Abstract
BALB/c Fechm1Pas mice have a mutated ferrochelatase gene resulting in protoporphyria that models the hepatic injury occurring sporadically in human erythropoietic protoporphyria. We used this mouse model to study the development of the injury and to compare the dysfunction of heme synthesis with hepatic gene expression of liver metabolism, oxidative stress, and cellular injury/inflammation. From an early age expression of total cytochrome P450 and many of its isoforms was significantly lower than in wild-type mice. However, despite massive accumulation of protoporphyrin in the liver, expression of the main genes controlling heme synthesis and catabolism (Alas1 and Hmox1, respectively) were only modestly affected even in the presence of the cytochrome P450-inducing CAR agonist 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene. In contrast, in BALB/c mice exhibiting griseofulvin-induced hepatic protoporphyria with induction and destruction of cytochrome P450, both Alas1 and Hmox1 genes were markedly up-regulated. Other expression profiles in BALB/c Fechm1Pas mice identified roles for oxidative mechanisms in liver injury while modulated gene expression of hepatocyte transport proteins and cholesterol and bile acid synthesis illustrated the development of cholestasis. Subsequent inflammation and cirrhosis were also shown by the up-regulation of cytokine, cell cycling, and procollagen genes. Thus, gene expression profiles studied in Fechm1Pas mice may provide candidates for human polymorphisms that explain the sporadic hepatic consequences of erythropoietic protoporphyria.
Heme is vital to the transport and utilization of oxygen in oxidative and signaling pathways. A large proportion of heme usage in the liver is accounted for by cytochrome P450 isoforms, many of which are associated with drug metabolism. In the final step of heme synthesis ferrous iron is inserted into the precursor porphyrin protoporphyrin IX by the mitochondrial enzyme ferrochelatase. The structure and regulation of ferrochelatase have been extensively studied.1 In humans, mutations in the genes of heme synthesis may lead to clinical symptoms associated with toxic accumulation of precursor molecules.2 These may occur in both erythroid and nonerythroid tissue especially the liver. In patients with erythropoietic protoporphyria (EPP), that have mutations of the ferrochelatase gene, leakage of protoporphyrin IX into plasma and its accumulation in tissues may lead to toxic actions in the skin because of light activation and free radical generation.3–5 In a minority of patients there is hepatic injury, cirrhosis, and jaundice, which in extreme circumstances requires liver transplantation.6–8 This might be because of the primary disturbance of hepatic heme synthesis or a secondary consequence of deposition in the liver of protoporphyrin formed in the erythropoietic system. How protoporphyrin in a dark environment could cause toxicity is not understood or why only some patients develop progressive liver failure.6
The Fechm1Pas mutant BALB/c mouse contains a point mutation in the ferrochelatase gene.9,10 The resulting enzyme exhibits less than 5% of normal ferrochelatase activity in liver and spleen and 6% of the wild-type activity when expressed as a recombinant protein. This leads to insufficiency in erythropoietic heme synthesis, hemolytic anemia, and splenic enlargement. The mice suffer from phototoxicity that can be alleviated by bone marrow transplantation and gene therapy.11,12 Protoporphyrin accumulates in the liver and plasma of mice with associated elevated plasma transaminases and bilirubin and hyperlipidemia.9,13,14 Progressive hepatobiliary injury leads to biliary fibrosis and neoplastic development13,15 but whether the liver is suffering from heme deficiency and how the toxicity arises are not clear. A proposal has been made for the involvement of cytotoxic bile containing high concentrations of bile salts and protoporphyrin that might damage bile duct epithelium and cause biliary fibrosis.13
Although initiated by a ferrochelatase mutation, the pathological consequences of protoporphyria are likely to be the result of a complex interaction between metabolic systems. To understand mechanisms of liver injury and demonstrate possible candidate genes that could explain the sporadic incidence of liver injury in EPP, we investigated hepatic gene expression in the mutant mouse using cDNA microarray analysis. We focused on expression of cytochrome P450 hemoproteins and of genes associated with bile acid and lipid metabolism, oxidative stress, and inflammation that might be affected by possible disturbance of heme synthesis and deposition of protoporphyrin (Figure 1). Results were compared to those obtained with mice exposed to the drug griseofulvin that causes protoporphyria by initiating formation in the liver of an inhibitor of ferrochelatase.16
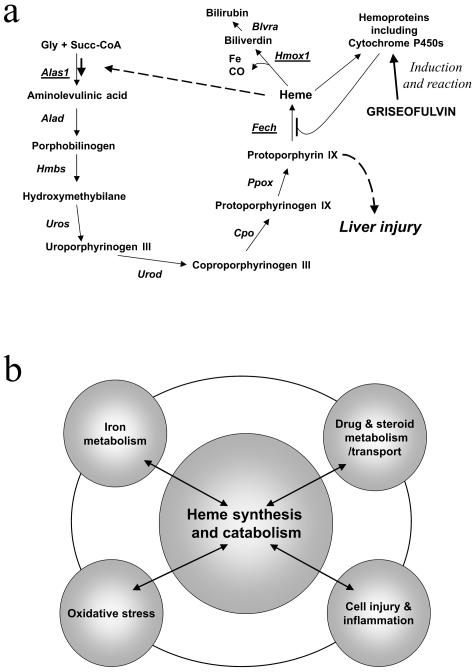
Figure 1.
Hepatic heme synthesis leading to protoporphyria and possible impact with other metabolic systems. a: Scheme of heme metabolism showing negative feedback deregulation of ALA synthase 1 and cytochrome P450 induction and inhibition of ferrochelatase after griseofulvin treatment. b: Scheme of possible metabolic pathways and systems that might impact on hepatic heme metabolism.
Materials and Methods
Animals and Treatments
The BALB/c-Fechm1Pas mouse strain (abbreviated to Fech mice) was originally produced by ethylnitrosourea mutagenesis and backcrossed on to BALB/c mice.9 The mutation appears to be inherited as a single autosomal recessive gene. The strain was obtained from the Jackson Laboratories, Bar Harbor, ME, and bred according to Home Office regulations under project licenses 80/1329 and 40/2571 by homozygous mating while maintained at 21°C under reduced light to protect from skin lesions and fed RM1 diet (SDS, Witham, UK). The data shown in Tables 1 and 2 were obtained by crossing F1 mice from mating female BALB/c mice with male homozygous Fechm1Pas mutants to give male BALB/c mice −/−, −/+, and +/+ for the Fechm1Pas mutation (wild types, heterozygotes, and homozygotes, respectively). For studies with older mice, age-matched control BALB/c mice were obtained from Harlan Ltd., Bicester, UK, or bred in-house. Fech mice were culled within a year as a consequence of hepatic adenomas or carcinomas. All mice analyzed were males. Mice were administered 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) by intraperitoneal injection (3 mg/kg) dissolved in corn oil (2.5 ml/kg), which was also used as a vehicle control, at 8 weeks of age and analyzed after 3 days. BALB/c mice (6 to 8 weeks old) were administered griseofulvin (Sigma, Poole, UK) (as 1% of the diet containing 2% arachis oil) for 3 weeks.17
Table 1.
Phenotypes of F2 Mice Generated from BALB/c × Fechm1Pas Mutation
| Fech mutation | n | Body wt (g) | Liver wt (g) | Hepatic protoporphyrin (nmol/g) | Plasma ALT (U/L) | Plasma bilirubin
|
|
|---|---|---|---|---|---|---|---|
| Direct (μmol/L) | Total (μmol/L) | ||||||
| Wild type | 6 | 26.1 ± 1.8 | 1.37 ± 0.23 | 1.3 ± 0.5 | 28 ± 8 | 0.7 ± 0.7 | 0.8 ± 1.2 |
| Heterozygotes | 14 | 26.0 ± 2.1 | 1.36 ± 0.21 | 1.1 ± 0.3 | 28 ± 9 | 1.4 ± 1.6 | 1.1 ± 0.8 |
| Homozygotes | 5 | 19.1 ± 3* | 2.05 ± 0.51* | 902 ± 366* | 522 ± 67* | 94 ± 59* | 82 ± 16* |
Female BALB/c mice were mated with male BALB/c Fechm1Pas mutant mice to give a F1 generation. From these a batch of F2 mice was produced and the 25 males (8 weeks old) were assessed for the Fech mutation as described in Materials and Methods.
Significantly different from heterozygote and homozygote mice (P < 0.05).
Table 2.
Influence of the Homozygous Fechm1Pas Mutation on the Hepatic Expression of Genes in Systems of Metabolism and Injury
| Gene | Gene symbol | 4 Weeks Fech/wild-type | P value | 8 Weeks Fech/wild-type | P value |
|---|---|---|---|---|---|
| Heme synthesis/catabolism | |||||
| Aminolevulinic acid synthase 1 | Alas1 | 1.11 | 1.86 | 0.042 | |
| Aminolevulinic acid synthase 2 | Alas2 | 0.54 | 0.039 | 0.16 | 0.006 |
| Aminolevulinate acid dehydratase | Alad | 1.06 | 0.81 | ||
| Hydroxymethylbilane synthase | Hmbs | 1.02 | 0.92 | ||
| Uroporphyrinogen III synthase | Uros | 0.89 | 1.18 | ||
| Uroporphyrinogen decarboxylase | Urod | 1.19 | 0.88 | ||
| Coproporphyrinogen oxidase | Cpo | 0.96 | 0.71 | ||
| Protoporphyrinogen oxidase | Ppox | 1.01 | 1.25 | ||
| Ferrochelatase | Fech | 0.92 | 0.76 | ||
| Heme oxygenase 1 | Hmox1 | 1.95 | 0.014 | 0.94 | |
| Heme oxygenase 2 | Hmox2 | 1.22 | 0.80 | ||
| Biliverdin reductase A | Blvra | 1.36 | 0.60 | ||
| UDP-glucuronosyltransferase 1a6 | Ugt1a6 | 0.57 | 0.70 | 0.035 | |
| Cytochrome P450/steroid metabolism | |||||
| Cytochrome P450 reductase | Por | 1.41 | 0.99 | ||
| Cytochrome b5 | Cyb5 | 0.98 | 0.92 | ||
| Cytochrome P450 1A2 | Cypla2 | 0.53 | 0.026 | 0.35 | 0.025 |
| Cytochrome P450 2A12 | Cyp2a12 | 0.97 | 0.62 | ||
| Cytochrome P450 2A5* | Cyp2a5 | 1.3 | 1.65 | ||
| Cytochrome P450 2B10† | Cyp2b10 | 0.59 | 0.72 | 0.037 | |
| Cytochrome P450 2B13 | Cyp2b13 | 0.48 | 0.006 | 0.65 | |
| Cytochrome P450 2C29 | Cyp2c29 | 0.22 | 0.028 | 0.18 | 0.006 |
| Cytochrome P450 2D10 | Cyp2d10 | 0.7 | 0.48 | 0.042 | |
| Cytochrome P450 2D26 | Cyp2d26 | 1.08 | 0.51 | 0.030 | |
| Cytochrome P450 2E1 | Cyp2e1 | 0.3 | 0.05 | 0.18 | 0.007 |
| Cytochrome P450 2F2 | Cyp2f2 | 0.25 | 0.011 | 0.09 | 0.009 |
| Cytochrome P450 2J6 | Cyp2j6 | 0.72 | 0.73 | 0.011 | |
| Cytochrome P450 3A11 | Cyp3a11 | 1.07 | 1.54 | 0.002 | |
| Cytochrome P450 3A25 | Cyp3a25 | 0.55 | 0.41 | 0.04 | |
| Cytochrome P450 4A10 | Cyp4a10 | 0.45 | 0.006 | 0.17 | 0.012 |
| Cytochrome P450 7A1 | Cyp7a1 | 1.15 | 1.58 | 0.017 | |
| Cytochrome P450 7B1 | Cyp7b1 | 0.8 | 0.08 | 0.002 | |
| Cytochrome P450 27A1 | Cyp27a1 | 0.81 | 0.24 | 0.034 | |
| Cytochrome P450 51 | Cyp51 | 1.55 | 3.07 | 0.006 | |
| 3-OH-Me glutaryl CoA synthase 1 | Hmgcs1 | 1.24 | 3.54 | 0.013 | |
| 3-OH-Me glutaryl CoA synthase 2 | Hmgcs2 | ND | 0.64 | 0.005 | |
| 3-OH-Me glutaryl CoA reductase | Hmgcr | 1.45 | 1.76 | ||
| 3β-OH steroid 5-dehydrogenase | Hsd3b4 | ND | 0.17 | 0.001 | |
| 11β-OH steroid dehydrogenase | Hsd11b1 | 0.38 | 0.048 | 0.34 | 0.011 |
| Nuclear receptors | |||||
| AHR | Ahr | 1.35 | 1.27 | ||
| HNF-1α | Tcf1 | 1.23 | 1.02 | ||
| HNF-1β | Tcf2 | 0.98 | 1.48 | ||
| HNF-4 | Hnf4 | 1.23 | 1.01 | ||
| LXR | Nr1h3 | 1.15 | 0.57 | ||
| FXR | Nr1h4 | 1.58 | 0.91 | ||
| PXR/SXR | Nr1i2 | 0.93 | 0.76 | ||
| CAR | Nr1i3 | 0.83 | 0.22 | 0.048 | |
| RXR α | Rxra | 0.9 | 1.08 | ||
| PPAR α | Ppara | ND | 0.45 | ||
| Organic transporters | |||||
| ABC‡ B1A (MDR1A) | Abcb1a | 1.64 | 1.95 | 0.010 | |
| ABC‡ B1B (MDR1B) | Abcb1b | 1.41 | 0.030 | 1.24 | |
| ABC‡ B4 (phospholipid transporter) (MDR2) | Abcb4 | 1.58 | 1.37 | ||
| ABC‡ B11 (bile acid transporter) (BSEP) | Abcb11 | 1.09 | 0.70 | ||
| ABC‡ C2 (organic ion transporter) (MRP2) | Abcc2 | 0.99 | 0.46 | ||
| Na taurocholate cotransporter (NTCP) | Slc10a1 | 0.37 | 0.002 | 0.23 | 0.000 |
| Organic anion transporter (OATP1) | Slco1a1 | 1.12 | 0.03 | 0.010 | |
| Organic anion transporter | Slco1a6 | 1.2 | 0.19 | 0.000 | |
| Apolipoprotein A-IV | Apoa4 | 1.75 | 5.38 | 0.000 | |
| Apolipoprotein B editing complex 1 | Apobec1 | 2.51 | 0.000 | 5.56 | 0.020 |
| Fatty acid-binding protein 1 | Fabp1 | 0.35 | 0.36 | 0.000 |
Table 2.
(Continued)
| Gene | Gene symbol | 4 Weeks Fech/wild-type | P value | 8 Weeks Fech/wild-type | P value |
|---|---|---|---|---|---|
| Iron metabolism | |||||
| Transferrin | Trf | 1.38 | 1.69 | 0.043 | |
| Transferrin receptor | Trfr | 0.77 | 0.89 | ||
| Transferrin receptor 2 | Trfr2 | 0.77 | 0.67 | 0.040 | |
| HFE | Hfe | 1.21 | 1.01 | ||
| Ferritin light chain 1 | Ftl1 | 1.09 | 1.62 | 0.000 | |
| Ferritin heavy chain | Fth | 1.59 | 1.52 | 0.009 | |
| Aconitase 1 | Aco1 | 1.11 | 0.95 | ||
| ABC‡ B7 | Abcb7 | 1.14 | 1.10 | ||
| Ferroportin | Slc40a1 | 1.18 | 1.37 | ||
| NRAMP1 | Slc11a1 | 1.43 | 3.21 | 0.037 | |
| β2-microglobulin | B2m | 0.97 | 1.45 | 0.011 | |
| Hepcidin | Hamp | 1.49 | 1.20 | ||
| Hephaestin | Heph | 1.53 | 0.001 | 2.74 | 0.003 |
| Ceruloplasmin | Cp | 2.19 | 3.53 | 0.001 | |
| Hemopexin | Hpxn | 2.45 | 4.54 | 0.003 | |
| Oxidative stress | |||||
| Jun oncogene | Jun | 1.16 | 3.38 | 0.016 | |
| Nuclear factor E2-derived 2, like 2 | Nfe2l2 | 1.63 | 0.006 | 3.92 | 0.003 |
| Metallothionein 1 | Mt1 | 1.19 | 2.52 | 0.008 | |
| Glutathione synthetase | Gss | 1.55 | 0.053 | 4.05 | 0.005 |
| Glutathione peroxidase 4 | Gpx4 | 2.78 | 0.025 | 3.85 | 0.000 |
| Glutathione S-transferase α2 | Gsta2 | 1.79 | 3.58 | 0.000 | |
| Glutathione S-transferase μ1 | Gstm1 | 2.25 | 2.42 | 0.002 | |
| Glutathione S-transferase μ5 | Gstm5 | 1.83 | 1.97 | 0.011 | |
| Fumarylacetoacetate hydrolase | Fah | 0.83 | 0.57 | 0.037 | |
| Catalase | Cat | 0.61 | 0.50 | 0.052 | |
| Superoxide dismutase 2 | Sod2 | 0.87 | 0.43 | 0.032 | |
| Glutamine synthase | Glul | 0.93 | 0.30 | 0.013 | |
| Carbonic anhydrase 3 | Car3 | 0.17 | 0.033 | 0.36 | 0.017 |
| Uncoupling protein 2 | Ucp2 | 2.34 | 0.005 | 3.92 | |
| Inflammation and wound healing | |||||
| TGF-β induced | Tgfbi | 2.02 | 0.013 | 2.58 | 0.003 |
| Transglutaminase 2 | Tgm2 | 2.84 | 0.009 | 4.64 | 0.002 |
| Osteopontin | Spp1 | 6.28 | 0.005 | 10.82 | 0.000 |
| Osteonectin | Sparc | 1.89 | 0.008 | 2.77 | 0.012 |
| Procollagen, type III, α1 | Col3a1 | 4.1 | 0.012 | 5.82 | 0.000 |
| Procollagen, type IV, α1 | Col4a1 | 2.49 | 0.036 | 5.85 | 0.038 |
| Procollagen, type VI, α3 | Col6a3 | 1.97 | 2.94 | 0.008 | |
| Annexin A2 | Anxa2 | 4.1 | 0.013 | 10.17 | 0.007 |
| Annexin A5 | Anxa5 | 4.18 | 0.001 | 6.63 | 0.000 |
| Biglycan | Bgn | 1.8 | 0.017 | 4.12 | 0.001 |
| Chemokine ligand 2 | Ccl2 | 2.83 | 0.003 | 7.94 | 0.011 |
| Cyclin D1 | Ccnd1 | 1.61 | 3.08 | 0.005 | |
| CD14 antigen | Cd14 | 2.82 | 0.002 | 3.87 | 0.003 |
| Lymphocyte antigen 6 complex | Ly6 days | 6.9 | 0.000 | 10.33 | 0.005 |
| Lymphocyte antigen 86 | Ly86 | 2.29 | 0.001 | 7.69 | 0.009 |
Results are the mean of three to four mice per group. Changes in expression are given as normal ratios relative to comparison with the wild-type mice. Degrees of significance (P values) were calculated from the original log2 data that were then converted to the normal values for ease of appreciation. Other genes in the groups were present on the array but have been omitted from the table for the sake of brevity or because the abundance of mRNA was too low for reliable quantitation. Symbols are from Genebank data base (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = unigene).
Also would probably detect Cyp2a4 (cytochrome P450 2A4).
Described as Cyp2b20 in Unigene but categorized as Cyp2b10 by Nelson and co-workers.57
ABC is ATP-binding cassette protein super family.
ND, not determined.
Histological and Biochemical Analyses
Tissues were fixed in neutral-buffered formalin and embedded in paraffin wax. Standard histological sections (5 μm thick) were stained with hematoxylin and eosin. Collagen was demonstrated with van Gieson’s stain. Tissue for electron microscopy was processed and examined as previously reported.18 Plasma alanine aminotransferase (ALT) and bilirubin levels were estimated with Sigma kits. Protoporphyrin levels were determined by fluorescence spectroscopy.19 RNA and microsomes were obtained as reported in previous publications.17,20 Total cytochrome P450 was assessed as described.21
Immunoblotting
Sodium dodecyl sulfate electrophoresis and Western blotting of microsomal proteins were performed as reported previously20 using chemiluminescence detection and primary antibodies from the following sources: heme oxygenase (HMOX1) with a rabbit antibody anti-rat HMOX1 peptide (Stressgen, Victoria, Canada), rat HMOX1 was a positive control; CYP1A2 with a goat antibody against rat enzyme (Gentest, Woburn, MA); CYP2A5 with a rabbit antibody against human CYP2A6 peptide (Chemicon, Temecula, CA) that recognizes the mouse ortholog22 with human CYP2A6 (Gentest) as a positive control; CYP2B10 with rabbit anti-human CYP2B6 (Chemicon). Rat microsomes expressing human CYP2B6 (74.4% homology with mouse CYP2B10) gave a single band but with a lower molecular weight. CYP2E1 and cytochrome P450 reductase were detected using rabbit antibodies (Chemicon) to the human forms and compared with human and rat proteins, respectively, from Gentest. CYP3A11 was detected with a rabbit antibody against rat CYP3A2 that may also recognize CYP2C29 (Gentest). Quantitation was achieved by densitometry and changes expressed relative to wild-type mice.20
Determination of Fech Genotype
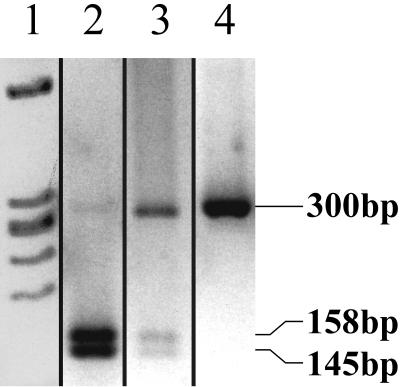
The mutated ferrochelatase gene in BALB/c Fechm1Pas mice carries a T→A transversion at nucleotide 293 leading to a methionine to lysine substitution at position 98.10To confirm the mutation, cDNA clones encoding the appropriate region of the ferrochelatase gene were isolated by amplification of cDNA from total liver RNA from wild-type and Fechm1Pas BALB/c mice using touchdown polymerase chain reaction (PCR) with primers 5′-GGTACATCATGCCAAGACCA-3′ and 5′-ACGACCTACTCAATAGGGGA-3′. A fragment of ∼300 bp was observed by 2.5% agarose gel electrophoresis for both strains. After amplification by PCR, purification and sequencing the T→A transversion were confirmed occurring at nucleotide 137 of the product from the Fechm1Pas mouse. The Fech heterozygotes and Fech homozygotes have been distinguished previously by phenotypic characteristics or dot blotting of cDNA fragments followed by hybridization with allele-specific oligonucleotides;9,10,13 neither method has been entirely satisfactory. Instead therefore, the wild-type 300-bp fragment was digested with the restriction enzyme Pag1, to two fragments of 158 and 145 bp. The Pag1 site is not present in the Fechm1Pas mutation and thus wild-type, heterozygous, and homozygous Fech mice were genotyped at the termination of experiments using liver cDNA and this procedure (Figure 2).
Figure 2.
Genotyping of Fechm1Pas mutation by cleavage of cDNA fragment of ferrochelatase. A 300-bp product amplified by PCR from ferrochelatase cDNA of wild-type or mutant mice was digested with Pag1 restriction enzyme. Lane 1, molecular weight markers; lane 2, homozygous wild-type BALB/c (−/−); lane 3, heterozygote; lane 4, homozygous mutant.
RNA Extraction, Labeling, and cDNA Gene Arrays
The mouse cDNA microarray consisted of ∼4000 candidate-expressed sequence tag clones obtained from Research Genetics, now Invitrogen, Carlsbad, CA and the IMAGE collection held originally at MRC Human Geneome Mapping Project. These clones represented many genes encompassing heme, iron, and drug metabolism; liver function; oxidative stress; apoptosis; cell cycling; inflammation; and wound-healing responses. Total RNA was extracted from liver tissues using Tri-Reagent (Sigma, UK). The RNA labeling, hybridization, analysis of fluorescence, and data processing were performed as previously described.17,23,24 The cDNAs of four mice were compared against cDNAs from four wild-type mice as appropriate for Fech or TCPOBOP studies. Clones for all genes reported were verified by sequence analysis.24 Data has been deposited via MIAMExpress in ArrayExpress at the European Bioinformatics Institute, Hinxton, UK (http://www.ebi.ac.uk/arrayexpress). ArrayExpress is a public repository for microarray data in accordance with Microarray Gene Expression Data Society’s MIAME recommendations. The accession numbers are as follows: experiment accession numbers, E-MEXP-189 and E-MEXP-194; array design accession number, A-MEXP-113 and A-MEXP-116.
Statistical Analyses
The microarray data were normalized using a program developed at the MRC Toxicology Unit, version NorTTNov2003.exe using the LOWESS option.25 Statistical analysis of the data were performed using the two-tailed paired t-test on the logarithm (base 2) of the ratio of the data. Statistical significance of phenotype and Western blotting data were performed using two-tailed Student’s t-test.
Real-Time Reverse Transcriptase (RT)-PCR
mRNA expressions from Alas1 and Hmox1 genes were determined by real-time RT-PCR with primers designed to cross exon-exon boundaries to avoid interference from genomic DNA. Primers were designed using Primer Express Software v2.0, Applied Biosystems. Primers were as follows: Hmox1 forward primer, 5′-CACTTCGTCAGAGGCCTGCTA-3′ and reverse primer, 5′-GTCTGGGATGAGCTAGTGCTGAT-3′; Alas1 forward primer, 5′-TCTTCCGCAAGGCCAGTCT-3′ and reverse primer, 5′-TGGGCTTGAGCAGCCTCTT-3′. Results were normalized against β2-microglobulin (β2m). B2m forward primer, 5′-CATACGCCTGCAGAGTTAAGCA-3′; reverse primer, 5′-GATCACATGTCTCGATCCCAGTAG-3′.
cDNA was synthesized from total RNA using random hexamers and SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) as per the manufacturer’s protocols. Product cDNAs (from 10 ng RNA), or a nontemplate control, were incubated with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) containing 900 nmol/L forward primer and 300 nmol/L reverse primer in an ABI Prism 7700 sequence detection system. The thermal-cycler protocol was: 50°C for 2 minutes, 95°C for 10 minutes, 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute.
Results
Phenotypic Characteristics of Fech Mutation
Male wild-type, heterozygous, and homozygous Fech mice bred as a F2 cross from BALB/c and BALB/c Fechm1Pas parent strains at 8 weeks of age representing early adulthood mice were phenotyped for hepatic porphyria and plasma levels of bilirubin and ALT (Table 1). Only the homozygous Fech mice showed hepatic porphyria and elevated plasma levels of ALT and bilirubin. All homozygous Fech mice had enlarged livers and spleens. Livers contained moderate amounts of protoporphyrin as fine droplets of red-brown pigment in hepatocytes as well as variable-sized deposits in bile canaliculi, bile ducts, and interstitial cells of both hepatic portal tracts and parenchyma.9,13 Portal tracts also showed varying degrees of inflammation, fibrosis, bile duct, and oval cell proliferation. The enlarged spleens showed active hematopoiesis in the zones of red pulp. Only 1 heterozygote of 14 exhibited slight hepatic portal tract proliferation, the remainder were identical to the wild-type group.
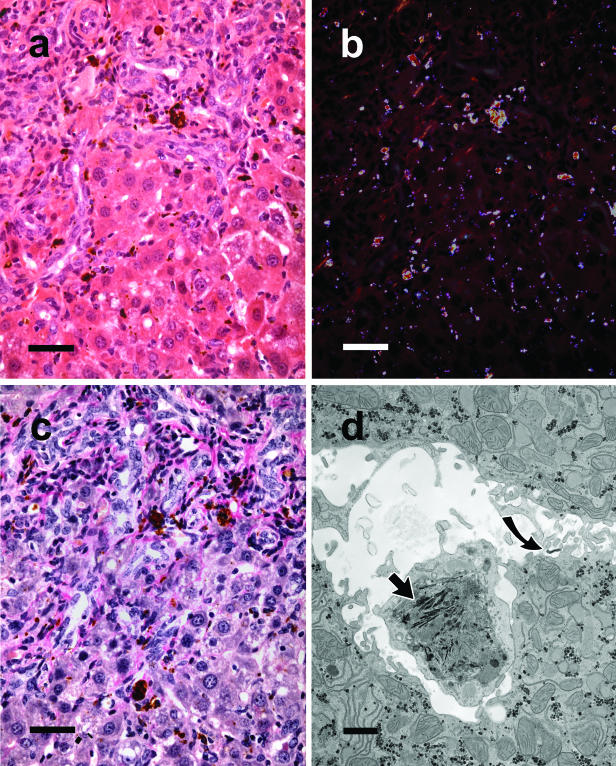
Histological findings in 4-week-old homozygous Fech mice (1 week after weaning) were qualitatively similar to those that were 8 weeks old, but there was obviously less pigment within hepatic parenchyma and less florid proliferative alterations and collagen around portal tracts (data not shown). In comparison, in older Fech mice at 13 weeks and 26 weeks although protoporphyrin continued to be widely distributed, showing red birefringence under polarized light,5 it tended to aggregate into larger deposits around portal tracts with greater inflammation, fibrosis, and proliferation of bile ducts and oval cells than seen at 8 weeks (Figure 3). Cytological changes in hepatocytes became marked with foci of chronic inflammation. By electron microscopy most hepatocytes appeared normal with only a few signs of lipid accumulation. The marked distension and inflammation of portal regions was accompanied by ingress of numerous macrophages containing lipofuscin and crystalline needles of protoporphyrin that were rarely seen in Kupffer cells. Small amounts of densely staining material were seen in the lumen of bile canaliculi and between hepatocytes (Figure 3). By 32 to 52 weeks livers became so enlarged that they accounted for up to 25% of body weight with large eosinophilic and clear cell foci, adenomas, and adenocarcinomas as reported by Libbrecht and colleagues.15
Figure 3.
Microscopic examination of 13-week-old homozygous male mouse Fech liver. a: Shows a portal zone from liver with bile duct proliferation and a scattering of red-brown pigment mainly located in and around portal tracts as well as in interstitial cells of the hepatic parenchyma. b: Shows the same microscopic field under crossed polarizing filters revealing the wide distribution of the pigment. H&E stain. c: Same zone as in a and b stained for collagen. Pink collagen bands can be seen spreading from the portal zones into the hepatic parenchyma as shown by van Gieson’s stain. d: Electron microscopy showing a macrophage containing needle-shaped crystals (arrowhead) in the dilated space between two hepatocytes exhibiting normal ultrastructure. Traces of a densely staining material are also evident (curved arrow) in the extracellular space. Scale bars: 50 μm (a–c); 1 μm (d).
Impact of Fech Mutation on Gene Expression
The expression of a wide range of genes was investigated by cDNA array analysis focusing on metabolic systems that we hypothesized might show interaction (Figure 1). As with histological assessment, few significant changes were detected in heterozygous mice carrying one copy of the Fech mutation and are not discussed further. A summary of the main changes in 4-week-old Fech homozygous mice compared with young adults (8 weeks of age) is shown in Table 2. Most changes were greater at 8 weeks than 4 weeks.
Heme and Hemoprotein Metabolism
Few consistent changes in the regulation of genes of the nonerythroid pathway of heme synthesis were detected in homozygous Fech mice. Thus despite the fact that hepatic ferrochelatase activity is <5% of normal in the livers of homozygous Fech mice,9 this did not lead to a regulatory hepatic heme pool sufficiently lowered to cause transcriptional activation of the Alas1 gene or increased stability mRNA.26,27 Curiously, down-regulation of Alas2, which is associated with erythroid heme synthesis, was observed. The mutated Fech gene was not up-regulated to compensate for the low enzyme activity. There was no evidence that heme catabolism was stimulated because expression of heme oxygenase1 (Hmox1) was not markedly elevated.
Cytochrome P450 isoforms account for a considerable proportion of the total liver hemoprotein content. Many of the genes for major constitutively expressed isoforms such as CYP1A2, CYP2C29, and CYP2E1, were down-regulated in Fech mice even at 4 weeks of age (Table 2). However, the decreased expression of cytochrome P450 genes was not uniform. Cyp3a11, the isoform that is involved in the hydroxylation of bile acids was significantly induced by 8 weeks. The genes for cytochrome P450 reductase and cytochrome b5 were unaffected. The Cyp3a11 gene is a target gene of the pregnane X receptor (PXR) as are a number of other genes involved in liver metabolism whose expression may be mediated in part by bile acid ligands for example Abcb1b (MDR1B).
Pertinent to the established disturbance of bile acid excretion in Fech mice was the effect on the expression of the genes for CYP7 sterol 7α-hydroxylases in bile acid synthesis. CYP7A1 enzyme catalyzes the rate-limiting initial 7α-hydroxylation step in metabolism of cholesterol in the neutral pathway with PXR, LXR, and FXR receptors implicated in the regulation of the gene.28 A small significant increase in expression of Cyp7a1 was observed in Fech mice at 8 weeks. In contrast, the Cyp7b1 gene that codes for a CYP isoform with oxysterol 7α-hydroxylase activity that is not regulated by these receptors was >90% down-regulated in 8-week-old mice. CYP7B1 enzyme acts in the acidic pathway of bile acid synthesis that is initiated by 27-hydroxylation of cholesterol.29 Expression of the Cyp27a1 gene also became depressed (Table 2). Additionally, the mRNA level from Cyp51, a controlling gene of cholesterol synthesis, became significantly increased (Table 2) compatible with the elevated biliary excretion of cholesterol and total bile acids in these mice.13
Expression of Transport Proteins
The proposal that disruption of bile acid metabolism contributed to the mechanism of liver injury was strengthened by expression of organic anion transport proteins concerned with influx and efflux from hepatocytes and chondriocytes and consistent with bile acid-mediated genes in cholestasis.30,31 Basolateral bile salt uptake genes Slc10a1(sodium taurocholate transporter or NTCP) and especially Slco1a1 and Slcola6 (organic anion transporters) became markedly down-regulated in Fech mice. In contrast, the genes Abcb11 and Abcc2 (BSEP and MRP2, respectively) associated with canalicular bile salt and bilirubin efflux showed only a statistically nonsignificant decrease. The Abcb1a gene (MDR1A) was up-regulated and the Abcb4 (MDR2) gene unchanged, the products of which are associated with phospholipid transport and cholestasis.32 Genes for CYP4A10 and fatty acid-binding protein are potentially co-ordinately expressed with Abcb4 via the nuclear receptor PPARα, but were both significantly down-regulated (Table 2). Transcript profiles of apolipoprotein A- and apolipoprotein B-editing complex genes (Table 2) were markedly elevated in the livers of Fech mice but other apolipoprotein clones on our arrays showed no significant changes from wild-type mice (data not shown).
Oxidative Stress and Iron Metabolism
The hypothesis that oxidative stress might play a role in Fech liver injury (Figure 1) was supported by the array data. In particular, expression of genes for Jun, anti-oxidant transcription factor NFE2, glutathione peroxidase 4, glutathione transferases, glutathione synthetase, and metallothionine 1 were increased and other genes decreased as occurs in oxidative stress (Table 2). On the other hand, although heme oxygenase 1 (Hmox1 gene) can be regulated by the anti-oxidant transcription factor NFE2, it showed little increase in Fech mice.
Modest effects on the hepatic expression of iron metabolism genes were detected (Table 2). In particular, synthesis of the plasma heme transporter hemopexin and ceruloplasmin, haephaestin, and NRAMP1, believed to be involved in plasma iron regulation, were significantly induced, possibly as a response to hemolysis and inflammation.
Liver Injury and Inflammation
Whatever the precise mechanism of liver dysfunction as a consequence of protoporphyrin deposition, up-regulation of many genes associated with cytokines and collagens and with lymphocytes and macrophage infiltration (such as Tgm2, Spp1, Col4a1, Anxa5, Cd14, and Ly6d) was consistent with observations of subsequent bile tract inflammation and proliferation (Table 2).
Modulation of Expression with Age
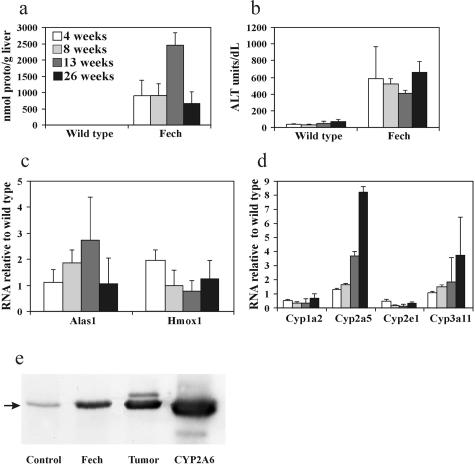
Previous biochemical changes in Fech mice have been examined in mice of an age range of 3 to 6 months.9,13,14 Here hepatic protoporphyrin and plasma ALT levels were compared with gene expression in mice from 4 weeks (1 week after weaning) up to 26 weeks of age. Porphyrin and ALT levels were already markedly increased by 4 weeks although protoporphyrin levels were maximum at 13 weeks whereas ALT levels were sustained throughout the period (Figure 4, a and b). Despite porphyria only a small increase in Alas1 expression was detected at even 13 weeks and little in Hmox1 at any time (Figure 4c). In contrast, genes for major cytochrome P450 isoforms, such Cyp2e1, Cyp1a2, and Cyp2c29 were markedly down-regulated from the earliest time and throughout the studies. Two exceptions were expressions of Cyp3a11 and Cyp2a5 that increased steadily in older mice (Figure 4d). The latter has been associated with porphyria, liver injury, and hepatic tumors.33–35 Western blotting confirmed the marked presence of CYP2A5 protein in livers of 26-week mice and hepatic adenoma tissue (Figure 4e). Changes in gene expressions associated with hepatocyte bile acid up-take, lymphocyte infiltration, remodeling, cirrhosis, and proliferation in older mice were mainly significantly less or absent in the 4-week-old mice compared with 8 weeks (Table 2) and continued to be greatly altered throughout the study period consistent with the more severe pathology.
Figure 4.
Phenotypic and gene expression changes in male Fech mice with increased age from 4 to 26 weeks compared to wild-type mice. a: Hepatic protoporphyrin. b: Plasma ALT. c: Comparison of Alas1 and Hmox1 gene expressions. d: Comparison of Cyp1a2, Cy2a5, Cyp2e1, and Cyp3a11 gene expressions in Fech mice relative to wild-type mice. Values represent means ± of three to five mice. Cyp1a2 and Cy2e1 in Fech were significantly (P < 0.05) less than wild type at all ages. e: Detection of CYP2A5 protein by Western blot in liver from wild-type and Fech mice after 26 weeks and an example of hepatic adenoma (taken from a 42-week-old mouse) using antibody against human CYP2A6 to detect the mouse ortholog.22 Mean increased level for Fech liver was 3.2 of wild-type (n = 3).
Further Studies of Heme Supply
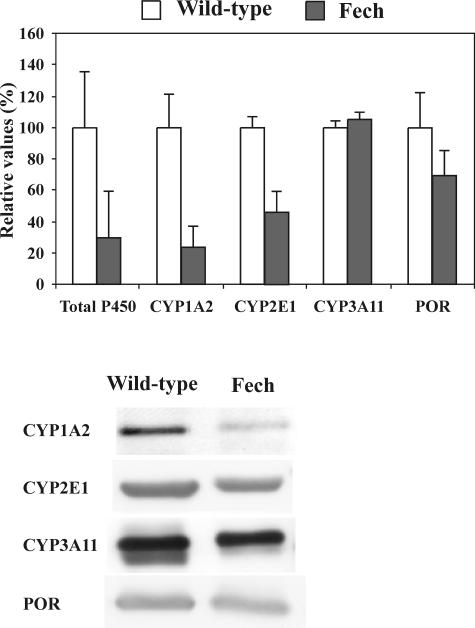
That Alas1 and Hmox1 were not greatly up-regulated suggested that the supply of heme in the protoporphyric Fech mutant mouse was sufficient to obviate major regulatory changes in Alas1 transcription or RNA stability27 (Figure 1) as a consequence of early down-regulation of major cytochrome P450 isoforms. The total hepatic cytochrome P450 level of 6-week-old Fech mice was ∼30% of that in age-matched controls (Figure 5). Western blotting confirmed significantly lower protein levels of the major hepatic isoforms CYP1A2 and CY2E1 in Fech mice compared with wild type. That this was not a simple sequela of general microsomal damage after liver injury was illustrated by the lack of significant effects on expression of Cyp3a11 and cytochrome P450 reductase (Table 2 and Figure 5).
Figure 5.
Comparison of levels of total cytochrome P450 and some specific isoforms in liver microsomes from 6-week-old Fech and wild-type BALB/c mice as assessed by Western blotting. Antibodies, sources, and standards (not shown) were as described in Materials and Methods. Anti-CYP3A11 may also detect CYP2C29. Total cytochrome P450, CYP1A2, and CYP2E1 were significantly (P < 0.05) different between the strains. CYP3A11 and cytochrome P450 reductase (POR) were not different.
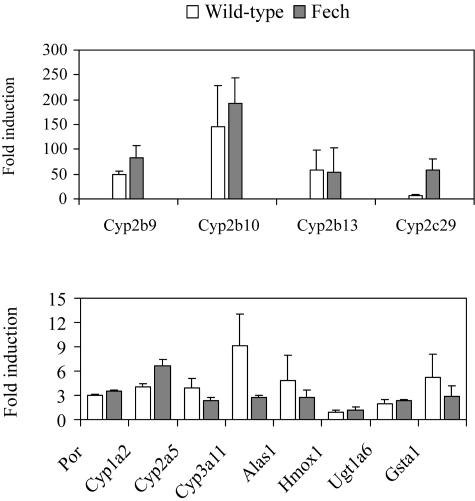
Induction of Cyp2b and Alas1 genes by phenobarbital-type drugs can be effected through the CAR/PXR transcription factors independent of heme status.36,37 After administration of TCPOBOP, a potent CAR agonist and cytochrome P450 inducer, Cyp2b isoforms were markedly up-regulated in control and Fech mice (Figure 6). Alas1 was modestly increased (but not Hmox1) in both strains demonstrating that in Fech mice the potential for up-regulation of the Alas1 gene by this mechanism is present. The induction was lower than in the wild type possibly as a consequence of reduced CAR expression (Table 2).
Figure 6.
Induction of expressions of hepatic cytochrome P450 2B and 5-aminolevulinate synthase 1 genes in BALB/c and Fech mice after exposure to the CAR ligand TCPOBOP. Mice (8 weeks old) were administered TCPOBOP (3 mg/kg) and after 3 days hepatic gene expression was determined by cDNA array analysis. No induction of heme oxygenase 1 (Hmox1) was detected. Full names of genes corresponding to gene symbols can be found in Table 2.
Comparison of Fech and Griseofulvin Porphyric Mice
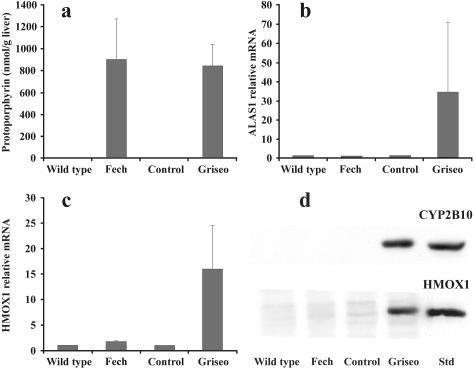
The drug griseofulvin causes protoporphyria in mice by formation of an inhibitor of ferrochelatase in the liver after cytochrome P450 induction (Figure 1).16 When griseofulvin-treated BALB/c mice with porphyria were compared with Fech mice the hepatic levels of protoporphyrin (Figure 7a) and the intra- and intercellular distributions were similar17 as were the depressed ferrochelatase activities (B. Clothier and A.G. Smith, unpublished data). However, comparison of array data from 8-week Fech mice (Table 2) with that generated from griseofulvin-treated BALB/c mice17 indicated that in these two types of protoporphyria Alas1 and Hmox1 showed different responses. Further analysis using real-time RT-PCR demonstrated that expressions of Alas1 and Hmox1 in the livers of Fech mice were not significantly changed whereas both genes were massively induced in the livers of mice treated with griseofulvin (Figure 7, b and c). Western blotting confirmed that the level of HMOX1 protein was greatly increased after drug treatment. In addition, the level of cytochrome P450 protein of the phenobarbital-inducible type CYP2B10, was highly elevated (Figure 7) in line with gene array analysis in our previous study.17 As in other studies38 and the Fech mouse (Table 2) no marked compensatory increase in ferrochelase RNA was detected (data not shown).
Figure 7.
Comparison of hepatic 5-aminolevulinate synthase 1 and heme oxygenase 1 expression, using real-time RT-PCR, in protoporphyric homozygous 8-week-old Fech mice and BALB/c mice treated with griseofulvin for 3 weeks. a: Hepatic protoporphyrin levels. b: RT-PCR of Alas1 expression. c: RT-PCR of Hmox1 expression. Expression of β2-microblobulin was used as a reference. d: Western blotting of proteins for HMOX1 and CYP2B10 using rabbit anti-rat HMOX1 and rabbit anti-human CYP2B6 polyclonal antibodies. HMOX1 mean value was 0.96-fold of wild type for Fech mice and 6.6-fold of controls for griseofulvin-treated mice (n = 3). CYP2B10 mean value was1.2-fold of wild type for Fech mice and 224-fold of controls for griseofulvin-treated mice (n = 3). Rat protein was used as a positive control for HMOX1 and microsomes from TCPOBOP-treated mice as a standard for CYP2B isoforms, particularly CYPB10. Rat cell microsomes expressing human CYP2B6 (CYP2B10 in the mouse is one of the orthologs of this human gene57 with a 74% protein homology) gave a single band but with a lower molecular weight (data not shown).
Discussion
Demonstration of Cholestatic Gene Expression
To our knowledge this is the first example of gene array analysis applied to a model of human porphyrias that is purely genetically derived. The development of cholestasis and liver failure in human EPP is not an automatic progression, although liver disease may develop in patients before minor clinical manifestations.5 The precise mechanism of uptake of protoporphyrin into the liver is not understood if, as is likely, a major part originates from the hemopoietic system. Study of the Fech mouse should increase our understanding. After plasma transport on albumin and hemopexin,39 uptake of protoporphyrin to the liver is thought to depend on affinity for fatty acid-binding protein in hepatocytes,39 the expression of which is markedly down-regulated in Fech (Table 2) and griseofulvin-treated mice.40 This may reflect attempts by hepatocytes to limit protoporphyrin uptake. Similarly, genes of basolateral transport proteins (Slc10a1 and Slco1a1) for import of anions and bile acids from the blood into the hepatocytes became markedly down-regulated. There is evidence that export of protoporphyrin from cells is mediated by ABC protein MDR2 (ABCB4)3 and expression of this was increased in our studies (Table 2). Other evidence implicates ABCG2.41 These patterns of transport protein expressions are characteristic of cholestatic experimental models with down-regulation of hepatocyte systems associated with uptake from portal blood flow, for bile acids mediated by HNF-1α and HNF-4α,31 and less reduction or even up-regulation of transport systems associated with export.30,42,43 How protoporphyrin causes this effect and whether it is purely physical or a result of interference in a specific signaling pathway remains to be elucidated. The marked rise in apolipoprotein IV-A expression may be an adaptive response to the elevated plasma porphyrin levels and has been reported for protoporphyric drugs.44 It may also be linked to the rise in cholesterol synthesis. A manifestation of the homozygous Fech mutation mouse is hyperlipidemia and atherosclerotic lesions.14
Consistent with previous work13 we found no major disturbances of the neutral pathway of bile acid synthesis from cholesterol in Fech mice; in fact there was a small increase in Cyp7a1 expression. In contrast, the Cyp7b1 gene became markedly down-regulated after 8 weeks. CYP7A1 and CYP7B1 proteins have identical sterol 7α-hydroxylase activity, but the latter acts in the acidic pathway of bile acid metabolism after 27-hydroxylation of cholesterol. In humans, a mutation of the equivalent gene is associated with accumulation of toxic sterols.45 Down-regulation of the preceding gene of this path, Cyp27a1 (Table 2) might be a protective mechanism, but null Cyp27a1 (−/−) mice also show significant down-regulation of Cyp7b1 and disturbance of lipid metabolism.46 Similar changes occur after rat bile duct ligation.47 In Fech mice, total bile acid excretion and bile acid levels in plasma are greatly increased.13 Thus part of the toxic response in the protoporphyric livers is probably a consequence of disturbed bile acid homeostasis with toxic bile acids and oxysterols perhaps acting in combination with protoporphyrin.13 If oxidized porphyrins besides protoporphyrin are formed in Fech livers they might contribute to the toxicity. The up-regulation of Cyp3a11 is probably an adaptive response in cholestasis to detoxify bile salts by 6β-hydroxylation to less toxic products such as β-muricholic acid, the excretion of which by Fech mice rises significantly.13,48
The expression data also demonstrate that oxidative processes49 could contribute to cell injury in Fech livers but it is not possible to conclude whether this would be a consequence of cholestasis or is initiated by protoporphyrin. The net result is inflammation with expression of genes to be expected from invading cells as well as injured hepatocytes (Table 2). The marked up-regulation of hemopexin, ceruloplasmin, NRAMP1, and haephaestin are probably part of such mechanisms to minimize heme-catalyzed free radical production because of intravascular hemolysis that leads to splenomegaly and liver inflammation.50
Potential for Individual Variation
One reason for the sporadic incidence of liver injury in EPP might be because of individual differences in the mechanism of protoporphyrin uptake and export since most EPP patients have normal protoporphyrin levels and liver function.5 Preliminary data suggest that the severity of the effects of the Fechm1Pas mutation on liver injury may depend on the genetic background of the mice.5 The present studies demonstrate a number of metabolic systems associated with protoporphyria, bile acid transport, and metabolism as well as subsequent fibrosis51 in which genetic polymorphisms (for instance genes responsible for hepatocyte protoporphyrin import or export) might have phenotypic influence in the development of cholestasis and liver injury in different Fech mice and some EPP patients.
Response of Heme Metabolism
The results of this study also demonstrate that although highly porphyric, heme supply seemed sufficient in the Fech mouse liver to require only modest up-regulation of Alas1 or other enzymes of the heme synthetic pathway. This might be explained by reduced heme usage for cytochrome P450 as an early response in this mutant. Decreased Cyp1a2, Cyp2e1, and Cyp2c29 occurred before response of other genes such as Slco1a1 and Slco1a6 (Table 2). In addition, induction of heme oxygenase that would exacerbate heme shortage was not observed. Only modest elevation of Alas1 in both BALB/c and the Fech mutant in response to the phenobarbital type cytochrome P450 inducer TCPOBOP occurred, despite large increases in Cyp2b expression, supporting the view that basal heme enzyme levels might be sufficient to cope with general cell function and could be up-regulated by drugs inducing cytochrome P450. In contrast, mice heterozygous for the null Hmbs gene (hydroxymethylbilane synthase or porphobilinogen deaminase) in heme synthesis exhibit super induction of Alas1 expression in response to phenobarbital.22 The difference may reflect relative activities of hydroxymethylbilane synthase and ferrochelatase in some circumstances of heme synthesis.52 That the liver in Fech mice does not show marked Alas1 and Hmox1 induction but contains large quantities of protoporphyrin is also consistent with a significant proportion of the hepatic porphyrin originating from an erythropoietic source as is likely for EPP patients.5,11 The detection and down-regulation of Alas2, the erythroid-isoform of aminolevulinate synthase, in hepatic tissue is not simple to explain but may reflect an inability of the gene in a particular cell type to be up-regulated in these mice.53
Comparison with Griseofulvin Protoporphyria
The present finding demonstrates that heme metabolism and catabolism in Fech mice differ from that seen when protoporphyria in mice is induced by griseofulvin. Hepatic ferrochelatase activities would be expected to be decreased by >90% in both Fech mice and those treated with griseofulvin for 3 weeks.9,54,55 Despite equivalent levels of hepatic protoporphyrin and cholestatic injury in the two models there was a marked stimulation of cytochrome P450 isoforms, Hmox1 and Alas1 expression after drug treatment that was not seen in the mutant mouse (Figure 7).17,38 This induction of Alas1 and Hmox1 was much greater than the modest change observed with the phenobarbital-like CAR agonist TCPOBOP. Griseofulvin not only induces cytochrome P450 but also stimulates turnover of its heme moiety in a process that induces Hmox1.56 Thus with griseofulvin, protoporphyria is a hepatic consequence of not only ferrochelatase inhibition but the induction and destruction of cytochrome P450 with accompanying Alas1 derepression.16 Furthermore in the longer term, differences between Fech- and griseofulvin-treated hepatic tissue may reflect adaptation of cells throughout their lifetime in the mutant compared to the more acute exposure of mice to the drug.
Summary
In conclusion, hepatic gene expression in protoporphyric Fechm1Pas mice is consistent with deposition of protoporphyrin causing complex gene expressions associated with cholestasis but not with a marked depletion of the heme regulatory pool as observed with the protoporphyric drug griseofulvin.
Acknowledgments
We thank Susan Robinson for advice; and Colin Travis, Judy McWilliams, Shu-Dong Zhang, Jin-Li Luo, Joan Riley, and Jennifer Edwards and colleagues for assistance.
Footnotes
Address reprint requests to A.G. Smith, MRC Toxicology Unit, Hodgkin Bldg., University of Leicester, Lancaster Rd, Leicester, LE1 9HN, UK. E-mail: ags5@le.ac.uk.
References
- Dailey HA, Dailey TA. Ferrochelatase. Kadish KM, Smith KM, Guilard RG, editors. Amsterdam: Elsevier,; The Porphyrin Handbook. 2003:pp 93–121. [Google Scholar]
- Anderson KE, Sassa S, Bishop DF, Desnick RJ. Disorders of heme biosynthesis: X-linked sideroblastic anemia and the porphyrias. Scriver CR, Beudet AL, Sly WS, Valle D, Vogelstein B, Childs MD, editors. New York: McGraw-Hill; The Metabolic and Molecular Basis of Inherited Disease. 2001:pp 2991–3062. [Google Scholar]
- Beukeveld GJ, In’t Veld G, Havinga R, Groen AK, Wolthers BG, Kuipers F. Relationship between biliary lipid and protoporphyrin secretion; potential role of mdr2 P-glycoprotein in hepatobiliary organic anion transport. J Hepatol. 1996;24:343–352. doi: 10.1016/s0168-8278(96)80015-8. [DOI] [PubMed] [Google Scholar]
- Meerman L. Erythropoietic protoporphyria. An overview with emphasis on the liver. Scand J Gastroenterol. 2000;35(Suppl 232):79–85. [PubMed] [Google Scholar]
- Cox TM. Protoporphyria. Kadish KM, Smith KM, Guilard RG, editors. Amsterdam: Elsevier,; The Porphyrin Handbook. 2003:pp 121–149. [Google Scholar]
- Bloomer JR, Poh-Fitzpatrick MB. Theodore Woodward Award. Pathogenesis of biochemical abnormalities in protoporphyria. Trans Am Clin Climatol Assoc. 2000;111:245–256. [PMC free article] [PubMed] [Google Scholar]
- Ishibashi A, Ogata R, Sakisaka S, Kumashiro R, Koga Y, Mitsuyama K, Kuromatsu R, Uchimura Y, Ijyuin H, Tanaka K, Iwao T, Ishii K, Sata M, Inoue Y, Kin Y, Oizumi K, Nishida H, Imaizumi T, Tanikawa K. Erythropoietic protoporphyria with fatal liver failure. J Gastroenterol. 1999;34:405–409. doi: 10.1007/s005350050284. [DOI] [PubMed] [Google Scholar]
- Meerman L, Haagsma EB, Gouw AS, Slooff MJ, Jansen PL. Long-term follow-up after liver transplantation for erythropoietic protoporphyria. Eur J Gastroenterol Hepatol. 1999;11:431–438. doi: 10.1097/00042737-199904000-00012. [DOI] [PubMed] [Google Scholar]
- Tutois S, Montagutelli X, Da Silva V, Jouault H, Rouyer-Fessard P, Leroy-Viard K, Guenet JL, Nordmann Y, Beuzard Y, Deybach JC. Erythropoietic protoporphyria in the house mouse. A recessive inherited ferrochelatase deficiency with anemia, photosensitivity, and liver disease. J Clin Invest. 1991;88:1730–1736. doi: 10.1172/JCI115491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulechfar S, Lamoril J, Montagutelli X, Guenet JL, Deybach JC, Nordmann Y, Dailey H, Grandchamp B, de Verneuil H. Ferrochelatase structural mutant (Fechm1Pas) in the house mouse. Genomics. 1993;16:645–648. doi: 10.1006/geno.1993.1242. [DOI] [PubMed] [Google Scholar]
- Pawliuk R, Bachelot T, Wise RJ, Mathews-Roth MM, Leboulch P. Long-term cure of the photosensitivity of murine erythropoietic protoporphyria by preselective gene therapy. Nat Med. 1999;5:768–773. doi: 10.1038/10488. [DOI] [PubMed] [Google Scholar]
- Fontanellas A, Mazurier F, Landry M, Taine L, Morel C, Larou M, Daniel JY, Montagutelli X, de Salamanca RE, de Verneuil H. Reversion of hepatobiliary alterations by bone marrow transplantation in a murine model of erythropoietic protoporphyria. Hepatology. 2000;32:73–81. doi: 10.1053/jhep.2000.8531. [DOI] [PubMed] [Google Scholar]
- Meerman L, Koopen NR, Bloks V, Van Goor H, Havinga R, Wolthers BG, Kramer W, Stengelin S, Muller M, Kuipers F, Jansen PL. Biliary fibrosis associated with altered bile composition in a mouse model of erythropoietic protoporphyria. Gastroenterology. 1999;117:696–705. doi: 10.1016/s0016-5085(99)70464-6. [DOI] [PubMed] [Google Scholar]
- Bloks VW, Plosch T, van Goor H, Roelofsen H, Baller J, Havinga R, Verkade HJ, van Tol A, Jansen PL, Kuipers F. Hyperlipidemia and atherosclerosis associated with liver disease in ferrochelatase-deficient mice. J Lipid Res. 2001;42:41–50. [PubMed] [Google Scholar]
- Libbrecht L, Meerman L, Kuipers F, Roskams T, Desmet V, Jansen P. Liver pathology and hepatocarcinogenesis in a long-term mouse model of erythropoietic protoporphyria. J Pathol. 2003;199:191–200. doi: 10.1002/path.1257. [DOI] [PubMed] [Google Scholar]
- De Matteis F, Marks GS. Cytochrome P450 and its interactions with the heme biosynthetic pathway. Can J Physiol Pharmacol. 1996;74:1–8. [PubMed] [Google Scholar]
- Gant TW, Baus PR, Clothier B, Riley J, Davies R, Judah DJ, Edwards RE, George E, Greaves P, Smith AG. Gene expression profiles associated with inflammation, fibrosis, and cholestasis in mouse liver after griseofulvin. Environ Health Perspect. 2003;111:847–853. [PubMed] [Google Scholar]
- Smith AG, Carthew P, Francis JE, Edwards RE, Dinsdale D. Characterization and accumulation of ferritin in hepatocyte nuclei of mice with iron overload. Hepatology. 1990;12:1399–1405. doi: 10.1002/hep.1840120622. [DOI] [PubMed] [Google Scholar]
- Grandchamp B, Deybach JC, Grelier M, de Verneuil H, Nordmann Y. Studies of porphyrin synthesis in fibroblasts of patients with congenital erythropoietic porphyria and one patient with homozygous coproporphyria. Biochim Biophys Acta. 1980;629:577–586. doi: 10.1016/0304-4165(80)90163-4. [DOI] [PubMed] [Google Scholar]
- Smith AG, Clothier B, Carthew P, Childs NL, Sinclair PR, Nebert DW, Dalton TP. Protection of the Cyp1a2(−/−) null mouse against uroporphyria and hepatic injury following exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Appl Pharmacol. 2001;173:89–98. doi: 10.1006/taap.2001.9167. [DOI] [PubMed] [Google Scholar]
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- Jover R, Hoffmann F, Scheffler-Koch V, Lindberg RL. Limited heme synthesis in porphobilinogen deaminase-deficient mice impairs transcriptional activation of specific cytochrome P450 genes by phenobarbital. Eur J Biochem. 2000;267:7128–7137. doi: 10.1046/j.1432-1327.2000.01815.x. [DOI] [PubMed] [Google Scholar]
- Turton NJ, Judah DJ, Riley J, Davies R, Lipson D, Styles JA, Smith AG, Gant TW. Gene expression and amplification in breast carcinoma cells with intrinsic and acquired doxorubicin resistance. Oncogene. 2001;20:1300–1306. doi: 10.1038/sj.onc.1204235. [DOI] [PubMed] [Google Scholar]
- Smith AG, Davies R, Dalton TP, Miller ML, Judah D, Riley J, Gant T, Nebert DW. Intrinsic hepatic phenotype associated with the cyp1a2 gene as shown by cDNA expression microarray analysis of the knockout mouse. Environ Health Perspect. 2003;111:855–861. [PubMed] [Google Scholar]
- Zhang S-D, Gant TW. A statistical framework for the design of microarray experiments and effective detection of differential gene expression. Bioinformatics. 2004;20:2821–2828. doi: 10.1093/bioinformatics/bth336. [DOI] [PubMed] [Google Scholar]
- Hamilton JW, Bement WJ, Sinclair PR, Sinclair JF, Alcedo JA, Wetterhahn KE. Heme regulates hepatic 5-aminolevulinate synthase mRNA expression by decreasing mRNA half-life and not by altering its rate of transcription. Arch Biochem Biophys. 1991;289:387–392. doi: 10.1016/0003-9861(91)90428-l. [DOI] [PubMed] [Google Scholar]
- Cable EE, Miller TG, Isom HC. Regulation of heme metabolism in rat hepatocytes and hepatocyte cell lines: delta-aminolevulinic acid synthase and heme oxygenase are regulated by different heme-dependent mechanisms. Arch Biochem Biophys. 2000;384:280–295. doi: 10.1006/abbi.2000.2117. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Gauthier KC, Umetani M, Watson MA, Lochansky MI, Collins JL, Leitersdorf E, Mangelsdorf DJ, Kliewer SA, Repa JJ. Identification of bile acid precursors as endogenous ligands for the nuclear xenobiotic pregnane X receptor. Proc Natl Acad Sci USA. 2003;100:223–228. doi: 10.1073/pnas.0237082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Hawkins J, Lund EG, Turley SD, Russell DW. Disruption of the oxysterol 7alpha-hydroxylase gene in mice. J Biol Chem. 2000;275:16536–16542. doi: 10.1074/jbc.M001811200. [DOI] [PubMed] [Google Scholar]
- Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- Jung D, Kullak-Ublick GA. Hepatocyte nuclear factor 1 alpha: a key mediator of the effect of bile acids on gene expression. Hepatology. 2003;37:622–631. doi: 10.1053/jhep.2003.50100. [DOI] [PubMed] [Google Scholar]
- Borst P, Elferink RO. Mammalian ABC transporters in health and disease. Annu Rev Biochem. 2002;71:537–592. doi: 10.1146/annurev.biochem.71.102301.093055. [DOI] [PubMed] [Google Scholar]
- Camus-Randon AM, Raffalli F, Bereziat JC, McGregor D, Konstandi M, Lang MA. Liver injury and expression of cytochromes P450: evidence that regulation of CYP2A5 is different from that of other major xenobiotic metabolizing CYP enzymes. Toxicol Appl Pharmacol. 1996;138:140–148. doi: 10.1006/taap.1996.0107. [DOI] [PubMed] [Google Scholar]
- Kobliakov V, Kulikova L, Samoilov D, Lang MA. High expression of cytochrome P450 2a-5 (coumarin 7-hydroxylase) in mouse hepatomas. Mol Carcinog. 1993;7:276–280. doi: 10.1002/mc.2940070411. [DOI] [PubMed] [Google Scholar]
- Salonpaa P, Krause K, Pelkonen O, Raunio H. Up-regulation of CYP2A5 expression by porphyrinogenic agents in mouse liver. Naunyn Schmiedebergs Arch Pharmacol. 1995;351:446–452. doi: 10.1007/BF00169087. [DOI] [PubMed] [Google Scholar]
- Kakizaki S, Yamamoto Y, Ueda A, Moore R, Sueyoshi T, Negishi M. Phenobarbital induction of drug/steroid-metabolizing enzymes and nuclear receptor CAR. Biochim Biophys Acta. 2003;1619:239–242. doi: 10.1016/s0304-4165(02)00482-8. [DOI] [PubMed] [Google Scholar]
- Fraser DJ, Zumsteg A, Meyer UA. Nuclear receptors constitutive androstane receptor and pregnane X receptor activate a drug-responsive enhancer of the murine 5-aminolevulinic acid synthase gene. J Biol Chem. 2003;278:39392–39401. doi: 10.1074/jbc.M306148200. [DOI] [PubMed] [Google Scholar]
- Inafuku K, Takamiyagi A, Oshiro M, Kinjo T, Nakashima Y, Nonaka S. Alteration of mRNA levels of delta-aminolevulinic acid synthase, ferrochelatase and heme oxygenase-1 in griseofulvin induced protoporphyria mice. J Dermatol Sci. 1999;19:189–198. doi: 10.1016/s0923-1811(98)00073-5. [DOI] [PubMed] [Google Scholar]
- Knobler E, Poh-Fitzpatrick MB, Kravetz D, Vincent WR, Muller-Eberhard U, Vincent SH. Interaction of hemopexin, albumin and liver fatty acid-binding protein with protoporphyrin. Hepatology. 1989;10:995–997. doi: 10.1002/hep.1840100617. [DOI] [PubMed] [Google Scholar]
- Vincent SH, Smith AG, Muller-Eberhard U. Modulation of hepatic heme-binding Z protein in mice by the porphyrogenic carcinogens griseofulvin and hexachlorobenzene. Cancer Lett. 1989;45:109–114. doi: 10.1016/0304-3835(89)90144-4. [DOI] [PubMed] [Google Scholar]
- Jonker JW, Buitelaar M, Wagenaar E, Van Der Valk MA, Scheffer GL, Scheper RJ, Plosch T, Kuipers F, Elferink RP, Rosing H, Beijnen JH, Schinkel AH. The breast cancer resistance protein protects against a major chlorophyll-derived dietary phototoxin and protoporphyria. Proc Natl Acad Sci USA. 2002;99:15649–15654. doi: 10.1073/pnas.202607599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos TA, Hooiveld GJ, Koning H, Childs S, Meijer DK, Moshage H, Jansen PL, Muller M. Up-regulation of the multidrug resistance genes, Mrp1 and Mdr1b, and down-regulation of the organic anion transporter, Mrp2, and the bile salt transporter, Spgp, in endotoxemic rat liver. Hepatology. 1998;28:1637–1644. doi: 10.1002/hep.510280625. [DOI] [PubMed] [Google Scholar]
- Schrenk D, Gant TW, Preisegger KH, Silverman JA, Marino PA, Thorgeirsson SS. Induction of multidrug resistance gene expression during cholestasis in rats and nonhuman primates. Hepatology. 1993;17:854–860. [PubMed] [Google Scholar]
- Buchberg AM, Kinniburgh AJ. Induction of liver apolipoprotein A-IV mRNA in porphyric mice. Nucleic Acids Res. 1985;13:1953–1963. doi: 10.1093/nar/13.6.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setchell KD, Schwarz M, O’Connell NC, Lund EG, Davis DL, Lathe R, Thompson HR, Weslie Tyson R, Sokol RJ, Russell DW. Identification of a new inborn error in bile acid synthesis: mutation of the oxysterol 7alpha-hydroxylase gene causes severe neonatal liver disease. J Clin Invest. 1998;102:1690–1703. doi: 10.1172/JCI2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Lund EG, Horton JD, Leitersdorf E, Russell DW, Dietschy JM, Turley SD. Disruption of the sterol 27-hydroxylase gene in mice results in hepatomegaly and hypertriglyceridemia. Reversal by cholic acid feeding. J Biol Chem. 2000;275:39685–39692. doi: 10.1074/jbc.M007653200. [DOI] [PubMed] [Google Scholar]
- Setchell KD, Rodrigues CM, Clerici C, Solinas A, Morelli A, Gartung C, Boyer J. Bile acid concentrations in human and rat liver tissue and in hepatocyte nuclei. Gastroenterology. 1997;112:226–235. doi: 10.1016/s0016-5085(97)70239-7. [DOI] [PubMed] [Google Scholar]
- Stedman C, Robertson G, Coulter S, Liddle C. Feed-forward regulation of bile acid detoxification by CYP3A4: studies in humanized transgenic mice. J Biol Chem. 2004;279:11336–11343. doi: 10.1074/jbc.M310258200. [DOI] [PubMed] [Google Scholar]
- Dalton TP, Shertzer HG, Puga A. Regulation of gene expression by reactive oxygen. Annu Rev Pharmacol Toxicol. 1999;39:67–101. doi: 10.1146/annurev.pharmtox.39.1.67. [DOI] [PubMed] [Google Scholar]
- Tolosano E, Fagoonee S, Hirsch E, Berger FG, Baumann H, Silengo L, Altruda F. Enhanced splenomegaly and severe liver inflammation in haptoglobin/hemopexin double-null mice after acute hemolysis. Blood. 2002;100:4201–4208. doi: 10.1182/blood-2002-04-1270. [DOI] [PubMed] [Google Scholar]
- Bataller R, North KE, Brenner DA. Genetic polymorphisms and the progression of liver fibrosis: a critical appraisal. Hepatology. 2003;37:493–503. doi: 10.1053/jhep.2003.50127. [DOI] [PubMed] [Google Scholar]
- Brodie MJ, Moore MR, Thompson GG, Campbell BC, Goldberg A. Is porphobilinogen deaminase activity a secondary control mechanism in heme biosynthesis in humans? Biochem Soc Trans. 1977;5:1466–1468. doi: 10.1042/bst0051466. [DOI] [PubMed] [Google Scholar]
- Sassa S, Nagai T. The role of heme in gene expression. Int J Hematol. 1996;63:167–178. doi: 10.1016/0925-5710(96)00449-5. [DOI] [PubMed] [Google Scholar]
- De Matteis F, Gibbs AH. Stimulation of the pathway of porphyrin synthesis in the liver of rats and mice by griseofulvin, 3,5-diethoxycarbonyl-1,4-dihydrocollidine and related drugs: evidence for two basically different mechanisms. Biochem J. 1975;146:285–287. doi: 10.1042/bj1460285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley A, King LJ, Gibbs AH, De Matteis F. Strain and sex differences in the response of mice to drugs that induce protoporphyria: role of porphyrin biosynthesis and removal. J Biochem Toxicol. 1990;5:175–182. doi: 10.1002/jbt.2570050307. [DOI] [PubMed] [Google Scholar]
- Denk H, Eckerstorfer R. Turnover of cytochrome P-450 and cytochrome b5 hemes in griseofulvin-induced murine porphyria. FEBS Lett. 1977;76:67–70. doi: 10.1016/0014-5793(77)80122-1. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, psuedogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]