility to periodontitis in tobacco smokers. J Periodontal Res. 1999;34:363–369. doi: 10.1111/j.1600-0765.1999.tb02267.x. [DOI] [PubMed] [Google Scholar]
- Taybos G. Oral changes associated with tobacco use. Am J Med Sci. 2003;326:179–182. doi: 10.1097/00000441-200310000-00005. [DOI] [PubMed] [Google Scholar]
- Obeid P, Bercy P. Effects of smoking on periodontal health: a review. Adv Ther. 2000;17:230–237. doi: 10.1007/BF02853162. [DOI] [PubMed] [Google Scholar]
- Chowdhury P, MacLeod S, Udupa KB, Rayford PL. Pathophysiological effects of nicotine on the pancreas: an update. Exp Biol Med (Maywood) 2002;227:445–454. doi: 10.1177/153537020222700708. [DOI] [PubMed] [Google Scholar]
- Weir HK, Thun MJ, Hankey BF, Ries LA, Howe HL, Wingo PA, Jemal A, Ward E, Anderson RN, Edwards BK. Annual report to the nation on the status of cancer, 1975–2000, featuring the uses of surveillance data for cancer prevention and control. J Natl Cancer Inst. 2003;95:1276–1299. doi: 10.1093/jnci/djg040. [DOI] [PubMed] [Google Scholar]
- Casiglia J, Woo SB. A comprehensive review of oral cancer. Gen Dent. 2001;49:72–82. [PubMed] [Google Scholar]
- Birrer MJ, Alani R, Cuttitta F, Preis LH, Sabich AL, Sanders DA, Siegfried JM, Szabo E, Brown PH. Early events in the neoplastic transformation of respiratory epithelium. J Natl Cancer Inst Monogr. 1992;13:31–37. [PubMed] [Google Scholar]
- Anto RJ, Mukhopadhyay A, Shishodia S, Gairola CG, Aggarwal BB. Cigarette smoke condensate activates nuclear transcription factor-kappaB through phosphorylation and degradation of IkappaB(alpha): correlation with induction of cyclooxygenase-2. Carcinogenesis. 2002;23:1511–1518. doi: 10.1093/carcin/23.9.1511. [DOI] [PubMed] [Google Scholar]
- Muller T. Expression of c-fos in quiescent Swiss 3T3 cells exposed to aqueous cigarette smoke fractions. Cancer Res. 1995;55:1927–1932. [PubMed] [Google Scholar]
- Benowitz NL. Clinical pharmacology of nicotine. Annu Rev Med. 1986;37:21–32. doi: 10.1146/annurev.me.37.020186.000321. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Gourlay SG. Cardiovascular toxicity of nicotine: implications for nicotine replacement therapy. J Am Coll Cardiol. 1997;29:1422–1431. doi: 10.1016/s0735-1097(97)00079-x. [DOI] [PubMed] [Google Scholar]
- Benowitz NL. Systemic absorption and effects of nicotine from smokeless tobacco. Adv Dent Res. 1997;11:336–341. doi: 10.1177/08959374970110030501. [DOI] [PubMed] [Google Scholar]
- Macklin KD, Maus AD, Pereira EF, Albuquerque EX, Conti-Fine BM. Human vascular endothelial cells express functional nicotinic acetylcholine receptors. J Pharmacol Exp Ther. 1998;287:435–439. [PubMed] [Google Scholar]
- Maus ADJ, Pereira EFR, Karachunski PI, Horton RM, Navaneetham D, Macklin K, Cortes WS, Albuquerque EX, Conti-Fine BM. Human and rodent bronchial epithelial cells express functional nicotinic acetylcholine receptors. Mol Pharmacol. 1998;54:779–788. doi: 10.1124/mol.54.5.779. [DOI] [PubMed] [Google Scholar]
- Zevin S, Gourlay SG, Benowitz NL. Clinical pharmacology of nicotine. Clin Dermatol. 1998;16:557–564. doi: 10.1016/s0738-081x(98)00038-8. [DOI] [PubMed] [Google Scholar]
- Lavelle C, Birek C, Scott DA. Are nicotine replacement strategies to facilitate smoking cessation safe? J Can Dent Assoc. 2003;69:592–597. [PubMed] [Google Scholar]
- Wallstrom M, Sand L, Nilsson F, Hirsch JM. The long-term effect of nicotine on the oral mucosa. Addiction. 1999;94:417–423. doi: 10.1046/j.1360-0443.1999.94341711.x. [DOI] [PubMed] [Google Scholar]
- Steinbach JH. Mechanism of action of the nicotinic acetylcholine receptor. Bock G, Marsh J, editors. New York: John Wiley and Sons Ltd.,; The Biology of Nicotine Dependence (Meeting, London, UK, November 7–9, 1989). 1990:pp 53–61. [Google Scholar]
- Grando SA. Biological functions of keratinocyte cholinergic receptors. J Invest Dermatol Symp Proc. 1997;2:41–48. doi: 10.1038/jidsymp.1997.10. [DOI] [PubMed] [Google Scholar]
- Grando SA. Receptor-mediated action of nicotine in human skin. Int J Dermatol. 2001;40:691–693. doi: 10.1046/j.1365-4362.2001.01293.x. [DOI] [PubMed] [Google Scholar]
- Kim YH, Won JS, Won MH, Lee JK, Suh HW. Role of proto-oncogenes in the regulation of proenkephalin mRNA expression induced by repeated nicotine injections in rat adrenal medulla. Life Sci. 2002;70:2915–2929. doi: 10.1016/s0024-3205(02)01539-4. [DOI] [PubMed] [Google Scholar]
- Pandey SC, Xu T, Zhang D. Regulation of AP-1 gene transcription factor binding activity in the rat brain during nicotine dependence. Neurosci Lett. 1999;264:21–24. doi: 10.1016/s0304-3940(99)00172-x. [DOI] [PubMed] [Google Scholar]
- Sugano N, Shimada K, Ito K, Murai S. Nicotine inhibits the production of inflammatory mediators in U937 cells through modulation of nuclear factor-kappaB activation. Biochem Biophys Res Commun. 1998;252:25–28. doi: 10.1006/bbrc.1998.9599. [DOI] [PubMed] [Google Scholar]
- Wonnacott S. Presynaptic nicotinic ACh receptors. Trends Neurosci. 1997;20:92–98. doi: 10.1016/s0166-2236(96)10073-4. [DOI] [PubMed] [Google Scholar]
- Walker LM, Preston MR, Magnay JL, Thomas PB, El Haj AJ. Nicotinic regulation of c-fos and osteopontin expression in human-derived osteoblast-like cells and human trabecular bone organ culture. Bone. 2001;28:603–608. doi: 10.1016/s8756-3282(01)00427-6. [DOI] [PubMed] [Google Scholar]
- Dunckley T, Lukas RJ. Nicotine modulates the expression of a diverse set of genes in the neuronal SH-SY5Y cell line. J Biol Chem. 2003;278:15633–15640. doi: 10.1074/jbc.M210389200. [DOI] [PubMed] [Google Scholar]
- Belluardo N, Mudo G, Blum M, Cheng Q, Caniglia G, Dell’Albani P, Fuxe K. The nicotinic acetylcholine receptor agonist (+/−)-epibatidine increases FGF-2 mRNA and protein levels in the rat brain. Brain Res Mol Brain Res. 1999;74:98–110. doi: 10.1016/s0169-328x(99)00266-1. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Hall LL, Gallacher G, Ndoye A, Jolkovsky DL, Webber RJ, Buchli R, Grando SA. Choline acetyltransferase, acetylcholinesterase, and nicotinic acetylcholine receptors of human gingival and esophageal epithelia. J Dent Res. 2000;79:939–949. doi: 10.1177/00220345000790040901. [DOI] [PubMed] [Google Scholar]
- Arredondo J, Nguyen VT, Chernyavsky AI, Jolkovsky DL, Pinkerton KE, Grando SA. A receptor-mediated mechanism of nicotine toxicity in oral keratinocytes. Lab Invest. 2001;81:1653–1668. doi: 10.1038/labinvest.3780379. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Ndoye A, Grando SA. Novel human α9 acetylcholine receptor regulating keratinocyte adhesion is targeted by pemphigus vulgaris autoimmunity. Am J Pathol. 2000;157:1377–1391. doi: 10.1016/s0002-9440(10)64651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredondo J, Hall LL, Ndoye A, Chernyavsky AI, Jolkovsky DL, Grando SA. Muscarinic acetylcholine receptors regulating cell cycle progression are expressed in human gingival keratinocytes. J Periodont Res. 2003;38:79–89. doi: 10.1034/j.1600-0765.2003.01006.x. [DOI] [PubMed] [Google Scholar]
- Lax P, Fucile S, Eusebi F. Ca(2+) permeability of human heteromeric nAChRs expressed by transfection in human cells. Cell Calcium. 2002;32:53–58. doi: 10.1016/s0143-4160(02)00076-3. [DOI] [PubMed] [Google Scholar]
- Shankaran K, Kandarkar SV, Contractor QQ, Kalro RH, Desai HG. Ultrastructural changes in esophageal mucosa of chronic tobacco chewers. Ind J Med Res. 1993;98:15–19. [PubMed] [Google Scholar]
- Elliott KJ, Ellis SB, Berckhan KJ, Urrutia A, Chavez-Noriega LE, Johnson EC, Velicelebi G, Harpold MM. Comparative structure of human neuronal α2-α7 and β2-β4 nicotinic acetylcholine receptor subunits and functional expression of the α2, α3, α4, α7, β2, and β4 subunits. J Mol Neurosci. 1996;7:217–228. doi: 10.1007/BF02736842. [DOI] [PubMed] [Google Scholar]
- Mulle C, Choquet D, Korn H, Changeux JP. Calcium influx through nicotinic receptor in rat central neurons: its relevance to cellular regulation. Neuron. 1992;8:135–143. doi: 10.1016/0896-6273(92)90115-t. [DOI] [PubMed] [Google Scholar]
- Heeschen C, Jang JJ, Weis M, Pathak A, Kaji S, Hu RS, Tsao PS, Johnson FL, Cooke JP. Nicotine stimulates angiogenesis and promotes tumor growth and atherosclerosis. Nat Med. 2001;7:833–839. doi: 10.1038/89961. [DOI] [PubMed] [Google Scholar]
- Arredondo J, Hall LH, Ndoye A, Nguyen VT, Chernyavsky AI, Bercovich D, Orr-Urtreger A, Beaudet AL, Grando SA. Central role of fibroblast α3 nicotinic acetylcholine receptor in mediating cutaneous effects of nicotine. Lab Invest. 2003;83:207–225. doi: 10.1097/01.lab.0000053917.46614.12. [DOI] [PubMed] [Google Scholar]
- Xu W, Gelber S, Orr-Urtreger A, Armstrong D, Lewis RA, Ou CN, Patrick J, Role L, De Biasi M, Beaudet AL. Megacystis, mydriasis, and ion channel defect in mice lacking the α3 neuronal nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 1999;96:5746–5751. doi: 10.1073/pnas.96.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredondo J, Nguyen VT, Chernyavsky AI, Bercovich D, Orr-Urtreger A, Vetter DE, Grando SA. Functional role of α7 nicotinic receptor in physiological control of cutaneous homeostasis. Life Sci. 2003;72:2063–2067. doi: 10.1016/s0024-3205(03)00084-5. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dlugosz AA, McKay R, Dean NM, Yuspa SH. Definition by specific antisense oligonucleotides of a role for protein kinase C alpha in expression of differentiation markers in normal and neoplastic mouse epidermal keratinocytes. Mol Carcinog. 1997;18:44–53. [PubMed] [Google Scholar]
- Li L, Tucker RW, Hennings H, Yuspa SH. Chelation of intracellular Ca2+ inhibits murine keratinocyte differentiation in vitro. J Cell Physiol. 1995;163:105–114. doi: 10.1002/jcp.1041630112. [DOI] [PubMed] [Google Scholar]
- Teague SV, Pinkerton KE, Goldsmith M, Gebremichael A, Chang S, Jenkins RA, Moneyhun JH. Sidestream cigarette smoke generation and exposure system for environmental tobacco smoke studies. Inhal Toxicol. 1994;6:79–93. [Google Scholar]
- Joad JP, Ji C, Kott KS, Bric JM, Pinkerton KE. In utero and postnatal effects of sidestream cigarette smoke exposure on lung function, hyperresponsiveness, and neuroendocrine cells in rats. Toxicol Appl Pharmacol. 1995;132:63–71. doi: 10.1006/taap.1995.1087. [DOI] [PubMed] [Google Scholar]
- Joad JP, Pinkerton KE, Bric JM. Effects of sidestream smoke exposure and age on pulmonary function and airway reactivity in developing rats. Pediatr Pulmonol. 1993;16:281–288. doi: 10.1002/ppul.1950160503. [DOI] [PubMed] [Google Scholar]
- Ji CM, Royce FH, Truong U, Plopper CG, Singh G, Pinkerton KE. Maternal exposure to environmental tobacco smoke alters Clara cell secretory protein expression in fetal rat lung. Am J Physiol. 1998;275:L870–L876. doi: 10.1152/ajplung.1998.275.5.L870. [DOI] [PubMed] [Google Scholar]
- Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM. A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors. J Biol Chem. 1996;271:7522–7528. doi: 10.1074/jbc.271.13.7522. [DOI] [PubMed] [Google Scholar]
- Chernyavsky AI, Arredondo J, Wess J, Karlsson E, Grando SA. Novel signaling pathways mediating reciprocal control of keratinocyte migration and wound epithelialization by M3 and M4 muscarinic receptors. J Cell Biol. 2004;166:261–272. doi: 10.1083/jcb.200401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Bieche I, Onody P, Laurendeau I, Olivi M, Vidaud D, Lidereau R, Vidaud M. Real-time reverse transcription-PCR assay for future management of ERBB2-based clinical applications. Clin Chem. 1999;45:1148–1156. [PubMed] [Google Scholar]
- Bieche I, Parfait B, Le Doussal V, Olivi M, Rio MC, Lidereau R, Vidaud M. Identification of CGA as a novel estrogen receptor-responsive gene in breast cancer: an outstanding candidate marker to predict the response to endocrine therapy. Cancer Res. 2001;61:1652–1658. [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Zia S, Ndoye A, Nguyen VT, Grando SA. Nicotine enhances expression of the α3, α4, α5, and α7 nicotinic receptors modulating calcium metabolism and regulating adhesion and motility of respiratory epithelial cells. Res Commun Mol Pathol Pharmacol. 1997;97:243–262. [PubMed] [Google Scholar]
- Ndoye A, Buchli R, Greenberg B, Nguyen VT, Zia S, Rodriguez JG, Webber RJ, Lawry MA, Grando SA. Identification and mapping of keratinocyte muscarinic acetylcholine receptor subtypes in human epidermis. J Invest Dermatol. 1998;111:410–416. doi: 10.1046/j.1523-1747.1998.00299.x. [DOI] [PubMed] [Google Scholar]
- Li MD, Konu O, Kane JK, Becker KG. Microarray technology and its application on nicotine research. Mol Neurobiol. 2002;25:265–285. doi: 10.1385/MN:25:3:265. [DOI] [PubMed] [Google Scholar]
- Konu O, Xu X, Ma JZ, Kane J, Wang J, Shi SJ, Li MD. Application of a customized pathway-focused microarray for gene expression profiling of cellular homeostasis upon exposure to nicotine in PC12 cells. Brain Res Mol Brain Res. 2004;121:102–113. doi: 10.1016/j.molbrainres.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Harvey SC, McIntosh JM, Cartier GE, Maddox FN, Luetje CW. Determinants of specificity for alpha-conotoxin MII on alpha3beta2 neuronal nicotinic receptors. Mol Pharmacol. 1997;51:336–342. doi: 10.1124/mol.51.2.336. [DOI] [PubMed] [Google Scholar]
- Nayak SV, Dougherty JJ, McIntosh JM, Nichols RA. Ca(2+) changes induced by different presynaptic nicotinic receptors in separate populations of individual striatal nerve terminals. J Neurochem. 2001;76:1860–1870. doi: 10.1046/j.1471-4159.2001.00197.x. [DOI] [PubMed] [Google Scholar]
- Timoteo MA, Faria M, Correia-de-Sa P. Endogenous adenosine prevents post-tetanic release facilitation mediated by alpha3beta2 nicotinic autoreceptors. Eur J Pharmacol. 2003;464:115–125. doi: 10.1016/s0014-2999(03)01374-8. [DOI] [PubMed] [Google Scholar]
- Grinevich VP, Letchworth SR, Lendenberger KA, Menager J, Mary V, Sadieva KA, Buhlman L, Bohme GA, Pradier L, Benavides J, Lukas RJ, Bencherif M. Heterologous expression of human {alpha}6{beta}4{beta}3{alpha}5 nicotinic acetylcholine receptors: binding properties consistent with their natural expression require quaternary subunit assembly including the {alpha}5 subunit. J Pharmacol Exp Ther. 2004 doi: 10.1124/jpet.104.075069. [DOI] [PubMed] [Google Scholar]
- Chang YC, Huang FM, Tai KW, Yang LC, Chou MY. Mechanisms of cytotoxicity of nicotine in human periodontal ligament fibroblast cultures in vitro. J Periodontal Res. 2002;37:279–285. doi: 10.1034/j.1600-0765.2002.01612.x. [DOI] [PubMed] [Google Scholar]
- Bergstrom J. Cigarette smoking as risk factor in chronic periodontal disease. Commun Dent Oral Epidemiol. 1989;17:245–247. doi: 10.1111/j.1600-0528.1989.tb00626.x. [DOI] [PubMed] [Google Scholar]
- Ismail AI, Morrison EC, Burt BA, Caffesse RG, Kavanagh MT. Natural history of periodontal disease in adults: findings from the Tecumseh Periodontal Disease Study, 1959–87. J Dent Res. 1990;69:430–435. doi: 10.1177/00220345900690020201. [DOI] [PubMed] [Google Scholar]
- Haber J. Cigarette smoking: a major risk factor for periodontitis. Compendium. 1994;15:1002–1008. [PubMed] [Google Scholar]
- Mandel I. Smoke signals: an alert for oral disease. J Am Dent Assoc. 1994;125:872–878. doi: 10.14219/jada.archive.1994.0192. [DOI] [PubMed] [Google Scholar]
- Berger F, Gage FH, Vijayaraghavan S. Nicotinic receptor-induced apoptotic cell death of hippocampal progenitor cells. J Neurosci. 1998;18:6871–6881. doi: 10.1523/JNEUROSCI.18-17-06871.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Zhu YK, Umino T, Spurzem JR, Romberger DJ, Wang H, Reed E, Rennard SI. Cigarette smoke inhibits osteogenic differentiation and proliferation of human osteoprogenitor cells in monolayer and three-dimensional collagen gel culture. J Lab Clin Med. 2001;137:208–219. doi: 10.1067/mlc.2001.113066. [DOI] [PubMed] [Google Scholar]
- Chen YP, Johnson GK, Squier CA. Effects of nicotine and tobacco-specific nitrosamines on hamster cheek pouch and gastric mucosa. J Oral Pathol Med. 1994;23:251–255. doi: 10.1111/j.1600-0714.1994.tb00054.x. [DOI] [PubMed] [Google Scholar]
- Ringdahl BE, Johnson GK, Ali RB, Organ CC. Effect of nicotine on arachidonic acid metabolites and epithelial parameters in rat oral mucosa. J Oral Pathol Med. 1997;26:40–45. doi: 10.1111/j.1600-0714.1997.tb00008.x. [DOI] [PubMed] [Google Scholar]
- Arredondo J, Nguyen VT, Chernyavsky AI, Bercovich D, Orr-Urtreger A, Kummer W, Lips K, Vetter DE, Grando SA. Central role of α7 nicotinic receptor in differentiation of the stratified squamous epithelium. J Cell Biol. 2002;159:325–336. doi: 10.1083/jcb.200206096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann D, Adams JD. Carcinogenic tobacco-specific N-nitrosamines in snuff and in the saliva of snuff dippers. Cancer Res. 1981;41:4305–4308. [PubMed] [Google Scholar]
- Lindell G, Farnebo LO, Chen D, Nexo E, Rask Madsen J, Bukhave K, Graffner H. Acute effects of smoking during modified sham feeding in duodenal ulcer patients an analysis of nicotine acid secretion gastrin catecholamines epidermal growth factor prostaglandin E-2 and bile acids. Scand J Gastroenterol. 1993;28:487–494. doi: 10.3109/00365529309098254. [DOI] [PubMed] [Google Scholar]
- Russell MA, Jarvis M, Iyer R, Feyerabend C. Relation of nicotine yield of cigarettes to blood nicotine concentrations in smokers. Br Med J. 1980;280:972–976. doi: 10.1136/bmj.280.6219.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilig C, Bernd A, Ramirez-Bosca A, Gormar FF, Bereiter-Hahn J, Keller-Stanislawski B, Sewell AC, Rietbrock N, Holzmann H. Reactions of human keratinocytes in vitro after application of nicotine. Skin Pharmacol. 1994;7:307–315. doi: 10.1159/000211311. [DOI] [PubMed] [Google Scholar]
- Kwon OS, Chung JH, Cho KH, Suh DH, Park KC, Kim KH, Eun HE. Nicotine-enhanced epithelial differentiation in reconstructed human oral mucosa in vitro. Skin Pharmacol Appl Skin Physiol. 1999;12:227–234. doi: 10.1159/000066247. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hershey JWB, Mathews MB. New York: Cold Spring Harbor Laboratory Press,; Translational Control of Gene Expression. 2000:pp 1020. [Google Scholar]
- Cucina A, Sapienza P, Corvino V, Borrelli V, Mariani V, Randone B, Santoro D’Angelo L, Cavallaro A. Nicotine-induced smooth muscle cell proliferation is mediated through bFGF and TGF-beta 1. Surgery. 2000;127:316–322. doi: 10.1067/msy.2000.104249. [DOI] [PubMed] [Google Scholar]
- Pausova Z, Paus T, Sedova L, Berube J. Prenatal exposure to nicotine modifies kidney weight and blood pressure in genetically susceptible rats: a case of gene-environment interaction. Kidney Int. 2003;64:829–835. doi: 10.1046/j.1523-1755.2003.00172.x. [DOI] [PubMed] [Google Scholar]
- Hu M, Liu QS, Chang KT, Berg DK. Nicotinic regulation of CREB activation in hippocampal neurons by glutamatergic and nonglutamatergic pathways. Mol Cell Neurosci. 2002;21:616–625. doi: 10.1006/mcne.2002.1202. [DOI] [PubMed] [Google Scholar]
- Chang YC, Hsieh YS, Lii CK, Huang FM, Tai KW, Chou MY. Induction of c-fos expression by nicotine in human periodontal ligament fibroblasts is related to cellular thiol levels. J Periodontal Res. 2003;38:44–50. doi: 10.1034/j.1600-0765.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- Shin VY, Wang HY, Liu ES, Koo MW, Cho CH. Differential effects of cigarette smoke extracts on cell proliferation in gastric and colon cells. Cancer Invest. 2003;21:200–207. doi: 10.1081/cnv-120016416. [DOI] [PubMed] [Google Scholar]
- Mizobuchi S, Furihata M, Sonobe H, Ohtsuki Y, Ishikawa T, Murakami H, Kurabayashi A, Ogoshi S, Sasaguri S. Association between p53 immunostaining and cigarette smoking in squamous cell carcinoma of the esophagus. Jpn J Clin Oncol. 2000;30:423–428. doi: 10.1093/jjco/hyd114. [DOI] [PubMed] [Google Scholar]
- Nordskog BK, Blixt AD, Morgan WT, Fields WR, Hellmann GM. Matrix-degrading and pro-inflammatory changes in human vascular endothelial cells exposed to cigarette smoke condensate. Cardiovasc Toxicol. 2003;3:101–117. doi: 10.1385/ct:3:2:101. [DOI] [PubMed] [Google Scholar]
- Garrido R, King-Pospisil K, Son KW, Hennig B, Toborek M. Nicotine upregulates nerve growth factor expression and prevents apoptosis of cultured spinal cord neurons. Neurosci Res. 2003;47:349–355. doi: 10.1016/s0168-0102(03)00222-0. [DOI] [PubMed] [Google Scholar]
- Lee SD, Lee DS, Chun YG, Shim TS, Lim CM, Koh Y, Kim WS, Kim DS, Kim WD. Cigarette smoke extract induces endothelin-1 via protein kinase C in pulmonary artery endothelial cells. Am J Physiol. 2001;281:L403–L411. doi: 10.1152/ajplung.2001.281.2.L403. [DOI] [PubMed] [Google Scholar]
- Coleman DT, Bancroft C. Nicotine acts directly on pituitary GH3 cells to inhibit prolactin promoter activity. J Neuroendocrinol. 1995;7:785–789. doi: 10.1111/j.1365-2826.1995.tb00715.x. [DOI] [PubMed] [Google Scholar]
- Li MD, Kane JK, Parker SL, McAllen K, Matta SG, Sharp BM. Nicotine administration enhances NPY expression in the rat hypothalamus. Brain Res. 2000;867:157–164. doi: 10.1016/s0006-8993(00)02283-6. [DOI] [PubMed] [Google Scholar]
- Zhang S, Day I, Ye S. Nicotine induced changes in gene expression by human coronary artery endothelial cells. Atherosclerosis. 2001;154:277–283. doi: 10.1016/s0021-9150(00)00475-5. [DOI] [PubMed] [Google Scholar]
- Kenny PJ, File SE, Rattray M. Nicotine regulates 5-HT(1A) receptor gene expression in the cerebral cortex and dorsal hippocampus. Eur J Neurosci. 2001;13:1267–1271. doi: 10.1046/j.0953-816x.2001.01501.x. [DOI] [PubMed] [Google Scholar]
- Serova L, Danailov E, Chamas F, Sabban EL. Nicotine infusion modulates immobilization stress-triggered induction of gene expression of rat catecholamine biosynthetic enzymes. J Pharmacol Exp Ther. 1999;291:884–892. [PubMed] [Google Scholar]
- Chong IW, Lin SR, Hwang JJ, Huang MS, Wang TH, Hung JY, Paulauskis JD. Expression and regulation of the macrophage inflammatory protein-1 alpha gene by nicotine in rat alveolar macrophages. Eur Cytokine Netw. 2002;13:242–249. [PubMed] [Google Scholar]
- Chang YC, Tsai CH, Yang SH, Liu CM, Chou MY. Induction of cyclooxygenase-2 mRNA and protein expression in human gingival fibroblasts stimulated with nicotine. J Periodontal Res. 2003;38:496–501. doi: 10.1034/j.1600-0765.2003.00681.x. [DOI] [PubMed] [Google Scholar]
- Gebel S, Gerstmayer B, Bosio A, Haussmann HJ, Van Miert E, Muller T. Gene expression profiling in respiratory tissues from rats exposed to mainstream cigarette smoke. Carcinogenesis. 2004;25:169–178. doi: 10.1093/carcin/bgg193. [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Ndoye A, Hall LL, Zia S, Arredondo J, Chernyavsky AI, Kist DA, Zelickson BD, Lawry MA, Grando SA. Programmed cell death of keratinocytes culminates in apoptotic secretion of a humectant upon secretagogue action of acetylcholine. J Cell Sci. 2001;114:1189–1204. doi: 10.1242/jcs.114.6.1189. [DOI] [PubMed] [Google Scholar]
- Heusch WL, Maneckjee R. Signalling pathways involved in nicotine regulation of apoptosis of human lung cancer cells. Carcinogenesis. 1998;19:551–556. doi: 10.1093/carcin/19.4.551. [DOI] [PubMed] [Google Scholar]
- Wang HY, Shin VY, Leung SY, Yuen ST, Cho CH. Involvement of bcl-2 and caspase-3 in apoptosis induced by cigarette smoke extract in the gastric epithelial cell. Toxicol Pathol. 2003;31:220–226. doi: 10.1080/01926230390183715. [DOI] [PubMed] [Google Scholar]
- Mai H, May WS, Gao F, Jin Z, Deng X. A functional role for nicotine in Bcl2 phosphorylation and suppression of apoptosis. J Biol Chem. 2003;278:1886–1891. doi: 10.1074/jbc.M209044200. [DOI] [PubMed] [Google Scholar]
- Sugano N, Minegishi T, Kawamoto K, Ito K. Nicotine inhibits UV-induced activation of the apoptotic pathway. Toxicol Lett. 2001;125:61–65. doi: 10.1016/s0378-4274(01)00416-7. [DOI] [PubMed] [Google Scholar]
- West KA, Brognard J, Clark AS, Linnoila IR, Yang X, Swain SM, Harris C, Belinsky S, Dennis PA. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest. 2003;111:81–90. doi: 10.1172/JCI16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoshiba K, Nagai A, Yasui S, Konno K. Nicotine prolongs neutrophil survival by suppressing apoptosis. J Lab Clin Med. 1996;127:186–194. doi: 10.1016/s0022-2143(96)90077-3. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Bayna E, Dalle Molle E, Lew WY. Nicotine inhibits cardiac apoptosis induced by lipopolysaccharide in rats. J Am Coll Cardiol. 2003;41:482–488. doi: 10.1016/s0735-1097(02)02820-6. [DOI] [PubMed] [Google Scholar]
- Hakki A, Pennypacker K, Eidizadeh S, Friedman H, Pross S. Nicotine inhibition of apoptosis in murine immune cells. Exp Biol Med (Maywood) 2001;226:947–953. doi: 10.1177/153537020122601011. [DOI] [PubMed] [Google Scholar]
- Pugh PC, Margiotta JF. Nicotinic acetylcholine receptor agonists promote survival and reduce apoptosis of chick ciliary ganglion neurons. Mol Cell Neurosci. 2000;15:113–122. doi: 10.1006/mcne.1999.0810. [DOI] [PubMed] [Google Scholar]
- Maneckjee R, Minna JD. Opioids induce while nicotine suppresses apoptosis in human lung cancer cells. Cell Growth Differ. 1994;5:1033–1040. [PubMed] [Google Scholar]
- Wright SC, Zhong J, Zheng H, Larrick JW. Nicotine inhibition of apoptosis suggests a role in tumor promotion. FASEB J. 1993;7:1045–1051. [PubMed] [Google Scholar]
- Eves EM, Xiong W, Bellacosa A, Kennedy SG, Tsichlis PN, Rosner MR, Hay N. Akt, a target of phosphatidylinositol 3-kinase, inhibits apoptosis in a differentiating neuronal cell line. Mol Cell Biol. 1998;18:2143–2152. doi: 10.1128/mcb.18.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickenden JA, Clarke MC, Rossi AG, Rahman I, Faux SP, Donaldson K, MacNee W. Cigarette smoke prevents apoptosis through inhibition of caspase activation and induces necrosis. Am J Respir Cell Mol Biol. 2003;29:562–570. doi: 10.1165/rcmb.2002-0235OC. [DOI] [PubMed] [Google Scholar]
- Wang J, Wilcken DE, Wang XL. Cigarette smoke activates caspase-3 to induce apoptosis of human umbilical venous endothelial cells. Mol Genet Metab. 2001;72:82–88. doi: 10.1006/mgme.2000.3115. [DOI] [PubMed] [Google Scholar]
- Aoshiba K, Tamaoki J, Nagai A. Acute cigarette smoke exposure induces apoptosis of alveolar macrophages. Am J Physiol. 2001;281:L1392–L1401. doi: 10.1152/ajplung.2001.281.6.L1392. [DOI] [PubMed] [Google Scholar]
- Ma L, Wang HY, Chow JY, Cho CH. Cigarette smoke increases apoptosis in the gastric mucosa: role of epidermal growth factor. Digestion. 1999;60:461–468. doi: 10.1159/000007692. [DOI] [PubMed] [Google Scholar]
- Rajpurkar A, Jiang Y, Dhabuwala CB, Dunbar JC, Li H. Cigarette smoking induces apoptosis in rat testis. J Environ Pathol Toxicol Oncol. 2002;21:243–248. [PubMed] [Google Scholar]
- Suzuki N, Wakisaka S, Takeba Y, Mihara S, Sakane T. Effects of cigarette smoking on Fas/Fas ligand expression of human lymphocytes. Cell Immunol. 1999;192:48–53. doi: 10.1006/cimm.1998.1432. [DOI] [PubMed] [Google Scholar]
- Wang H, Ma L, Li Y, Cho CH. Exposure to cigarette smoke increases apoptosis in the rat gastric mucosa through a reactive oxygen species-mediated and p53-independent pathway. Free Radic Biol Med. 2000;28:1125–1131. doi: 10.1016/s0891-5849(00)00207-0. [DOI] [PubMed] [Google Scholar]
- Crowley-Weber CL, Dvorakova K, Crowley C, Bernstein H, Bernstein C, Garewal H, Payne CM. Nicotine increases oxidative stress, activates NF-kappaB and GRP78, induces apoptosis and sensitizes cells to genotoxic/xenobiotic stresses by a multiple stress inducer, deoxycholate: relevance to colon carcinogenesis. Chem Biol Interact. 2003;145:53–66. doi: 10.1016/s0009-2797(02)00162-x. [DOI] [PubMed] [Google Scholar]
- Fucile S. Ca(2+) permeability of nicotinic acetylcholine receptors. Cell Calcium. 2004;35:1–8. doi: 10.1016/j.ceca.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Toth E, Vizi ES, Lajtha A. Effect of nicotine on levels of extracellular amino acids in regions of the rat brain in-vivo. Neuropharmacology. 1993;32:827–832. doi: 10.1016/0028-3908(93)90192-6. [DOI] [PubMed] [Google Scholar]
- Sastry BVR. Neuropharmacology of nicotine: effects on the autoregulation of acetylcholine release by substance P and methionine enkephalin in rodent cerebral slices and toxicological implications. Clin Exp Pharmacol Physiol. 1995;22:288–290. doi: 10.1111/j.1440-1681.1995.tb01997.x. [DOI] [PubMed] [Google Scholar]
- Ogle CW, Glavin GB. Chronic nicotine treatment upregulates muscarinic M1-receptors in the rat stomach. Med Sci Res. 1996;24:83–84. [Google Scholar]
- Wang H. Modulation by nicotine on muscarinic receptor-effector systems. Chung Kuo Yao Li Hsueh Pao. 1997;18:193–197. [PubMed] [Google Scholar]
- Flores CM, Rogers SW, Pabreza LA, Wolfe BB, Kellar KJ. A subtype of nicotinic cholinergic receptor in rat brain is composed of α4 and β2 subunits and is up-regulated by chronic nicotine treatment. Mol Pharmacol. 1992;41:31–37. [PubMed] [Google Scholar]
- James JR, Nordberg A. Genetic and environmental aspects of the role of nicotinic receptors in neurodegenerative disorders: emphasis on Alzheimer’s disease and Parkinson’s disease. Behav Genet. 1995;25:149–159. doi: 10.1007/BF02196924. [DOI] [PubMed] [Google Scholar]
- Miao H, Liu C, Bishop K, Gong ZH, Nordberg A, Zhang X. Nicotine exposure during a critical period of development leads to persistent changes in nicotinic acetylcholine receptors of adult rat brain. J Neurochem. 1998;70:752–762. doi: 10.1046/j.1471-4159.1998.70020752.x. [DOI] [PubMed] [Google Scholar]
- Zia S, Ndoye A, Lee TX, Webber RJ, Grando SA. Receptor-mediated inhibition of keratinocyte migration by nicotine involves modulations of calcium influx and intracellular concentration. J Pharmacol Exp Ther. 2000;293:973–981. [PubMed] [Google Scholar]
- Lebargy F, Benhammou K, Morin D, Zini R, Urien S, Bree F, Bignon J, Branellec A, Lagrue G. Tobacco smoking induces expression of very-high-affinity nicotine binding sites on blood polymorphonuclear cells. Am J Respir Crit Care Med. 1996;153:1056–1063. doi: 10.1164/ajrccm.153.3.8630545. [DOI] [PubMed] [Google Scholar]
- Kimura R, Ushiyama N, Fujii T, Kawashima K. Nicotine-induced Ca2+ signaling and down-regulation of nicotinic acetylcholine receptor subunit expression in the CEM human leukemic T-cell line. Life Sci. 2003;72:2155–2158. doi: 10.1016/s0024-3205(03)00077-8. [DOI] [PubMed] [Google Scholar]