Abstract
Heterogeneity of the microvasculature in different organs has been well documented by multiple methods including in vivo phage display. However, less is known about the diversity of blood vessels within functionally distinct regions of organs. Here, we combined in vivo phage display with laser pressure catapult microdissection to identify peptide ligands for vascular receptors in the islets of Langerhans in the murine pancreas. Protein database analyses of the peptides, CVSNPRWKC and CHVLWSTRC, showed sequence identity to two ephrin A-type ligand homologues, A2 and A4. Confocal microscopy confirmed that most immunoreactivity of CVSNPRWKC and CHVLWSTRC phage was associated with blood vessels in pancreatic islets. Antibodies recognizing EphA4, a receptor for ephrin-A ligands, were similarly associated with islet blood vessels. Importantly, binding of both islet-homing phage and anti-EphA4 antibody was strikingly increased in blood vessels of pancreatic islet tumors in RIP-Tag2 transgenic mice. These results indicate that endothelial cells of blood vessels in pancreatic islets preferentially express EphA4 receptors, and this expression is increased in tumors. Our findings show in vivo phage display and laser pressure catapult microdissection can be combined to reveal endothelial cell specialization within focal regions of the microvasculature.
Peptides that target vascular receptors by in vivo phage display have been successfully identified in the mouse and in a human subject.1–6 The resultant peptide sequences illustrate the heterogeneous nature of vascular receptors from organ to organ.1,7,8 In addition to this difference, many organs such as the adrenal gland, kidney, pancreas, and brain contain functionally distinct regions that are embedded within the tissue and exhibit a unique vascular organization that suggests differential molecular vascular addresses within the same organ. Extraction of such functionally specialized regions from excised organs using conventional biochemical methods may fail to recover ligand-receptor pairs. For example, physical separation of a specific site from the surrounding tissues after in vivo phage library biopanning may disrupt the complex between the receptor and the bound peptide-phage by mechanical manipulations and/or nonphysiological buffer conditions. Moreover, protease inhibitor cocktails may alleviate phage degradation by endogenous proteases in crude tissue homogenates, however the cost effectiveness of their use may be prohibitive in a complex subcellular fractionation scheme that may ultimately diminish final yields.
We set out to determine whether phage that home to a functionally specialized region within an organ could be isolated using a combination of in vivo phage display with fluorescence laser microbeam microdissection and laser pressure catapulting, hereafter referred to as laser pressure catapult microdissection (LPCM).9 We investigated the vascular heterogeneity10 of the murine pancreatic islet for the following rationale: islets perform specialized functions that are biologically and clinically relevant, and the islet vasculature is readily identifiable from that of the surrounding acinar pancreas.11 Applications of this study include development of specific peptide-based targeting therapies that recognize functionally distinct intravascular networks that encompass a broad range of human diseases.
Materials and Methods
Animals
Eight-week-old wild-type C57BL/6 male mice were purchased from Harlan (Indianapolis, IN). Male C57BL/6/RIP-Tag2 transgenic mice produce spontaneous pancreatic islet tumors12 and were genotyped using tail-tip DNA by the polymerase chain reaction (PCR). Mice were housed under barrier conditions at the animal care facility at either The University of Texas, M. D. Anderson Cancer Center (MDACC) or the University of California, San Francisco (UCSF). All experimental procedures were approved by the Institutional Animal Care and Use Committees at MDACC and UCSF.
In Vivo Phage Display
109 transforming units of a cyclic CX7C peptide phage library1 was intravenously injected via the tail vein of Avertin (2,2,2-tribromoethanol, 0.015 to .017 mg/g injected intraperitoneally; Sigma-Aldrich Corp., St. Louis, MO) anesthetized C57BL/6 male mice,13 and allowed to circulate for 6 minutes while maintaining the body temperature of the mice at 37°C with a heating pad. The pancreas was removed, weighed, and homogenized in ice-cold Dulbecco’s modified Eagle’s medium containing Earle salts (MDACC cell culture facility) supplemented with 1% bovine serum albumin, 1 mmol/L 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), 10 μg/ml aprotinin, 1 μg/ml leupeptin (Calbiochem, San Diego, CA). The pancreas homogenate was resuspended, rinsed 3× in the same buffer and phage were recovered by infection of the bacterial host, K91.14 Phage-infected K91 recovered in 10 ml of Luria-Bretani (LB)/kanamycin (Kan, 100 mg/L)/tetracycline (Tet, 0.2 mg/L) for 20 minutes in the dark at room temperature. Aliquots from each culture were plated onto LB/Kan/Tet (40 mg/L) agar plates and incubated overnight in the dark at 37°C. Two hundred and forty-six bacterial colonies were each grown overnight in 5 ml of LB/Kan (100 mg/L)/Tet (40 mg/L) at 37°C in the dark with agitation. Phage were purified from pooled overnight cultures and infective titers were determined using established protocols.15 Three hundred colonies were picked from the second round, of which 96 colonies were randomly selected for colony PCR and automated sequencing of the peptide insert at the MDACC Sequencing Core Facility. For the third panning round, intravenous injection of 109 transforming units of purified, amplified round II phage was followed by an injection of 50 μg of fluorescein isothiocyanate (FITC)-lectin (Vector Laboratories, Inc., Burlingame, CA). Mice were infused through the right ventricle with 3 ml of 37°C Dulbecco’s modified Eagle’s medium. The inferior vena cava was cut for the outlet. The pancreas was harvested, frozen at −80°C in Tissue Tek O.C.T. compound (Sakura, Torrance, CA), and 14-μm sections were cut and placed onto prepared microscope slides for LPCM (MDACC Veterinary Technical Services). The total amount of phage recovered from the round III pancreas was determined by extrapolating the number of bacterial colonies recovered from four, 14-μm thawed round III pancreatic sections to the final thickness (5 mm) of the frozen tissue block.
Fluorescence LPCM
One hundred and twenty islet sections were microdissected and catapulted from 14-μm frozen pancreas sections using the positioning and ablation with laser microbeams system (P.A.L.M. Mikrolaser Technologie GmbH, Bernried, Germany) fitted to a Zeiss Axiovert 135 microscope with Fluar objectives and a fluorescence module using the manufacturer’s suggested laser-cutting and focus energy. Excised islet sections were catapulted into 30 μl of either 1 mmol/L ethylenediaminetetraacetic acid, pH 8.0 (Sigma-Aldrich Corp.) for PCR amplification, or a protease inhibitor cocktail consisting of 1 mmol/L AEBSF 20 μg/ml aprotinin, 10 μg/L leupeptin, 1 mmol/L elastase inhibitor I, 0.1 mmol/L Na-Tosyl-Phe chloromethyl ketone, 1 nmol/L pepstatin A in phosphate buffered saline (PBS), pH 7.4, for phage recovery by bacterial infection (see below). All protease inhibitors were purchased from Calbiochem.
Phage Recovery by Bacterial Infection
Catapulted islet sections were incubated with 1 ml of stationary phase K91 for 2 hours at room temperature with gentle agitation. Each culture was transferred to 1.2 ml of LB/Kan (100 mg/L)/Tet (0.2 mg/L) and incubated in the dark at room temperature for 40 minutes. The tetracycline concentration was increased to 40 mg/L, and the cultures were incubated overnight at 37°C with agitation. Cultures were plated out onto LB/Kan/Tet agar plates to obtain single colonies. Peptide insert sequences were amplified by colony PCR and sequenced at the MDACC Sequencing Core Facility.
Phage Recovery by DNA Amplification
Peptide insert sequences were initially amplified by PCR from catapulted islet sections using the fUSE5 phage forward primer,16 and reverse primer, 5′-CCCTCATAGTTAGCGTAACGATCT-3′. Flanking SfiI restriction sites were added to each peptide insert sequence by a second PCR amplification by using the forward Sfi library primer 5′-CACTCGGCCGACGGGC-3′, and reverse SfiI primer: 5′-CAGTTTCGGCCCCAGCGGCCC-3′ for ligation into SfiI-digested CsCl2-purified fUSE5 genomic DNA.17 All primers were purchased from Genosys (The Woodlands, TX). Ligation products were electroporated into MC1061 bacterial hosts,14 and plated onto LB/streptomycin (100 mg/L)/Tet (40 mg/L) agar plates. Single colonies were subjected to colony PCR and sequenced at the MDACC Sequencing Core Facility.
Phage Preparation, Phage and Antibody Injections, and Tissue Preparation
Single-stranded CVSNPRWKC, CHVLWSTRC, and fd-tet phage DNA were electroporated into electrocompetent MC1061 bacterial hosts, amplified phage were purified from overnight cultures and titered using established protocols.15 Peptide insert sequences were verified by automated sequencing (ELIM Biopharmaceuticals, Hayward, CA). Phage preparations were used within 24 hours of preparation. Before injection, 109 transforming units of CVSNPRWKC, CHVLWSTRC, and fd-tet phage preparations were diluted into sterile PBS to a final volume of 200 μl. Fifty μg of polyclonal goat anti-mouse EphA4 antibodies (R&D Systems, Minneapolis, MN) and normal goat IgG (Jackson ImmunoResearch, West Grove, PA) were diluted with sterile PBS and filtered through a 0.22-μm filter. Phage, EphA4 antibodies, or goat IgGs were injected intravenously into male C57BL/6 or RIP-Tag2 mice, allowed to circulate for 6 minutes, and then the mice were systemically perfused with 1% paraformaldehyde in PBS, pH 7.4 (PFA/PBS) through the left ventricle for 3 minutes at 120 mmHg. The inferior vena cava was cut as an outlet for the fixative. Organs were removed, incubated in 1% PFA/PBS for 1 to 2 hours, embedded overnight at 4°C in 30% sucrose/PBS/0.01% Thimerosol (Sigma-Aldrich Corp.), and frozen in O.C.T. on dry ice.
Immunohistochemistry
All steps were performed at room temperature unless otherwise indicated. Eighty-μm frozen tissue sections were air dried, rinsed 2× for 5 minutes each with PBS, 1× with PBS/1% Triton X-100, and then blocked with 5% normal mouse serum (Jackson ImmunoResearch) in PBS/1% Triton X-100 for 1 hour. For phage immunostaining, sections were incubated in a 0.22-μm filtered primary antibody solution containing monoclonal rat anti-mouse CD31 antibody (1:500; Pharmingen, San Diego, CA), polyclonal rabbit fd-bacteriophage antibody (1:5000; Sigma-Aldrich Corp.), and 5% normal mouse serum in PBS/1% Triton X-100 for at least 14 hours. Sections were rinsed 3× for 5 minutes each with PBS/1% Triton X-100. Rinsed sections were incubated in a 0.22-μm filtered solution containing mouse FITC-conjugated anti-rat IgG (1:200, Jackson ImmunoResearch), mouse Cy3-conjugated anti-rabbit IgG (1:400, Jackson ImmunoResearch), and 5% normal mouse serum in PBS/1% Triton X-100 for 2 hours. Prepared sections containing EphA4 antibody were incubated with monoclonal rat anti-mouse CD31 in 5% normal mouse serum in PBS/1% Triton X-100, rinsed, and incubated with mouse FITC-conjugated anti-rat IgG, mouse Cy3-conjugated anti-goat IgG (1:400, Jackson ImmunoResearch), and 5% normal mouse serum in PBS/1% Triton X-100. After immunostaining, all sections were rinsed 3×, 10 minutes each, with PBS/1% Triton X-100, fixed for 5 minutes with 4% PFA/PBS, rinsed 3× with PBS for 10 minutes each, and mounted with Vectashield (Vector Laboratories, Inc.).
Microscopy
Fluorescence images were acquired using an externally coded, three-chip charge-couple device camera (CoolCam; SciMeasure Analytical Systems, Atlanta, GA) fitted on a Zeiss Axiophot fluorescence microscope with Fluar objectives or with a Zeiss LSM 510 laser scanning confocal microscope with krypton-argon and helium-neon lasers (Carl Zeiss, Jena, Germany) and analyzed with the LSM 510 software (version 3.2.2).
Results
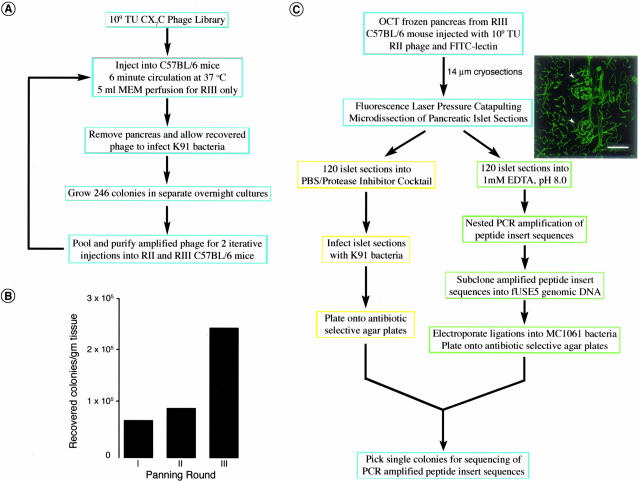
To prepare tissues for LPCM, we performed three rounds of in vivo phage display starting with a cyclic CX7C phage library (Figure 1A).15 Colony counts of phage recovered from excised pancreata showed increased selectivity of targeting peptides homing to the pancreas with each successive panning round (Figure 1B). Islets were readily distinguished from the surrounding acinar pancreas by their characteristic vascular organization as revealed by fluorescein isothiocyanate-conjugated Lycopersicon esculentum lectin (FITC-lectin)-labeled blood vessels (Figure 1C).18 Phage recovery from LPCM islet sections was achieved by two routes, bacterial infection15 and subcloning nested PCR-amplified peptide insert sequences into the fUSE5 phage genome (Figure 1C).
Figure 1.
In vivo phage display and fluorescence LPCM. A: Isolation of islet-homing phage from a cyclic CX7C phage library by fluorescence LPCM proceeded by in vivo phage display with B, an increased selectivity of pancreas-targeting phage after two iterative rounds of panning in the wild-type C57BL/6 murine pancreas. C: Peptide-phage and phage DNA were isolated from microdissected normal mouse pancreatic islet sections from the round III pancreas by K91 infection (yellow) and DNA amplification/subcloning of peptide insert sequences (green), respectively. Islets were visually identified by fluorescence microscopy in C57BL/6 pancreatic sections by their distinctive vascular organization. Scale bar, 18 μm (C).
Bacterial infection of phage recovered from catapulted islet sections yielded eight bacterial clones from which seven unique peptide sequences were obtained (Table 1). Conversely, subcloning PCR-amplified peptide insert sequences from catapulted islets into the fUSE5 phage genome yielded hundreds of transformed bacterial clones, however, sequence analyses of 80 randomly selected clones yielded 11 unique sequences. Identified peptide sequences were analyzed using the Basic Local Alignment Search Tool (BLAST) on the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/BLAST) to search for putative homologous proteins (Table 1). Peptides CVPRRWDVC and CQHTSGRGC exhibited partial sequence identity to the laminin β-2 chain at positions 197 to 201 and 1088 to 1092, respectively.19 Peptides CVSNPRWKC and CHVLWSTRC exhibited partial sequence identity to ephrin-A2 and ephrin-A4 which are glycosylphosphatidylinositol-anchored proteins that bind to EphA-type receptors.20
Table 1.
Peptide Sequences Expressed by Phage Recovered from Pancreatic Islet Sections
| Islet-homing peptide sequence* | Colonies recovered by K91 infection | Colonies recovered by PCR/subcloning | Murine protein sharing complete/partial sequence identity | SWISSPROT accession number |
|---|---|---|---|---|
| CVSNPRWKC | 1 | 0 | Ephrin-A2, Ephrin-A4 | P52801, O08542 |
| CQHTSGRGC | 1 | 0 | S-laminin β2 chain | Q61292 |
| CRARGWLLC | 1 | 0 | α3 integrin | Q62470 |
| CGGVHALRC | 1 | 0 | PDGF receptor β | P05622 |
| CFNRTWIGC | 1 | 0 | Frizzled-1 | O70421 |
| CSRGPAWGC | 1 | 0 | Bone morphogenetic protein 3b | P97737 |
| CVPRRWDVC | 2 | 0 | S-laminin β2 chain | Q61292 |
| CWSRGQGGC | 0 | 21 | Endothelin-1 receptor | Q61614 |
| CHVLWSTRC | 0 | 11 | Ephrin-A2, Eprhrin-A4 | P52801, O08542 |
| CLGLLMAGC | 0 | 11 | VE-cadherin | P55284 |
| CMSSPGVAC | 0 | 9 | Insulin receptor substrate 1 | P35569 |
| CLASGMDAC | 0 | 8 | Tumor necrosis factor-α | P06804 |
| CHDERTGRC | 0 | 6 | Angio-associated migratory protein | Q8K2C1 |
| CAHHALMEC | 0 | 5 | Adiponectin receptor 1, 2 | Q91VH1, Q8BQS5 |
| CMQGAATSC | 0 | 4 | Grb10 | Q60760 |
| CVRDLLTGC | 0 | 3 | α4 integrin | Q00651 |
| CMQGARTSC | 0 | 1 | α2C-adrenergic receptor | Q01337 |
| CGGLVAVGC | 0 | 1 | β7 integrin | P26011 |
Amino acid residues in bold share sequence identity, including conservative replacements, to the listed murine proteins.
Precedence for vascular expression of both classes of Eph receptors and ephrin ligands has been documented. Mouse knockout studies have shown interactions between the EphB2, B3, and B4 receptors, and their corresponding ligands, ephrinB1 and B2, are necessary for vasculogenesis and angiogenesis.21 Ephrin-A1 expression is induced by tumor necrosis factor-α in human umbilical vein endothelial cells and its binding to the EphA2 receptor results in receptor autophosphorylation.22 Ephrin-A1 induces angiogenesis in rat corneal pocket assays via endothelial cell migration rather than by endothelial cell proliferation.22 The EphA2 receptor and cognate ephrin-A1 ligand are expressed in RIP-Tag2 murine pancreatic islet tumor endothelium suggesting a role for these proteins in cancer.23 In vitro phage display identified two peptides, SWLAYPGAVSYR and YSAYPDSVPMMS, that mimic ephrin A-type ligands because both peptides bind to immobilized EphA2-Fc protein and to the EphA2 receptor expressed on human umbilical vein endothelial cells and MDA-MB-435 cultured human breast carcinoma cells.24
We mapped the location of the CVSNPRWKC and CHVLWSTRC peptides to the aligned primary structures of murine ephrin-A2 and ephrin-A4, and human ephrin-A2 using DIALIGN 2.2.125 (Figure 2). Murine and human ephrin-A2 share 89% sequence identity, whereas the murine ephrins A2 and A4 share 43% sequence identity. Secondary structure predictions of the glycosylphosphatidylinositol-linked A-type ephrins were extrapolated from the crystal structures of the ephrin-B2 extracellular domain26 and the EphB2-ephrin-B2 complex.27 Peptide CVSNPRWKC mapped to the murine ephrin A2 N-terminus at amino acid residues 41 to 44 with a conservative replacement of a Trp residue in the peptide sequence for Phe45 in ephrin-A2. Structurally, the SNPRW peptide sequence lies between the A′ and B anti-parallel β-strands and appears to be well conserved among the A-type ephrins from mice, rats, and humans.26,27 In addition, this sequence is C-terminal from a putative glycosylation site at Asn38 that is analogous to Asn39 in ephrins B2 and B3, and is conserved in ephrins A1, A2, A4, and A5.26,27 Asn38 has been proposed to be involved in a weaker affinity ligand-receptor tetramerization interface.28 Peptide CHVLWSTRC mapped to the unstructured C-terminus of murine ephrinA2 at residues 202 to 205. The VLWS peptide sequence also mapped to the signal sequence of murine ephrin-A4 at positions 10 to 12, and showed a weaker sequence identity in the D β-strand at positions 78 to 81.
Figure 2.
Mapping peptide sequences to murine ephrin A2 and A4, and human ephrin A2. Capitalized amino acid residues of the aligned ephrin sequences delineate identical or conservative replacements. Four conserved cysteine residues are shown in green. Blue-highlighted residues are glycosylphosphatidylinositol-anchor sites. Olive-highlighted amino acids correspond to potential glycosylated Asn residues. The predicted secondary structure of the A-type ephrins shows the sequence of the β-strands (gray) and two α-helices (magenta) that were extrapolated from the crystallographic structure of the murine ephrin B2 extracellular domain. Amino acid residues that are identical between the CVSNPRWKC and CHVLWSTRC peptides and ephrin A2 and A4 are highlighted in yellow whereas conserved residues are shown in red. Weaker sequence identities of the CHVLWSTRC peptide to murine ephrin A4 are highlighted in pink. Sequences from various species are abbreviated as follows: Mm, Mus musculus; Hs, Homo sapiens.
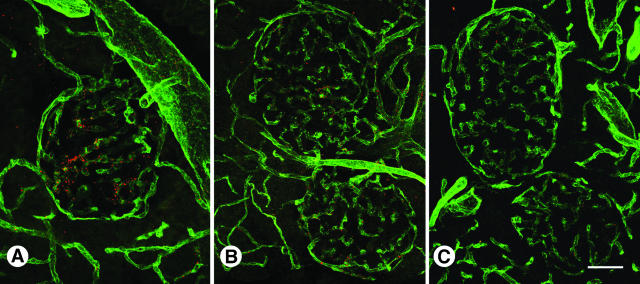
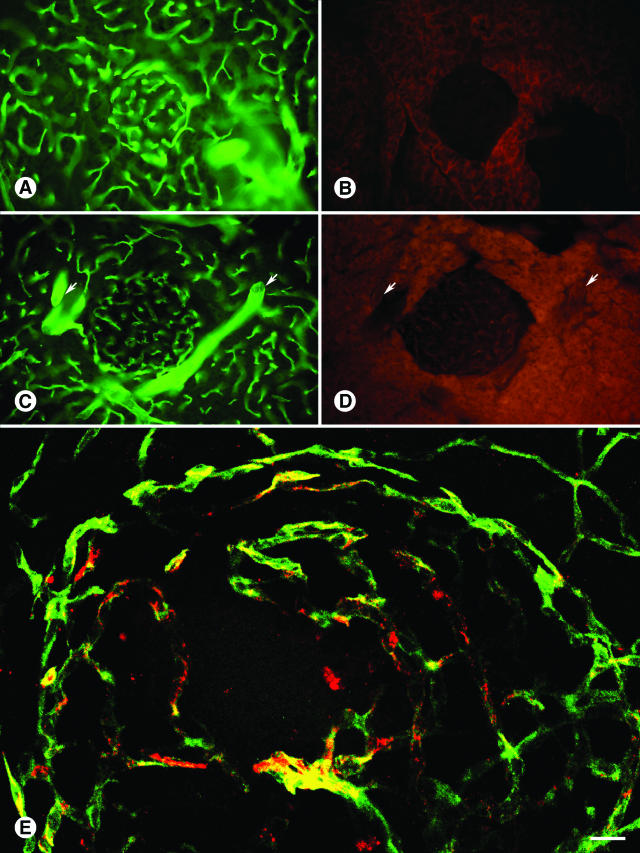
We verified CVSNPRWKC and CHVLWSTRC phage binding to the C57BL/6 pancreatic islet vasculature by immunohistochemistry (Figure 3). Intravenously injected CVSNPRWKC phage (Figure 3A), and CHVLWSTRC phage (Figure 3B) localized predominantly to normal pancreatic islet blood vessels, whereas the insertless control phage, fd-tet, did not bind to either the acinar or islet vasculature (Figure 3C).
Figure 3.
Distribution of CVSNPRWKC and CHVLWSTRC phage in wild-type islet blood vessels. CVSNPRWKC phage (A, red) and CHVLWSTRC phage (B, red) localize to C57BL/6 islet blood vessels immunostained with CD31 (green) whereas insertless fd-tet phage (C, red) do not. Scale bar: 53 μm (A, C); 27 μm (B).
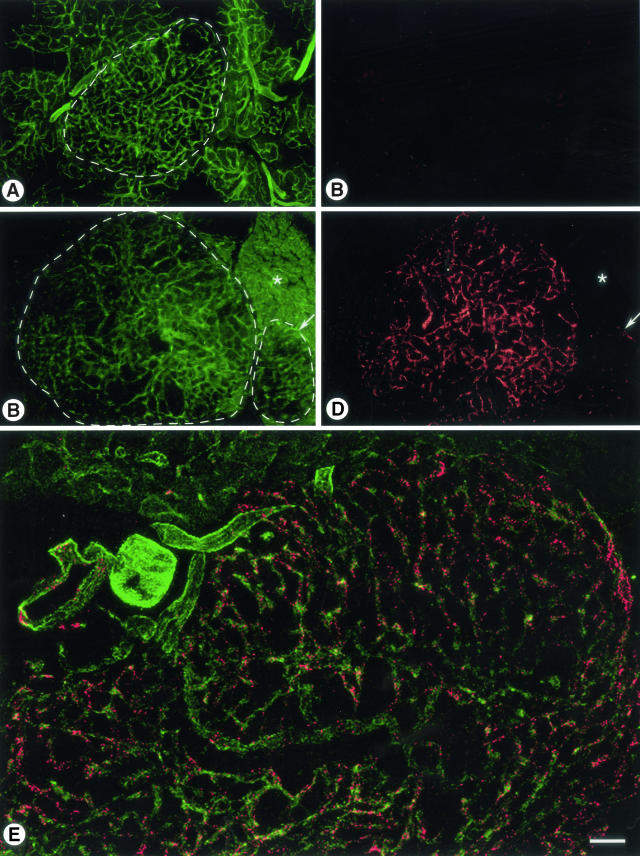
Because unregulated overexpression of proteins is a common feature in tumors,29 we wanted to determine whether localization of the islet homing peptide-phage, CVSNPRWKC and CHVLWSTRC, was increased in angiogenic tumor blood vessels. We compared the binding of CVSNPRWKC and CHVLWSTRC phage to the negative control phage, fd-tet, in pancreatic islet tumors from RIP-Tag2 transgenic mice12 (Figure 4). Insertless fd-tet phage neither accumulated in islet tumor blood vessels nor in blood vessels of the surrounding normal acinar pancreas (Figure 4, A and B). Conversely, CHVLWSTRC phage were abundant in tumor blood vessels that were delineated by phage immunoreactivity (Figure 4, C and D; encircled large tumor). Binding of CHVLWSTRC phage was reduced in blood vessels of a smaller premalignant angiogenic islet (Figure 4C, encircled right islet; and Figure 4D, arrow), whereas phage binding was absent in acinar blood vessels (Figure 4, C and D; asterisk). Similarly, CVSNPRWKC phage immunoreactivity was robust in blood vessels of large islet tumor with insignificant phage immunoreactivity in acinar blood vessels (Figure 4E). CVSNPRWKC or CHVLWSTRC phage did not bind to blood vessels of the brain, lung, or mesenteric lymph nodes, whereas the liver and spleen contained both peptide-expressing phage and fd-tet phage as expected (data not shown).6,15
Figure 4.
Immunoreactivity of fd-tet, CVSNPRWKC, and CHVLWSTRC phage in islet tumor blood vessels from RIP-Tag2 transgenic mice. A and B: CD31-immunoreactive blood vessels (green) do not show the insertless control, fd-tet phage (red) in either the blood vessels of the islet tumor (encircled in A) or surrounding acinar pancreas. C and D: Blood vessels in a large islet tumor (C, encircled large tumor) show significant amounts of CHVLWSTRC phage immunoreactivity (D, red), whereas CHVLWSTRC phage immunoreactivity is less in a small premalignant angiogenic islet (C, encircled small islet; D, arrow). In contrast, CHVLWSTRC phage do not bind to acinar pancreas blood vessels (asterisk). E: A large RIP-Tag2 islet tumor shows that the blood vessels (CD31, green) contain significant amounts of CVSNPRWKC phage immunoreactivity (red). Scale bar, 140 μm (A–D); 106 μm (E).
The distribution of CHVLWSTRC and CVSNPRWKC phage was also evaluated in smaller, hyperplastic angiogenic RIP-Tag2 islets (Figure 5). CHVLWSTRC phage localized to angiogenic blood vessels in hyperplastic islets, however the distribution of phage immunoreactivity was negative in some islet blood vessels (Figure 5, A and B; arrows). Similarly, binding of CVSNPRWKC phage to blood vessels in a hyperplastic islet was sporadic (Figure 5C), considerably less than that found in tumor blood vessels (compare Figure 5C with Figure 4, C and D, large tumor), and consistent with the amount of CVSNPRWKC phage found in a smaller premalignant angiogenic islet (compare Figure 5C with Figure 4, C and D, small islet).
Figure 5.
CHVLWSTRC and CVSNPRWKC phage bind to blood vessels in hyperplastic angiogenic islets. A and B: CD31-immunoreactive blood vessels (green) in a RIP-Tag2 pancreas section contain CHVLWSTRC phage (red) in the vasculature of a hyperplastic angiogenic islet but not in the surrounding acinar pancreas blood vessels. Some islet blood vessels (A and B, arrows), however, do not contain phage. C: Immunoreactivity of CVSNPRWKC phage (red) in blood vessels (green) of a hyperplastic angiogenic islet show a nonuniform distribution. Scale bar, 80 μm (A, B); 27 μm (C).
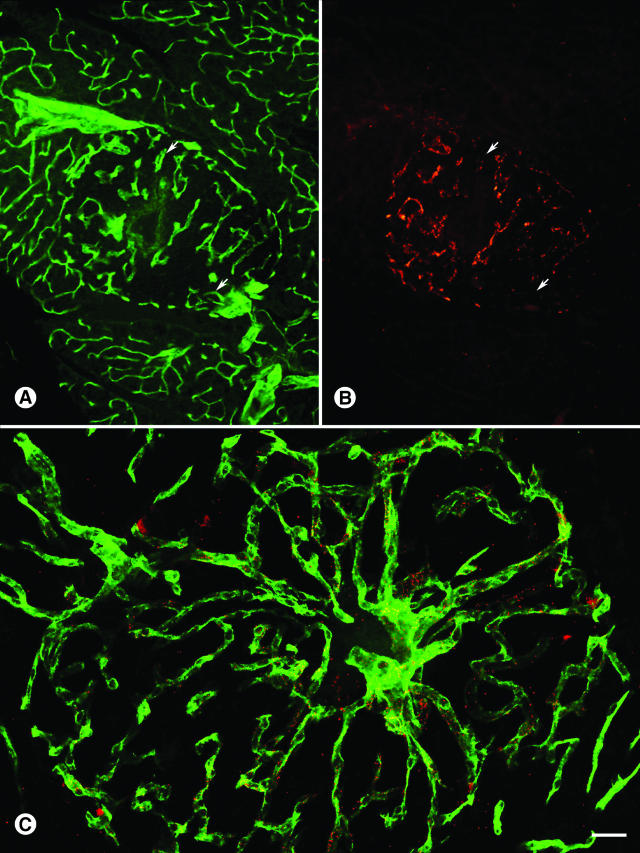
Because the CHVLWSTRC and CVSNPRWKC sequences mapped to the primary structure of ephrin A2 and A4, we chose to investigate the expression of the EphA4 receptor in normal and RIP-Tag2 islet blood vessels (Figure 6). Our rationale to evaluate the EphA4 receptor in pancreatic islets was twofold. First, both A- and B-type ephrin ligands and receptors are present and exhibit functional roles in blood vessels. Second, the EphA4 receptor is the only Eph receptor that binds to both ephrin A- and B-type ligands.30 Although injected control goat IgG did not bind to normal C57BL/6 islet blood vessels (Figure 6, A and B), injected EphA4 antibodies bound to the islet vasculature and to arterioles in the acinar pancreas (Figure 6, C and D; arrows). Immunoreactivity of EphA4 antibodies was greater in RIP-Tag2 islet tumor blood vessels (Figure 6E), but did not bind to resident arterioles in the RIP-Tag2 acinar pancreas (data not shown). Expression of the EphA4 receptor was also negative in the liver, spleen, lungs and mesenteric lymph nodes (data not shown).
Figure 6.
EphA4 expression in wild-type and islet tumor blood vessels. A and B: Binding of control goat IgGs (red) is negative in C57BL/6 islet and acinar pancreas blood vessels (CD31, green). C and D: Goat EphA4 antibody (red) localizes to C57BL/6 islet blood vessels (green) and to arterioles (arrows) in the acinar pancreas. E: Goat EphA4 antibody (red) is strongly immunoreactive in RIP-Tag2 tumor blood vessels. Scale bar, 70 μm (A–D); 8 μm (E).
Discussion
In the study described here, we have identified unique peptide ligands that were isolated from wild-type murine pancreatic islet blood vessels by using in vivo phage display in combination with fluorescence LPCM. Evidence that CVSNPRWKC and CHVLWSTRC phage homed preferentially to blood vessels in normal islets with increased binding to angiogenic blood vessels of solid islet tumors was shown by immunohistochemistry to be mediated by the peptide insert sequences. Increased accumulation of CVSNPRWKC and CHVLWSTRC phage in tumor blood vessels than in premalignant angiogenic islets and normal islets suggests increased expression of the cognate receptor in the tumor vasculature. The two-peptide sequences, CVSNPRWKC and CHVLWSTRC, demonstrate partial sequence identity with ephrin-A2 and ephrin-A4, and suggest an EphA-type receptor to be the putative receptor candidate. Similarity of the EphA4 receptor expression patterns in blood vessels of wild-type and tumor islets to that observed for the CVSNPRWKC and CHVLWSTRC phage suggests EphA4 may be the binding receptor.
The expression of EphA-type receptors and ligands in pancreatic islet tumor angiogenesis is consistent with the overexpression of EphA2 in melanoma cell lines,31 and malignant transformation of cultured MCF-10A mammary epithelial cells.32 Activation of the EphA2 receptor has been shown in tumor angiogenesis of RIP-Tag2 mice, 4T1 mouse mammary adenocarcinomas, and human xenograft tumors in nude mice.23,24,33,34 We tested the binding of CVSNPRWKC and CHVLWSTRC phage to the recombinant extracellular domain of the EphA4 receptor using an enzyme-linked immunosorbent assay-based in vitro binding assay. Islet-homing phage showed 2× greater binding relative to fd-tet phage (data not shown), despite the fact that the Eph receptor binding requires receptor clustering.35 Structural analysis of Eph receptors and their ligands proposes the SNPRW sequence to be adjacent to a lower affinity site tetramer interface that forms when two dimer receptor/ligand pairs form the active tetramer complex.28 The expression of EphA4 antibody in the arterioles of the normal acinar pancreas is consistent with the arteriolar expression of ephrin B-type ligands,21 and the ability of the EphA4 receptor to bind to both ephrin A- and B-type ligands.30 The absence of the islet-homing phage to the acinar pancreas arterioles may reflect functional differences between ephrin A- and B-type binding affinities.
Given the promiscuous binding of ephrin ligands to Ephrin receptors and that the EphA2 receptor is expressed in the RIP-Tag2 tumor vasculature,23,33 it is also possible that the differential binding patterns of CVSNPRWKC and CHVLWSTRC phage in the normal versus tumor vasculature reflects preferential binding of the islet-homing phage for either the EphA2 or EphA4 receptor or both receptors. In addition, because the peptide sequences exhibit sequence identity to different regions of the ephrin A2 and A4 ligands, the binding affinities exhibited by the two islet homing phage to a single EphA receptor may also illustrate this difference. Formation of new binding surfaces deduced by the Eph receptor-ephrin crystal structure26–28 may also account for differential binding of the two islet-homing phage by influencing the accessibility of multibinding domains of the membrane-bound activated dimeric or tetrameric receptor/ligand complex.
Our working hypothesis was that we would be able to identify pancreatic islet homing phage however, given the levels of vascular diversity, we also expected a fraction of the peptides might indeed home to the pancreas without discriminating between the blood vessels of the islets and the acinar pancreas. LPCM on the acinar pancreas showed that ∼95% of the peptides that were isolated from the acinar pancreas were unique and not found in the peptide sequences identified from microdissected islet sections (V.J. Yao, unpublished results). In contrast, ∼84% of islet-targeting phage were unique to the islet vasculature. These differences reflect the heterogeneity of vascular proteins expressed in the two functionally distinct regions of the pancreas. The two islet-homing phage evaluated in this study clearly demonstrate the intraorgan diversity of receptor expression in the pancreas by unequivocal binding to the islet blood vessels.
Localization of CVSNPRWKC and CHVLWSTRC phage to the islet vasculature is similar to binding of peptide ligands to a membrane-bound receptor. Like other ligands binding to their cognate receptors, CVSNPRWKC and CHVLWSTRC phage do not co-localize with the CD31 receptor (PECAM-1), nor do we suggest that the bound phage are internalized by the endothelial cells during the short 6-minute in vivo circulation time. We have always observed a punctate pattern for phage immunoreactivity in the mouse vasculature. We interpret this immunostaining pattern as peptide phage binding to receptors on the luminal surface of endothelial cells. Optical sectioning by confocal microscopy illustrates the detailed localization of peptide phage immunofluorescence in normal and tumor blood vessels (Figures 3 and 4E, respectively). The punctate pattern of phage immunoreactivity is unlike the smooth brushstroke-like quality of antibody immunoreactivity with the EphA4 antibody (Figure 6E). Whether each fluorescent dot represents 1, 10, or 100 phage particles is beyond the scope of our study.
Peptide sequences isolated by either bacterial infection or PCR amplification of catapulted wild-type islet sections indicates three rounds of panning were sufficient for selection of islet targeting phage. The paucity of infective phage from catapulted islet sections suggests the isolation of islet-homing phage from whole pancreas homogenates may be technically challenging. Our results also suggest that recovery of phage DNA may be possible from fixed stained tissues.
Recently, another group reported using in vivo phage display in RIP-Tag2 mice to isolate peptides that home to angiogenic tumor blood vessels or pericytes, or to both in premalignant hyperplastic islets and islet tumors.36 Detailed comparison of the similarities and differences between the approaches used in this earlier study and our work is worthwhile. In the study by Joyce and colleagues,36 in vivo phage display using a CX7C peptide library expressed on T7 phage identified a number of linear peptides. The T7 cyclic peptide phage library was precleared twice for 12 hours each on collagenase-digested RIP-Tag2 angiogenic islets before two to three in vivo panning rounds in RIP-Tag2 mice. In lieu of showing the distribution of injected homing phage in the islet vasculature, the authors showed the distribution of injected FITC-conjugates of their recovered peptides bound to both endothelial cells and/or pericytes of premalignant and/or solid tumors. Although cyclic peptide sequences were fused to the C-terminus of the T7 phage major coat protein, all of the reported peptide sequences were linear. Whether this resulted from fortuitous stop codons as the authors stated or recombination events with the bacterial host during phage amplification is ambiguous.
We used a CX7C peptide library inserted near the N-terminus of the pIII minor coat protein in fd bacteriophage,15 and isolated cyclic islet-targeting peptide phage from the normal pancreas after three rounds of in vivo biopanning by LPCM. Unlike linear peptides, cyclic peptides show increased binding affinities presumably because of a constrained secondary structure that accommodates conformational access to active binding sites.37 Two islet-homing phage recovered from normal islet sections by LPCM in round 3 localized to blood vessels of normal islets and islet tumors. We did not have, a priori, a select subset of receptors from which we sought peptide ligands. The identification of two ephrin A-type ligands by using a combination of in vivo phage display/LPCM and corroboration of this information with emerging studies from other laboratories clearly demonstrates an important proof of principle.
As tumor blood vessels are inherently leaky,38 some fd phage (diameter, 7 to 9 nm) would be expected to extravasate. However, the close association of CVSNPRWKC and CHVLWSTRC phage with CD31 immunoreactivity of endothelial cells indicated that most of the cognate receptor was expressed on endothelial cells and that any extravasated phage remained close to the vessel wall for at least 6 minutes after injection. No immunoreactivity for CVSNPRWKC or CHVLWSTRC phage co-localized with pancreatic islet cells and neither phage bound to β-cells isolated from normal islets (R. Giordano, unpublished results) using the BRASIL method.39 Also, the rapid attachment and generally uniform distribution of CVSNPRWKC and CHVLWSTRC phage fit with binding to the luminal surface of endothelial cells and differed from the patchy leakiness of blood vessels in RIP-Tag2 tumors.
The distribution of CVSNPRWKC or CHVLWSTRC phage in blood vessels of RIP-Tag2 tumors differed in some ways from that of the tumor-homing phage, RGD-4C and NGR.40–43 Although most of RGD-4C and NGR phage bind to the luminal surface of endothelial cells, some phage co-localize with pericytes (mural cells)44 or leukocytes in focal regions (V. Yao, unpublished results). Although leakage of T7 phage from the vasculature of RIP-Tag2 tumors has not been directly addressed, other studies have shown that, after intravenous injection, 50-nm fluorescent microspheres, which are approximately the size of the T7 capsid head, extravasate from focal regions of blood vessels in RIP-Tag2 tumors and remain close to vessel walls (T. Nakahara, unpublished results). Leakage of peptide ligands bound to either fd and T7 bacteriophage may reflect differences between using wild-type or transgenic mice in the original screening assay, and may also be a function of inherent parameters and engineered peptides that induce changes in the net electrostatic charge and effect specific receptor/ligand binding affinities, respectively.
Comparison of the vasculature-associated proteins from our studies and those from Joyce and colleagues36 revealed many different protein homologues as well as several common homologous proteins, such as platelet-derived growth factor-related proteins, basement membrane proteins such as laminin and collagen, and endothelial proteins such as endothelin-1 receptor, FGFR1, and Tie-1. Although we elected to focus the validation steps on a limited set of peptides, we have tested a few of the other candidate ligands listed in Table 1 and found they bound exclusively to the islet vasculature (data not shown). As for the acinar binding peptides, we have not pursued these here.
Although much clinical interest in pancreatic islets focuses on glucose homeostasis, the findings presented in our study as well as others illustrate the diversity of islet-specific vascular targeting ligands that underlie the heterogeneity and biological specialization within the pancreas. Indeed, others have shown the expression of proteins in the islet vasculature influence the development of the pancreas.45–47 Furthermore, the function and integrity of the pancreas may be maintained by islet cells via their expression of enzymes that influence drug metabolism and disease progression.48,49
The vascular heterogeneity that exists in the pancreas is most likely to be recapitulated in other organs that have anatomically distinct regions with functional specialization. Identification of novel peptide ligands by in vivo phage display/LCPM that target vascular receptors within a discrete functional domain in an organ presents novel strategies for directed delivery of therapeutics, genes, and molecular probes such as imaging agents for future clinical applications. Detailed understanding of specific cellular targets and properties of targeting peptides may lead to broad spectrum therapies that simultaneously challenge endothelial cells, perivascular cells, and/or tumor cells. A number of recent reports50,51 and clinical trials are exploring these types of combinatorial therapies.
Acknowledgments
We thank Carlotta Cavazos, Tina Poseno, Yan Sun, Johanna Lahdenranta (M. D. Anderson Cancer Center), and Yvan Chanthery (University of California, San Francisco) for technical assistance and advice; Erin Ator and Michael Mancuso (University of California, San Francisco) for overseeing the care of the RIP-Tag2 colony; Gyulnar Baimekanova for genotyping the mice (University of California, San Francisco); and Mikhail Kolonin (M. D. Anderson Cancer Center) for critical reading of this manuscript.
Footnotes
Address reprint requests to Renata Pasqualini, Departments of Genitourinary Medical Oncology and Cancer Biology, The University of Texas, M.D. Anderson Cancer Center, 1515 Holcombe Blvd., Houston, Texas, 77030. E-mail: rpasqual@mdanderson.org.
Supported by the National Institutes of Health (grants HL-24136 and HL-59157 from the National, Heart, Lung, and Blood Institute to D.M.; the Vascular Mapping Project to D.M.; P50-CA90270 from the National Cancer Institute to D.M., R.P., and W.A.; and National Institutes of Health grants CA78512 and CA88106 to R.P. and CA90810 to R.P. and W.A.), the Gillson-Longenbaugh Foundation (to R.P. and W.A.), the V Foundation (to R.P. and W.A.), and the AngelWorks Foundation (to D.M., R.P., and W.A.).
References
- Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardó-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- Kolonin MG, Pasqualini R, Arap W. Teratogenicity induced by targeting a placental immunoglobulin transporter. Proc Natl Acad Sci USA. 2002;99:13055–13060. doi: 10.1073/pnas.162468499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essler M, Ruoslahti E. Molecular specialization of breast vasculature: a breast-homing phage-displayed peptide binds to aminopeptidase P in breast vasculature. Proc Natl Acad Sci USA. 2002;99:2252–2257. doi: 10.1073/pnas.251687998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajotte D, Ruoslahti E. Membrane dipeptidase is the receptor for a lung-targeting peptide identified by in vivo phage display. J Biol Chem. 1999;274:11593–11598. doi: 10.1074/jbc.274.17.11593. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Koivunen E, Ruoslahti E. Alpha v integrins as receptors for tumor targeting by circulating ligands. Nat Biotechnol. 1997;15:542–546. doi: 10.1038/nbt0697-542. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Arap W, McDonald DM. Probing the structural and molecular diversity of tumor vasculature. Trends Mol Med. 2002;8:563–571. doi: 10.1016/s1471-4914(02)02429-2. [DOI] [PubMed] [Google Scholar]
- Trepel M, Arap W, Pasqualini R. In vivo phage display and vascular heterogeneity: implications for targeted medicine. Curr Opin Chem Biol. 2002;6:399–404. doi: 10.1016/s1367-5931(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Schutze K, Lahr G. Identification of expressed genes by laser-mediated manipulation of single cells. Nat Biotechnol. 1998;16:737–742. doi: 10.1038/nbt0898-737. [DOI] [PubMed] [Google Scholar]
- Dib SA, Vardi P, Bonner-Weir S, Eisenbarth GS. Selective localization of factor VIII antigenicity to islet endothelial cells and expression of class II antigens by normal human pancreatic ductal epithelium. Diabetes. 1988;37:482–487. doi: 10.2337/diab.37.4.482. [DOI] [PubMed] [Google Scholar]
- Beck JSP, Berg BN. The circulatory pattern in the islands of Langerhans. Am J Pathol. 1931;7:31–35. [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE, Fox JG. Efficacy of tribromoethanol anesthesia in mice. Lab Anim Sci. 1993;43:189–192. [PubMed] [Google Scholar]
- Smith GP, Scott JK. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Arap W, Rajotte D, Ruoslahti E. In vivo selection of phage display libraries. Barbas CF III, Burton DR, Scott JK, Silverman GJ, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press,; Phage Display A Laboratory Manual. 2001:pp 22–29. [Google Scholar]
- Koivunen E, Arap W, Rajotte D, Lahdenranta J, Pasqualini R. Identification of receptor ligands with phage display peptide libraries. J Nucl Med. 1999;40:883–888. [PubMed] [Google Scholar]
- Bonnycastle LLC, Menendez A, Scott JK. General phage methods, phage preparation on CsCl gradients. Barbas CF III, Burton DR, Scott JK, Silverman GJ, editors. Cold Spring Harbor: Cold Spring Harbor Laboratory Press,; Phage Display A Laboratory Manual. 2001:pp 15.11–15.13. [Google Scholar]
- Thurston G, McLean JW, Rizen M, Baluk P, Haskell A, Murphy TJ, Hanahan D, McDonald DM. Cationic liposomes target angiogenic endothelial cells in tumors and chronic inflammation in mice. J Clin Invest. 1998;101:1401–1413. doi: 10.1172/JCI965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin ME, Gautam M, Loechel F, Sanes JR, Merlie JP, Albrechtsen R, Wewer UM. Structural organization of the human and mouse laminin beta2 chain genes, and alternative splicing at the 5′ end of the human transcript. J Biol Chem. 1996;271:13407–13416. doi: 10.1074/jbc.271.23.13407. [DOI] [PubMed] [Google Scholar]
- Gale NW, Yancopoulos GD. Ephrins and their receptors: a repulsive topic? Cell Tissue Res. 1997;290:227–241. doi: 10.1007/s004410050927. [DOI] [PubMed] [Google Scholar]
- Gale NW, Baluk P, Pan L, Kwan M, Holash J, DeChiara TM, McDonald DM, Yancopoulos GD. Ephrin-B2 selectively marks arterial vessels and neovascularization sites in the adult, with expression in both endothelial and smooth-muscle cells. Dev Biol. 2001;230:151–160. doi: 10.1006/dbio.2000.0112. [DOI] [PubMed] [Google Scholar]
- Pandey A, Shao H, Marks RM, Polverini PJ, Dixit VM. Role of B61, the ligand for the Eck receptor tyrosine kinase, in TNF-alpha-induced angiogenesis. Science. 1995;268:567–569. doi: 10.1126/science.7536959. [DOI] [PubMed] [Google Scholar]
- Brantley DM, Cheng N, Thompson EJ, Lin Q, Brekken RA, Thorpe PE, Muraoka RS, Cerretti DP, Pozzi A, Jackson D, Lin C, Chen J. Soluble Eph A receptors inhibit tumor angiogenesis and progression in vivo. Oncogene. 2002;21:7011–7026. doi: 10.1038/sj.onc.1205679. [DOI] [PubMed] [Google Scholar]
- Koolpe M, Dail M, Pasquale EB. An ephrin mimetic peptide that selectively targets the EphA2 receptor. J Biol Chem. 2002;277:46974–46979. doi: 10.1074/jbc.M208495200. [DOI] [PubMed] [Google Scholar]
- Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- Toth J, Cutforth T, Gelinas AD, Bethoney KA, Bard J, Harrison CJ. Crystal structure of an ephrin ectodomain. Dev Cell. 2001;1:83–92. doi: 10.1016/s1534-5807(01)00002-8. [DOI] [PubMed] [Google Scholar]
- Himanen JP, Rajashankar KR, Lackmann M, Cowan CA, Henkemeyer M, Nikolov DB. Crystal structure of an Eph receptor-ephrin complex. Nature. 2001;414:933–938. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- Himanen JP, Nikolov DB. Eph signaling: a structural view. Trends Neurosci. 2003;26:46–51. doi: 10.1016/s0166-2236(02)00005-x. [DOI] [PubMed] [Google Scholar]
- Bennasroune A, Gardin A, Aunis D, Cremel G, Hubert P. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit Rev Oncol Hematol. 2004;50:23–38. doi: 10.1016/j.critrevonc.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Gale NW, Holland SJ, Valenzuela DM, Flenniken A, Pan L, Ryan TE, Henkemeyer M, Strebhardt K, Hirai H, Wilkinson DG, Pawson T, Davis S, Yancopoulos GD. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron. 1996;17:9–19. doi: 10.1016/s0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- Dodelet VC, Pasquale EB. Eph receptors and ephrin ligands: embryogenesis to tumorigenesis. Oncogene. 2000;19:5614–5619. doi: 10.1038/sj.onc.1203856. [DOI] [PubMed] [Google Scholar]
- Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61:2301–2306. [PubMed] [Google Scholar]
- Cheng N, Brantley D, Fang WB, Liu H, Fanslow W, Cerretti DP, Bussell KN, Reith A, Jackson D, Chen J. Inhibition of VEGF-dependent multistage carcinogenesis by soluble EphA receptors. Neoplasia. 2003;5:445–456. doi: 10.1016/s1476-5586(03)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Pasqualini R, Lindberg RA, Kain R, Freeman AL, Pasquale EB. The ephrin-A1 ligand and its receptor, EphA2, are expressed during tumor neovascularization. Oncogene. 2000;19:6043–6052. doi: 10.1038/sj.onc.1204004. [DOI] [PubMed] [Google Scholar]
- Davis S, Gale NW, Aldrich TH, Maisonpierre PC, Lhotak V, Pawson T, Goldfarb M, Yancopoulos GD. Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science. 1994;266:816–819. doi: 10.1126/science.7973638. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Laakkonen P, Bernasconi M, Bergers G, Ruoslahti E, Hanahan D. Stage-specific vascular markers revealed by phage display in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2003;4:393–403. doi: 10.1016/s1535-6108(03)00271-x. [DOI] [PubMed] [Google Scholar]
- Koivunen E, Restel BH, Rajotte D, Lahdenranta J, Hagedorn M, Arap W, Pasqualini R. Integrin-binding peptides derived from phage display libraries. Methods Mol Biol. 1999;129:3–17. doi: 10.1385/1-59259-249-X:3. [DOI] [PubMed] [Google Scholar]
- Hashizume H, Baluk P, Morikawa S, McLean JW, Thurston G, Roberge S, Jain RK, McDonald DM. Openings between defective endothelial cells explain tumor vessel leakiness. Am J Pathol. 2000;156:1363–1380. doi: 10.1016/S0002-9440(10)65006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano RJ, Cardó-Vila M, Lahdenranta J, Pasqualini R, Arap W. Biopanning and rapid analysis of selective interactive ligands. Nat Med. 2001;7:1249–1253. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]
- Koivunen E, Wang B, Ruoslahti E. Phage libraries displaying cyclic peptides with different ring sizes: ligand specificities of the RGD-directed integrins. Biotechnology. 1995;13:265–270. doi: 10.1038/nbt0395-265. [DOI] [PubMed] [Google Scholar]
- Koivunen E, Wang B, Ruoslahti E. Isolation of a highly specific ligand for the alpha 5 beta 1 integrin from a phage display library. J Cell Biol. 1994;124:373–380. doi: 10.1083/jcb.124.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- Morikawa S, Baluk P, Kaidoh T, Haskell A, Jain RK, McDonald DM. Abnormalities in pericytes on blood vessels and endothelial sprouts in tumors. Am J Pathol. 2002;160:985–1000. doi: 10.1016/S0002-9440(10)64920-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Pour PM, Standop J, Batra SK. Are islet cells the gatekeepers of the pancreas? Pancreatology. 2002;2:440–448. doi: 10.1159/000064718. [DOI] [PubMed] [Google Scholar]
- Pour PM, Schmied BM, Ulrich AB, Friess H, Andren-Sandberg A, Buchler MW. Abnormal differentiation of islet cells in pancreatic cancer. Pancreatology. 2001;1:110–116. doi: 10.1159/000055802. [DOI] [PubMed] [Google Scholar]
- Erber R, Thurnher A, Katsen AD, Groth G, Kerger H, Hammes HP, Menger MD, Ullrich A, Vajkoczy P. Combined inhibition of VEGF and PDGF signaling enforces tumor vessel regression by interfering with pericyte-mediated endothelial cell survival mechanisms. FASEB J. 2004;18:338–340. doi: 10.1096/fj.03-0271fje. [DOI] [PubMed] [Google Scholar]
- Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J Clin Invest. 2003;111:1287–1295. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]