Abstract
In this study we addressed the role of the nuclear factor (NF)-κB1/p50 subunit in chronic injury of the liver by determining the inflammatory and fibrotic responses of nfκb1-null mice in an experimental model that mimics chronic liver disease. Mice received repeated hepatic injuries throughout 12 weeks by intraperitoneal injection of the hepatotoxin carbon tetrachloride. In response nfκb1−/− mice developed more severe neutrophilic inflammation and fibrosis compared to nfκb1+/+ mice. This phenotype was associated with elevated hepatic expression of tumor necrosis factor (TNF)-α, which was localized to regions of the liver associated with inflammation and fibrosis. Hepatic stellate cells are important regulators of hepatic inflammatory and fibrogenic events but normally do not express TNF-α. Hepatic stellate cells derived from nfκb1−/− mice expressed TNF-α promoter activity, mRNA, and protein. By contrast the expression of other NF-κB-responsive genes (ICAM1 and interleukin-6) was similar between nfκb1−/− and nfκb1+/+ cells. We provide experimental evidence that the inappropriate expression of TNF-α by nfκb1−/− cells is because of lack of a p50-dependent histone deacetylase 1 (HDAC1)-mediated repression of TNF-α gene transcription. Taken together these data indicate that the p50 NF-κB subunit plays a critical protective role in the injured liver by limiting the expression of TNF-α and its recruitment of inflammatory cells.
Nuclear factor (NF)-κB is a cardinal regulator of immune and inflammatory responses, controlling the expression of genes encoding cytokines, chemokines, adhesion molecules, and regulators of the cell cycle and apoptosis.1 Prolonged activation of NF-κB leads to perpetuated inflammatory responses and as such the transcription factor is a target for a range of anti-inflammatory drugs used to treat diseases including rheumatoid arthritis, asthma, and inflammatory bowel disease.2 NF-κB is a dimeric transcription factor generated from combinations of Rel (c-Rel), p65 (RelA), RelB, p50/p105 (NFκB1), and p52/p100 (NFκB2) all of which contain an evolutionary conserved Rel homology domain that includes the DNA-binding and dimerization motifs as well as the nuclear localization signal.1–3 The prototypic NF-κB consists of a dimer of p50 and p65 and is classically held in an inhibitory complex with IκBα.3 In response to stimulation, IκBα is phosphorylated via activation of the IKK complex and is subsequently polyubiquitinated and targeted for degradation by the 26S proteosome. This leads to release of the p50/p65 dimer that translocates to the nucleus to activate transcription of genes containing their DNA-binding motif, known as a κB-binding site.
This classical mechanism of activation and operation of NF-κB is however complicated by the fact there are at least 10 potential homodimeric and heterodimeric combinations of Rel factors. An additional level of complication is that there is emerging evidence from both in vitro and in vivo studies that the individual Rel proteins have distinct biological activities, however the precise physiological role of each protein is not fully understood.4 Of particular interest, transcription repression functions have been attributed to homodimers of p50 as well as to NF-κB heterodimers containing RelB.5–7 These repressive properties may serve to fine tune NF-κB-directed immune and inflammatory responses. Studies using mice carrying targeted disruptions of the Rel family genes are providing important evidence for distinct biological functions for these factors. Nfκb1−/− mice effectively lack two proteins, a 105-kd non-DNA-binding cytoplasmic protein (p105) and the p50 NF-κB subunit that is generated from p105 by proteolytic processing. Nfκb1−/− mice develop normally but have multifocal defects in immune responses and B cell function.8,9 Mice deficient in relB have a variety of pathological conditions including multiorgan inflammatory cell infiltrates, myeloid hyperplasia, and splenomegaly because of extramedullary hyperplasia.10 However, mice lacking both relB and nfκb1 spontaneously develop severe multiorgan inflammation, which suggests that these factors may operate in concert to limit inflammatory events.11
In this study we have investigated the role played by nfκb1 in the injury/wound-healing response by using a well-established model of iterative liver injury that mimics a repetitive inflammation-driven injury and wound-healing process. Importantly, this model of chronic liver disease significantly differs from the acute injury models that have previously been used to conclude lack of a role for p50 in liver disease.12,13 By comparing the response of nfκb1+/+ and nfκb1−/− to iterative injury with the hepatotoxin carbon tetrachloride (CCl4) we show that lack of p50 results in an exacerbated neutrophilic inflammatory response and development of severe fibrosis. We describe a mechanism underlying this previously undescribed phenotype of nfκb1 mice that involves the absence of a p50-dependent transcriptional repression of tumor necrosis factor (TNF)-α expression by the histone deacetylase HDAC1.
Materials and Methods
Nfkb1−/−/Nfkb1+/+ Mice and 12-Week CCl4 Injury Model
Experiments were performed on C57Bl/6;129PF2/J nfkb1−/− and F2 hybrid nfkb1+/+ mice. Because F2 hybrids carry random combinations of alleles derived from both the C57BL/6 and 129 genetic backgrounds they are more appropriate physiological controls than inbred mice of either parental strain (http://jaxmice.jax.org/info/hybrid.html). Breeding pairs were purchased from Jackson Laboratories (Bar Harbor, ME) and were housed in pathogen-free conditions. CCl4 and olive oil were purchased from Sigma Chemical Co., Dorset, UK. Age-matched male nfkb1−/− and nfkb1+/+ mice were treated with CCl4 at 1 μl [CCl4:olive oil (1:3 {v/v})/g body weight by intraperitoneal injection twice weekly for 12 weeks]. At days 1, 3, 5, 7, and 14 after the last CCl4 injection animals were killed by cervical dislocation and livers harvested. To determine age-associated differences in liver pathology mice were maintained in pathogen-free conditions for 12 months before harvesting of livers.
Cell Culture
Hepatic stellate cells (HSCs) were isolated from normal livers of nfkb1−/− and nfkb1+/+ male mice. Livers were washed in Hanks’ buffered saline minus calcium and then cut into small pieces and digested with collagenase and pronase. Enzymatic digestion was followed by discontinuous density centrifugation in 11.5% Optiprep (Life Technologies, UK). HSCs were cultured on plastic in Dulbecco’s modified Eagle’s medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mmol/L l-glutamine, and 16% fetal calf serum and maintained at 37°C at an atmosphere of 5% CO2. All experiments using HSC-derived myofibroblasts were performed on cells passaged between two and four times.
Plasmid DNA, Cell Transfections, and Reporter Assays
All plasmid DNA was prepared using a commercial DNA extraction and isolation kit (Maxiprep; Qiagen). Expression vectors for HDAC1 and p50 were gifts from Dr. Neil Perkins (University of Dundee, Dundee, Scotland) and Professor Ron Hay (University of St. Andrews, Fife, Scotland), respectively. Reporter constructs pxp-TNF-α-luc (referred to here as pTNF-α-Luc) was kindly provided by Dr. Peter Johnson (NCI, Frederick, MD). Culture-activated nfkb1−/− and nfkb1+/+ mouse HSCs were transfected by the nonliposomal Effectene protocol (Qiagen) using 1 μg of reporter plasmid DNA, ±2 μg of expression plasmid/control vector, and 10 ng of control Renilla plasmid pRLTK according to the manufacturer’s instructions. For experiments using trichostatin A (TSA), 24 hours after transfection, nfkb1−/− and nfkb1+/+ mouse HSCs were treated with TSA (500 nmol/L) for 24 hours and reporter gene activity assay performed. Luciferase assays were performed using a dual luciferase kit (Promega) according to the manufacturer’s instructions. Firefly luciferase activities were normalized for differences in transfection efficiency by measurement of the activity of co-transfected pRLTK.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis and Immunoblotting
Whole cell extracts were prepared, and protein concentration of samples was determined using a Bradford DC assay kit (Bio-Rad). Whole cell extracts (30 or 20 μg) from activated nfkb1−/− and nfkb1+/+ mouse HSCs, respectively, were then fractionated by electrophoresis through a 9% sodium dodecyl sulfate-polyacrylamide gel. Gels were run at a 100 V for 1.5 hours before transfer onto nitrocellulose. After blockade of nonspecific protein binding, nitrocellulose blots were incubated for 1 hour with primary antibodies diluted in Tris-buffered saline (TBS)/Tween 20 (0.075%) containing 3% Marvel. Mouse monoclonal horseradish peroxidase-conjugated antibody recognizing HDAC1 (Upstate Biotechnology, Lake Placid, NY) was used at 1 μg/ml. Mouse monoclonal antibodies directed against α-smooth muscle actin (SMA) (1:2000), desmin (1:2000), and β-actin (1:1000) were obtained from Sigma. Rabbit polyclonal directed against glial fibrillary acidic protein (1:1000) was obtained from Sigma. Rabbit polyclonal directed against p75 (1:2000) was obtained from Promega. After incubation with primary antibodies, blots were washed three times in TBS/Tween 20 before incubation for 1 hour in goat anti-mouse or mouse anti-rabbit horseradish peroxidase conjugate antibody at 1:2000 dilution in TBS/Tween 20 (0.075%) containing 3% Marvel. After extensive washing in TBS/Tween 20, the blots were processed with distilled water for detection of antigen using the enhanced chemiluminescence system (Amersham Biosciences).
Semiquantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was purified from isolated cells or frozen livers using the Total RNA purification kit (Qiagen, UK) following the manufacturer’s instructions and was used to generate first strand cDNA using a random hexamer primer [p(dN)6] and MMLV reverse transcriptase. Oligonucleotide primers for PCR amplification of murine β-actin, interleukin (IL)-6, p65, p50, p52, RelB, c-Rel, and IκBα have been previously published.11,14 Primers for mouse TNF-α 5′-cca acg gca tgg atc tca-3′ (sense) and 5′-tac ttg ggc aga ttg acc-3′ (anti-sense), for mouse ICAM 1, 5′-acc cca agg acc cca agg aga-3′ (sense) and 5′-aga gcg gca gag caa aag aag-3′ (anti-sense). Primers for mouse HDAC1 were 5′-cga gac ggg att gat gac-3′ (sense) and 5′-tcg tgt tct ggt tag tca-3′ (anti-sense). PCR reactions comprised of 1 μl of cDNA template, 100 ng each of sense and anti-sense oligonucleotide primers, 2.5 μl of optimized TaqPCR buffer (Promega), 0.4 mmol/L dNTP mixture, and 2 U of Taq polymerase in a total reaction volume of 25 μl. After an initial 5-minute incubation at 94°C, PCRs were performed using a 1-minute annealing step (TNF-α at 55°C, ICAM1 at 59.3°C, HDAC1 at 51.6°C), followed by a 2-minute elongation step at 72°C and a 45-second denaturation step at 94°C. Thirty-five PCR cycles were performed for amplification of all cDNAs, followed by a final elongation for 10 minutes at 72.0°C. PCR products were separated by electrophoresis through a 1% agarose gel and detected by ethidium bromide staining.
Quantitative TaqMan RT-PCR
18s rRNA TaqMan primers and probe were purchased from Applied Biosystems (UK) and the reaction was conducted according to the manufacturer’s instructions using 10 ng of cDNA for each reaction. Procollagen I mRNA expression was determined using forward primer 5′-ttc acc tac agc acg ctt gtg-3′, reverse primer 5′-gat gac tgt ctt gcc cca agt t-3′ and probe 5′-atg gct gca cga gtc aca-3′ quencher, TAMRA and fluorophore, FAM. For α-SMA forward primer 5′-tca gcg cct cca gtt cct-3′, reverse primer 5′-aaa aaa aac cac gag taa caa atc aa-3′ and probe 5′-tcc aaa tca ttc ctg ccc a-3′ quencher, TAMRA and fluorophore, FAM. For TIMP-1, forward primer 5′-gca tgg aca ttt att ctc cac tgt-3′, reverse primer 5′-tct cta gga gcc ccg atc tg-3′ and probe 5′-cag ccc ctg ccg cca tca-3′ quencher, TAMRA and fluorophore, FAM. Relative level of transcriptional difference between nfkb1−/− and nfkb1+/+ was calculated using the following equation: [1/(2A)] ×100, where A = average nfkb1+/+ ct − nfkb1−/− ct after the 18s RNA value had been deducted from the target gene for each animal. As the polymerase chain reaction follows logarithmic scale kinetics it is most accurate to measure the ct value in the exponential phase of the reaction. The ct (cycle threshold) value is calculated by measuring the cycle number at the threshold point where amplification (fluorescence) is at the start of the exponential phase of the reaction, ie, just above background fluorescence. Statistical analysis was performed using a paired t-test on both the nfkb1−/− and nfkb1+/+ using the following equation; 2(A). This equation allows statistical analysis on data before normalization but accounts for the logarithmic kinetics of the PCR amplification.
Histology and Immunohistochemistry
Sirius Red and hematoxylin and eosin staining was performed as previously described.15,16 For immunohistochemical staining for SMA-α, slides were dewaxed in xylene and dehydrated in alcohol. Antigen retrieval was achieved by microwaving in citric saline for 15 minutes. Endogenous peroxidase activity was blocked by hydrogen peroxide pretreatment for 10 minutes then further blocked using Vector Laboratories avidin/biotin blocking kit, 3 drops/section, with TBS washes between each stage. Slides were incubated with complete culture medium for 20 minutes, then in mouse anti-human α-SMA-fluorescein isothiocyanate (Sigma) diluted 1:30,000 in TBS, and applied to the slides and incubated overnight at 4°C. Slides were washed and rabbit anti-fluorescein isothiocyanate antibody (DAKO) diluted 1:4000 and incubated at room temperature for 30 minutes. Slides were then washed and incubated at room temperature for 30 minutes with a 1:200 dilution of biotin swine anti-rabbit (DAKO). Slides were washed in TBS and a streptavidin biotin-peroxidase complex at 1:200 dilution (DAKO) was incubated at room temperature for 30 minutes. Cells positive for α-SMA were visualized by diaminobenzidine (DAB) staining. Slides were counterstained with Mayer’s hematoxylin for 30 seconds, dehydrated, cleared in xylene, and mounted in DPX. Immunohistochemical staining for neutrophils was performed using the neutrophil antibody clone 7/4 (Serotec). Slides were initially processed in the manner described for α-SMA immunostaining. The primary antibody (Serotec, UK) was diluted 1:100 and incubated overnight at 4°C, secondary and anti-IgG horseradish peroxidase-conjugated tertiary antibodies were incubated for 20 minutes (Vector Laboratories, UK) and neutrophil recruitment was visualized by DAB staining. Immunohistochemical staining for TNF-α was performed using the goat anti-mouse TNF-α antibody (Serotec). Dewaxed and microwave-treated slides were initially blocked for endogenous peroxidase activity by hydrogen peroxide pretreatment for 10 minutes then further blocked using Vector Laboratories avidin/biotin blocking kit, 3 drops/section, with phosphate-buffered saline (PBS) washes between each stage. The primary antibody (Serotec, UK) was diluted 1:500 and incubated overnight at 4°C, secondary and anti-IgG horseradish peroxidase-conjugated tertiary antibodies were incubated for 20 minutes (Vector Laboratories, UK) and TNF-α was visualized by DAB staining. Slides were counterstained with Mayer’s hematoxylin for 30 seconds, dehydrated, and mounted in DPX. For TNF-α and α-SMA dual immunostaining the methods are as described above for the individual antibody stains with the exception of an additional serum-blocking step between the DAB visualization of α-SMA and the incubation of goat anti-mouse TNF-α antibody. To distinguish between TNF-α and α-SMA DAB plus nickel (black) was used to visualize TNF-α.
Cell Counts and Pathology Scoring
All cell counts were performed in a blinded manner and were representative of an average number of positive cells/field counted in 20 high-power fields from four to five mice per time point of recovery. Statistical differences between nfκb1−/− and nfκb1+/+ cell counts were calculated using a two-tailed unpaired t-test, assuming equal variation in SD between the two populations using Graph Pad Prism computer software. Histopathological analysis and grading of liver sections for degree of fibrosis and inflammation was performed by an expert pathologist (H.M.-S.) in a blinded manner.
TNF-α Enzyme-Linked Immunosorbent Assay (ELISA)
Livers or HSCs were lysed in Dignam buffer A supplemented with a cocktail of protease and phosphatase inhibitors [300 nmol/L 4-(2-aminoethyl) benzenesulfonyl fluoride, 0.2 mmol/L ethylenediaminetetraacetic acid, 250 μg/ml leupeptin, 250 μg/ml pepstatin A, 10 mmol/L aprotinin, 10 mmol/L NaV, and 10 mmol/L NaF]. Lysates were passed through a 19- and 21-gauge needle and then through a Qiashredder that was centrifuged for 2 minute at 13,000 rpm. Whole cell extracts were transferred to fresh Eppendorf tubes, and their protein content was determined using the Bradford DC assay kit (Bio-Rad). A 96-well ELISA plate was coated with capture antibody; rat monoclonal anti-mouse TNF-α (Biosource) at 4 μm/ml in diluted in PBS and incubated overnight at 4°C. Wells were washed three times in wash buffer (0.5% Tween in PBS), then incubated at room temperature for 1 hour with blocking buffer (1% fetal calf serum, 5% sucrose in PBS). Wells were washed as before and 100 μl of standard (0 to 250 ng/ml of recombinant mouse TNF-α; Cambridge Bioscience) or sample (10 to 25 mg whole cell or whole liver extract) was added to each well and incubated at room temperature for 2 hours. Wells were washed as before and 100 μl of biotinylated rabbit anti-mouse TNF-α polyclonal antibody (Peprotech EC Ltd.) at 100 ng/ml was added and incubated at room temperature for 2 hours followed by further washes and incubation at room temperature for 20 minutes with 0.2 μg/ml of streptavidin-horseradish peroxidase (Biosource). After final washing, 100 μl of substrate solution (1:1 mix H2O2 and 40 μmol/L tetramethylbenzidine diluted in 0.1 mol/L sodium citrate buffer) was added to each well and incubated for 20 minutes at room temperature. Fifty μl of stop solution (1 mol/L sulfuric acid) and was then added to each well and the optical density of reactions was measured at 405 nm and the amount of TNF-α in samples was determined using the standard curve.
Results
Severe Injury-Induced Hepatic Inflammation in Nfκb1−/− Mice
A histological comparison revealed mild to severe spontaneous inflammatory changes in the livers of nfκb1−/− mice that were absent in nfκb1+/+ mice (Figure 1 and Table 1). However these changes were not progressive or associated with a disease state because the severity of inflammation was not altered with aging (comparing 12-week-old to 1-year-old mice) and we were unable to detect any signs of fibrosis. The discovery of a reproducible inflammatory phenotype in the liver of nfκb1−/− mice suggested that the animals may be susceptible to a more severe response to chronic injury of the liver. To test this idea we compared the effects of chronic chemical injury of the liver between nfκb1−/− and nfκb1+/+ mice. Chronic injury of the rodent liver by repeated administration of CCl4 causes an inflammation-driven wound-healing response and fibrosis.15 Cessation of injury leads to spontaneous recovery and resolution of fibrosis.15,16 The responses of nfκb1−/− and nfκb1+/+ mice to CCl4 injury throughout 12 weeks and their subsequent spontaneous recovery were evaluated. Firstly, we confirmed a differential expression of p50 between nfκb1−/− and nfκb1+/+ livers and also determined the expression of RelB and p52 because these factors can have overlapping functions with p50.4,11 Both p50 and RelB transcripts were expressed in wild-type sham (olive oil)-injured animals, however p52 mRNA was undetectable (Figure 2). As expected p50 transcript was not detected in nfκb1−/− livers, whereas levels of RelB and absence of p52 was similar to wild type. Interestingly, although injury and recovery had no effect on expression of p50, a dramatic loss of RelB expression and weak induction of p52 was observed in both nfκb1−/− and nfκb1+/+ livers. In the case of RelB, expression was replenished during recovery (days 5 and 7) while there appeared to be a consistent but low-level induction of p52. The main conclusion from these observations is that CCl4-injured nfκb1−/− livers lack expression of both p50 and RelB which have been described to have compensatory regulatory functions in inflammatory responses.11
Figure 1.
Stable inflammatory changes in nfκb1−/− liver. H&E staining of liver sections from 12-week-old and 1-year-old nfκb1+/+ and nfκb1−/− mice (numbering refers to precise animal from which liver section was derived as given in Table 1). Black arrows denote minor or major inflammatory changes in nfκb1−/− liver. Photomicrographs are representative of five nfκb1+/+ and four 12-week-old or five 1-year-old nfκb1−/− mice. Original magnifications, ×200.
Table 1.
Stable Inflammatory Changes Associated with Lack of nfκb1
| Mouse genotype | Inflammatory change
|
|
|---|---|---|
| 12 weeks | 1 year | |
| nfκ b1−/− 1 | Major change | Major change |
| nfκ b1−/− 2 | Minor change | Major change |
| nfκ b1−/− 3 | Minor change | Minor change |
| nfκ b1−/− 4 | Minor change | Minor change |
| nfκ b1−/− 5 | Minor change | |
| nfκ b1+/+ 1 | Normal | Normal |
| nfκ b1+/+ 2 | Normal | Normal |
| nfκ b1+/+ 3 | Normal | Normal |
| nfκ b1+/+ 4 | Normal | Normal |
| nfκ b1+/+ 5 | Normal | Normal |
Five (1 to 5) nfκb1−/− and five (1 to 5) nfκb1+/+ mice were aged for 1 year and liver pathology (normal, minor, or major inflammatory changes) compared with 12-week-old mice.
Figure 2.
Expression of hepatic p50, RelB, and p52 during injury and recovery. RT-PCR for p50, RelB, and p52 transcripts was performed using RNA isolated from nfκb1+/+ and nfκb1−/− livers at days 1, 5, and 7 of recovery after 12 weeks of carbon tetrachloride injury or olive oil (sham). The gels shown are at least two experimental repeats from a minimum of three animals/treatment group/genotype. A 1-kb DNA ladder (not shown) was run alongside the PCR products to confirm correct sizes of the amplified cDNA fragments.
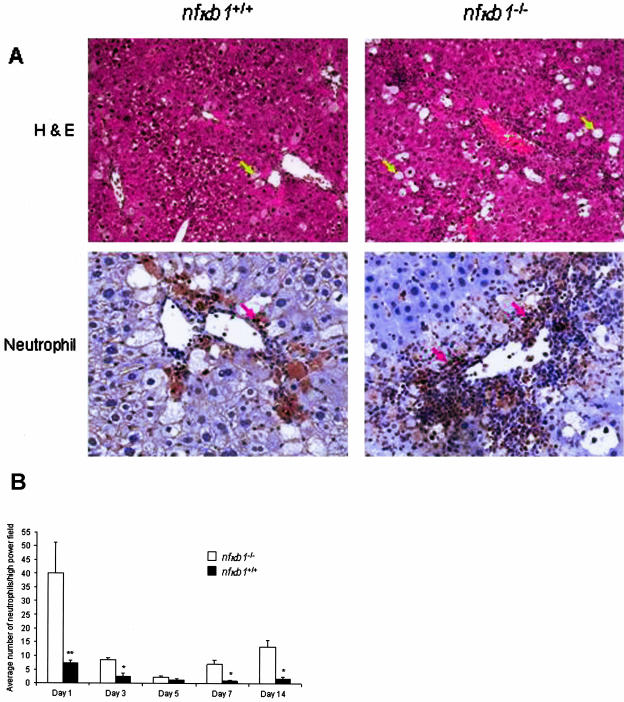
We next compared histological parameters of liver injury and disease. The most obvious and dramatic difference was a severe periportal/venular inflammatory infiltrate present in the livers of injured nfκb1−/− mice that was not seen in injured nfκb1+/+ mice (Figure 3A). Histological analysis showed that the cellularity of the infiltrate in nfκb1−/− livers was mainly neutrophilic. By counting numbers of stained neutrophils we found that at peak injury (day 1) there were a total of 40 (±11 SEM) neutrophils/field in nfκb1−/− livers compared with 7 (±1 SEM) neutrophils/field in nfκb1+/+ livers (Figure 3B). Although numbers of recruited neutrophils dropped by day 3 after injury in both nfκb1+/+ and nfκb1−/− livers there remained an elevated neutrophil count in the latter until at least day 14 after injury. By contrast, numbers of hepatic T cells and macrophages were not significantly different between the two genotypes (data not shown). These data indicate that a severe and persistent neutrophilic inflammatory response to injury occurs in the absence of nfκb1. In addition, we observed higher numbers of ballooned hepatocytes in nfκb1−/−-injured livers indicative of increased hepatocyte damage (Figure 3A). Since a previous study reported no significant differences in the acute response to CCl4-induced liver injury,12 the greater degree of hepatocyte damage in nfκb1−/− livers cannot simply be explained by inherent differences in CCl4 metabolism or the mechanism of injury between nfκb1−/− and nfκb1+/+ animals.
Figure 3.
Severe inflammation in chronically injured nfκb1−/− livers. A: H&E (top left and right, yellow arrows denote ballooned hepatocytes) and neutrophil immunostaining (bottom left and right, red arrows denote neutrophils) staining on nfκb1+/+ and nfκb1−/− liver sections at day 1 of recovery after 12 weeks of CCl4 injury. Photomicrographs are representative of five nfκb1+/+ and four nfκb1−/− mice. B: Numbers of neutrophils were counted in 15 high-power fields and expressed as average number of neutrophils/high-power field ± SEM. Day 1, P = 0.0016; day 3, P = 0.0185; day 5, not significant; day 7, P = 0.0039; and day 14, P = 0.0117; P values calculated using a paired t-test. Original magnifications, ×100.
Greater Numbers of Activated Hepatic Stellate Cells and Fibrosis in Injured Nfκb1−/− Mice
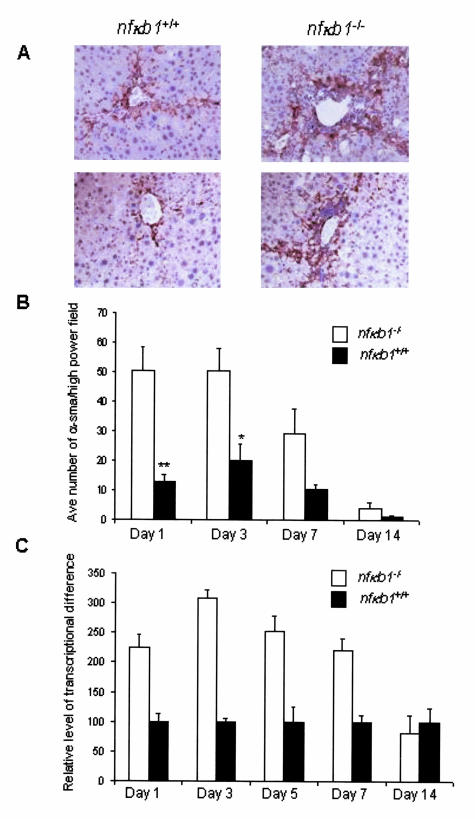
CCl4-induced chronic liver injury leads to a fibrogenic response characterized by trans-differentiation (or activation) of quiescent hepatic stellate cells (HSCs) into α-SMA-positive, proliferating myofibroblast-like cells.16 The increased synthesis and secretion of collagen I/III and tissue inhibitor of metalloproteinase 1 (TIMP-1) by these cells leads to a net deposition of fibril-forming collagens and eventually liver fibrosis and cirrhosis.17 Examination of the extent of fibrosis in nfκb1−/− and nfκb1+/+ livers of 12-week CCl4-injured mice was performed by histopathological analysis and grading of liver sections stained with Sirius Red that stains collagen I/III containing fibers (Figure 4A). This analysis showed a significantly worse fibrotic response in nfκb1−/− mice evident at both days 1 and 7 after injury (Figure 4, A and B). Underlying the more severe fibrosis in nfκb1−/− mice was elevated hepatic expression of transcripts for procollagen α1 (I) and TIMP-1 (Figure 4, C and D). Furthermore, numbers of profibrogenic α-SMA-positive cells were dramatically elevated in nfκb1−/− livers both at peak injury and throughout the recovery process consistent with increased fibrosis in these animals (Figure 5, A and B). In support of these findings, we observed a trend toward elevated levels of transcript for α-SMA in nfκb1−/− livers at peak injury and up to day 7 of recovery although this effect did not reach statistical significance (Figure 5C).
Figure 4.
Liver fibrosis is worse in nfκb1−/− mice. A: Sirius red staining of liver sections at days 1 and 7 of recovery after 12 weeks of CCl4 injury. Black arrows denote collagen fibers and are representative of five nfκb1+/+ and four nfκb1−/− mice. B: Fibrosis scores expressed as average grade ± SEM of five nfκb1+/+ and four nfκb1−/− mice at days 1 and 7 of recovery. C: TaqMan analysis of procollagen I mRNA measured in 20 ng of cDNA from whole liver extracts at days 1, 3, 5, 7, and 14 after 12 weeks of CCl4 injury. The relative level of transcriptional difference was calculated and expressed as an average ± SEM from four nfκb1−/− and five nfκb1+/+ animals. Average cycle numbers were: day 1, 24.60 and 27.21; day 3, 25.63 and 27.32; day 5, 26.33 and 27.10; day 7, 27.0 and 27.87; and day 14, 31.87 and 29.88 for nfκb1−/− and nfκb1+/+, respectively. The differences were statistically significant, P = 0.0329, and were calculated using a paired t-test. D: TaqMan analysis of TIMP1 mRNA measured in 20 ng of cDNA from whole liver extracts. The relative level of transcriptional difference was calculated and expressed as an average ± SEM from four nfκb1−/− and five nfκb1+/+ animals. Average cycle numbers were: day 1, 31.42 and 33.47; day 3, 34.09 and 35.15; day 5, 34.19 and 35.14; day 7, 34.27 and 34.23; and day 14, 36.68 and 36.09 for nfκb1−/− and nfκb1+/+, respectively. The differences were statistically significant, P = 0.0302, and were calculated using a paired t-test. Original magnifications, ×100.
Figure 5.
Elevated numbers of wound-healing hepatic myofibroblasts in nfκb1−/− livers. A: Hepatic α-SMA immunostaining at days 1 and 7 after injury. Photomicrographs are representative of four nfκb1−/− and five nfκb1+/+ mice at each time point. B: Number of α-SMA-positive cells were counted in 15 high-power fields and expressed as average number of α-SMA-positive cells/high-power field ± SEM from four nfκb1−/− and five nfκb1+/+ animals at each time point. Day 1, P = 0.0017; day 3, P = 0.035; day 7, P = 0.048; and day 14 is not significant; P values calculated using a paired t-test. C: TaqMan quantification of α-SMA mRNA measured in 20 ng of cDNA from whole liver at days 1, 3, 5, 7, and 14 of recovery. The relative level of transcriptional difference was expressed as an average ± SEM from four nfκb1−/− and five nfκb1+/+ animals per time point. Average cycle numbers were: day 1, 32.42 and 33.57; day 3, 33.66 and 35.16; day 7, 35.29 and 36.69; day 14, 35.62 and 37.08 for nfκb1−/− and nfκb1+/+, respectively. However, differences were not found to be statistically significant, P = 0.16, using the paired t-test. Original magnifications, ×200.
Selective Elevated Expression of TNF-α by Activated Nfκb1−/− HSCs
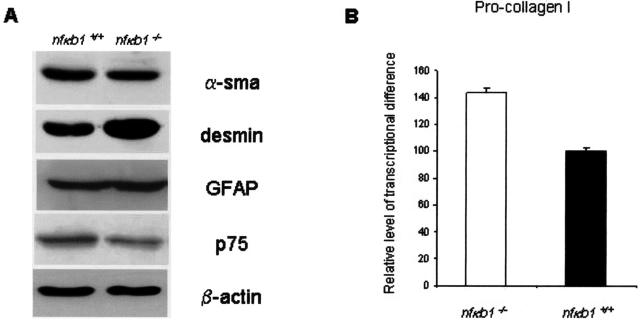
The more pronounced inflammatory and fibrogenic responses of nfκb1−/− mice to chronic injury raised the possibility that the phenotype of HSCs may be attenuated in these animals. HSCs were therefore isolated from uninjured mouse livers and then activated to a myofibroblast-like phenotype by culturing on plastic, this being a widely accepted and used model of HSC activation.17 Expression of the p75 low-affinity nerve growth factor receptor and high levels of GFAP expression in the cultured myofibroblasts confirmed that the cells originally isolated were of HSC origin.18 We did not observe any significant differences in the rate of culture growth or expression of classic markers (α-SMA and desmin) of activation between nfκb1−/− and nfκb1+/+ HSCs (Figure 6A). A weak, although not statistically significant, increase in procollagen α1 (I) mRNA expression was observed in nfκb1−/− HSCs (Figure 6B). HSC activation is accompanied by reprogramming of their constitutive NF-κB activity to persistently elevated levels by a mechanism involving CBF-1-mediated repression of IκB-α that results in the induction of NF-κB-dependent genes such as ICAM-1 and IL-6.19–22 As shown in Figure 7A, the expression of ICAM-1 and IL-6 was similar between nfκb1−/− and nfκb1+/+ HSCs indicating no general attenuation of NF-κB-directed gene expression. By contrast, levels of transcript for TNF-α were dramatically higher in nfκb1−/− HSCs. To confirm that nfκb1−/− HSCs produced elevated levels of TNF-α we additionally showed by ELISA that these cells produce 10-fold higher amounts of the cytokine than nfκb1+/+ HSCs (Figure 7B). If this elevated production of TNF-α by HSCs results in a general increase in hepatic expression of the cytokine it would be of relevance to the pathology observed in nfκb1−/− mice. We therefore compared TNF-α expression and found both the level of mRNA (Figure 8A) and protein (Figure 8B) to be elevated in injured nfκb1−/− compared to nfκb1+/+ livers. As shown in Figure 8C, immunohistochemical analysis for TNF-α in nfκb1−/− livers demonstrated that expression was mainly located in cells surrounding the inflammatory infiltrate and was of a similar pattern to that for α-SMA staining (Figure 5A). Dual staining for α-SMA and TNF-α confirmed in situ expression of TNF-α by nfκb1−/− myofibroblasts (Figure 8D). This supports the conclusion of the in vitro studies that nfκb1−/− hepatic myofibroblasts are one source of the elevated levels of TNF-α. These are highly relevant findings because TNF-α is proinflammatory, functions as a recruitment factor for neutrophils to the liver, and has hepatotoxic properties.23,24
Figure 6.
Lack of differences in markers of activation between nfκb1−/− and nfκb1+/+ HSCs. A: Whole cell extracts were isolated from culture-activated HSCs and 25 μg were used to immunoblot for α-SMA, desmin, p75, glial fibrillary acid protein (GFAP), and β-actin. All gels were representative of at least two independent experiments. B: TaqMan analysis of procollagen I mRNA measured in 20 ng of cDNA from activated HSCs. The relative level of transcriptional difference was calculated and expressed as an average ± SEM from three independent cell preparations. Average cycle numbers were 18.45 and 20.55 for collagen and 18.68 and 20.45 for 18s RNA in nfκb1−/− and nfκb1+/+, respectively. Use of paired t-test revealed difference not to be significant (P = 0.4).
Figure 7.
Selective overexpression of TNF-α by nfκb1−/− HSCs. A: Three nfκb1−/− and three nfκb1+/+ mice were used to generate six independent lines of activated HSCs (−/−1 to 3 and +/+ 1 to 3). Total RNA was isolated from all six lines and used as a template in RT-PCR reactions for detection of ICAM-1, IL-6, TNF-α, and β-actin transcripts using protocols described in the Materials and Methods. The gels shown are presented as three sets of genotype pairs and are representative of two repeat RT-PCRs for each RNA sample. A 1-kb DNA ladder was run alongside the PCR products to confirm correct sizes of the amplified cDNA fragments (not shown). B: Whole cell extracts were isolated from nfκb1−/−- and nfκb1+/+-activated HSCs and sandwich ELISA was used to quantify TNF-α levels as average ng of TNF-α/μg whole cell extract ± SEM, n = 3 nfκb1−/− and nfκb1+/+-independent isolations.
Figure 8.
Elevated TNF-α in chronic injured nfκb1−/− liver. A: Total RNA was isolated from the livers of three nfκb1−/− and three nfκb1+/+ mice at day 1 after 12 weeks after injury and were used as templates in RT-PCR reactions for TNF-α and β-actin transcripts. The gels show RT-PCR products for the three livers/genotype and were representative of two repeat RT-PCRs for each RNA sample. B: Whole liver protein extracts were at day 1 after injury and sandwich ELISA was used to quantify TNF-α (as average ng of TNF-α/μg whole liver protein ± SEM, n = 4 nfκb1−/− and 5 nfκb1+/+). C: TNF-α immunostaining on liver sections at day1 after injury, arrows denote TNF-α-expressing cells mainly localized to regions surrounding inflammatory infiltrates in nfκb1−/− livers. Photomicrographs are representative of four nfκb1−/− and five nfκb1+/+ animals. D: TNF-α and α-SMA dual immunostaining on nfκb1−/− liver sections at day1 after injury, yellow arrows denote TNF-α and α-SMA co-localization. Original magnifications: ×100 (C, top); ×200 (C, bottom); ×1000 (D).
Repression of TNF-α Gene Transcription by p50 and HDAC1
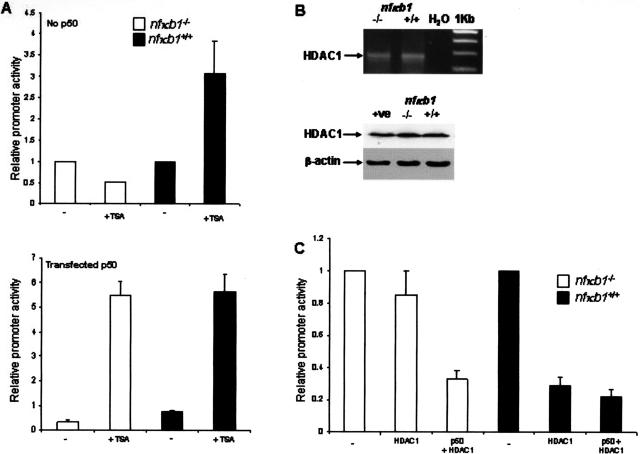
We next investigated the mechanism by which absence of the nfκb1 gene leads to elevation of TNF-α expression by HSC-derived myofibroblasts and wanted to confirm that absence of p50 is mechanistically the critical factor. A murine TNF-α promoter-luciferase construct (pTNF-α-Luc) was transfected into nfκb1−/−- and nfκb1+/+-activated HSCs either alone or together with an expression vector for p50. The TNF-α promoter was 10-fold more active in nfκb1−/− than nfκb1+/+ HSCs (Figure 9A) and activity of the promoter in both cell types was suppressed by co-transfection of p50 (Figure 9B). Hence, p50 functions in wild-type cells to suppress TNF-α gene transcription, we therefore examined the mechanism underlying this function. It has recently emerged that homodimeric p50 can associate with the transcriptional co-repressor histone deacetylase 1 (HDAC1) and that this complex is stable when p50 homodimers bind to κB DNA-binding sites.25 It is proposed that recruitment of p50-HDAC1 complexes to NF-κB-responsive genes results in deacetylation of histones and modifies local chromatin structure into a condensed and transcriptionally repressed state.25 To determine whether there is a role for deacetylases in p50-mediated repression of TNF-α promoter activity we initially treated pTNF-α-Luc-transfected nfκb1−/− and nfκb1+/+ HSCs with the broad specific HDAC inhibitor TSA. Figure 10A shows that TSA stimulated TNF-α promoter activity by threefold in nfκb1+/+ HSCs but had no effect on activity of the TNF-α promoter in nfκb1−/− HSCs. However, nfκb1−/− HSCs transfected with a p50 expression vector responded to TSA treatment with a 10-fold stimulation of TNF-α promoter activity. Ability of the TNF-α promoter to respond to TSA therefore critically depends on expression of p50, this observation implicates p50 as an essential component of HDAC-mediated repression of TNF-α expression. We next investigated a specific role for HDAC1. Firstly we showed that both nfκb1−/− and nfkb1+/+ HSCs express HDAC1 (Figure 10B), hence the differences in TSA responses for the TNF-α promoter between the two genotypes cannot be simply explained by differences in HDAC1 expression. Overexpression of HDAC1 in nfκb1+/+ repressed TNF-α promoter activity, by contrast this effect was not observed in nfκb1−/− HSCs (Figure 10C). However, transfection of p50 into nfκb1−/− HSCs rescued the ability of exogenous HDAC1 to repress TNF-α promoter activity. We therefore conclude that HDAC1 is a repressor of TNF-α gene transcription that requires p50 to perform this function. We suggest that in wild-type cells p50 homodimers act to recruit HDAC1 to the TNF-α promoter and that absence of p50 in nfκb1−/− cells disables the repressive function of HDAC1 resulting in aberrant overexpression of TNF-α. This provides a molecular mechanism to explain the elevated expression of TNF-α and severe neutrophilic inflammation in injured nfκb1−/− livers.
Figure 9.
TNF-α promoter activity is increased in nfκb1−/− HSCs. A: Activated HSCs isolated from nfκb1−/− and nfκb1+/+ mice were transfected with 10 ng of pRLTK (Renilla control) and 1 μg of pTNF-α-Luc. After 48 hours the luciferase activities were determined, normalized to pRLTK, and promoter activity in nfκb1−/− cells expressed as promoter activity relative to the activity measured in nfκb1+/+ cells (mean ± SE of triplicate transfections) B: HSCs were transfected with 10 ng of pRLTK, 1 μg of pTNF-α-Luc, and 2 μg of pCDNA3-p50 (+p50) or pCDNA3 (−). After 48 hours, luciferase activities were determined, normalized to pRLTK activity, and expressed as promoter activity relative to the activity measured in pCDNA3-transfected cells (mean ± SE of triplicate transfections).
Figure 10.
HDAC1-mediated repression of TNF-α transcription is p50-dependent. A (top): Activated HSCs were transfected with 10 ng of pRLTK and 1 μg of pTNF-α-Luc for 24 hours before 24 hours of treatment with or without 500 nmol/L TSA. Luciferase activities were determined, normalized to pRLTK activity, and expressed as promoter activities relative to activity measured in cells lacking TSA treatment. A (bottom): As above but with the inclusion of pCDNA3p50 in the transfection mixture. All transfections were performed in triplicate. B (top): Total RNA was isolated from nfκb1−/− and nfκb1+/+ HSCs and used as a template for RT-PCR detection of HDAC1. B (bottom): Whole cell extracts were isolated from nfκb1−/−- and nfκb1+/+-activated HSCs and 25 μg of protein was used to immunoblot for HDAC1 and β-actin. C: Activated HSCs were transfected with 10 ng of pRLTK and 0.5 μg of pTNF-α-Luc alone (−) or together with 1 μg of pCDNA3 and 2 μg of pHDAC1 (HDAC1) or 1 μg of pCDNA3-p50 and 2 μg of pHDAC1 (p50 + HDAC1). After 48 hours, luciferase activities were determined, normalized to pRLTK activity, and expressed as promoter activities relative to activity in cells transfected with pTNF-α-Luc alone (mean ± SE of triplicate transfections).
Discussion
Now that there is unequivocal and direct evidence that NF-κB drives inflammation,26 there is a need to understand the specific physiological role played by each individual Rel protein in the inflammatory response. In addition, the emerging function of transcriptional co-activators and co-repressors as regulators of Rel activities27 necessitate studies that determine how these factors interplay with each Rel protein to achieve appropriate physiological responses of tissues to stress, injury, and infections. The data presented in this study define a protective role for p50 that operates to limit the inflammatory and fibrogenic responses in the chronically injured liver. We further define a transcriptional mechanism to explain this anti-inflammatory property of p50, which involves HDAC-dependent repression of TNF-α gene transcription.
The nfκb1−/− mouse has previously been used in attempts to determine a function for p50 in acute liver injury and regeneration, however these studies concluded a lack of a significant role in either process.12,13 The fundamental difference between these earlier studies and the work presented here is our use of a model that addresses the potential for p50 to influence the physiological response to iterative injury. The chronic CCl4-injury model generates repeated and overlapping rounds of injury, inflammation, and wound-healing processes that mimic the pathological events underlying the development of fibrosis of the chronically diseased liver. Using this model we show that absence of nfκb1 is associated with more profound hepatic inflammation and development of a more severe fibrogenic response. Of note nfκb1−/− mice are also susceptible to more severe lung inflammation and damage when challenged by pulmonary infection with Escherichia coli.28 Taken together these observations indicate that p50 is an important negative regulator of tissue inflammation in multiple organs responding to injury and infections. It was therefore of interest to determine a mechanism through which p50 may exert its protective anti-inflammatory mode of action.
The most dramatic difference between CCl4-injured nfκb1−/− and nfκb1+/+ mice was the massive accumulation of neutrophils in periportal/venular regions of nfκb1−/− livers. Neutrophils accumulate in and adhere to sinusoids and postsinusoidal venules during hepatic inflammation.29 Neutrophils have been strongly implicated in the development of liver damage and HSC activation via their production of reactive oxygen species and various hydrolytic enzymes.24,29,30 Disease states in which neutrophil-induced liver damage is associated include hepatic ischemic-reperfusion, alcoholic hepatitis, and drug-induced liver injuries.31 Again of relevance to our findings the inflammation generated in the lungs of E. coli-infected nfκb1−/− mice mainly consisted of neutrophils suggesting that absence of p50 renders tissues more susceptible to neutrophil infiltration.28 One of the most potent mediators of the hepatic accumulation and activation of neutrophils is TNF-α.32 The livers of CCl4-injured nfκb1−/− mice expressed higher levels of TNF-α than injured nfκb+/+ mice and immunohistochemical analysis showed that TNF-α expression was mainly associated with α-SMA-positive cells surrounding inflammatory infiltrates. There are currently believed to be at least two main sources of α-SMA-positive myofibroblasts in the injured liver, these being the periportal myofibroblast and the activated HSCs.18 Importantly, both cell types have a similar profibrogenic and proinflammatory phenotype and if their persistence in the chronically injured liver leads to progressive deposition of cross-linked collagen and ultimately hepatic fibrosis. By isolating HSCs and subsequently activating them in culture we were able to show that although there were no gross phenotypic differences between nfκb1−/− and nfκb1+/+ HSCs, the former cell type produced highly elevated levels of TNF-α. There have been no previous reports of HSCs expressing TNF-α and we measured only low barely detectable levels of the cytokine in nfκb1+/+ HSCs. We therefore propose that aberrant production of TNF-α by myofibroblasts probably of both periportal and perisinusoidal (HSCs) origin provides an explanation for the severe neutrophilic inflammatory infiltrate observed in injured nfκb1−/− mice.
There is both clinical and experimental evidence in support for a pathophysiological role for TNF-α in liver fibrosis. Elevated serum TNF-α concentrations in patients with alcoholic hepatitis have by been reported by several groups and the values correlate with severity of cirrhosis and mortality.33 Similarly, there is a strong correlation between level of serum TNF-α and development of periportal fibrosis in human Schistosoma mansoni infection, with higher concentrations of TNF-α being associated with a higher risk of fibrosis.34 Liver fibrosis resulting from chronic CCl4 exposure is almost completely absent by histological analysis in TNF receptor-deficient mice and is accompanied by marked reductions in hepatic collagen and transforming growth factor-β expression.35 Furthermore, neutralizing antibodies to TNF-α prevent the development of rodent liver fibrosis induced by concanavalin A.36 However, it is not yet clear as to whether TNF-α has a direct influence on the fibrogenic process and given its highly pleiotropic function in the liver it is more likely that it exerts its influence indirectly via enhancing the inflammatory response to liver damage.
TNF-α expression is tightly regulated at the transcriptional level and is under the repressive influence of two NF-κB proteins, p50 and RelB.5–7 Knockout mouse studies show that p50 and RelB have overlapping compensatory functions with respect to inflammation.11 Although RelB−/− mice spontaneously develop a mild inflammatory infiltrate in several organs, a combined deficiency of p50 and RelB leads to a severe multiorgan inflammatory phenotype involving recruitment of T cells, macrophages, and neutrophils. Of note in the current study we observed a loss of hepatic RelB at peak injury that was replenished on recovery, the physiological significance of this injury-induced attenuation of RelB is currently not understood. However, it is significant that peak liver injury in nfκb1−/− mice was effectively associated with a combined deficiency of p50 and RelB. RelB−/− fibroblasts overexpress TNF-α in response to stimulation with LPS although the precise mechanism by which RelB negatively regulates TNF-α gene transcription remains poorly defined.6,7 The repressive function of p50 on TNF-α is also incompletely described although it is known that the distal region of both the human and murine TNF-α promoters carry κB sites that preferentially associate with p50 homodimers.5,37 Murine macrophages have been shown to secrete a TNF-α-inhibitory factor that stimulates their enhanced expression of p50 and repression of TNF-α promoter activity.7 This effect and the ability of transfected p50 to repress TNF-α promoter activity was lost if three upstream κB sites with preference for p50 were removed from the promoter. Our study not only provides a physiological explanation for the requirement for tight inhibitory regulation of the TNF-α locus by p50 (ie, the protection of tissues against excessive inflammation and damage resulting from overexpression of TNF-α) but also describes how p50 mediates this repression. The murine TNF-α promoter not only displayed highly elevated activity in nfκb1−/− HSCs, it was also unresponsive to the stimulatory effects of TSA and the repressive effects of HDAC1. Importantly we demonstrated that ectopic expression of p50 in nfκb1−/− HSCs repressed activity of the TNF-α promoter and restored the sensitivity to TSA and HDAC1 observed in wild-type HSCs. Overexpression of TNF-α by nfκb1−/− HSCs is therefore because of absence of p50 (rather than a function attributed to the precursor protein p105) and specifically arises as a consequence of the loss of p50-dependent HDAC-mediated transcriptional repression of the TNF-α gene.
Zhong and colleagues25 described how a complex of homodimeric p50 and HDAC1 binds to κB sites and functions as a general repressor of NF-κB-dependent transcription in resting cells. On cell stimulation, the phosphorylated p65 subunit of newly formed nuclear p50/p65 heterodimers associates with CBP/p300 and this complex then replaces the repressive p50/p50-HDAC1 complex at κB sites to activate transcription. Interestingly we did not observe a general difference in the expression of NF-κB-responsive genes between nfκb1−/− and nfκb+/+ HSCs, including IL-6 that Zhong and colleagues25 showed to be a target for p50/p50-HDAC1-mediated repression in resting cells. The most obvious explanation for this difference between the two studies is that the HSC-derived myofibroblasts used in the present study are in an activated state in which the majority of NF-κB-responsive genes will be under the influence of p50/p65-CBP complexes. This idea is supported by the fact that p50/p65 dimers are the major κB-binding proteins detected by electrophoretic mobility shift assay in activated HSCs.20–22 However, genes such as TNF-α that are under the control of κB sites with high-binding affinity for p50 homodimers will preferentially associate with p50/p50-HDAC1 complexes over p50/p65-CBP complexes. This is supported by experimental evidence that TNF-α transcription is highly sensitive to the repressive effects of p50 even when in the presence of high levels of p65.5 Our findings therefore complement and advance those of Zhong and colleagues25 by showing that p50/p50-HDAC1 complexes remain functional in the stimulated cell to enable the selective repression of a subset of NF-κB-responsive genes. Additional and possibly signal- or/and cell-dependent regulatory mechanisms are presumably then required to overcome p50/p50-HDAC1 repression for appropriate induction of TNF-α. The severity of tissue inflammation and damage observed in the livers of the CCl4-injured nfκb1−/− mice serves to illustrate how this complexity of transcriptional regulation operates to prevent the pathological consequences of inappropriate overexpression of the cytokine. An intriguing observation was that treatment of p50-transfected nfκb1−/− HSCs with TSA resulted in the powerful activation of the TNF-α promoter indicating that the deacetylase inhibitor converts p50 from a repressor to an activator of TNF-α transcription (Figure 10A). One way in which p50/p50 dimers can function as transcriptional activators is by associating with Bcl3 that itself binds to CBP/p300 and can interact with components of the basal transcription machinery.37 HSC activation is accompanied by induction of Bcl3.21 This raises the possibility that p50/p50-Bcl3 and p50/p50-HDAC1 complexes may co-exist in HSCs and compete for κB sites or alternatively that association of Bcl3 and HDAC1 with p50 may be mutually exclusive events. Regulation of these interactions will help dictate the level of TNF-α-driven inflammation in infected and injured tissues.
In summary, the p50 subunit of NF-κB is protective in the chronically injured liver and we have described a plausible mechanism to explain this effect involving repression of TNF-α expression by p50/p50-HDAC1 complexes. Further studies on the function of p50 in inflammatory states and the role played by its co-activators and co-repressors may lead to new anti-inflammatory drugs that specifically target p50 activities.
Acknowledgments
We thank Miss Shona Mathery for technical assistance with the TNF-α ELISA.
Footnotes
Address reprint requests to Prof. Derek A. Mann, Liver Group, IIR Division, Level D, South Block, Southampton General Hospital, Southampton SO16 6YD UK. E-mail: dam2@soton.ac.uk.
Supported by the Wellcome Trust (grants 050443/Z and 068524/Z/02/Z), the UK Medical Research Council (Co-operative Group component grant G9900279), and the Childrens Liver Disease Foundation.
J.P.I. is a Medical Research Council senior clinician fellow.
References
- Pahl HL. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Gaynor RB. Therapeutic potential of inhibition of the NF-κB pathway in the treatment of inflammation and cancer. J Clin Invest. 2001;107:135–142. doi: 10.1172/JCI11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionary conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Gerondakis S, Grossmann M, Nakamura Y, Pohl T, Grumont R. Genetic approaches in mice to understand Rel/NF-κB and IκB function: transgenics and knockouts. Oncogene. 1999;18:6888–6895. doi: 10.1038/sj.onc.1203236. [DOI] [PubMed] [Google Scholar]
- Baer M, Dillner A, Schwartz RC, Sedon C, Nedospasov S, Johnson PF. Tumor necrosis factor alpha transcription in macrophages is attenuated by an autocrine factor that preferentially induces NF-κB p50. Mol Cell Biol. 1998;18:5678–5689. doi: 10.1128/mcb.18.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Chen S, Wang Y, Mackman N, Ku G, Lo D, Feng L. RelB modulation of IκBα stability as a mechanism of transcription suppression of interleukin-1α (IL-1α), IL-1β and tumor necrosis factor alpha in fibroblasts. Mol Cell Biol. 1999;19:7688–7696. doi: 10.1128/mcb.19.11.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld R, May MJ, Berberich I, Serfling E, Ghosh S, Neumann M. RelB forms transcriptionally inactive complexes with RelA/p65. J Biol Chem. 2003;278:19852–19860. doi: 10.1074/jbc.M301945200. [DOI] [PubMed] [Google Scholar]
- Sha WC, Liou H-C, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- Snapper CM, Zelazowski P, Rosas FR, Kehry FR, Tian M, Baltimore D, Sha WC. B cells from p50/NF-κB knockout mice have selective defects in proliferation, differentiation, germ line CH transcription, and Ig class switching. J Immunol. 1996;156:183–191. [PubMed] [Google Scholar]
- Weih F, Carrasco D, Durham SK, Barton DS, Rizzo CA, Ryseck R-P, Lira SA, Bravo R. Multi-organ inflammation and hematopoietic abnormalities in mice with targeted disruption of RelB, a member of the NF-κB/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- Weih F, Durham SK, Barton DS, Sha WC, Baltimore D, Bravo R. p50-NF-κB complexes partially compensate for the absence of RelB: severely increased pathology in p50−/−relB−/− double-knockout mice. J Exp Med. 1997;185:1359–1370. doi: 10.1084/jem.185.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis RA, Kovalovich K, Cressman DE, Taub R. Normal liver regeneration in p50/nuclear factor κB1 knockout mice. Hepatology. 2001;33:915–924. doi: 10.1053/jhep.2001.23192. [DOI] [PubMed] [Google Scholar]
- Kato A, Edwards MJ, Lentsch AB. Gene deletion of NF-κB p50 does not alter the hepatic inflammatory response to ischemia/reperfusion. J Hepatol. 37:48–55. doi: 10.1016/s0168-8278(02)00068-5. [DOI] [PubMed] [Google Scholar]
- Mann J, Oakley F, Johnson PW, Mann DA. CD40 induces interleukin-6 gene transcription in dendritic cells: regulation by TRAF2, AP-1, NF-kappaB and CBF1. J Biol Chem. 2002;277:17125–17138. doi: 10.1074/jbc.M109250200. [DOI] [PubMed] [Google Scholar]
- Iredale JP, Benyon RC, Pickering J, McCullen M, Northrop M, Pawley S, Arthur MJP. Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J Clin Invest. 1998;102:538–549. doi: 10.1172/JCI1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MC, Issa R, Smart DE, Trim N, Murray GI, Primrose JN, Arthur MJP, Iredale JP, Mann DA. Gliotoxin stimulates the apoptosis of human and rat hepatic stellate cells and enhances the resolution of liver fibrosis in rats. Gastroenterology. 2001;121:685–698. doi: 10.1053/gast.2001.27188. [DOI] [PubMed] [Google Scholar]
- Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem. 2000;275:2247–2250. doi: 10.1074/jbc.275.4.2247. [DOI] [PubMed] [Google Scholar]
- Cassiman D, Libbrecht L, Desmet V, Denef C, Roskams T. Hepatic stellate cell/myofibroblast subpopulations in fibrotic human and rat livers. J Hepatol. 2002;36:200–209. doi: 10.1016/s0168-8278(01)00260-4. [DOI] [PubMed] [Google Scholar]
- Hellerbrand C, Jobin C, Licato LL, Sartor RB, Brenner DA. Cytokines induce NF-κB in activated but not in quiescent rat hepatic stellate cells. Am J Physiol. 1998;275:G269–G278. doi: 10.1152/ajpgi.1998.275.2.G269. [DOI] [PubMed] [Google Scholar]
- Hellerbrand C, Jobin C, Limuro Y, Licato L, Sartor RB, Brenner DA. Inhibition of NF-κB in activated rat hepatic stellate cells by proteasome inhibitors and an IκB super-repressor. Hepatology. 1998;27:1285–1295. doi: 10.1002/hep.510270514. [DOI] [PubMed] [Google Scholar]
- Elsharkawy AM, Wright MC, Hay RT, Arthur MJ, Hughes T, Bahr MJ, Degitz K, Mann DA. Persistent activation of nuclear factor-kappa B in cultured rat hepatic stellate cells involves the induction of potentially novel Rel-like factors and prolonged changes in the expression of IκB family proteins. Hepatology. 1999;30:761–769. doi: 10.1002/hep.510300327. [DOI] [PubMed] [Google Scholar]
- Oakley F, Mann J, Ruddell R, Pickford J, Weinmaster G, Mann DA. Basal expression of IκBα is controlled by the mammalian transcriptional repressor RBP-J (CBF1) and its activator Notch1. J Biol Chem. 2003;278:24359–24370. doi: 10.1074/jbc.M211051200. [DOI] [PubMed] [Google Scholar]
- Bajt ML, Farhood A, Jaeschke H. Effects of CXC chemokines on neutrophil activation and sequestration in hepatic vasculature. Am J Physiol. 2001;281:G1188–G1195. doi: 10.1152/ajpgi.2001.281.5.G1188. [DOI] [PubMed] [Google Scholar]
- Jaeschke H, Smith CW. Mechanisms of neutrophil-induced parenchymal cell injury. J Leukoc Biol. 1997;61:647–653. doi: 10.1002/jlb.61.6.647. [DOI] [PubMed] [Google Scholar]
- Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- Chen LW, Egan L, Li ZW, Greten FR, Kagnoff MF, Karin M. The two faces of IKK and NF-κB inhibition: prevention of systemic inflammation but increased local injury following intestinal ischemia-reperfusion. Nat Med. 2003;9:575–581. doi: 10.1038/nm849. [DOI] [PubMed] [Google Scholar]
- Chen LF, Greene WC. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Mizgerd JP, Lupa MM, Kogan MS, Warren HB, Kobzik L, Topulos GP. NF-κB p50 limits inflammation and lung injury during Escherichia coli pneumonia. Am J Respir Crit Care Med. 2003;168:810–817. doi: 10.1164/rccm.200303-412OC. [DOI] [PubMed] [Google Scholar]
- Chosay JG, Essani NA, Dunn CJ, Jaeschke H. Neutrophil margination and extravasation in sinusoids and venules of liver during endotoxin-induced injury. Am J Physiol. 1997;272:G1195–G1200. doi: 10.1152/ajpgi.1997.272.5.G1195. [DOI] [PubMed] [Google Scholar]
- Maher JJ. Interactions between hepatic stellate cells and the immune system. Semin Liver Dis. 2001;21:417–426. doi: 10.1055/s-2001-17555. [DOI] [PubMed] [Google Scholar]
- Jaeschke H. Neutrophil-mediated tissue injury in alcoholic hepatitis. Alcohol. 2002;27:23–27. doi: 10.1016/s0741-8329(02)00200-8. [DOI] [PubMed] [Google Scholar]
- Bajt ML, Farhood A, Jaeschke H. Effects of CXC chemokines on neutrophil activation and sequestration in hepatic vasculature. Am J Physiol. 2001;281:G1188–G1195. doi: 10.1152/ajpgi.2001.281.5.G1188. [DOI] [PubMed] [Google Scholar]
- McClain CJ, Song Z, Barve SS, Hill DB, Deaciuc I. Recent advances in alcoholic liver disease IV. Dysregulated cytokine metabolism in alcoholic liver disease. Am J Physiol. 2004;287:G497–G502. doi: 10.1152/ajpgi.00171.2004. [DOI] [PubMed] [Google Scholar]
- Booth M, Mwatha JK, Joseph FM, Kadzo H, Ireri E, Kazibwe F, Kemijumbi J, Kariuki C, Kimani G, Ouma JH, Kabatereine NB, Vennervald BJ, Dunne DW. Periportal fibrosis in human Schistosoma mansoni infection is associated with low IL-10, low IFN-gamma, high TNF-alpha, or low RANTES, depending on age and gender. J Immunol. 2004;172:1295–1303. doi: 10.4049/jimmunol.172.2.1295. [DOI] [PubMed] [Google Scholar]
- Simeonova PP, Gallucci RM, Hulderman T, Wilson R, Kommineni C, Rao M, Luster MI. The role of tumor necrosis factor-α in liver toxicity, inflammation and fibrosis induced by carbon tetrachloride. Toxicol Appl Pharmacol. 2001;177:112–120. doi: 10.1006/taap.2001.9304. [DOI] [PubMed] [Google Scholar]
- Kimura K, Ando K, Ohnishi H, Ishikawa T, Kakumu S, Takemura M, Muto Y, Moriwaki H. Immunopathogenesis of hepatic fibrosis in chronic liver injury induced by repeatedly administered concanavalin A. Int Immunol. 1999;11:1491–1500. doi: 10.1093/intimm/11.9.1491. [DOI] [PubMed] [Google Scholar]
- Kuprash DV, Udalova IA, Turetskaya RL, Kwiatkowski D, Rice NR, Nedospasov SA. Similarities and differences between human and murine TNF promoters in their response to lipopolysaccharide. J Immunol. 1999;162:4045–4052. [PubMed] [Google Scholar]
- Na SY, Choi JE, Kim HJ, Jhun BH, Lee YC, Lee JW. Bcl3, and IκB protein, stimulates activating protein-1 transactivation and cellular proliferation. J Biol Chem. 1999;274:28491–28496. doi: 10.1074/jbc.274.40.28491. [DOI] [PubMed] [Google Scholar]