Abstract
The molecular basis for bacterial responses to host signals during natural infections is poorly understood. The gram-positive bacterial pathogen group A Streptococcus (GAS) causes human mucosal, skin, and life-threatening systemic infections. During the transition from a throat or skin infection to an invasive infection, GAS must adapt to changing environments and host factors. To better understand how GAS adapts, we used transcript profiling and functional analysis to investigate the transcriptome of a wild-type serotype M1 GAS strain in human blood. Global changes in GAS gene expression occur rapidly in response to human blood exposure. Increased transcription was observed for many genes that likely enhance bacterial survival, including those encoding superantigens and host-evasion proteins regulated by a multiple gene activator called Mga. GAS also coordinately expressed genes involved in proteolysis, transport, and catabolism of oligopeptides to obtain amino acids in this protein-rich host environment. Comparison of the transcriptome of the wild-type strain to that of an isogenic deletion mutant (ΔcovR) mutated in the two-component regulatory system designated CovR-CovS reinforced the hypothesis that CovR-CovS has an important role linking key biosynthetic, catabolic, and virulence functions during transcriptome restructuring. Taken together, the data provide crucial insights into strategies used by pathogenic bacteria for thwarting host defenses and surviving in human blood.
Little is known about how pathogenic bacteria adapt to permit growth in human blood. A model organism to address this issue is group A Streptococcus (GAS), which causes a broad spectrum of human diseases ranging from relatively mild throat and skin infections to fulminant, life-threatening invasive diseases such as puerperal sepsis, myositis, necrotizing fasciitis, and streptococcal toxic shock syndrome.1–3 GAS has long been known to be capable of replicating in nonopsonizing human blood.2 However, despite years of study, the molecular mechanisms mediating GAS-host interactions remain poorly understood. Several bacterially encoded molecules contribute to GAS immune evasion by interfering with opsonophagocytosis and killing by polymorphonuclear lymphocytes.2,3 Others protect GAS by disrupting important innate host defenses such as complement activation and complement-mediated cell lysis.2,3 However, additional bacterial proteins likely are involved.
Recently, Gryllos and colleagues4 demonstrated that the expression of the hyaluronic acid capsule biosynthesis (has) operon is stimulated in the bloodstream of infected mice. In addition, two transcriptome studies have demonstrated GAS adaptive transcription after in vitro exposure to human polymorphonuclear lymphocytes and iron limitation.5,6 However, no studies have assessed GAS global transcription or the regulatory networks that govern GAS adaptive responses during growth in human blood.
A two-component regulatory system (TCS) designated CovR-CovS (Cov, control of virulence; also known as CsrR-CsrS) plays an important role in GAS virulence by negatively regulating the has operon and other genes encoding secreted and membrane-anchored factors that promote survival and virulence in humans.7–10 Isogenic Δcov mutant strains are hypervirulent in mouse skin infections and have enhanced resistance in vitro to complement-mediated opsonophagocytic killing by human polymorphonuclear lymphocytes,7–9,11 consistent with increased virulence gene transcription and extracellular capsule production. Frameshift mutations in the covRS locus also arise spontaneously in vivo, and synergistically enhance the virulence of wild-type (WT) bacteria,12 and hyperencapsulated GAS variants have been isolated after in vitro passage in human blood.13 Taken together, these observations suggest that the CovR-CovS TCS responds to molecular signals in human blood.
We directly analyzed GAS global transcription during ex vivo culture in human whole blood using a high-density oligonucleotide array. We hypothesized that the CovR-CovS TCS is involved in GAS adaptation allowing growth in blood and that virulence gene expression would be augmented. To test this hypothesis, we compared the transcriptomes of a WT, serotype M1 GAS strain and its isogenic covR-deletion mutant (ΔcovR). The data provide important new insights into the early stages of GAS survival in blood and evidence that the CovR-CovS TCS functions to coordinate bacterial fitness attributes during disseminated host infections.
Materials and Methods
Bacterial Strains and Growth Conditions
Serotype M1 strain MGAS5005 and the isogenic MGAS5005 ΔcovR derivative (JRS950) have been described.9 GAS was cultured on Trypticase soy agar containing 5% sheep blood agar (Becton Dickinson, Cockeysville, MD), or in Todd-Hewitt (TH) broth (Becton Dickinson) containing 0.2% (w/v) yeast extract (THY; Difco Laboratories, Detroit, MI), at 37°C in 5% CO2. Bacteria were grown in THY broth to late-exponential phase (OD600 = 0.8), harvested by centrifugation at 6000 × g at 37°C for 8 minutes, suspended in an equal volume of human whole blood maintained at 37°C with 5% CO2, and then incubated. Aliquots were removed at 0, 30, 60, and 90 minutes, and added to 2 vol of RNAProtect bacteria reagent (Qiagen, Valencia, CA). Cells were harvested by centrifugation and stored at −80°C before bacterial RNA isolation. Viable counts were obtained for GAS cultures immediately before time course initiation and after 4-hour co-culture in human blood.
Human Study Participants and Whole Blood Collection
Owing to inherent interindividual and gender-related variability of human peripheral blood specimens, 12 human blood donors were used to provide generalizability and sufficient statistical power. Clinical data measurements showed subject and gender-associated variability, so that six donor patients of each gender were used. All blood donors were within normal parameters for 24 tested analytes (data not shown). Heparinized human venous blood (125-ml) was collected from the 12 healthy individuals in accordance with a protocol approved by the Institutional Review Board for Human Subjects, National Institute of Allergy and Infectious Diseases. Informed consent was obtained from all study participants. GAS clinical disease history was not assessed. Blood donors (six females, six males) were from many ethnic backgrounds and their ages ranged from 26 to 54 years; (mean age: females, 37.2 years; males, 36.2 years). Heparin was used in preference to ethylenediaminetetraacetic acid as an anti-coagulant because ethylenediaminetetraacetic acid chelates divalent cations, which would influence cellular functions during GAS-blood cell interactions. On collection, venous blood was divided into aliquots for antibody (Ab) testing (1 ml), cytokine analysis (1 ml), and blood analysis (1 ml; Alpha Veterinary Laboratories, Hamilton, MT). The remaining blood was maintained at 37°C with 5% CO2 until the time course was initiated.
RNA Isolation
Bacterial cell pellets were suspended in 5 vol of EL buffer (Qiagen), incubated for 20 minutes on ice, and separated from lysed erythrocytes by centrifugation at 4500 × g at 4°C for 6 minutes. Cells were rinsed with 2 vol of EL buffer. RNA was isolated from the bacterial pellets as described,9 except that 0.8 μg of bacteriophage MS2 carrier RNA (Roche Bioscience, Indianapolis, IN) and 250 μg of glycogen (Roche) were added. RNA was purified further using the RNeasy 96 kit (Qiagen), with on-column RNase-free DNase I treatment and after treatment with DNAFree (Ambion, Austin, TX). Electrophoretic analysis with an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA) and A260/A280 ratios were used to assess RNA integrity. TaqMan polymerase chain reaction (PCR) assays were performed with RNA templates to ensure contaminating genomic DNA was absent as described.9
cDNA Labeling
RML GeneChip targets were prepared according to the protocol supplied by the manufacturer (Affymetrix Inc., Santa Clara, CA), with modifications. Control spike transcript mixes (containing 0.025 to 0.000025 pmol each of DAP, LYS, THR, and TRP spike transcript cRNAs) (1 μl) were added to each RNA aliquot, and 4.5 μg of random primers (Invitrogen, Carlsbad, CA) were annealed (10 minutes at 70°C, 10 minutes at 25°C). First-strand cDNA was synthesized with 25 U/μl SuperScript III (Invitrogen) in the presence of 0.5 mmol/L dNTPs, 0.5 U/μl SUPERaseIn RNase inhibitor (Ambion), and 10 mmol/L dithiothreitol (10 minutes at 25°C, 60 minutes at 37°C, 60 minutes at 42°C, 10 minutes at 70°C). RNA was removed by hydrolysis in 1 N NaOH (30 minutes at 65°C), and neutralized with 1 N HCl before cDNA purification using the QIAquick 96 kit (Qiagen) according to the manufacturer’s recommendations, except that an extra 10-minute centrifugation was used to remove trace phycoerythrin-ethanol buffer. For cDNA fragmentation, 10.5 μg of cDNA and 1.75 U (0.35U/μg) of DNase I (Roche) were used (10 minutes at 37°C, 10 minutes at 98°C). The fragmented cDNA (averaging 50 to 100 bases) was 3′ end-labeled with biotin-ddUTP using the BioArray terminal labeling kit (Enzo Life Sciences, Inc., Farmingdale, NY) (60 minutes at 37°C) according to the manufacturer’s instructions. The fragmented and end-labeled cDNA was added to the hybridization solution without further purification.
RML GeneChip Composition and Performance
An anti-sense oligonucleotide array (18-μm feature size) representing ∼249,690 25-mer probe pairs (16 pairs per probe set) was manufactured by Affymetrix Inc.14 The custom GeneChip (RMLChip herein) contains 2636 probe sets (42,351 probe pairs) for 2636 predicted GAS open reading frames (ORFs). These features represent a composite superset of six GAS genomic sequences representative of serotypes M1, M3, M5, M12, M18, and M49 (sequenced strains are designated SF370, MGAS315, Manfredo, MGAS9429, MGAS8232, and CS101, respectively). To facilitate the analysis of GAS samples in the presence of host cells, all probe set sequences were pruned during the design process to exclude cross-hybridizing sequences (those exhibiting sequence similarity) with human, rat, and mouse genome ORFs represented on Affymetrix Inc. arrays, and 12 additional bacterial genome sequences. Although the RMLChip was not designed based on the genome sequence of strain MGAS5005, the genome sequence has since been obtained and annotated for this strain under GenBank accession no. CP000017 (Sumby PA, Madrigal A, Kent KD, Porcella SF, Ricklefs SM, Virtaneva K, Sturdevant D, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM, submitted), and the composite RMLChip contains 1692 redundant probe sets (high BLAST score match to MGAS5005) that represent more than 90% coverage of the total number of predicted coding regions (1869 ORFs) encoded by this M1 GAS genome.
GeneChip Hybridization
Target hybridizations, washing, staining, and scanning were performed by the National Institute of Allergy and Infectious Diseases Affymetrix core facility (Science Applications International Corporation (SAIC) Frederick, MD), following the manufacturer’s recommendations (Affymetrix).
Experimental Design and Statistical Analysis
For each of the 12 human blood donors, arrays were hybridized in a complete two-factor experimental design with two treatment levels (WT or mutant GAS strain) and four time points (0, 30, 60, and 90 minutes). To minimize experimental variability, all 12 blood samples were collected within a 2-hour time period and GAS culturing was conducted in parallel. Cultured samples were randomized before all preparation procedures were performed.
Expression estimates for each gene were obtained using the PM-MM difference model of dCHIP version 1.3 software available at http://www.dchip.org/.15 The gene expression estimates were further normalized across samples by simple quadratic scaling on all genes with the median expression for each gene as a baseline.16 Two-dimensional scatterplots were generated for all pairs of samples within a factor level to examine the uniformity of the normalized expression values across donors; five samples with low correlation to the other within-factor samples were removed as outliers (data not shown). Principal component analyses were performed using all MGAS5005 genome-specific probe sets (n = 1925). Hierarchical clustering also was performed to explore single gene effects.
A mixed-effects analysis of variance model was applied to an absolute square root transform of the dCHIP expression estimates, with time, treatment, and gender as fixed effects, and subject as a random effect using Partek Pro 5.1 (Partek Inc., St. Louis, MO). In reporting the significance of effects, both the nominal P values and the false discovery rate (FDR) Q-value17 were reported (Supplementary Table 1; supplementary data available at http://ajp.amjpathol.org) because it is important to account for multiple testing. The experimental design permitted differentially expressed genes to be identified with very high confidence, corresponding to FDRs of 0.06% for time, strain, and subject effects. FDR levels of 0.06% are equivalent to ∼1 false positive in a genome encoding ∼1900 ORFs (approximately the size of the GAS genome).
To elucidate the biology underlying the GAS transcriptional response, we looked for evidence that sets of genes belonging to a functional category showed a coordinated response to experimental factors. Functional annotation for the MGAS5005 genome was generated through in-house compilation. All probe sets were assigned to 1 of 17 functional categories (including unknown), and further classified into 1 of 52 subcategories. The differential expression of a functional category was assessed across both time and treatment by an approach first used in Virtaneva and colleagues,18 that is similar to recent efforts, such as Mootha and colleagues19 For each gene, F-statistics were obtained for the time and treatment effects from a fixed-effects analysis of variance model. A two-sample Wilcoxon ranked sum statistic was then computed for the rank statistics of genes belonging to the functional category relative to the ranks of all remaining genes. Empirical P values for each functional category were obtained by recomputing Wilcoxon statistics across 10,000 permutations of the array assignments. In each permutation, the subject assignment of each array was held constant, whereas the treatment and time assignments were randomized. This approach allowed computation of permutation-based estimates for the FDR20 to account for the multiple testing of functional categories. Nominal P values and FDR estimates are reported for the set of 17 categories and 52 subcategories (Supplementary Table 2).
Reverse Transcriptase (RT)-PCR
Real-time PCR assays were conducted to confirm a subset of the microarray data as described,6 except that Platinum Quantitative PCR SuperMix (Invitrogen) was used and each PCR reaction was performed in quadruplicate.
Flow Cytometry
Strains MGAS5005 and JRS950 were grown in vitro in THY broth to late-exponential phase. Bacteria were collected by centrifugation, rinsed once with Dulbecco’s phosphate-buffered saline (PBS) (Sigma-Aldrich, St. Louis, MO), suspended in Dulbecco’s PBS in 96-well plates, and maintained at 4°C throughout the staining procedure. Cells were blocked with 2% human serum in Dulbecco’s PBS (staining buffer, SB) for 10 minutes before immunostaining for 30 minutes with GAS-specific, affinity-purified primary Abs (Bethyl Laboratories, Montgomery, TX) in SB.21 GAS antigens that were more highly expressed at the RNA level in the ΔcovR mutant strain were selected for the analysis. Control Abs were raised in rabbits against rSLA (spyM3_1204), an ORF that is not encoded in the GAS serotype M1 MGAS5005 genome. Detection was achieved using phycoerythrin-conjugated polyclonal donkey anti-rabbit IgG (1:500; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) in SB for 30 minutes, and subsequent flow cytometric analysis performed with a FACscalibur flow cytometer (Becton Dickinson, Mountain View, CA).21
Results
GAS Molecular Signature during Growth in Human Blood
To model bacteria proliferating rapidly during host sepsis, GAS cells in the late-exponential phase of growth were recultured at approximately the same cell density in freshly heparinized human whole blood for 0 to 90 minutes, and their transcribed cDNAs used to prepare microarray hybridization targets. Evaluation of scatterplots, spike-in control transcripts (data not shown), and histograms of the ex vivo expression data (Supplementary Figure 1) indicated high quality for the resultant data set (comprised of 91 RMLChips). The resultant principal component analyses plots and clustering dendrogram discriminated by treatment within time (Supplementary Figure 2).
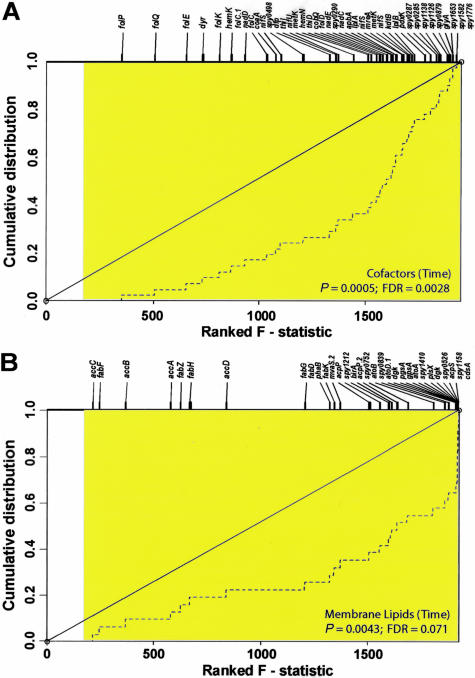
The expression data revealed that extensive remodeling of the transcript profile occurred in both strains during ex vivo blood culture. Within 30 minutes, 716 transcripts were more abundant (up-regulated) and 425 transcripts were less abundant (down-regulated) in the WT strain (Supplementary Table 1). Time affected transcription of the greatest number of GAS genes (n = 1467, or 76.2% of the genome) belonging to functional categories expected to be important for growth adaptation in blood, such as de novo synthesis of macromolecular precursors (including co-factors and nucleotides), carbohydrate metabolism, membrane transport, and transcriptional regulation (Supplementary Figure 3A and Supplementary Table 1). To investigate the ex vivo expression data in more detail, we identified functional categories that were overrepresented within the differentially expressed GAS genes and again, temporal effects were most significant. Transcription of 41 genes involved in co-factor biosynthesis (particularly, chorismate and folate) was significantly reduced with time (nominal P value = 0.0005; FDR estimate = 0.0028) (Figure 1A and Supplementary Table 2). Reduced transcription of genes functioning in cell envelope biogenesis, including biosynthesis of peptidoglycan (n = 47), and of membrane lipids (n = 31; nominal P < 0.005; FDR <0.10) (Figure 1B and Supplementary Table 2), also occurred in both strains throughout time in whole blood. Significant expression changes (nominal P < 0.05; FDR < 0.10) also were observed throughout time in cellular shape and division, carbohydrate metabolism, and nucleotide metabolism. Additional noteworthy functional categories in our time analysis showing respectable FDR estimates when controlling for multiple testing include heat shock proteins (nominal P < 0.05; FDR = 0.14), amino acid catabolism (nominal P < 0.05; FDR = 0.14), and stress adaptation (nominal P = 0.087; FDR = 0.16) (Supplementary Table 2).
Figure 1.
Statistical analysis of GAS functional categories for time effects during ex vivo blood culture. A and B: Plots show the cumulative distribution for ranked F statistics of genes within the selected functional categories when testing for the effect of time in blood (hatched lines). If the degree of differential expression of genes within a category is the same as that of all other GAS genes, the distribution will trace the identity line (black diagonal line). If small P values occur with greater frequency among genes in a category, then the distribution curve diverges from the diagonal line, with the most differentially regulated genes shifted to the right. Tick marks at the top of plots show the ranks of genes within the functional category, indicated by their assigned gene name or spy ORF numbers. All genes exhibiting significant differential expression at the nominal 0.05 level (F >2.71) are shaded in the plot, demonstrating the extensive transcriptional changes observed throughout time. The effect of time on GAS shows significant differential expression in co-factors (A, nominal P < 0.0005; FDR <0.005) and membrane lipids (B, nominal P < 0.005; FDR <0.10), during blood culture.
Adaptive Metabolic Shift in WT GAS during Whole Blood Exposure
Human blood is relatively poor in free amino acids (AAs), but rich in carbon and energy sources such as carbohydrates and peptides.22 Within 30 minutes of blood culturing in the WT strain, transcript levels of genes involved in glycolysis were reduced, suggesting a depletion of bacterial intracellular glucose levels. We also observed massive transcriptional changes that reflect temporal adjustment of GAS cellular metabolism, firstly to augment transport and fermentation of alternate metabolizable carbohydrates, and subsequently to transport and catabolize oligopeptides, aminosugars, and AAs (Supplementary Table 1). In light of the temporal derepression observed in the WT during blood culture of GAS transcripts encoding nutrient-releasing proteins, including the potent cytolysin streptolysin S (SLS: sagA), streptokinase (ska), and the Ig-endopeptidase designated Mac (spy0861), carbon- and nitrogen-containing host substrates would be expected to become increasingly available throughout time for bacterial use. Consistent with this idea, increased transcription after 30 minutes of oligopeptidase F (pepF), oligopeptidase (pepB), and the oligopeptide permease (opp) operon suggested a concerted strategy to acquire oligopeptides. Temporal reduction of transcripts encoding RelA (spy1981), encoding the bifunctional enzyme involved in the synthesis and hydrolysis of (p)ppGpp during amino acid starvation, and CodY (spy1777), encoding a pleiotropic transcriptional repressor responsive to branched chain AAs, supports the notion that GAS intracellular AA levels likely accumulated in the WT strain within 30 minutes in human blood (Supplementary Table 1). Temporal reduction of transcripts encoding the transcriptional regulator Rgg, which represses arginine and serine degrading enzymes when glucose is present,23 coincided with temporal up-regulation of transcripts encoding arginine (arcCBA: spy1543-44, and 1547) and serine (sdhBA: spy2189-90) catabolic enzymes, contributing to a metabolic transition from carbohydrate to AA fermentation.
GAS Virulence Gene Transcription during Blood Adaptation
On blood exposure, we observed accumulation of transcripts belonging to the multigene activator (mga) regulon in both GAS strains.24,25 Maximum levels of mga (spy2019) regulator transcripts were detected 30 minutes after blood exposure (Supplementary Table 1). In concordance, Mga-regulated transcripts emm1 and sic, encoding the surface-associated Emm1 (or M1) protein and secreted streptococcal inhibitor of complement (SIC), subsequently accumulated (Supplementary Table 1). These virulence determinants promote bacterial adherence and subversion of Fc- and complement-mediated immune defenses.2,3 Similar patterns of increased transcription were observed for other genes located upstream of mga indicating possible co-regulation, including those encoding a putative metal-transporting ABC-transporter TptA-D (spy2033-39), a TCS designated Irr-Ihk (spy2027-26), and secreted antigens Isp (spy2025) and SPy2023. After blood exposure, levels for transcripts encoding Mga-independent virulence determinants such as capsular polysaccharide (hasABC; spy2000-02), streptolysin S (sagA; spy0738) and accessory genes, and Ig-endopeptidase Mac (spy0861) also increased in both strains. In contrast, the levels of transcripts encoding the cysteine protease (speB; spy2039) and transcriptional repressor cathelicidin-resistance gene regulator (crgR; spy1870) were temporally reduced in both strains during blood culture.26 However, reduction of speB and crgR transcripts is predicted to (indirectly) enhance bacterial resistance to phagocytosis and host antimicrobial peptides, respectively.
Transcripts encoding ribosomal protein subunits, molecular chaperones GroEL, DnaJ, and DnaK and translation elongation factor Tu (TufA), as well as others involved in glycolysis (such as Fba and Eno) or anaerobic respiration (Ldh) were among the most abundant GAS transcripts detected in the WT strain (Supplementary Figure 4 and Supplementary Table 3), thereby supporting recent theoretical predictions for highly expressed GAS genes.27 Interestingly, transcripts encoding a transcriptional regulator (RofA: spy0124) and several proven virulence factors (Emm1, SIC, SLO, and ScpA) also were among the most abundant GAS transcripts; the latter remained highly abundant throughout the time course of the experiment.
Importantly, we also observed an immediate and robust increase of transcript levels for the pyrogenic toxin superantigen (PTSAg)-encoding genes speA (spyM18_0393) and speJ (spy0436) in both GAS strains throughout time in blood (Supplementary Table 1). More modest increases of speG (spy0212) and smeZ (spy1998) transcript levels occurred. Increased levels of PTSAg transcripts were verified by real-time RT-PCR analysis (Supplementary Figure 5). Overstimulation of proinflammatory host responses will disturb innate host defenses, increase host cell damage, and concomitantly release carbon and energy sources for bacterial use.
Temporal Expression of Hypothetical Genes in Blood
Forty-three percent of the genes in strain MGAS5005 are hypothetical ORFs of unknown function. Although many of these genes have homologues in other bacteria, little or no functional information is known in these other systems. Blood exposure significantly affected the level of 531 GAS transcripts encoding proteins of unknown function. Of the 531 affected genes, 40 genes have been identified as essential for growth in Bacillus subtilis28 and 27 in Staphylococcus aureus.29,30 Transcripts encoding a basic, hydrophilic, GAS surface antigen (SPy0843) increased approximately twofold within 30 minutes of blood exposure, and then gradually declined. SPy0843 is homologous to BspA of Bacteroides forsythus, and is known to be made during GAS infections.31
CovR-CovS Regulatory Influence on GAS Gene Transcription in Blood
Transcription of the covRS operon, which is negatively autoregulated, and of the covRS activator gene rocA (spy1605),32 increased within 30 minutes after transfer of the WT strain from THY culture broth into whole blood (Supplementary Table 1). covRS transcripts returned to pre-existing levels by 60 minutes, whereas rocA levels remained elevated throughout the time course. After derepression of covRS at 30 minutes, derepression occurred thereafter of many known virulence-associated genes and other genes under CovR-regulation, such as sagA (spy0738), mac (spy0861), isp2 (spy1801), and ska (spy1979), to name a few (Supplementary Table 1).9
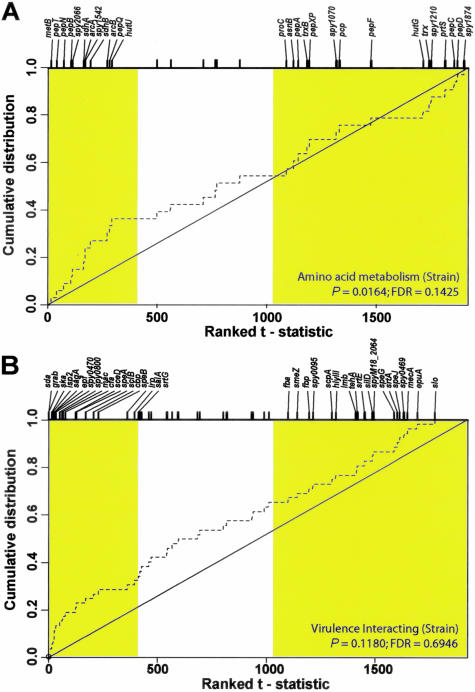
Strain genotype (ΔcovR versus WT) affected a large proportion of GAS transcripts (n = 1075 genes, or 55.8% of the genome) (Supplementary Figure 3B). Transcripts involved in protein folding and/or modification (nominal P < 0.005; FDR = 0.028) (Supplementary Figure 6a), amino acid metabolism (nominal P < 0.05; FDR = 0.14) (Figure 2A), and amino acid catabolism (nominal P < 0.05; FDR = 0.49) were consistently more abundant in the ΔcovR mutant in our functional analysis (Supplementary Table 2). We also observed up-regulation of 36 GAS transcripts involved in proven or putative virulence interactions in the ΔcovR mutant strain compared to WT (Figure 2B), including cfa (spy1273) encoding the CAMP factor hemolysin, and sceD (spy2191). Derepression of the capsule biosynthesis operon (has; spy2200-02) in the ΔcovR strain was observed and verified by RT-PCR analysis (Supplementary Figure 4). In addition, the cathelicidin resistance transcriptional repressor crgR (spy1870)26 transcripts were more abundant in the ΔcovR strain (Q < 0.0003). The category “virulence-interacting” did not achieve statistical significance after multiplicity correction in our strain comparison (Supplementary Table 2), primarily owing to the presence of numerous covR-independent virulence-associated transcripts, such as the mga-regulated emm1 and isp (spy2025), and others. However, derepression of 18 of 52 virulence-associated transcripts (Q <0.0006) concurs with in vitro data previously reported for Δcov strains,9,33 and emphasizes the impact of this regulatory locus.
Figure 2.
Statistical analysis of GAS functional categories for treatment effects during ex vivo blood culture. A and B: Plots show the cumulative distributions for ranked test statistics of genes within the selected functional categories for treatment effects in blood (hatched lines). Genes differentially expressed with treatment that exhibit a leftward shift indicate up-regulation in the ΔcovR strain and those showing a rightward shift indicate down-regulation. All genes exhibiting significant differential expression at the nominal 0.05 level ( t > 1.99) are shaded in the plot. The effect of treatment in blood on GAS amino acid metabolism (A) showing up-regulation in the ΔcovR strain (nominal P value <0.05; FDR = 0.14), and virulence interacting (B), showing up-regulation in the ΔcovR strain, but not in a statistically significant manner (nominal P value = 0.11; FDR = 0.69).
Transcripts of the following genes were down-regulated in the ΔcovR mutant relative to the WT strain: spy0425-0427 and spy2105-10 encoding purine nucleoside biosynthesis genes; spy1785-1798 encoding the heme-binding iron transporter system Shp (shp/siaA; spy1796) and (htsABC/sia; spy1795-93); and dppA-E (spy2000-04) encoding the dipeptide permease transporter, as seen previously in vitro.9 Transcription of the genes encoding stringent factor RelA (spy1981) and some AA-tRNA synthetases also was reduced in the ΔcovR strain relative to the WT strain (Supplementary Table 1). The effect of time on oxidative stress, a functional category that was not statistically overrepresented in the expression data (P value = 0.600), is depicted for illustrative purposes (Supplementary Figure 6b).
The CovR-CovS TCS plays an important role in modulating GAS virulence capacity.9,34 Our transcriptome analysis supports the hypothesis that CovR-CovS mediates GAS compensatory metabolism in blood. For example, transcripts encoding the pleiotropic regulator carbon catabolite control protein (CcpA; spy0514),35 were higher in the ΔcovR mutant than in the WT strain throughout the time course (Q < 0.0001). Transcripts of genes encoding proteins involved in transport of cations (spy1240-1249) and carbohydrates (spy1591-95), as well as folate (spy1096-1100) and chorismate (spy1350-1358) biosynthesis also were higher in the ΔcovR mutant (Supplementary Table 1). Strain-dependent patterns also were seen for transcripts encoding arginine catabolic enzymes (ArcCBA: spy1543-44-47), serine dehydratase (sdhBA; spy2189-90), putative phosphotransferase systems (PTS) for galactitol (spy1709-12) and N-acetylglucosamine (spy629-34), carbohydrate assimilatory functions (GlpFOK: spy1682-4; spy1586; spy1704-12), and those putatively involved in aminosugar metabolism (spy0716; spy1399; spy1694). These strain-dependent transcript differences indicate that CovR-CovS is involved in substantial restructuring of GAS metabolism in whole blood.
Validation of Oligonucleotide-Array Gene Expression Results
We used real-time RT-PCR to validate the microarray data. A subset of five targets was selected to encompass a range of expression estimates from within the RMLChip data set. Relative expression estimates initially determined with RMLChip hybridization also were correlated (slope, m = 1.0044) with RT-PCR results in both strains for all five tested virulence genes (hasA, speA, speG, speJ, and smeZ) (Supplementary Figure 5; R2 = 0.755 for all time points; range, 0.72 to 0.88). As others have observed,9,36 we obtained positive regression slopes indicating that higher relative transcript estimates were obtained by real-time RT-PCR determination than by microarray hybridizations.
Confirmation of GAS Surface Antigen Expression by Flow Cytometry
We used flow cytometry to corroborate the microarray gene expression data at the protein level. The tested surface antigens were detected at higher levels on the hyperencapsulated ΔcovR mutant relative to the WT (Table 1), a result concordant with the increased transcript levels in the ΔcovR strain detected by RNA profiling at time 0 for the in vitro grown WT and ΔcovR strains (Supplementary Table 1). After a conservative (Bonferroni) correction for multiplicity testing (P < 0.005), four GAS differentially expressed proteins (Spy0319, OppA, SceD, and MtsA) were detected with significance (P = 0.0001, 0.0006, 0.0007, and 0.004, respectively) on the GAS strains (Table 1), which is consistent with RMLChip expression estimates (Supplementary Table 1). Three of the four antigens are substrate-binding, ABC-type transporter lipoproteins involved in membrane transport,37,38 whereas the function of the secreted SceD protein is unknown.
Table 1.
Flow Cytometric Analysis of Group A Streptococcus Surface Proteins
| Designation | Protein | Description | Mutant | WT | P value |
|---|---|---|---|---|---|
| SPy0319 | Surface lipoprotein | 22.43 ± 0.60 | 15.00 ± 0.68 | 0.0001 | |
| SPyM18_0281 | OppA | Surface lipoprotein | 23.31 ± 1.05 | 15.03 ± 0.98 | 0.0006 |
| SPy2191 | SceD | Secreted protein | 22.90 ± 0.70 | 18.20 ± 0.50 | 0.0007 |
| SPy0453 | MtsA | Surface lipoprotein | 12.74 ± 0.80 | 9.60 ± 0.45 | 0.0040 |
| SPy1592 | Surface lipoprotein | 69.87 ± 5.08 | 50.02 ± 4.20 | 0.0064 | |
| SPy1245 | PstS | Surface lipoprotein | 25.70 ± 0.95 | 19.73 ± 1.97 | 0.0090 |
| SPyM3_1204 | SLA | Secreted protein | 13.75 ± 0.78 | 10.58 ± 0.95 | 0.0111 |
| Auto-fluorescence | FACs control | 4.14 ± 0.05 | 4.29 ± 0.04 | 0.0127 | |
| SPy0252 | Surface lipoprotein | 10.78 ± 0.82 | 8.93 ± 0.43 | 0.0258 | |
| Secondary Ab | FACs control | 4.07 ± 0.08 | 4.15 ± 0.12 | 0.3414 |
MGAS5005 (WT) and JRS950 (ΔcovR) cells were harvested at OD600 = 0.8 after in vitro growth in THY broth at 37°C with 5% CO2. Immunostaining was performed with affinity-purified rabbit polyclonal antibodies, or control rabbit α-SLA antibodies. GAS surface antigens were detected with a phycoerythrin-conjugated donkey anti-rabbit IgG secondary antibody, and analyzed by flow cytometry.
Listed are mean fluorescence within the analysis gate ± SD from two independent experiments comprised of triplicate measurements. Minimums of 17,900 gated events (representing GAS cells) were analyzed for each replicate. Statistical significance was assessed at the P < 0.05 level after Bonferroni correction for 10 comparisons (adjusted P < 0.005).
Discussion
Adaptive gene expression determines whether bacteria successfully persist and disseminate during encounters with host defenses and diverse microenvironments. Identification of GAS genes uniquely expressed in blood is important both for understanding infection processes and for the development of new approaches to control GAS infections. Unfortunately, there is no current experimental approach that accurately recapitulates the human blood environment, and inclusion of all host variables influencing bacterial transcription is presently unachievable. For example, a patient’s genetic makeup, innate defenses, cytokine levels, and changing clinical conditions (such as dehydration, fever, changing electrolyte or blood sugar levels, and anemia) likely influence bacterial responses and infection course. As a first step toward understanding this complex host-bacterium interaction, we aimed to identify GAS transcripts that correlate with ex vivo exposure to human blood supplied from a diverse set of donors. To allow for bacterial losses during isolation from host cells, GAS was inoculated into whole blood at ∼108 CFU/ml (or ∼10 CFU per white blood cell). Although in vitro phagocytosis assays and experimental septicemia in animal models are routinely conducted in this GAS cell density range, lower cell densities are observed during natural human infections. Human whole blood was used without previous heat treatment (to denature host complement components), thereby enabling us to measure GAS transcript expression during full interaction of host cellular and bacterial factors. Although an alternative model species (eg, inbred animal) blood could have been used, it would have been difficult to obtain appropriate volumes of blood, and may have introduced bias owing to species-specific differences in responding cell types, hemodynamic properties, PTSAg susceptibilities, proinflammatory cytokine responses, and host defense mechanisms that likely impact on bacterial responses. Moreover, GAS is a human-specific pathogen, which clearly also justifies the use of human blood. Randomly selected human blood donations were preferable because this allowed us to account for individual and gender-based variation. Knowing that GAS must evade innate host defenses in nutrient-depleted host environments, late exponential phase GAS cells were chosen for our experimental design because this is considered a transitional growth phase when bacterial cell densities are high and nutrient sources (such as carbon, nitrogen, and the like) become limiting.
Our goal was to comprehensively investigate the GAS transcriptional response to human blood (as culture medium) and to evaluate the involvement of the CovR-CovS TCS during blood adaptation. Although the ex vivo growth technique used in this study successfully mimics some of the conditions existing within infected hosts, we must acknowledge it cannot accurately replicate the highly complex environment that pathogens encounter during natural infections. Consequently, the salient points of this ex vivo study are provided within this context. This being said, our strategy is more clinically relevant than the analyses of transcriptomes derived from bacteria cultured exclusively in laboratory media, which are artificially nutrient-rich and lack host factors and cells. Moreover, the study demonstrated strong evidence for consistent bacterial adaptive responses to blood components. For example, on blood exposure we observed increased expression of GAS genes that interact with host cell surfaces (adhesins such as M1, collagen-binding proteins SclB and Cbp, and capsule),39 and that contribute to the evasion of host innate defenses (SIC, Mac, and SPEA). Increased transcription of has genes (encoding capsule) concurs with data previously obtained for GAS in mouse blood after intraperitoneal inoculation.4 Human serological data have shown that the proteins encoded by many transcripts we observed were up-regulated during ex vivo blood culture, such as sic, emm1, ska, and speA, are also expressed in vivo during disseminated GAS infections.40–42
Our analysis revealed that many genes involved in key metabolic functions were substantially affected by blood exposure. Being auxotrophic for most amino acids, GAS must rely on host sources for anabolic and catabolic substrates.43 In human blood, which is low in free amino acids, our data support that GAS coordinates the proteolysis, transport, and catabolism of oligopeptides to obtain essential free AAs. Recent discoveries that the pleiotropic transcriptional repressor CodY senses intracellular levels of branched-chain AAs in Bacillus subtilis44 suggest that CodY (Spy1777) may be involved in mediating some of the GAS dynamic responses. Importantly, we discovered that the CovR-CovS TCS also plays an important role during the remodeling of GAS metabolism in response to human blood.
Elevated systemic cytokines are thought to contribute to the pathogenesis of streptococcal toxic shock syndrome by activating host complement and coagulation cascades, increasing plasminogen activator inhibitor type 1, and reducing fibrinolysis. In extreme cases, elevated cytokines result in hypotension, disseminated intravascular coagulation, multiple organ failure, and death because of lethal shock.45 In concordance, PTSAg activity is found in acute-phase serum samples from patients with severe GAS disease, and lack of neutralizing anti-PTSAg antibodies appears to be a key risk factor for the development of invasive streptococcal disease.42,46,47 Although regulation of PTSAg production is poorly understood, up-regulation of PTSAg-encoding transcripts during blood culture concurs with recent studies indicating that host factors enhance production of particular PTSAgs.48–50 Worldwide resurgence of streptococcal toxic shock syndrome has been associated with circulation of novel speA+ strains.51 Our direct measurement of GAS PTSAg transcripts in human blood supports that SPEG, SPEJ, SMEZ, but especially SPEA, likely contribute to GAS-disseminated infections more than previously appreciated. Increased detection of GAS emm1 transcripts in response to human blood is also notable in light of recent evidence that soluble M1 protein (released from GAS surfaces by SPEB) complexes with soluble fibrinogen to activate polymorphonuclear lymphocytes, resulting in a proinflammatory cascade that promotes vascular leakage, tissue damage, and streptococcal toxic shock syndrome development.52
In vitro, we previously observed that CovR plays a central role in gene regulatory networks by influencing expression of genes encoding other transcriptional regulators, including other TCSs, and many GAS genes encoding surface and secreted proteins mediating host-pathogen interactions.9 Although the spectrum of molecular signals to which GAS is responsive remains poorly understood, Gryllos and colleagues53 recently reported that CovR-regulated genes are repressed in vitro under high environmental Mg+2 concentrations. Lower concentrations of Mg+2 present in mucosal and extracellular body fluids would predict that CovR repressor activity is low in these host environments. In keeping with this postulate, we observed temporary ablation of CovR-mediated autoregulation and derepression of covRS transcription within 30 minutes in blood. Our data support temporary CovR-CovS inactivity and enhanced GAS virulence factor production immediately after human blood exposure. Surprisingly, transcription of speB (spy2039), shown in vitro to be CovR-regulated and encoding the major secreted protease, was decreased throughout the 90-minute time course in both the ΔcovR and WT GAS strains (Supplementary Table 1). This is a significant observation in the context of reports that this protease can degrade most of the GAS-secreted proteome, including virulence factors such as Emm, SIC, and PTSAgs.54 This study revealed that in the complex milieu of human blood and cells, unknown signals (or regulatory systems) apparently override CovR regulation of speB. Importantly, such a prompt reduction of SPEB proteolytic functionality after blood exposure may be advantageous to GAS by enhancing (for example) resistance to phagocytosis (mediated by intact Emm protein), and disrupting host innate defenses through the proliferative and T-cell Vβ clonal responses of host cells (mediated by full-length PTSAgs).
Many regulatory genes (n = 97) were differentially expressed throughout time in blood. Some directly modulate GAS gene expression to elude host defenses, thereby facilitating persistence and/or dissemination. For example, transcripts encoding the multiple gene activator Mga (spy2019) accumulated within 30 minutes and subsequently declined, whereas transcripts encoding the TCS Irr-Ihk accumulated within 60 minutes of whole blood exposure and were sustained. These regulatory systems play crucial roles in protecting GAS against immune and innate host defenses.5,55,56 However, additional transcriptional regulators likely play significant downstream roles in mediating GAS blood persistence and adaptation mechanisms.
The data argue that a GAS stress response occurred during the 90-minute time course and accounts for some of the observed gene expression changes. For example, genes functioning in cell envelope biogenesis were significantly down-regulated in both strains throughout time in whole blood. Some degree of cellular shock may be expected, especially during the first 30 minutes of blood exposure, owing to GAS harvest by centrifugation, cell resuspension in iron- and nutrient-limiting whole blood (after growth in a nutrient-rich, THY medium), increased osmolality of blood relative to THY broth, and in light of host inflammatory responses in whole blood. In this context, some temporal changes could be interpreted as sequential transcriptional remodeling events aimed at facilitating GAS adaptation and shock recovery on blood exposure. However, as extracellular physiological fluids are considered more nutrient-deprived than human blood, our findings are representative of the kinds of GAS transcriptional responses that occur during natural infections, albeit throughout a longer time frame. Transcripts encoding peptidases, AA-acyl tRNA synthetases, and protein chaperones were more abundant in the ΔcovR mutant. Interestingly, the CovR-CovS system was derepressed within 30 minutes and then returned to pre-existing levels; supporting the notion that temporary inactivation of the CovR repressor allows expression of GAS gene products that contribute to its cellular adaptation response during mild stress (culture shock) recovery.9 This is supported by a recent report that growth of GAS, which lacks a sigB ortholog, under mild stress conditions requires inactivation of CovR to relieve repression of many GAS genes.57 Consistent with these observations, GAS viable cell counts after ex vivo culture indicated that approximately two doublings had occurred in the WT strain and slightly more in the ΔcovR mutant, but the strain count difference did not achieve significance (data not shown).
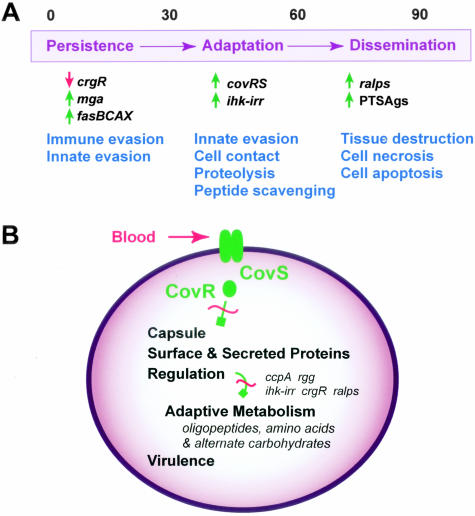
Our study has provided the first comprehensive analysis of how GAS adapts to growth in human blood. On blood exposure, GAS immediately remodels transcription through coordinated expression of regulatory, metabolic, and virulence genes. Transcriptional regulators designated Mga and Ihk-Irr, and the three-component regulatory system called FasBCA (Spy0243-0245) (Figure 3A), are up-regulated. Increased expression of these regulatory systems in turn promotes bacterial fitness and survival in this host environment by enabling the evasion of immune and innate host defenses.5,55 Our data also support the idea that human blood stimuli evoke GAS responses that facilitate pathogen adaptation in this free AA-limiting host environment (Figure 3A). Interference with CovR repressor activity mobilizes metabolic machinery for peptide scavenging, and enhances host cell contact through derepression of capsule and bacterial surface adhesin expression. With time, localized buildup within the extracellular milieu of nutrient-releasing proteins, such as cytolysins, as well as the proinflammatory PTSAgs and M protein will disrupt host tissue barriers and cause host damage that enhances the potential for bacterial dissemination (Figure 3A). In keeping with models put forth previously,9,34,58 the CovR-CovS TCS apparently serves to couple as-yet-unidentified environmental cues to multiple effector outputs, including stress adaptation and virulence-associated functions such as capsule, surface adhesins, and secreted proteins (Figure 3B). It is unlikely that CovR is directly responsible for all of the extensive metabolic remodeling we observed. Rather, we believe that most effects are because of the downstream impact of CovR derepression on numerous other regulatory loci (n = 97). This interpretation is exemplified by a disparate number of down-regulated transcripts (n = 532) relative to up-regulated transcripts (n = 270) in the ΔcovR versus WT strain comparison throughout the entire time course. In our simplified model, indirect effects are implied but primarily omitted (Figure 3B).
Figure 3.
Time effect and CovR-CovS two-component system involvement during GAS adaptation to human blood. A: A model for GAS adaptation throughout time during blood exposure. Numbers at top of figure represent minutes. On blood entry, GAS immediately remodels transcription to promote bacterial survival. Successfully persistent, GAS subsequently promotes host cell contact through surface adhesins and mobilizes metabolic machinery for peptide and AA scavenging in this protein-rich host environment. Buildup of proteases and proinflammatory molecules secreted into the extracellular milieu disrupts host tissue barriers and causes host cell damage in a manner that synergistically enhances bacterial dissemination. B: CovR-CovS involvement in coordinating fundamental GAS cellular processes. Normally, phosphoryl group transfer from the membrane-associated histidine kinase component (CovS) to CovR, its cognate DNA-binding response regulator, enhances CovR DNA-binding activity. However, human blood exposure temporarily ablates this transfer event (red hashed line) and CovR-binding activity is decreased, resulting in derepression of virulence-associated genes such as capsule, surface adhesins, and secreted proteins. Through direct and indirect means, GAS adaptively regulates a vast array of genes involved in cellular metabolism and virulence functions that enable GAS to persist and replicate successfully in human blood.
To summarize, GAS undergoes a rapid, adaptive response to host molecular signals in human blood. As a consequence, cellular metabolism and virulence pathways are remodeled, resulting in enhanced survival and pathogenesis. The CovR-CovS TCS plays a critical role in directly and indirectly coordinating diverse regulatory networks in blood and during fulminant GAS infections. Increased understanding of how GAS and other bacteria respond to blood could be helpful in developing rapid diagnostic or therapeutic strategies for disseminated bacterial infections.
Supplementary Material
Acknowledgments
We thank the blood donors for making this study possible; J. Yang and R. Lempicki of Science Applications International Corporation (SAIC) Frederick Affymetrix Core facility for hybridizations; T. Downey, J. Lin, and X. Wang (Partek Inc.) for experimental design and statistical assistance; F. DeLeo, S. Kobayashi, and K. Braughton for collecting whole blood; B. Doughty and L. Parkins for expert technical assistance; N. Hoe and G. Somerville for critical review of the manuscript; and J. Scott for providing the ΔcovR mutant strain JRS950.
Footnotes
Address reprint requests to James M. Musser, M.D. Ph.D., Center for Human Bacterial Pathogenesis, Department of Pathology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030. E-mail: musser@bcm.tmc.edu.
Supported by the National Institutes of Health (U01-AI-60595) to J.M.M. and (T32-ES 07018) to W.T.B.
References
- Musser JM, Krause RM. The revival of group A streptococcal diseases, with a commentary on staphylococcal toxic shock syndrome. Krause RM, editor. New York: Academic Press,; Emerging Infections. 1998:pp 185–218. [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- Gryllos I, Cywes C, Shearer MH, Cary M, Kennedy RC, Wessels MR. Regulation of capsule gene expression by group A streptococcus during pharyngeal colonization and invasive infection. Mol Microbiol. 2001;42:61–74. doi: 10.1046/j.1365-2958.2001.02635.x. [DOI] [PubMed] [Google Scholar]
- Voyich JM, Sturdevant DE, Braughton KR, Kobayashi SD, Lei B, Virtaneva K, Dorward DW, Musser JM, DeLeo FR. Genome-wide protective response used by group A streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc Natl Acad Sci USA. 2003;100:1996–2001. doi: 10.1073/pnas.0337370100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot LM, Smoot JC, Graham MR, Somerville GA, Sturdevant DE, Migliaccio CA, Sylva GL, Musser JM. Global differential gene expression in response to growth temperature alteration in group A streptococcus. Proc Natl Acad Sci USA. 2001;98:10416–10421. doi: 10.1073/pnas.191267598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CAL, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. Virulence control in group A streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B, DeLeo FR, Reid SD, Voyich JM, Magoun L, Liu M, Braughton KR, Ricklefs S, Hoe NP, Cole RL, Leong JM, Musser JM. Opsonophagocytosis-inhibiting Mac protein of group A streptococcus: identification and characteristics of two genetic complexes. Infect Immun. 2002;70:6880–6890. doi: 10.1128/IAI.70.12.6880-6890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath A, DiRita VJ, Barg NL, Engleberg NC. A two-component regulatory system, CsrR-CsrS, represses expression of three streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleberg CN, Heath A, Miller A, Rivera C, DiRita VJ. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J Infect Dis. 2001;183:1043–1054. doi: 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- Raeder R, Harokopakis E, Hollingshead S, Boyle MD. Absence of SpeB production in virulent large capsular forms of group A streptococcal strain 64. Infect Immun. 2000;68:744–751. doi: 10.1128/iai.68.2.744-751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21(Suppl 1):20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Liyanarachchi S, Wright FA, Davuluri R, Lockman JC, de la Chapelle A, Pellegata NS. Gene expression profiling of isogenic cells with different TP53 gene dosage reveals numerous genes that are affected by TP53 dosage and identifies CSPG2 as a direct target of p53. Proc Natl Acad Sci USA. 2002;99:15632–15637. doi: 10.1073/pnas.242597299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva K, Wright FA, Tanner SM, Yuan B, Lemon WJ, Caligiuri MA, Bloomfield CD, de la Chapelle A, Krahe R. Expression profiling reveals fundamental biological differences in acute myeloid leukemia with isolated trisomy 8 and normal cytogenetics. Proc Natl Acad Sci USA. 2001;98:1124–1129. doi: 10.1073/pnas.98.3.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Yekutieli D, Benjamini Y. Resampling based FDR controlling multiple hypotheses testing. J Stat Plan Infer. 1999;82:171–196. [Google Scholar]
- Lei B, Smoot LM, Menning HM, Voyich JM, Kala SV, Deleo FR, Reid SD, Musser JM. Identification and characterization of a novel heme-associated cell surface protein made by Streptococcus pyogenes. Infect Immun. 2002;70:4494–4500. doi: 10.1128/IAI.70.8.4494-4500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman L, Bennet JC, editors. Philadelphia: W.B. Saunders; CecilTextbook of Medicine. 2000:p 2525. [Google Scholar]
- Chaussee MS, Somerville GA, Reitzer L, Musser JM. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J Bacteriol. 2003;185:6016–6024. doi: 10.1128/JB.185.20.6016-6024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon MG, Geist RT, Perez-Casal J, Scott JR. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J Bacteriol. 1992;174:5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver KS, Heath AS, Scott JR. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange and iron limitation on EMM transcription. Infect Immun. 1995;63:4540–4542. doi: 10.1128/iai.63.11.4540-4542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Karlin S, Theriot J, Mrazek J. Comparative analysis of gene expression among low G+C gram-positive genomes. Proc Natl Acad Sci USA. 2004;101:6182–6187. doi: 10.1073/pnas.0401504101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O’Reilly M, O’Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JFML, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Zhang B, Van SF, Horn, Warren P, Woodnutt G, Burnham MK, Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, Kedar GC, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu-Zy ZY, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- Reid SD, Green NM, Sylva GL, Voyich JM, Stenseth ET, DeLeo FR, Palzkill T, Low DE, Hill HR, Musser JM. Postgenomic analysis of four novel antigens of group A streptococcus: growth phase-dependent gene transcription and human serologic response. J Bacteriol. 2002;184:6316–6324. doi: 10.1128/JB.184.22.6316-6324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas I, Scott JR. Identification of rocA, a positive regulator of covR expression in the group A streptococcus. J Bacteriol. 2003;185:3081–3090. doi: 10.1128/JB.185.10.3081-3090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AA, Engleberg NC, DiRita VJ. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol Microbiol. 2001;40:976–990. doi: 10.1046/j.1365-2958.2001.02441.x. [DOI] [PubMed] [Google Scholar]
- Steiner K, Malke H. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol Microbiol. 2000;38:1005–1016. doi: 10.1046/j.1365-2958.2000.02203.x. [DOI] [PubMed] [Google Scholar]
- Titgemeyer F, Hillen W. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek. 2002;82:59–71. [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Podbielski A, Pohl B, Woischnik M, Korner C, Schmidt KH, Rozdzinski E, Leonard BA. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (opp) and its effect on cysteine protease production. Mol Microbiol. 1996;21:1087–1099. doi: 10.1046/j.1365-2958.1996.661421.x. [DOI] [PubMed] [Google Scholar]
- Janulczyk R, Ricci S, Bjorck L. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect Immun. 2003;71:2656–2664. doi: 10.1128/IAI.71.5.2656-2664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrager HM, Alberti S, Cywes C, Dougherty GJ, Wessels MR. Hyaluronic acid capsule modulates M protein-mediated adherence and acts as a ligand for attachment of group A streptococcus to CD44 on human keratinocytes. J Clin Invest. 1998;101:1708–1716. doi: 10.1172/JCI2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe NP, Kordari P, Cole R, Liu M, Palzkill T, Huang W, McLennan D, Adams GJ, Hu M, Vuopio-Varkila J, Cate TR, Pichichero ME, Edwards KM, Eskola J, Low DE, Musser JM. Human immune response to Streptococcal inhibitor of complement (Sic), a serotype M1 group A streptococcus extracellular protein involved in epidemic waves. J Infect Dis. 2000;182:1425–1436. doi: 10.1086/315882. [DOI] [PubMed] [Google Scholar]
- Batsford S, Brundiers M, Schweier O, Horbach E, Monting JS. Antibody to streptococcal cysteine proteinase as a seromarker of group A streptococcal (Streptococcus pyogenes) infections. Scand J Infect Dis. 2002;34:407–412. doi: 10.1080/00365540110080674. [DOI] [PubMed] [Google Scholar]
- Basma H, Norrby-Teglund A, Guedez Y, McGeer A, Low DE, El-Ahmedy O, Schwartz B, Kotb M. Risk factors in the pathogenesis of invasive group A streptococcal infections: role of protective humoral immunity. Infect Immun. 1999;67:1871–1877. doi: 10.1128/iai.67.4.1871-1877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade HD. The metabolism of amino acids by streptococci. McCarty M, editor. New York: Columbia University Press,; Streptococcal Infections. 1954 [Google Scholar]
- Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol. 2003;185:1911–1922. doi: 10.1128/JB.185.6.1911-1922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T, Sriskandan S, Yang L, Fraser JD. Superantigens and streptococcal toxic shock syndrome. Emerg Infect Dis. 2003;9:1211–1218. doi: 10.3201/eid0910.030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson BK, Andersson J, Holm SE, Norgren M. Invasive group A streptococcal infections: T1M1 isolates expressing pyrogenic exotoxins A and B in combination with selective lack of toxin-neutralizing antibodies are associated with increased risk of streptococcal toxic shock syndrome. J Infect Dis. 1999;180:410–418. doi: 10.1086/314872. [DOI] [PubMed] [Google Scholar]
- Kotb M, Norrby-Teglund A, McGeer A, Green K, Low DE. Association of human leukocyte antigen with outcomes of infectious diseases: the streptococcal experience. Scand J Infect Dis. 2003;35:665–669. doi: 10.1080/00365540310015962. [DOI] [PubMed] [Google Scholar]
- Kazmi SU, Kansal R, Aziz RK, Hooshdaran M, Norrby-Teglund A, Low DE, Halim AB, Kotb M. Reciprocal, temporal expression of SpeA and SpeB by invasive M1T1 group A streptococcal isolates in vivo. Infect Immun. 2001;69:4988–4995. doi: 10.1128/IAI.69.8.4988-4995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broudy TB, Pancholi V, Fischetti VA. The in vitro interaction of Streptococcus pyogenes with human pharyngeal cells induces a phage-encoded extracellular DNase. Infect Immun. 2002;70:2805–2811. doi: 10.1128/IAI.70.6.2805-2811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks DJ, Lei B, Musser JM. Prophage induction and expression of prophage-encoded virulence factors in group A streptococcus serotype M3 strain MGAS315. Infect Immun. 2003;71:7079–7086. doi: 10.1128/IAI.71.12.7079-7086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Kapur V, Kanjilal S, Shah U, Musher DM, Barg NL, Johnston KH, Schlievert PM, Henrichsen J, Gerlach D. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (Scarlet fever toxin). J Infect Dis. 1993;167:337–346. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- Herwald H, Cramer H, Morgelin M, Russell W, Sollenberg U, Norrby-Teglund A, Flodgaard H, Lindbom L, Bjorck L. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 2004;116:367–379. doi: 10.1016/s0092-8674(04)00057-1. [DOI] [PubMed] [Google Scholar]
- Gryllos I, Levin JC, Wessels MR. The CsrR/CsrS two-component system of group A streptococcus responds to environmental Mg2+. Proc Natl Acad Sci USA. 2003;100:4227–4232. doi: 10.1073/pnas.0636231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. Invasive M1T1 group A streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol. 2004;51:123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Voyich JM, Braughton KR, Sturdevant DE, Vuong C, Kobayashi SD, Porcella SF, Otto M, Musser JM, DeLeo FR. Engagement of the pathogen survival response used by group a streptococcus to avert destruction by innate host defense. J Immunol. 2004;173:1194–1201. doi: 10.4049/jimmunol.173.2.1194. [DOI] [PubMed] [Google Scholar]
- Dalton TL, Scott JR. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J Bacteriol. 2004;186:3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Malke H. relA-independent amino acid starvation response network of Streptococcus pyogenes. J Bacteriol. 2001;183:7354–7364. doi: 10.1128/JB.183.24.7354-7364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.