Abstract
Given that expression of many genes changes when cells become malignant or are placed in different microenvironments, we asked whether these changes were accompanied by global reorganization of chromatin. We reasoned that sequestration or exposure of chromatin-sensitive sites to restriction enzymes could be used to detect this reorganization. We found that AluI-sensitive sites of nonmalignant cells were relatively more exposed compared to their malignant counterparts in cultured cells and human tumor samples. Changes in exposure and sequestration of AluI-sensitive sites in normal fibroblasts versus fibrosarcoma or those transfected with oncogenes, nonmalignant breast cells versus carcinomas and poorly metastatic versus highly invasive melanoma were shown to be independent of the cell cycle and may be influenced by proteins rich in disulfide bonds. Remarkably, regardless of degree of malignancy, AluI-sensitive sites became profoundly sequestered when cells were incubated with laminin, Matrigel, or a circular RGD peptide (RGD-C), but became exposed when cells were placed on collagen I or in serum-containing medium. Disruption of the actin cytoskeleton led to exposure, whereas disruption of microtubules or intermediate filaments exerted a sequestering effect. Thus, AluI-sensitive sites are more sequestered with increasing malignant behavior, but the sequestration and exposure of these sites is exquisitely sensitive to information conferred to the cell by molecules and biomechanical forces that regulate cellular and tissue architecture.
It is well known from pathological specimens that normal and malignant cells differ in terms of nuclear structure. We have postulated that tissue organization depends on interactions between the extracellular matrix (ECM) and the nuclear structure and chromatin.1,2 Observations from in vivo and culture studies have provided evidence that the ECM may indeed influence higher order chromatin structure and gene expression.3–12 Furthermore, experiments on endothelial cells and fibroblasts have demonstrated that a mechanical tug to an integrin receptor at the cell surface could change the organization of both the cytoskeleton and the alignment of intranuclear molecules within 1 second, whereas force applied to non-integrin receptors such as LDL did not change nuclear organization.2
Using AluI restriction enzyme that cuts the widely distributed AG-CT sites, we asked five interrelated questions: 1) Do sensitivities to chromatin digestion differ in nonmalignant and malignant cells of different origins? 2) If so, can one use this information to distinguish between cells of low and high metastatic potential? 3) Does the microenvironment in general, and ECM in particular, affect chromatin structure regardless of degree of malignancy? 4) If so, is there some specificity with regards to the nature of the ECM molecules used? And finally, 5) Is the cytoskeleton involved? We find that answers to the above questions are affirmative.
Materials and Methods
Cell Culture
We used primary uveal melanoma cell lines of low (OCM1a) and high (M619) invasive potential, and a highly invasive metastatic uveal melanoma cell line (MUM2B); the characteristics of these cell lines have been described in detail previously.13 Determination of the modal chromosome number revealed the OCM1a and M619 lines to be in the tetraploid range while the MUM2B line was pseudo-diploid. These cell lines were also characterized by their differential ability to form laminin-rich patterns that appear to control melanoma morphogenesis.14 OCM1a uveal melanoma cells were a generous gift from Dr. June Kan-Mitchell, Karmanos Cancer Institute, Wayne State University, Detroit, MI, and UM54 normal uveal melanocytes were a generous gift from Dr. J. William Harbour, Washington University, St Louis, MO. Melanoma cells and uveal melanocytes were plated in EMEM (BioWhittaker, Inc., Walkersville, MD), and supplemented with heat inactivated 15% fetal bovine serum (Fisher, Ontario, Canada) without the addition of exogenous extracellular matrix molecules or growth factors.
Normal WI-38 human fibroblasts were obtained from the ATTC (Rockville, MD), and were maintained in 10% serum in DMEM. Telomerase, large-T, and ras transfected fibroblasts were developed by Hahn et al15 and maintained according to their protocols, and were a generous gift from Dr. Igor Roninson (University of Illinois at Chicago). HT1089 fibrosarcoma cells were obtained from the ATTC. No antibacterial or antifungal drugs were used in the maintenance of cell lines or in experiments, as their chronic use has been shown to interfere with the differentiation potential of other primary cell types.7 MCF10A breast epithelial cells and MDA-MB231 breast carcinoma cells were obtained from the ATTC, and were maintained on DMEM plus heat inactivated calf serum. The isolation and phenotypes of HMT-3522 human mammary epithelial cells16,17 both nonmalignant (S1) and their tumorigenic counterparts designated T4–218 were described previously. All cell cultures were determined to be free of mycoplasma contamination using the GenProbe rapid detection system (Fisher, Itasca, IL).
Restriction Enzyme Assays of Interphase Cells
We used three assays to compare sequestration and exposure of AluI binding sites in interphase cells of different invasive behaviors. In the permeabilized cell assay, we exposed cells in monolayer cultures to AluI restriction enzyme after permeabilization with Triton X-100. In the cell smear assay, we mechanically dislodged cells from culture, applied them to glass slides, allowed the cells to air dry, and exposed the cells to AluI restriction enzyme, thus simulating a diagnostic cytological examination performed in a clinical setting (eg, the examination of cells extracted by fine needle aspiration biopsy, brush biopsy, or scraping). In the flow cytometry assay, we quantified differential digestion of chromatin between cell lines by AluI restriction enzyme over time, again using methodology that could be easily adapted to a clinical diagnostic setting.
Permeabilized Cell Assay
Cells grown as monolayer cultures in the absence of exogenous matrix molecules were exposed to 0.1% Triton X-100 (Sigma, St. Louis, MO) for 90 seconds followed by a rinse of DMEM. The preparation was exposed to AluI restriction enzyme (0.5 μl in 40 μl DMEM; Promega, Madison, WI) for 60 minutes to 24 hours. The permeabilized cultures were then exposed to ethidium bromide (25 μl, 1 μg/ml; Sigma) and photographed with a Leica inverted microscope (Leica, Bannockburn, IL).
Cell Smear Assay
After mechanically dislodging cells grown under monolayer conditions, the slurry was suspended in 1X PBS or serum-free DMEM, and a drop containing 15 μl of the suspension was placed onto a glass slide. The drops were allowed to evaporate over 30 minutes to 1 hour. AluI restriction enzyme (0.5 μl in 40 μl) or MspI restriction enzyme (0.5 μl in 40 μl DMEM; Promega) was applied to the dried cells, and the preparation was placed in a humidified 37°C chamber to optimize enzyme activity and minimize enzyme evaporation. Endonuclease digestions were terminated at pre-designated time points (30 minutes and hourly increments thereafter up to 24 hours) to determine the optimum digestion time that would allow for discrimination of differential chromatin digestion between cell lines. Ethidium bromide was added to terminate the digestion, and the preparation was photographed immediately. Touch preparations of normal human tissue and human tumor tissue were made and air dried. The preparation was incubated with AluI restriction enzyme and the reaction was terminated with ethidium bromide at 5 and 24 hours.
To test the influence of different soluble ECM molecules on AluI sensitivity, we mechanically dislodged cells that had been growing under monolayer conditions from flasks and suspended the slurry of dislodged cells in 1X PBS or serum-free DMEM. Two rows of 15 μl drops of the suspension were applied to a glass slide, 3 drops per row. In some experiments as described below, we added soluble laminin, Matrigel, FBS, RGD-C, collagen Type I, bFGF, EGF (all reagents from Clontech, Palo Alto, CA), circularized RGD (RGD-C, a kind gift from Renata Pasquallini) to one of the two rows of drops. RGD-C is known to bind to integrin receptors.2 The test and control drops were then permitted to evaporate at room temperature for at least 1 hour, leaving “smears” of dried cells that had or had not been incubated with a test molecule. For each assay, cellular permeability was checked using the trypan blue exclusion method.
The AluI restriction enzyme (0.5 μl in 40 μl DMEM; Promega) was applied to each drop in a humidified 37° incubator for up to 24 hours. The buffers used in these assays did not contain either DTT or mercaptoethanol to avoid removing any proteins within the cytoplasm or nucleus that might sequester AluI binding sites from enzyme digestion. Incubation with AluI was terminated by adding ethidium bromide (Sigma; 250 μl, 100 ng/ml) to each drop per slide at 30 minutes and at hourly intervals thereafter up to 24 hours of incubation.
Scoring Results
In the cell smear assay and the permeabilized cell assay, samples were photographed with a Leica inverted microscope and were scored qualitatively from the micrographs as follows: 1) Nuclei in which no fluorescence was detected except for nucleoli were scored as complete digestion; 2) Nuclei in which fluorescence intensity was equal to that detected at time before application of restriction enzyme was scored as no digestion; and 3) Detection of non-nucleolar nuclear fluorescence that was less intense than detected before application of restriction enzyme was scored as partial digestion.
Flow Cytometry
To provide for a quantitative measure of DNA digestion, we performed flow cytometry. Cells from monolayer cultures of UM54 normal uveal melanocytes, poorly invasive OCM1a primary uveal melanoma, highly invasive M619 primary uveal melanoma, and highly invasive MUM2B metastatic uveal melanoma were suspended in DMEM and spun down at 1400 rpm for 5 minutes. The pellet was re-suspended in 0.1% Triton X-100, incubated for 1 minute at room temperature, and spun down again at 1400 rpm for 5 minutes. After re-suspension in DMEM, 0.5 μl of AluI restriction enzyme in 40 μl of DMEM was added and the preparation was incubated at following time points: 0 (baseline), 1, 3, and 5 hours at 37°C. Propidium iodide (10 μl/ml; Molecular Probes, Eugene, OR) was added at the conclusion of each digestion period. The cells were analyzed in a FACS Calibur (BD Bioscience, San Jose, CA) equipped with a 488 laser, detectors for forward and side scatter, and 520, 575, and 675 nm detectors. 10,000 cells were counted and the results were analyzed with FACS dot-plots and histograms. Three AluI digestions monitored by flow cytometry were performed for each of the three melanoma cell lines. The percentage of cells in M2–M4 (roughly corresponding to 102 to 104 fluorescence intensity) was calculated and compared between cell lines at 5 hours of digestion with the t-test. The PI signal, representing stoichometrically the amount of labeled undigested DNA,19 was recorded immediately after cell permeabilization and labeling with PI and used as a baseline (0 time point), and was then measured for each cell line after 1, 3, and, 5 hours of exposure to AluI restriction enzyme. CellQuest software (BD Bioscience) was used to generate overlays of the histograms for each cell line at each time point in the digestion experiment.
Isolated Chromosome Set Assay
We have previously described a method by which the entire chromatin contents of nuclei can be extracted, producing a complete chromosome set for each cell thus manipulated.20,21 We removed chromatin sets from cells that had not been pretreated with any agent. All chromosome extractions and digestions were carried out under isotonic culture conditions in DMEM at pH 7.4; similar results also were obtained using complete cell culture medium containing FBS.
Chromosome sets removed from metaphase cells of varying invasive potentials were studied under buffered media conditions that do not disturb the native structure of chromosomes.20–29 Cells from which chromosomes were to be removed were grown on small coverslips and placed in the center of a 35-mm plastic dish lid containing 2 ml of gassed DMEM. This preparation was allowed to equilibrate at 37°C in a 5% CO2 buffered incubator before micromanipulation. For optimal micromanipulation conditions, cells were grown to near confluence to produce strong attachments to each other and ECM needed to resist the pulling forces associated with rapid (within 1 second) micropuncture and removal of chromosomes without causing cell detachment or death of the manipulated cell. Chromosome sets were obtained either in the presence or absence of colchicine and cytochalasin-B as reported previously.20 All chromosome set extractions and digestions were carried out under isotonic culture conditions in DMEM at pH 7.4; similar results also were obtained using complete cell culture medium containing serum.
Chromosome sets were extracted from living metaphase cells by rapidly piercing the cell directly on the side of a mitotic plate with a glass microneedle and then laterally drawing out the chromosomes through the hole on the cell surface created by the micropuncture using a Leitz micromanipulator (Leica Microsystems). In certain studies which used treatments that cause cell detachment (mercaptoethanol, DTT, and proteinase K), chromosome sets were removed from the cell with a pipette and placed on culture substrata. The chromosome sets were then deposited to pre-designated areas defined by scratching the Petri dish lid with a fine needle. Narishige micropipettes (Narishige Scientific Instruments, Tokyo, Japan) were pulled with a Sutter micropipette puller (Sutter Instrument Company, Novato, CA) adjusted to produce long barrels approximately 1 to 5 μm wide along a length of 40 to 100 μm (tip widths were consistently less than 0.5 μm).
In this isolated chromosome set assay, it is important to use restriction enzymes and proteases in the range of specific ion concentrations if they are to function efficiently. We used buffered media conditions that preserve physiological ion concentrations to maintain the extent of chromatin compaction observed in living cells while providing the appropriate concentrations of NaCl and MgCl2 to permit enzymes to work.20 The restriction enzymes were used in the absence of DTT or β-mercaptoethanol (which are typically included in commercially available kits used to digest DNA) to avoid interfering with DNA-associated proteins.
Restriction enzymes were dissolved in 2 ml of DMEM applied directly to 35 mm tissue culture dish lids on which the chromosome preparations were attached in the concentrations listed below. Changes in chromosome morphology were photographed by time-lapse video microscopy in real time. All microsurgical procedures were observed at 63× by phase and fluorescence microscopy with a Leica inverted microscope, and captured using a time-lapse video recorder (Sony) and Pinnacle Image software (Pinnacle Systems, Modesto, CA).
We tested the following enzymes: AluI (5–50 units), Msp-I, (75 units, Promega), DNase I (1–20 units/droplet; Promega,), micrococcal nuclease (1–10 units; Promega), EcoRI (5–50 units; Promega), HindIII (5–10 units; Promega), BamHI (5–10 units; New England Biolabs, Beverly, MA), RNase A (1–100 units; Sigma), RNase I (1–100 units; Promega), EMBO (3 units; Promega), Sau-III (20 units; Promega), PstI (30 units; Promega), XHO-1 (40 units; Promega). Ethidium bromide (25 μl, 1 μg/ml; Sigma) was added to the medium after 1 to 3 hours of digestion with restriction enzyme and the preparations were photographed with a Leica inverted microscope.
Permeabilized Cell Assay with Polymerized ECM Molecules
We designed a method to place different polymerized ECM molecules on glass slides to screen for changes in the sensitivity of AluI-sensitive sites in normal cells and tumor cells or tumor cells with varying degrees of invasive behavior. First, a coating of FBS or Type I collagen was applied to the glass slide and allowed to dry completely. Then a stencil was positioned on top of the dried FBS or collagen and clamped so that it would not move. The stencil was fashioned in such a way that it allowed application of a second ECM molecule over the dried FBS or collagen in square patches. Laminin or Matrigel were then applied to areas of the slide exposed by the stencil and the entire assemblage was baked in an oven for 1 hour at 90°C. The stencil was gently removed under sterile conditions, and cells were then seeded at subconfluent density onto the entire surface. The cultures were then exposed to 0.1% Triton X-100 (Sigma) for 90 seconds followed by a rinse of DMEM. AluI restriction enzyme (0.5 μl in 40 μl DMEM) was applied for 30 to 60 minutes after which time ethidium bromide (250 μl, 100 ng/ml) was placed on the preparation. Nuclei were photographed immediately and scored for digestion as described above.
Three-Dimensional Cultures
As reported previously, highly invasive primary and metastatic melanoma cells grow in monolayers on tissue culture plastic but form looping patterns when placed in three-dimensional (3D) culture conditions identical to those seen in tissue sections of primary aggressive and metastatic lesions resected from patients.14 Three-dimensional cultures were established by growing cells on Matrigel30 (Collaborative Biomedical, Bedford, MA) or Type I collagen31 (Collaborative Biomedical) placed onto plastic tissue culture dishes to a depth of about 0.2 mm followed by polymerization for 1 hour at 37°C. The plates were then returned to the incubator and allowed to completely polymerize for several hours. Cells were then seeded at saturating densities (400 million cells/dish) on the polymerized 3D gel coatings, and the cultures were checked daily for the presence of looping patterns that are characteristic for highly invasive melanoma cells.13 Techniques for growing mammary cells on 3D gels have been described30,31 (see review by Schmiechle and Bissell32).
Transmission Electron Microscopy
Cells were fixed by adding in 4% buffered glutaraldehyde to the culture medium, followed by post fixation in osmium tetroxide. After dehydrations in ethanol, cultures were embedded in Epon. Ultra-thin sections were cut perpendicular and parallel to the culture, stained with uranyl acetate and lead citrate, and were examined with a JEOL 1011 transmission electron microscope (Peabody, MA).
Results
Differential Chromatin Digestion by AluI Restriction Enzyme in Different Interphase Cells with Varying Malignant Behavior
The sequestration and exposure of AluI binding sites was tested qualitatively by exposing permeabilized cultured cells on glass slides to restriction enzyme (permeabilized cell assay, Figure 1, A–F) to maintain the structural integrity of the nucleus and cytoplasm. We also smeared cultured cells on slides and allowed them to dry (cell smear assay, Figure 1, G–J) to rapidly assess a large number of samples. Each assay was repeated 18 times with identical results.
Figure 1.
Differential sensitivity of interphase cells to AluI restriction enzyme. A–F: Permeabilized cell assays. A: Phase micrograph of intact poorly invasive OCM1a uveal melanoma cells grown on monolayers after 2 minutes of exposure to Triton X-100 for permeabilization followed by 30 minutes of exposure to AluI restriction enzyme. Cells were labeled with ethidium bromide. B: Fluorescent micrograph of the same preparation illustrated in A. DNA has been digested leaving only the AluI enzyme-resistant nucleoli fluorescent. C: Phase micrograph of highly invasive M619 uveal melanoma cells prepared as in A. D: Fluorescent micrograph of the same preparation illustrated in C. DNA is resistant to digestion by AluI restriction enzyme. E: Phase micrograph of highly invasive MUM2B metastatic melanoma cells after preparation identical to cells illustrated in A. F: Fluorescent micrograph of same preparation illustrated in E labeled and with ethidium bromide. G–J: Cell smear assays comparing UM54 normal uveal melanocytes, OCM1a, M619, and MUM2B melanoma cells. After 5 hours of exposure to AluI restriction enzyme, only the nucleoli are detected in the UM54 and OCM1a cells, but the chromatin of M619 and MUM2B cells is undigested and therefore brightly fluorescent. K: Normal renal tubular epithelium. L: After 5 hours of digestion of normal renal tubular epithelial cells from the region depicted in K with AluI restriction enzyme: only the nucleoli fluoresce. M: Renal cell carcinoma. N: After 5 hours of digestion with AluI, the chromatin of cells from the adjacent renal cell carcinoma depicted in M has resisted digestion and is brightly fluorescent. Scale bars: A–F = 50 μm; H–J = 100 μm; K, L = 50 μm; M, N = 100 μm.
In both the permeabilized cell assay and the cell smear assay, we detected striking difference in the susceptibility of chromatin to digestion by AluI in cells of different invasive potential (Table 1). For example, the chromatin of UM54 normal uveal melanocytes and poorly invasive OCM1a melanoma cells digested completely within 60 minutes of exposure to AluI restriction enzyme. Bright fluorescence was detected in nucleoli, but no fluorescence was demonstrated elsewhere in the nucleus. By contrast, the nuclei of highly invasive M619 and MUM2B melanoma cells resisted digestion by AluI restriction enzyme and were brightly fluorescent (Figure 1). In a parallel experiment, chromatin of MCF10A breast epithelial cells digested completely within 60 minutes whereas chromatin from MDA-MB231 breast carcinoma cells resisted digestion (data not shown).
Table 1.
Summary of Response to DNA Digestions by AluI Restriction Enzyme Using Three Different Assays
| Cell Line | Interphase cell assays
|
Metaphase cell assay isolated chromosome sets | |
|---|---|---|---|
| Permeabilized | Smear | ||
| Melanoma | |||
| OCM1a poorly invasive primary melanoma13,71 | Sensitive | Sensitive | Sensitive |
| M619 highly invasive primary melanoma13 | Resistant | Resistant | Resistant |
| MUM2B highly invasive metastatic melanoma13 | Resistant | Resistant | Resistant |
| Fibroblast/fibrosarcoma | |||
| WI-38 fibroblasts | Sensitive | ||
| HT1080 fibrosarcoma | Resistant | ||
| Fibroblasts + transfected oncogenes15 | |||
| + Telomerase | Sensitive | ||
| + Telomerase + Large T | Sensitive | ||
| + Telomerase + Large T + ras | Resistant | ||
| Breast epithelium/breast carcinoma | |||
| MCF10A non-malignant breast epithelial cells | Sensitive | Sensitive | |
| MDA-MB231 metastatic breast carcinoma cells | Resistant | Resistant | |
| S1 non-malignant breast epithelial cells33 | Sensitive | Sensitive | |
| T4-2 tumorigenic breast cells derived from S133 | Resistant | Resistant | |
In preliminary experiments, we tested the feasibility of detecting differences in the susceptibility of chromatin to AluI between normal and neoplastic human tissue in touch preparations. For example, the chromatin of normal renal tubular epithelial cells digested completely within 5 hours, but chromatin from an adjacent focus of renal cell carcinoma resisted digestion by AluI restriction enzyme in the same time frame (Figure 1, K–N); the same differential digestion was detected between normal gastric epithelium and gastric carcinoma (data not shown).
To achieve quantitative assessment of chromatin digestion in the uveal melanocytic cell lines studied by cell smear assay (Figure 1, G–J), we measured DNA content in cell suspensions by flow cytometry after staining with propidium iodide (PI). Histograms comparing OCM1a, M619, and MUM2B melanoma cells with each other and the normal UM54 melanoma cell line are presented in Figure 2. There are striking differences in the percentage of cells in channels M2-M4 (corresponding to 102 − 104 fluorescence intensity on the x axis) over time. At 5 hours of digestion, only 1% of UM54 normal melanocytes are recorded in M2–4, compared with 52% of OCM1a cells, 73% of M619 cells, and 94% of MUM2B cells. The experiment illustrated in Figure 2 was repeated twice, and the percentage of cells in M2-M4 at 5 hours after digestion was compared between the three melanoma cell lines in three replicate assays using a t-test. There are significantly more MUM2B cells in M2-M4 than OCM1a cells (P = 0.006) and significantly more M619 cells than OCM1a cells in M2-M4 (P = 0.02). There were also more MUM2B cells than M619 cells in M2-M4 after 5 hours of digestion by AluI (P = 0.054). Thus, the quantitative flow cytometric assays showed that chromatin in highly invasive melanoma cells was more resistant to digestion with AluI restriction enzyme than poorly invasive melanoma cells and was consistent with the qualitative permeabilized cell and cell smear assays (Figure 1).
Figure 2.
Flow cytometric study of UM54 normal uveal melanocytes, OCM1a poorly invasive primary melanoma cells, M619 highly invasive primary melanoma cells, and MUM2B highly invasive metastatic melanoma cells displaying the propidium iodide (PI) fluorescence intensity signal in the x axis, and cell count in the y axis for each cell line at each of the four time points. A: Gating for M1-M4 is indicated in the histogram for UM54 at time 0. The percentage of cells in each gate is reported for all four cell lines at time 0 and 5 hours of digestion. Note the differences between the percentages of cells in M2–4 between the four cell lines at 5 hours. The histograms at 5 hours of digestion parallel the cell smear assay performed in these cell lines and illustrated in Figure 1, G–J. B: For each time-point (baseline, 1, 3, and 5 hours), the histogram outline of each cell line is superimposed, color coded according to cell line as indicated in the legend. Note that the PI signal for UM54 normal melanocytes degrades rapidly at 1 hour and continues to degrade throughout the time-course. The PI signal for OCM1a cells degrades rapidly at 1 hour and gradually throughout the 5-hour time course of digestion by AluI. The PI signal for M619 cells degrades at 1 hour but shows less degradation at 5 hours than OCM1a cells. The PI signal for MUM2B cells degrades the least at 1 hour of all cell lines and there is little signal degradation thereafter.
Differential Digestion of Chromatin by AluI Restriction Enzyme Is Independent of the Cell Cycle
The previous assays tested the digestion of chromatin by AluI in asynchronous populations of interphase cells. To test the susceptibility of chromatin digestion by AluI in cells in mitosis, we used a method to extract complete sets of mitotic chromosomes from cells.7,20–29 We studied WI-38 fibroblasts, oncogene transfected cells, and cancer cell lines of varying degrees of invasive behavior (Table 1).
Metaphase chromosome sets from the poorly invasive OCM1a, highly invasive M619, and the highly invasive and metastatic MUM2B were extracted and placed next to one another so that chromosomes from OCM1a were next to chromosomes from M619 and MUM2B cells (Figure 3A). After 30 minutes of digestion by AluI, the OCM1a chromosomes had digested to a much greater extent than the chromosome sets extracted from highly M619 or MUM2B cells (Figure 3B). After 60 minutes of digestion by AluI restriction enzyme, chromosomes were stained with ethidium bromide to visualize undigested DNA (Figure 3C). Chromosomes from the most highly invasive and metastatic cells digested the least, while the poorly invasive cell chromosomes digested nearly completely. The relative sensitivities of the chromosomes extracted from metaphase cells to AluI restriction enzyme were therefore identical to the sensitivities observed within the nuclei of corresponding asynchronous interphase cells (Figures 1 and 2).
Figure 3.
Differential sensitivity of isolated sets of chromosomes extracted from primary and metastatic uveal melanoma cells. A: Phase micrograph of chromosomes isolated from poorly invasive primary OCM1a uveal melanoma cells (labeled 1 and 2 in blue), highly invasive primary M619 uveal melanoma cells (labeled 3, 4, and 5 in yellow), and highly invasive MUM2B metastatic uveal melanoma cells (labeled 6 and 7 in red) are placed side-by-side on a glass slide. B: Same preparation as A after 30 minutes of digestion with AluI restriction enzyme. Chromosomes from the poorly invasive OCM1a primary uveal melanoma cell line are digesting whereas chromosomes from the more invasive cell lines remain intact. C: Fluorescence micrograph of same preparation as in A and B after 60 minutes of digestion with AluI restriction enzyme and labeling with ethidium bromide. Chromosomes extracted from the poorly invasive primary OCM1a uveal melanoma cells have almost completely digested leaving only trace amounts of ethidium-positive material whereas chromosome sets from the more invasive cell lines have largely resisted digestion. D–F: Differential effects of AluI restriction enzyme digestion on chromosomes extracted from metaphase fibroblasts, fibroblasts transfected with oncogenes, and fibrosarcoma cells. D: Phase micrograph of three sets of metaphase chromosomes extracted from fibrosarcoma cells (labeled 1–3 in red) and attached to charged plastic next to 4 sets of metaphase chromosomes from normal fibroblasts (labeled 4–7 in blue). E: Phase microscopy of the same preparation as in A after 30 minutes of digestion with AluI restriction enzyme. F: Phase microscopy of the same preparation as in A and B after 60 minutes of digestion with AluI restriction enzyme. Normal fibroblast chromosomes have dissolved while chromosomes from the fibrosarcoma cells are relatively intact. G: Fluorescence microscopy of two sets of chromosomes extracted from fibrosarcoma cells (labeled 1, 2) and 6 sets of chromosomes extracted from normal WI-38 fibroblasts (labeled 3–8). The preparation was exposed to AluI restriction enzyme for 2 hours and labeled with ethidium bromide. The chromosomes from the fibrosarcoma cells (labeled 1, 2) resist digestion. Note that individual chromosomes remain intact. By contrast, the chromosomes from the normal WI-38 fibroblasts (3–8) have digested. H: Phase video microscopy of chromosomes extracted from fibroblasts transfected with telomerase (labeled Tel in blue) and from fibroblasts transfected with telomerase plus SV-40 Large T antigen plus ras (labeled Tel+Large T+ras in red). I: Same preparation as in D after 38 minutes of exposure to AluI restriction enzyme. Chromosomes extracted from fibroblasts transfected with only telomerase have dissolved whereas chromosomes extracted from fibroblasts transfected with telomerase plus SV-40 plus Large T have largely resisted digestion. Scale bars: A–C = 30 μm; D–F = 40 μm; G = 20 μm; H, I = 20 μm.
Differential Sensitivity of Isolated Chromosome Sets from Normal Fibroblasts, Fibroblasts Transfected with Multiple Oncogenes, and Fibrosarcomas to AluI Restriction Enzyme
We compared the sensitivity to digestion by AluI for chromosomes extracted from WI-38 fibroblasts with chromosomes extracted from HT-1080 fibrosarcoma cells: the latter two resisted digestion by AluI, but chromosomes extracted from normal fibroblasts were digested rapidly by 30 minutes (Figure 3, D–G).
Normal fibroblasts and fibrosarcoma cells represent two extremes on a spectrum in tumor progression. To compare cells with similar genetic backgrounds, we therefore tested sensitivity to AluI restriction enzyme digestion on chromosomes of fibroblasts developed by Hahn et al15 that differ by single or multiple defined oncogene insertions: fibroblasts transfected only with telomerase, fibroblasts transfected with telomerase plus SV-40 Large T antigen, and fibroblasts transfected with telomerase plus Large T antigen plus ras. The chromosomes extracted from fibroblasts transfected with only the telomerase gene were digested by AluI within 38 minutes. Fibroblasts transfected with telomerase plus SV-40 Large-T digested within the same time frame. However, chromosomes from fibroblasts transfected with telomerase plus SV-40 plus ras (Figure 3, H and I) resisted digestion after the same time exposure to AluI restriction enzyme. Thus, in this fibroblast progression model, the insertion of three oncogenes induced a shift in the sensitivity of the entire genome to AluI digestion.
Differential Sensitivity of Isolated Chromosomes from a Breast Cancer Progression Series
To compare normal and spontaneously-induced malignant epithelial cells from the same genetic background, we used an extensively characterized series of breast and breast cancer cell lines (HMT3522). The nonmalignant cells referred to as S1 breast epithelial cells were isolated from reduction mammoplasty and a series of cell lines, including those malignant in nude mice, were derived in defined media after removal of EGFR as described previously.16–18 S1 cells form acini in three-dimensional cultures that reproduce the polarized structure of the breast acinus with high fidelity. The T4–2 line forms disorganized acini as do cells from primary human breast tumors33 Chromosomes were extracted from S1 and T4–2 cells grown in monolayer cultures and exposed to AluI restriction enzyme. After 60 minutes, there were clear differences in digestion rates between chromosomes extracted from S1 cells and those removed from T4–2 cells (Figure 4).
Figure 4.
Phase micrograph of differential effects of AluI restriction enzyme digestion of chromosomes extracted from S1 breast epithelial cell and T4 cells derived from this line. After 60 minutes of exposure to AluI restriction enzyme, T4 chromosomes are resistant to digestion compared with S1 chromosome sets. Scale bar = 20 μm.
Differential Digestion of Chromatin with Other Restriction Enzymes
To determine whether DNA digestion by enzymes other than AluI discriminated between cells of low and high invasive behavior, we tested EcoRI, SauI, PstI, XhoI, HindIII, and MspI in isolated chromosome digest assays using WI-38 fibroblasts, and melanoma cells of melanoma cells of increasing invasive potential (OCM1a, M619, and MUM2B). MspI (Figure 5) and DNase-I (not shown) also discriminated between cells of different invasive potential.
Figure 5.
Fibroblasts, poorly invasive melanoma cells, and highly invasive and metastatic melanoma cell nuclei were tested for their differential sensitivity to MspI applied for 24 hours in a humidified 37°C chamber. Note that the highly invasive cell nuclei do not digest to the extent that the poorly invasive or normal cell nuclei digest under the same conditions and in the same time period. Scale bar = 40 μm.
The Importance of Proteins in the Structural Sequestration of AluI Sites
We hypothesized that nuclear proteins are responsible for the sequestration and exposure of AluI sites. Therefore, we tested the sensitivity of chromosomes from non-invasive and highly invasive cells to proteases. We have previously shown that heparin and trypsin-sensitive proteins play key roles in chromosome length and compaction in normal cells.20,21 Exposure of chromosomes to heparin or trypsin did not discriminate between non-invasive and highly invasive cells (data not shown). However, these cells could be discriminated when chromatin from these cells was exposed to proteinase K. Under these conditions, chromosomes from highly invasive HT1080 cells dissolved within 6 minutes while chromosomes from non-invasive normal fibroblasts, normal human umbilical endothelial cells (not shown), and poorly invasive melanoma cells de-condensed but did not digest in the same time period (Figure 6, A–C). These data suggest a fundamental difference in the sensitivity of chromatin-associated proteinase K-sensitive proteins between normal and highly invasive cells.
Figure 6.
A: Phase micrograph of four sets of chromosomes extracted from WI-38 fibroblasts (labeled 1–4 in blue) and from two fibrosarcoma cells (labeled 5, 6 in red). B: Same preparation as in A after 2 minutes of exposure to proteinase K. C: Same preparation as in A and B after 6 minutes of exposure to proteinase K. Chromosomes from the fibrosarcoma cells have digested but chromosomes from the normal endothelial cells are relatively intact. D–K: Effects of disrupting protein disulfide bonds on sensitivity of chromatin to AluI restriction enzyme. D: Phase micrograph of two sets of chromosomes extracted from fibroblasts transfected with telomerase (labeled Tel in blue) are placed on the same slide as two sets of chromosomes extracted from fibroblasts transfected with telomerase plus Large T plus ras (labeled Tel+Large T+ras in red). Box identifies a portion of the Tel only transfected fibroblast chromosomes. E: Phase micrograph of same preparation illustrated in A after 80 minutes of exposure to β-mercaptoethanol and AluI restriction enzyme. Chromosomes from the fibroblasts transfected with telomerase plus Large T plus ras have digested completely but chromosomes transfected only with telomerase are only minimally digested, a sensitivity opposite that seen when AluI restriction enzyme is used alone (Figure 3, E and F). As the chromosomes from the Tel only transfected fibroblasts digest, they also shift position in the preparation. Note the relative positions of the boxed areas in D and E. F1: Phase microscopy high magnification of boxed region in D showing details of chromosomes. F2: phase microscopy of same chromosomes from the boxed area in E (after 80 minutes of digestion). Note the partial dissolution of the chromosomes. G: Phase micrograph of chromosomes extracted from poorly invasive OCM 1a primary melanoma cells (labeled 1–4 in blue) and highly invasive M619 primary melanoma cells (labeled 5–8 in red). H: fluorescence micrograph of same preparation as in G after 60 minutes of exposure to β-mercaptoethanol and AluI restriction enzyme, labeled with ethidium bromide. Chromosomes from the poorly invasive OCM 1a cell line (1–4) resist digestion, but chromosomes from the invasive M619 cell line are beginning to digest. Note specifically the fluorescent “cloud” of labeled DNA in chromosome set 8. I: Fluorescence micrograph of same preparation as in G and H after 80 minutes of exposure to β-mercaptoethanol and AluI restriction enzyme: Chromosomes from the poorly invasive OCM 1a cell line (1–4) still resist digestion while chromosomes from the aggressive M619 (5–8) cell line are completely digested. J: Fluorescence micrograph of smear preparation of invasive M619 melanoma cells after 30 minutes of digestion with AluI restriction enzyme in the presence of DTT. K: Fluorescence micrograph of smear preparation of poorly invasive OCM 1a melanoma cells after the same conditions and time point as in J. In the presence of DTT, the nuclei in highly invasive cells are more sensitive to digestion with AluI restriction enzyme, an effect opposite that seen when AluI is used alone (compare with Figure 1). Scale bars: A–C = 20 μm; D, E = 40 μm; F1, F2 = 10 μm; G, H = 20 μm; J, K = 20 μm.
It is not possible to study the role of proteins in the sequestration and exposure of AluI sensitive tissues using the nonspecific enzyme, proteinase K because this nonspecific protease would also digest the AluI restriction enzyme. Because there is evidence that disulfide-rich nuclear matrix-associated proteins play a key role in the sequestration and exposure of genes,34 we tested the digestion of chromatin by AluI in the presence of β-mercaptoethanol or DTT, agents that disrupt proteins rich in disulfide bonds but do not interfere with Alu enzyme activity. Additionally, commercially available restriction enzyme digest kits first remove proteins from DNA with reagents such as β-mercaptoethanol or DTT.
We therefore repeated the extraction of chromosomes from fibroblasts transfected with telomerase only and from fibroblasts transfected with telomerase plus Large T and ras. After exposure of these chromosomes to AluI and either β-mercaptoethanol or DTT, the chromosomes extracted from telomerase plus Large T and ras-transfected fibroblasts digested more rapidly than the chromosomes from fibroblasts transfected only with telomerase (Figure 6, D–F2). Thus, digestion by disulfide disrupting agents renders highly invasive cell chromatin more sensitive to AluI restriction enzyme.
Although chromosomes from M619 are relatively resistant to AluI restriction enzyme compared with chromosomes from OCM1a cells in the absence of β-mercaptoethanol or DTT (Figure 3), chromosomes from the former digested more rapidly than those from the OCM1a cells when these disulfide bond-disrupting agents were present (Figure 6, G–I). Interphase nuclei of highly invasive M619 uveal melanoma cells were also more sensitive to AluI restriction enzyme in the presence of DTT after 30 and 60 minutes of digestion than were nuclei of poorly invasive OCM1a cells (Figure 6, J and K). Thus, disulfide bond disrupting agents—β-mercaptoethanol or DTT—expose AluI binding sites in highly invasive cells.
Regulation of Susceptibility to AluI Restriction Enzyme by Matrix Composition in 2D Conditions by the Cell Smear Assay
There is now compelling evidence that the microenvironment including the ECM regulates gene expression.1,7,13,35–39 We therefore reasoned that different ECM molecules would induce changes in the sequestration and exposure of DNA.
We tested for differential digestion of chromatin in intact cells by AluI after cells were exposed to different soluble ECM molecules with a cell smear preparation that allowed high through-put analysis of cells dried onto glass slides. Given the results described above, the buffers used in these assays did not contain either DTT or mercaptoethanol to avoid removing any proteins within the cytoplasm or nucleus that might sequester AluI binding sites from enzyme digestion.
Remarkably, chromatin of both highly invasive (MUM2B) and low invasive (OCM1a) melanoma cells exposed to laminin for 30 to 60 minutes resisted digestion by AluI at 1 hour (Figure 7, A and C), whereas the chromatin from the same cells exposed to collagen 1 and FBS was completely digested (Figure 7, B and D) in the same time period (the same results were obtained with purified fibronectin, data not shown). Nucleoli, which are made of ribonucleoprotein and thus resistant to AluI, were not digested. The composition of ECM molecules affected the differential digestion of chromatin in the cell smear assay in multiple cell lines. Similar results were found when DNase was substituted for AluI restriction enzyme (data not shown).
Figure 7.
Laminin increases sequestration of AluI sensitive sites in cell smear assays. A: Highly invasive MUM2B cells pre-incubated with soluble laminin for 60 minutes, digested with AluI for 1 hour and labeled with ethidium bromide: chromatin resists digestion. B: MUM2B cells pre-incubated with FBS for 60 minutes, digested with AluI for 1 hour and labeled with ethidium bromide. Except for nucleoli, the chromatin is digested. C: Poorly invasive OCM1a cells pre-incubated with laminin for 60 minutes, digested for with AluI for 1 hour and labeled with ethidium bromide. D: OCM1a cells pre-incubated with serum for 60 minutes, digested for 1 hour with AluI and labeled with ethidium bromide. Except for nucleoli, the chromatin is digested. Scale bars: A—D = 50 μm.
The chromatin of highly invasive M619 melanoma cells when exposed for 30 minutes to soluble laminin and RGD-C peptide resisted digestion with AluI restriction enzyme. RGE had no effect on chromatin sequestration and exposure (data not shown). Chromatin of the same cells completely digested in the same time period after exposure to FBS, water (to ensure that exposure to ions did not affect chromatin structure), collagen I, EGF, and bFGF, a response identical to cells exposed only to PBS (Figure 8, A–N).
Figure 8.
Exposure of cells (invasive melanoma, invasive breast cancer, and breast epithelial cells) to different matrix molecules and growth factors on the cell smear assay. A–M: Paired comparison of highly invasive M619 primary uveal melanoma cells treated with matrix molecules, serum, H2O, or growth factors on upper row (A, C, E, G, I, K, M) and control incubations in PBS on lower row (B, D, F, H, J, L, N). Note resistance to digestion by AluI restriction enzyme on laminin (Lam) and RGD, but digestion on FBS, collagen Type I (coll), water, epithelial growth factor (EGF), and basic fibroblast growth factor (bFGF). Comparison of aggressive breast carcinoma cell line MDA-MB231 on different matrix molecules: O, Laminin; P, FBS; Q, RGD-C; and R, collagen Type I. Note the resistance to digestion on laminin and RGD, but susceptibility to digestion on FBS and collagen Type I (Col l). Comparison of MCF10A breast epithelial cells on matrix molecules: S, laminin; T, PBS control, U, serum; V, collagen Type I. Scale bars: A–N = 20 μm; O–U = 40 μm.
We then tested MCF10A nonmalignant breast cells and MDA-MB231, a metastatic breast cancer cell line with the cell smear assay. Similar to melanoma cells, 30 minutes of incubation with laminin or RGD-C before exposure to the AluI restriction enzyme rendered the chromatin of both MB321 and MCF10A cells resistant to digestion, whereas incubation of the same cells with collagen I and RGD-C rendered the chromatin susceptible to digestion (Figure 8, O–V).
Permeabilized Cell Assay with Polymerized ECM Molecules
In vivo, cells are exposed to both soluble and polymerized ECM molecules. Therefore, to test the exposure and sequestration of AluI sensitive sites on cells exposed to polymerized ECM molecules, we developed a permeabilized cell model system in which cells were situated on heat-denatured and localized thin ECM films of varying compositions. These heat-denatured ECM films resisted penetration and rapid digestion by highly invasive, matrix invading tumor cells, allowing the cells to rest on the ECM surface where they were accessible to various reagents after permeabilization with Triton X-100.
For this assay, we constructed a grid of different ECM molecules on glass slides: laminin or Matrigel deposited as squares, and dried FBS or Type I collagen in the rows between squares. Cells were then distributed evenly over the slide before exposure to the AluI restriction enzyme (see diagram in Figure 9, A and B).
Figure 9.
Effect of polymerized ECM molecules on sensitivity of Alu sites to AluI restriction enzyme with the permeabilized cell assay. A: Representation of a matrix chip on which squares of laminin or Matrigel are separated from each other by rows of pre-adsorbed FBS or Type I collagen (thick black lines). B: Phase-contrast micrograph of the matrix grid patterns. The squares in this grid are composed of laminin, and the intersquare rows are composed of FBS. C: Phase-contrast micrograph of highly invasive MDA-MB231 breast carcinoma cells seeded on laminin squares and FBS intersquare rows. D: Same field as C showing MDA-MB231 cells labeled with ethidium bromide after 3 hours of digestion with AluI restriction enzyme. The cells on the laminin square with brightly fluorescent nuclei have resisted digestion by AluI restriction enzyme. The small fluorescent dots in the intersquare rows are nucleoli. E: MDA-MB231 cells are easily dislodged from the grid surface with a microneedle, demonstrating that cells are on the surface of the laminin square and are therefore completely accessible to the AluI restriction enzyme. F: Three MCF10A breast epithelial cells at the interface between a laminin square and the FBS intersquare row. Two cells touch the edge of the laminin square, but one cell (black arrow) is situated within the FBS intersquare row. G: Corresponding fluorescence micrograph of the same field as in F. Only the two cells in direct contact with edge of the laminin square are not digested by AluI restriction enzyme. The nucleus of the one cell not in contact with the laminin square is digested (white arrow), and only its nucleolus (small dots) labels with ethidium bromide. H: Phase contrast of a cluster of MCF10A cells treated with AluI restriction enzyme for 60 minutes; the cluster of cells straddles the interface (arrows) between dried FBS (left) and laminin (lam; right). The cells on dried FBS (arrowheads) have flattened and spread, while the cells on laminin are round. I: Corresponding fluorescence micrograph of the same field as in H. The nuclei of three flattened cells on FBS (arrowheads) digest with AluI restriction enzyme, leaving only nucleoli undigested, while the cells on laminin resisted to AluI digestion. J: Phase contrast of a OCM1a cells treated with AluI restriction enzyme for 60 minutes; the cells are distributed both on the laminin square and on the FBS intersquare row. K: Corresponding fluorescence micrograph of the same field as in J. DNA of OCM1a melanoma cells on laminin resisted digestion by the AluI restriction enzyme, but DNA in the same cells situated on FBS digested completely: only the nucleoli are fluorescent. L: Fluorescence micrograph of metastatic MUM2B melanoma cells on laminin (top) collagen (bottom) grid. DNA in the melanoma cells on laminin resisted AluI enzyme digestion, while DNA from the same cells on collagen digested completely. M: Phase micrograph of same preparation illustrated in L showing MUM2B melanoma cells on laminin (top) and collagen (bottom) during digestion with AluI restriction enzyme. Note the horizontal orientation of the nucleolus marked by the arrow. N: Same field as in M but 3 minutes later. Note the vertical orientation of the nucleolus marked by the arrow indicating Brownian motion. Note that the nuclear membrane of the melanoma cells on laminin is sharply demarcated but the nuclei of the same cells on collagen appear to have no border: the chromatin of cells on collagen has been completely digested.
Polymerized laminin or Matrigel, but not FBS or Type I collagen, greatly increased the resistance of chromatin digestion by AluI restriction enzyme in MDA-MB231 cells (Figure 9, C and D). To ensure that cells had complete access to the enzymes and dyes added after the cells were permeabilized with Triton X-100, we dislodged the cells using a microneedle, and verified that they were indeed on the heat-denatured polymerized ECM surfaces instead of being buried within them (Figure 9E).
Identical ECM composition-dependent sensitivities to AluI digestion were observed with MCF10A breast epithelial cells (Figure 9, F–I). Small groups of these cells that were situated along a laminin-dried FBS interface provided the most compelling evidence that ECM contact itself, and not access to restriction enzyme or dye, influenced the digestion of chromatin by AluI. When MCF10A cells made contact with even the edge of a laminin square, DNA resisted digestion by AluI restriction enzyme for up to 60 minutes of exposure to the enzyme (Figure 9F), but cells not in contact with laminin digested in this time frame (Figure 9G). When clusters of MCF10A cells straddled the interface between laminin and dried FBS, the nuclei of cells in contact with laminin resisted digestion by AluI restriction enzyme within 60 minutes, but those cells situated entirely on dried FBS digested completely by 60 minutes (Figure 9, H and I). Matrix-dependent alterations in chromatin sensitivity to AluI restriction enzyme were also observed in melanoma cell lines (Figure 9, J–L).
In all permeabilized cell assays, nucleoli, which are made of ribonucleoprotein and thus resistant to AluI, were used as an internal control and were not digested (Figure 9L). Brownian motion by the ethidium-positive nucleolar residues (Figure 9, M and N) further confirmed digestion of DNA, suggesting disruption of the constraints that DNA imposes on the nucleoli.
To test if matrix composition on AluI site influences sequestration and exposure in mitotic cells, we extracted chromosomes20,21 from aggressive M619 primary uveal melanoma cells grown as a monolayer and placed them side by side with chromosomes extracted from the same cells grown on the surface of polymerized Matrigel. These chromosome sets were then exposed to the AluI restriction enzyme for 60 minutes, labeled with ethidium bromide, and photographed. Chromosomes from the aggressive melanoma cells grown as monolayers digested more rapidly that those extracted from the same cells grown on polymerized Matrigel (Figure 10).
Figure 10.
Comparing the sensitivity to AluI restriction enzyme of chromosomes from aggressive M619 uveal melanoma cells grown in monolayer conditions or on Matrigel. A: Chromosomes are extracted from an M619 cell grown in monolayer conditions. B: Chromosomes are extracted from an M619 uveal melanoma cell in a culture forming networks of cords on the surface of Matrigel. C: The chromosomes extracted in A and B are placed side by side on plastic. D: The chromosomes from M619 cells grown as monolayers after 60 minutes of exposure to AluI restriction enzyme, but the chromosomes from M619 cells grown on the surface of Matrigel resists digestion in this time period. Scale bars: A, B = 10 μm; C, D = 5 μm.
Regulation of Susceptibility to AluI Restriction Enzyme Digestion by Thickness of ECM Gels
The cell smear and polymerized ECM assays provide a limited approximation of the behavior of tumor cells in vivo where these cells are often embedded within the complex ECM of different compositions. Additionally, it is well known that there are large shifts in gene expression between 2D and 3D culture conditions,40–42 suggesting a possible rearrangement of chromatin structure that might be detectable to differential chromatin digestion to AluI. To determine whether the susceptibility and resistance of chromatin to AluI digestion is influenced by the thickness of ECM, we prepared cultures using 0.3 mm thick preparations of native Matrigel.
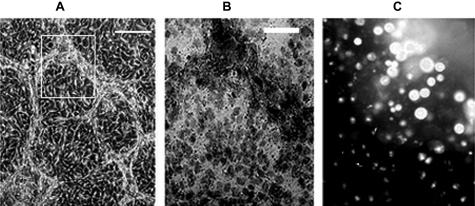
Highly invasive M619 and MUM2B uveal melanoma cells formed looping patterns in thick matrix culture conditions as described previously.13,14 The 3D cultures (Figure 11A) were treated with Triton X-100 and were exposed to Trypan for 10 minutes to ensure that cells buried within matrix were permeabilized (Figure 11B). These cultures were exposed to the AluI restriction enzyme for 1 hour, labeled with ethidium bromide, and immediately photographed. The chromatin of pattern-forming cells in thick ECM resisted digestion with AluI restriction enzyme as indicated by bright nuclear staining with ethidium bromide (Figure 11C). A comparison of gene expression by Affymetrix microarray between MUM619 and MUM2B cells in corresponding flat and 3D culture conditions produced a list of 990 transcripts expressed differentially (651 genes differentially expressed in cells grown in flat culture conditions, and 339 genes differentially expressed by cells in 3D cultures, not shown). A detailed listing and analysis of this differential gene expression will be reported separately.
Figure 11.
Resistance of AluI sites in lysed aggressive uveal melanoma cells to AluI digestion in three-dimensional cultures. A: Phase micrograph of highly invasive melanoma cells exhibiting looping vasculogenic mimicry patterns. B: Bright field photomicrograph taken of the same preparation as in A after 3 minutes of Triton X-100 treatment, and 10 minutes of Trypan blue exposure: all cells exhibit uptake of this dye. Note that the areas where the matrix patterns have formed vasculogenic mimicry patterns (box in A), Trypan blue dye appears to be most concentrated. C: Fluorescent micrograph of ethidium bromide uptake in the same area as shown in B after a 2 hour digestion with AluI: the tumor cell nuclei resist digestion with AluI restriction enzyme. Scale bars: A = 50 μm; B, C = 20 μm.
The Role of the Cytoskeleton in Resistance and Susceptibility to AluI Digestion
Both biochemical and mechanical signaling pathways have been implicated in the remodeling of chromatin.2,20,21,37,43–45 We have previously shown that specific cytoskeletal components alter nuclear shape, molecular alignment of structures in the nucleus, and nucleolar positioning in normal endothelial cells, fibroblasts, and breast cells.2,43 Therefore, we investigated the function of the cytoskeleton in modulating the sensitivity of chromatin digestion by AluI to matrix composition.
We studied normal fibroblasts instead of tumor cells because the cytoskeleton of tumor cells tends to be abnormal.46–48 Cell smear assays were used to assess the response of fibroblast nuclei to AluI digestion under different conditions because fibroblasts tend to spread when serum is present thus facilitating observation of specific cytoskeletal components. Most fibroblasts in the assay flattened and spread in 30 minutes, but rare cells remained round (Figure 12A). After 30 minutes of exposure to AluI restriction enzyme, the chromatin of the flat spreading fibroblasts was digested as detected by ethidium bromide labeling; the chromatin of the fibroblasts that did not spread resisted digestion (Figure 12B). However, after exposure to colchicine for 30 minutes, most of fibroblasts failed to flatten and spread (Figure 12, C and D), and the chromatin of these cells resisted 30 minutes of digestion by AluI restriction enzyme. The chromatin of colchicine-treated fibroblasts that did not flatten and spread remained resistant up to 3 hours of digestion with AluI (not shown). These findings were consistent with the role of the cytoskeleton in regulating cell and nuclear shape2,49 and suggested that microtubules participated in regulating the sequestration or exposure of AluI binding sites.
Figure 12.
Pharmacological inhibition of AluI sensitivity. A: Phase contrast micrograph of WI-38 fibroblasts dried and digested for 30 minutes with AluI restriction enzyme. Most cells are well spread. Note the two non-spreading cells (arrow) B: Fluorescence micrograph of the same cells shown in A after labeling with ethidium bromide. Note that all of the spread cells exhibit nuclei that completely digest after 30 minutes of treatment with AluI, but the nuclei in the cells that do not spread (arrow) resist digestion. C: Phase micrograph of fibroblasts treated with colchicine for 30 minutes, incubated with AluI for 30 minutes, and labeled with ethidium bromide. Note that most of the colchicine-treated cells have not spread (arrow points to a cell that has spread) D: Fluorescence micrograph of the same cells shown in C after labeling with ethidium bromide. Note that all of the nuclei of non-spread cells resist digestion, while the nuclei of the cells that spread (arrow) are completely digested. E: Fluorescence micrograph of WI-38 fibroblasts after drying, digestion with AluI restriction enzyme for 30 minutes, and labeling with ethidium bromide. The nuclei of all cells except for one (bright spot at left) are digested. F: Fluorescence micrograph of WI-38 fibroblasts after 30 minutes of treatment with cytochalasin B followed by 30 minutes of incubation with AluI restriction enzyme. Note that all of the nuclei are completely digested; only nucleoli are labeled. G: Fluorescence micrograph of WI-38 fibroblasts after 30 minutes of treatment with colchicine, followed by a 30-minute incubation with AluI and labeling with ethidium bromide. Note that every nucleus in each spread cell is completely digested while the nuclei in cells not spread completely resist digestion with AluI restriction enzyme. H: Fluorescence micrograph of WI-38 fibroblasts after 30 minutes of treatment with acrylamide, followed by 30 minutes incubation with AluI restriction enzyme and labeling with ethidium bromide. All cells failed to spread during the 30-minute drying period due to the presence of acrylamide, and the nucleus of each cell resists digestion with AluI. Scale bars: A–D = 20 μm; E–H = 20 μm.
To determine whether other components of the cytoskeleton—actin filaments or intermediate filaments—were involved, each component was disrupted in turn. Monolayer cultures of normal fibroblasts were either left untreated or treated for 30 minutes with cytochalasin B to disrupt actin filaments or acrylamide to disrupt intermediate filaments.50 Cells were prepared for a cell smear assay as described above and each preparation was exposed to AluI for 30 minutes before labeling with ethidium bromide. Compared with normal fibroblasts (Figure 12E), the chromatin of fibroblasts treated with cytochalasin B digested completely, whereas the chromatin of fibroblasts treated with either colchicine or acrylamide resisted digestion. Thus, disruption of the actin cytoskeleton exposes chromatin while disruption of intermediate filaments and microtubules sequesters the chromatin.
To investigate structural continuity between the ECM, the cytoskeleton, and the nuclear envelope, we attached 5-μm beads coated with RGD-C peptide to the surfaces of the fibroblasts in serum-free media to form a focal adhesion.2 Within 5 to 30 minutes of bead binding we documented an ultrastructural contiguity between the RGD-bead-induced focal adhesion complexes, microtubules, and the nuclear surface and its pore complexes (Figure 13A). Identical ultrastructural connections between the cell cortex, microtubules and intermediate filaments, and the nuclear surface were identified in normal fibroblasts exposed to laminin (Figure 13B). Whereas microtubules appeared to be predominantly associated with nuclear pore complexes, intermediate filaments in colchicine-treated fibroblasts appeared to associate with the nuclear envelope (Figure 13C).
Figure 13.
Ultrastructural characterization of structural contiguity between an RGD receptor site, the cytoskeleton, and the nucleus. A: RGD-C-coated 5 μm bead attached to a fibroblast for 5 minutes. Note microtubules (arrows) communicating between the focal adhesion forming around the bead and the nuclear pores. B: Normal fibroblast at higher magnification in the vicinity of nuclear pores reveals bowed microtubules apparently flanking the sides of nuclear pores (arrow identified a pore). The microtubular bowing may be the result of compression on these structures. C: Fibroblast photographed in the presence of colchicine to eliminate microtubules, showing intermediate filaments (arrow) associated with the nuclear envelope and possibly nuclear pores. Scale bars: A = 1 μm; B, C = 100 nm.
Discussion
There are significant precedents for the use of nucleases to study chromatin organization in normal cells and cancer cells. Weintraub and Groudine51 demonstrated that active genes in normal cells are preferentially digested by DNase-I. Puck et al52 demonstrated that the reversion of malignant cells to a normal phenotype by exposure to cyclic AMP derivatives was accompanied by differences in the sequestration and exposure of DNase I-sensitive sites. We tested the hypothesis that chromatin organization as measured by AluI restriction enzyme differs in benign and malignant cells and is regulated by the ECM and the cytoarchitecture.
We demonstrated that there are differences in chromatin digestion by AluI in between normal melanocytes and melanoma cells, normal breast epithelium and breast carcinoma cells, and fibroblasts and fibrosarcomas. These differences are independent of the cell cycle as demonstrated by the identical differential digestion of chromosomes extracted from cells and nuclei from intact cells of varying malignant behavior. The insertion of oncogenes into fibroblast DNA15 resulted in a decrease in the sensitivity of the entire genome to AluI restriction enzyme. Differential sensitivity of chromatin to AluI restriction enzyme is also independent of ploidy.
In performing AluI restriction enzyme chromatin assays, it is important to ensure that the reagents used are free of DTT or β-mercaptoethanol. Both of these reagents disrupt proteins rich in disulfide bonds that play key roles in maintaining the sequestration of AluI sensitive sites. Indeed, alterations in higher order chromatin structure may be mediated by the proteinaceous disulfide-rich nuclear matrix proteins.3,53–57 We further discovered that chromatin isolated from poorly invasive and highly invasive cells are digested at different rates by the non-specific protease, proteinase K, thus suggesting that proteins, in addition to DNA,20,29 play a critical role in maintaining the structural organization of the genome, especially in highly invasive cancer cells.
The differential response of chromatin to AluI restriction enzyme among normal cells and cancer cells of varying levels of invasive behavior suggests that it may be possible to extend this observation to a clinical setting. The cell smear assay used to discriminate between cell lines of different malignant behavior is similar to a diagnostic cytological smear or touch preparation. Preliminary studies indicate that differential chromatin digestion can be used to discriminate between cancer cells and the normal adjacent tissue in human diagnostic specimens (Figure 1), and additional studies are planned to test the sensitivity and specificity of this technique. Additionally, the quantitative discrimination between melanoma cells of different invasive potential in suspension by flow cytometry (Figure 2) suggests another possible and easily standardized translational approach to classification of patient diagnostic samples.
We investigated the possible effects of ECM on the sequestration and exposure of AluI sensitive sites in nonmalignant, benign, and malignant cells because there is considerable evidence that the ECM profoundly influences gene expression.37,45,58 The first evidence for the existence of an ECM-responsive element in the promoter of tissue-specific genes was produced more than a decade ago in one of our laboratories.59–62 In particular, we showed that activation of the ECM-response element could occur only in the context of chromatin and not by transient transfection, and that ECM (laminin, in particular) induced changes in histone acetylation of the ECM-response element (as reported previously62 and in unpublished data).
In this study, we demonstrate for the first time a complete and global reorganization of chromatin in response to ECM composition and thickness. This reorganization was accompanied by differential changes in the expression of 990 genes (data to be reported separately) and is consistent with reports of significant ECM-induced shifts in gene expression.37,45,58–62 In all degrees of malignancy, AluI-sensitive sites became profoundly sequestered when cells were incubated with laminin, Matrigel, or a circular RGD peptide (RGD-C), but became exposed when cells were placed on serum or collagen I.
The mechanism by which the ECM induces changes in AluI binding site sequestration and exposure appears to depend on the cytoskeleton. Disruption of selective elements of the cytoskeleton—microtubules, actin, and intermediate filaments—resulted in profound changes in cell shape as expected, but also in sequestration or exposure of Alu binding sites. We previously demonstrated that mechanical pulling on integrin receptors affected nuclear architecture, and that this effect was mediated differentially by microtubules, intermediate filaments, and actin: birefringence microscopy of living cells demonstrated that molecular alignment of structures within the nucleus changed in 1 second when integrin receptors, but not acetylated LDL receptors had a force applied to them.2 Over longer time frames than 1 second, it has been widely established that different types and quantities of ECM molecules can induce cells and nuclei to rapidly change their shape and differentiation programs simultaneously, and that these changes persist for months in culture.4–6,9–11,63–65
Pharmacological disruption of actin by cytochalasin B enhanced digestion of chromatin by the AluI restriction enzyme. Enhanced sensitivity of AluI-sensitive sites in the presence of cytochalasin may result from disruption of cytoplasmic organization, or because monomeric and polymeric actins are present in the nucleus, they may play a role in regulating chromatin organization and hence transcription.66
The function of intermediate filaments in mediating the susceptibility and resistance of chromatin to AluI restriction enzyme is less certain. In this study, we used acrylamide to solubilize the intermediate filament lattice completely in the cytoplasm while leaving the actin and microtubule lattices unaffected;2 it has been established in living cells that vimentin filaments help stabilize nuclear structure by acting as guy-wires.50,66–70 By solubilizing intermediate filaments, we may have also affected intermediate filaments within the nucleus.67,70
The disruption of microtubules resulted in a profound change in cell shape and the sequestration of Alu binding sites. The chromatin of colchicine-treated cells was resistant to digestion by the AluI restriction enzyme (Figure 12, G and H). The physical coupling of microtubules with nuclear pores observed at the ultrastructural level in this study (Figure 13) is consistent with biophysical findings that associate this component of the cytoskeleton with stabilizing nuclear structure. Microtubules together with cell-ECM adhesions are postulated to hold the cell in place against the inward pull of actin.9 Because of the strong effect of microtubule and intermediate filament inhibitors on Alu sequestration or exposure, and because of the association of microtubules with nuclear pores, the results here suggest that the mechanism for Alu exposure may involve the positioning of microtubules at the nuclear pores. Microtubules may act as compression-resistant structures,9 distorting the surface of the nucleus thereby allowing the nuclear pores to widen and permit the influx of chromatin remodeling enzymes and ions.2,21 After pharmacological disruption of microtubules with colchicine, the nucleus does not distort to the extent observed when microtubules are present.2 It is therefore possible that nuclear pores would assume a more closed status, thereby affecting the exposure of Alu binding sites.
ECM control of chromatin organization as detected by sequestration and exposure of AluI binding sites appears to be a mechanical signaling pathway mediated by the cell cortex, the cytoskeleton, and higher order chromatin structure consistent with a tensegrity model of the cell. This mechano-genomic response to ECM may operate alongside ECM-mediated chemical signaling pathways.1,45 Therefore, changes in the sequestration and exposure of AluI binding sites that are mediated by the cytoskeleton and induced by the ECM suggest novel mechanisms of drug resistance for those agents that bind to or disrupt DNA.
Acknowledgments
We are grateful to Dr. Valerie Lindgren for chromosome analyses of the uveal melanoma cell lines and to Dr. ShriHari Kadkol and Dr. Amy Lin for helpful discussions. RGD-C was a kind gift from Dr. Renata Pasquallini and UM54 melanoma cells were a generous gift from Dr. William Harbour.
Footnotes
Address reprint requests to Andrew J. Maniotis, Department of Pathology, University of Illinois at Chicago, 1819 W. Polk Street, 446 CMW (MC 847), Chicago, IL 60612. E-mail: amanioti@uic.edu.
Supported in part by grant EY10457 from the National Institutes of Health (to R.F.), and grant DEAC03–76SF0098 from the Department of Energy (to M.J.B.).
This article is featured in a commentary by G. Stein (Am J Pathol 2005, 166:959–962), published in their issue.
References
- Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- Maniotis A, Chen C, Ingber D. Demonstration of mechanical interconnections between integrins, cytoskeletal filaments, and nuclear scaffolds that stabilize nuclear structure. Proc Nat Acad Sci USA. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Nuclear protein matrix: association with newly synthesized DNA. Science. 1975;189:291–293. doi: 10.1126/science.1145202. [DOI] [PubMed] [Google Scholar]
- Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–349. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- Ingber DE, Madri JA, Folkman J. Endothelial growth factors and extracellular matrix regulate DNA synthesis through modulation of cell and nuclear expansion. J In Vitro Cell Dev Biol. 1987;23:387–394. doi: 10.1007/BF02620997. [DOI] [PubMed] [Google Scholar]
- Ingber DE, Folkman J. Mechanochemical switching between growth and differentiation during fibroblast growth factor-stimulated angiogenesis in vitro: role of extracellular matrix. J Cell Biol. 1989;198:317–330. doi: 10.1083/jcb.109.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohman RC, Bayne E, Spector D, Obinata T, Micou-Eastwood J, Maniotis A. Myogenesis and histogenesis of skeletal muscle on flexible membranes in vitro. In Vitro Cell Dev Biol. 1990;25:201–208. doi: 10.1007/BF02624113. [DOI] [PubMed] [Google Scholar]
- Sjakste NI, Sjakste TG. The characteristics of the molecular organization of the cell nucleus in leukocytes due to the degree of their differentiation. Vestn Ross Akad Med Nauk. 1993:56–63. [PubMed] [Google Scholar]
- Ingber DE, Dike L, Hansen L, Karp S, Liley H, Maniotis A, McNamee H, Mooney D, Plopper G, Sims J. Cellular tensegrity: exploring how mechanical changes in the cytoskeleton regulate cell growth, migration, and tissue pattern during morphogenesis. Int Rev Cytol. 1994;150:173–224. doi: 10.1016/s0074-7696(08)61542-9. [DOI] [PubMed] [Google Scholar]
- Singhvi R, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE. Engineering cell shape and function. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- Roskelley CD, Srebrow A, Bissell MJ. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr Opin Cell Biol. 1995;7:736–747. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LMG, Pe’er J, Trent JM, Meltzer PS, Hendrix MJC. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol. 1999;155:739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis AJ, Chen X, Garcia C, DeChristopher PJ, Wu D, Pe’er J, Folberg R. Control of melanoma morphogenesis, endothelial survival, and perfusion by extracellular matrix. Lab Invest. 2002;82:1031–1043. doi: 10.1097/01.lab.0000024362.12721.67. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Briand P, Petersen OW, van DB. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol. 1987;23:181–188. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- Briand P, Nielsen KV, Madsen MW, Petersen OW. Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Res. 1996;56:2039–2044. [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G. DNA content measurement for DNA ploidy and cell cycle analysis. Robinson JP, editor. New York: John Wiley & Sons, Inc.,; Current Protocols in Cytometry. 1997 doi: 10.1002/0471142956.cy0705s00. [DOI] [PubMed] [Google Scholar]
- Maniotis A, Bojanowski K, Ingber D. Mechanical continuity and reversible chromosome disassembly within intact genomes removed from living cells. J Cell Biochem. 1997;65:114–130. [PubMed] [Google Scholar]
- Bojanowski K, Maniotis AJ, Plisov S, Larsen AK, Ingber DE. DNA topoisomerase II can drive changes in higher order chromosome architecture without enzymatically modifying DNA. J Cell Biochem. 1998;69:127–142. doi: 10.1002/(sici)1097-4644(19980501)69:2<127::aid-jcb4>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Hoskins GC. Electron microscopic observations of human chromosomes isolated by micrurgy. Nature. 1965;207:1215–1216. doi: 10.1038/2071215a0. [DOI] [PubMed] [Google Scholar]
- Hoskins GC. Sensitivity of micrurgically removed chromosomal spindle fibres to enzyme disruption. Nature. 1968;217:748–750. doi: 10.1038/217748a0. [DOI] [PubMed] [Google Scholar]
- Diacumakos EG, Holland S, Pecora P. Chromosome displacement in and extraction from human cells at different mitotic stages. Nature. 1971;232:33–36. doi: 10.1038/232033a0. [DOI] [PubMed] [Google Scholar]
- Korf BR, Diacumakos EG. Random arrangement of mitotic chromosomes in radial metaphases of the Indian muntjac. Cytogenet Cell Genet. 1977;19:335–343. doi: 10.1159/000130827. [DOI] [PubMed] [Google Scholar]
- Korf BR, Diacumakos EG. Absence of true interchromosomal connectives in microsurgically isolated chromosomes. Exp Cell Res. 1980;130:377–385. doi: 10.1016/0014-4827(80)90015-4. [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B, Marko JF, Chatenay D, Libchaber A. Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. J Cell Biol. 1997;139:1–12. doi: 10.1083/jcb.139.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojanowski K, Ingber DE. Ionic control of chromosome architecture in living and permeabilized cells. Exp Cell Res. 1998;244:286–294. doi: 10.1006/excr.1998.4205. [DOI] [PubMed] [Google Scholar]
- Poirier MG, Marko JF. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc Natl Acad Sci USA. 2002;99:15393–15397. doi: 10.1073/pnas.232442599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AR, Bailey N, Damsky C, Petersen OW, Bissell MJ. Cellular growth and survival are mediated by beta 1 integrins in normal human breast epithelium but not in breast carcinoma. J Cell Sci. 1995;108:1945–1957. doi: 10.1242/jcs.108.5.1945. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci. 2003;116:2377–2388. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, Bissell MJ. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P, Siegel AJ, Berezney R. A comprehensive study on the isolation and characterization of the HeLa S3 nuclear matrix. J Cell Sci. 1991;98 (Pt 3):281–291. doi: 10.1242/jcs.98.3.281. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- Roskelley CD, Desprez PY, Bissell MJ. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci USA. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau N, Myers C, Bissell MJ. From laminin to lamin: regulation of tissue-specific gene expression by the ECM. Trends Cell Biol. 1995;5:1–4. doi: 10.1016/s0962-8924(00)88924-2. [DOI] [PubMed] [Google Scholar]
- Lin CQ, Dempsey PJ, Coffey RJ, Bissell MJ. Extracellular matrix regulates whey acidic protein gene expression by suppression of TGF-alpha in mouse mammary epithelial cells: studies in culture and in transgenic mice. J Cell Biol. 1995;129:1115–1126. doi: 10.1083/jcb.129.4.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srebrow A, Friedmann Y, Ravanpay A, Daniel CW, Bissell MJ. Expression of Hoxa-1 and Hoxb-7 is regulated by extracellular matrix-dependent signals in mammary epithelial cells. J Cell Biochem. 1998;69:377–391. doi: 10.1002/(sici)1097-4644(19980615)69:4<377::aid-jcb1>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Li D, Lelievre S, Bissell MJ, Brugge JS. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat Cell Biol. 2001;3:785–792. doi: 10.1038/ncb0901-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15:753–762. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli CH, Bailey N, Bissell MJ. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J Cell Biol. 1991;115:1383–1395. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holth LT, Chadee DN, Spencer VA, Samuel SK, Safneck JR, Davie JR. Chromatin, nuclear matrix and the cytoskeleton: role of cell structure in neoplastic transformation (review). Int J Oncol. 1998;13:827–837. doi: 10.3892/ijo.13.4.827. [DOI] [PubMed] [Google Scholar]
- Davie JR, Spencer VA. Signal transduction pathways and the modification of chromatin structure. Prog Nucleic Acid Res Mol Biol. 2001;65:299–340. doi: 10.1016/s0079-6603(00)65008-0. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Chu YW, Trevor KT, Seftor RE. Role of intermediate filaments in migration, invasion and metastasis. Cancer Metastasis Rev. 1996;15:507–525. doi: 10.1007/BF00054016. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Trevor KT. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am J Pathol. 1997;150:483–495. [PMC free article] [PubMed] [Google Scholar]
- Hendrix MJC, Seftor EA, Seftor REB, Gardner LM, Boldt HC, Meyer M, Pe’er J, Folberg R. Regulation of uveal melanoma interconverted phenotype by hepatocyte growth factor/scatter factor (HGF/SF). Am J Pathol. 1998;152:855–863. [PMC free article] [PubMed] [Google Scholar]
- Pourati J, Maniotis A, Spiegel D, Schaffer JL, Butler JP, Fredberg JJ, Ingber DE, Stamenovic D, Wang N. Is cytoskeletal tension a major determinant of cell deformability in adherent endothelial cells? Am J Physiol. 1998;274:C1283–C1289. doi: 10.1152/ajpcell.1998.274.5.C1283. [DOI] [PubMed] [Google Scholar]
- Hay M, De Boni U. Chromatin motion in neuronal interphase nuclei: changes induced by disruption of intermediate filaments. Cell Motil Cytoskeleton. 1991;18:63–75. doi: 10.1002/cm.970180107. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Puck TT, Krystosek A, Chan DC. Genome regulation in mammalian cells. Somat Cell Mol Genet. 1990;16:257–265. doi: 10.1007/BF01233362. [DOI] [PubMed] [Google Scholar]
- Capco DG, Wan KM, Penman S. The nuclear matrix: three-dimensional architecture and protein composition. Cell. 1982;29:847–858. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, Coffey DS. A structural analysis of the role of the nuclear matrix and DNA loops in the organization of the nucleus and chromosome. J Cell Sci Suppl. 1984;1:123–135. doi: 10.1242/jcs.1984.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- Nickerson JA, Krockmalnic G, Wan KM, Turner CD, Penman S. A normally masked nuclear matrix antigen that appears at mitosis on cytoskeleton filaments adjoining chromosomes, centrioles, and midbodies. J Cell Biol. 1992;116:977–987. doi: 10.1083/jcb.116.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini MA, He D, Ouspenski II, Brinkley BR. Dynamic continuity of nuclear and mitotic matrix proteins in the cell cycle. J Cell Biochem. 1996;62:158–164. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C158::AID-JCB3%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Roti Roti JL, Wright WD, VanderWaal R. The nuclear matrix: a target for heat shock effects and a determinant for stress response. Crit Rev Eukaryot Gene Expr. 1997;7:343–360. doi: 10.1615/critreveukargeneexpr.v7.i4.30. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Weaver VM, Lelievre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59:1757–1763s. [PubMed] [Google Scholar]
- Schmidhauser C, Bissell MJ, Myers CA, Casperson GF. Extracellular matrix and hormones transcriptionally regulate bovine beta-casein 5′ sequences in stably transfected mouse mammary cells. Proc Natl Acad Sci USA. 1990;87:9118–9122. doi: 10.1073/pnas.87.23.9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser C, Casperson GF, Myers CA, Sanzo KT, Bolten S, Bissell MJ. A novel transcriptional enhancer is involved in the prolactin- and extracellular matrix-dependent regulation of beta-casein gene expression. Mol Biol Cell. 1992;3:699–709. doi: 10.1091/mbc.3.6.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser C, Casperson GF, Bissell MJ. Transcriptional activation by viral enhancers: critical dependence on extracellular matrix-cell interactions in mammary epithelial cells. Mol Carcinog. 1994;10:66–71. doi: 10.1002/mc.2940100203. [DOI] [PubMed] [Google Scholar]
- Myers CA, Schmidhauser C, Mellentin-Michelotti J, Fragoso G, Roskelley CD, Casperson G, Mossi R, Pujuguet P, Hager G, Bissell MJ. Characterization of BCE-1, a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of histone acetylation. Mol Cell Biol. 1998;18:2184–2195. doi: 10.1128/mcb.18.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerman JT, Pitelka DR. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro. 1977;13:316–328. doi: 10.1007/BF02616178. [DOI] [PubMed] [Google Scholar]
- Lee EY, Lee WH, Kaetzel CS, Parry G, Bissell MJ. Interaction of mouse mammary epithelial cells with collagen substrata: regulation of casein gene expression and secretion. Proc Natl Acad Sci USA. 1985;82:1419–1423. doi: 10.1073/pnas.82.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ML, Aggeler J, Farson DA, Hatier C, Hassell J, Bissell MJ. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc Natl Acad Sci USA. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amankwah KS, De Boni U. Ultrastructural localization of filamentous actin within neuronal interphase nuclei in situ. Exp Cell Res. 1994;210:315–325. doi: 10.1006/excr.1994.1044. [DOI] [PubMed] [Google Scholar]
- Goldman AE, Maul G, Steinert PM, Yang HY, Goldman RD. Keratin-like proteins that coisolate with intermediate filaments of BHK-21 cells are nuclear lamins. Proc Natl Acad Sci USA. 1986;83:3839–3843. doi: 10.1073/pnas.83.11.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milankov K, De Boni U. Cytochemical localization of actin and myosin aggregates in interphase nuclei in situ. Exp Cell Res. 1993;209:189–199. doi: 10.1006/excr.1993.1301. [DOI] [PubMed] [Google Scholar]
- Sahlas DJ, Milankov K, Park PC, De Boni U. Distribution of snRNPs, splicing factor SC-35 and actin in interphase nuclei: immunocytochemical evidence for differential distribution during changes in functional states. J Cell Sci. 1993;105 (Pt 2):347–357. doi: 10.1242/jcs.105.2.347. [DOI] [PubMed] [Google Scholar]
- Tolstonog GV, Sabasch M, Traub P. Cytoplasmic intermediate filaments are stably associated with nuclear matrices and potentially modulate their DNA-binding function. DNA Cell Biol. 2002;21:213–239. doi: 10.1089/10445490252925459. [DOI] [PubMed] [Google Scholar]
- Kan-Mitchell J, Mitchell MS, Rao N, Liggett PE. Characterization of uveal melanoma cell lines that grow as xenografts in rabbit eyes. Invest Ophthalmol Vis Sci. 1989;30:829–834. [PubMed] [Google Scholar]