Abstract
Huntington's disease (HD), spinocerebellar ataxias types 1 and 3 (SCA1, SCA3), and spinobulbar muscular atrophy (SBMA) are caused by CAG/polyglutamine expansion mutations. A feature of these diseases is ubiquitinated intraneuronal inclusions derived from the mutant proteins, which colocalize with heat shock proteins (HSPs) in SCA1 and SBMA and proteasomal components in SCA1, SCA3, and SBMA. Previous studies suggested that HSPs might protect against inclusion formation, because overexpression of HDJ-2/HSDJ (a human HSP40 homologue) reduced ataxin-1 (SCA1) and androgen receptor (SBMA) aggregate formation in HeLa cells. We investigated these phenomena by transiently transfecting part of huntingtin exon 1 in COS-7, PC12, and SH-SY5Y cells. Inclusion formation was not seen with constructs expressing 23 glutamines but was repeat length and time dependent for mutant constructs with 43–74 repeats. HSP70, HSP40, the 20S proteasome and ubiquitin colocalized with inclusions. Treatment with heat shock and lactacystin, a proteasome inhibitor, increased the proportion of mutant huntingtin exon 1-expressing cells with inclusions. Thus, inclusion formation may be enhanced in polyglutamine diseases, if the pathological process results in proteasome inhibition or a heat-shock response. Overexpression of HDJ-2/HSDJ did not modify inclusion formation in PC12 and SH-SY5Y cells but increased inclusion formation in COS-7 cells. To our knowledge, this is the first report of an HSP increasing aggregation of an abnormally folded protein in mammalian cells and expands the current understanding of the roles of HDJ-2/HSDJ in protein folding.
Huntington's disease (HD) is an autosomal dominant neurodegenerative condition characterized by chorea, dementia, and psychiatric symptoms. The causative mutation is a CAG trinucleotide repeat expansion in exon 1 of huntingtin. Normal chromosomes have 35 or fewer repeats, whereas disease is associated with 36 or more repeats. The age at onset of symptoms is inversely correlated with the repeat number (reviewed in refs. 1 and 2).
HD is a member of a family of CAG trinucleotide repeat diseases, where the repeats are translated into an expanded polyglutamine (polyQ) that confers deleterious novel functions on the mutant proteins (reviewed in refs. 1 and 2). A hallmark of many of these diseases, including HD (3), spinobulbar muscular atrophy (SBMA) (4), dentatorubral-pallidoluysian atrophy (DRPLA) (5), and spinocerebellar ataxias (SCA) types 1 (6), 3 (7), and 7 (8), is the development of intracellular protein aggregates (inclusions) in the vulnerable neurons.
A pathological role for inclusions is suggested by the correlation of inclusion counts in the cortex of HD patients with CAG repeat number, which reflects disease severity (9). Inclusion formation precedes neurological dysfunction in some HD transgenic mice (10) and is associated with predisposition to cell death in in vitro models of HD (11, 12), DRPLA (13), SBMA (14), and SCA3 (7). The causal role for inclusions in these diseases has been challenged by experiments reporting a dissociation between cell death and inclusion formation in primary cell cultures (15). These findings were not straightforward and may be compatible with a pathogenic role for huntingtin polymerization (1, 2). Klement et al. deleted the self-association domain from a SCA1 transgene with expanded repeats preventing inclusion formation, but this transgene caused a SCA-like phenotype in transgenic mice (16). These mice do not show the progressive disease seen in mice expressing SCA1 transgenes with expanded repeats including the self-association domain (17). Because the latter mice develop inclusions, these aggregates may be necessary for disease progression.
Aggregates found in polyQ diseases are ubiquitinated in vivo and in vitro (7, 9, 10, 18, 19) and sequester proteasomal components in SCA1, SCA3, and SBMA (19–21). Ubiquitinated proteins are targeted to the 26S proteasome for degradation (22). Cummings et al. hypothesized that polyQ-containing inclusions may resist degradation, prevent ubiquitin recycling, and disrupt proteasome function (19). Because the ubiquitin–proteasome pathway is essential for the rapid degradation of many critical regulatory proteins (23), its disruption is fatal for a cell.
Heat shock proteins (HSPs) are expected to play protective roles in polyQ diseases, because they assist the folding of proteins into appropriate conformations, refold abnormally folded proteins, and rescue previously aggregated proteins (24–26). Ataxin-1 aggregates sequester HSP40 and HSP70 (in vitro) and inclusions formed by the androgen receptor with expanded repeats colocalize with HSP40, HSP70, and HSP90 (19, 20). Cummings et al. (19) and Stenoinen et al. (21) suggested that HSPs may play a protective role in SCA1 and SBMA, because the chaperone HDJ-2/HSDJ (a human homologue of the DnaJ or HSP40 family) reduced inclusion formation in HeLa cells. However, these experiments were not performed in other cell lines.
These findings suggest that it is necessary to understand the roles of HSPs and the proteasome in polyQ diseases. HDJ-2/HSDJ is a cochaperone and regulates the activity of members of the DnaK (HSP70) family (25, 26). Therefore, we investigated the effect of HDJ-2/HSDJ overexpression on polyQ-induced aggregation in models of HD by using three different cell types. We also examined the effects of nonspecific HSP induction (heat shock) and proteasome inhibition on inclusion formation.
Materials and Methods
Plasmid Construction.
HD exon 1 pEGFP-C1 (enhanced green fluorescent protein) constructs: genomic DNA from individuals with 21, 41, 51, 66, or 72 HD CAG repeats was used to amplify codons 8 to 57 inclusive of HD exon 1 (numbering applies to huntingtin with 23 Q residues—equivalent to 21 uninterrupted CAG repeats). We used primers HDF 5′-ATG AAG GCC TTC GAG TCC CTC AAG TCC TTC-3′ and HU3 5′-GGC GGC TGA GGA AGC TGA GAA-3′. Aliquots of these PCR products were reamplified with primers HM1 (5′-G CGC AGA TCT ATG AAG GCC TTC GAG TCC CTC AAG TCC TTC-3′) and HM3 (5′-G CGC GAA TTC GGC GGC TGA GGA AGC TGA GGA-3′), which have BglII and EcoRI restriction sites incorporated to allow cloning into pEGFP-C1 (CLONTECH).
HD pHM6 [hemagglutinin (HA)-tagged] constructs: the HD pEGFP constructs were used as templates for amplification with primers HDF-HindIII (5′-A CTG AAG CTT ATG AAG GCC TTC GAG TCC CTC AAG TCC TTC-3′) and HM3, to enable cloning into the HindIII and EcoRI sites of pHM6 (Roche Diagnostics, Lewes, U.K.). All constructs were validated by sequencing. DNA concentrations were determined spectrophotometrically and compared with gel estimates. HSP40/70 constructs were gifts from Huda Zoghbi and Chris Cummings (Baylor College of Medicine, Houston, TX) (19).
Cell Culture, Transfection Experiments, and FACS Analysis.
African green monkey kidney cells (COS-7) were grown in DMEM (Sigma) with 100 units/ml penicillin/streptomycin/2 mM l-glutamine/1 mM sodium pyruvate/10% FBS at 37°C, 5% CO2. Human neuroblastoma cells (SH-SY5Y) were grown in the same conditions, but in low glucose DMEM/HAM F-12 (1:1) (Sigma) with heat inactivated FBS. Rat pheochromocytoma cells (PC12 Tet-On) (CLONTECH) were cultured in DMEM with 100 units/ml penicillin/streptomycin/2 mM l-glutamine/10% heat-inactivated horse serum/5% FBS/100 μg/ml G418 at 37°C, 10% CO2. For transfection, cells were grown to 60–80% confluency on coverslips for 24 h, exposed to a mixture of 10 μl of LipofectAMINE Reagent (Life Technologies, Paisley, U.K.), and 2 μg of plasmid DNA for 5 h in serum-free medium, after which culture medium was added.
In cotransfection experiments, we used empty vector (pFLAG-CMV-2) (Sigma) and pFLAG-CMV-2 constructs expressing full-length human HDJ-2/HSDJ, the HDJ-2/HSDJ mutants Δ250 and Δ450 (see Results) (19) in combination with HD exon 1 constructs. We used a 3:1 ratio of HDJ-2/HSDJ construct to HD exon 1 construct DNA, to ensure that all cells expressing HD exon 1 constructs also expressed the appropriate HDJ-2/HSDJ construct. In all such experiments we used a total of 2 μg of DNA per 3.5-cm dish. At 24, 48, and 72 h after transfection, cells on coverslips were washed with 1× PBS, fixed with 4% paraformaldehyde in 1× PBS for 30 min, and mounted in antifadent supplemented with 4′,6-diamidino-2-phenylindole (3 μg/ml) to allow visualization of nuclear morphology.
Quantitative flow cytometry was performed by using a FACSort flow cytometer (Becton Dickinson). Cells, 10,000–20,000 per sample, were examined, and the data were analyzed by using lysis II software.
Heat Shock and Proteasome Inhibition Experiments.
Cells were heat shocked by incubation at 45°C for 30 min at 5, 20, and 30 h after transfection. Control cells were incubated at 37°C throughout. In proteasome inhibition experiments, we added 10 μM lactacystin (Affiniti, Exeter, U.K.) to the medium 24 h after transfection for 24 h, a concentration that effectively inhibits the proteasome (20).
Immunocytochemistry and Western Blotting.
Forty-eight hours after transfection, cells were washed and fixed with ice-cold methanol for 5–10 min. Alternatively, cells were fixed with 4% paraformaldehyde for 20 min, washed twice, and permeabilized (0.2% Triton X-100 in 1× PBS) for 15 min. After three wash steps, cells were incubated in blocking buffer (5% FBS) for 30 min at room temperature (RT), exposed to primary antibody (mouse anti-HA.11 monoclonal (Babco, Richmond, CA), rabbit anti-Hsp40 polyclonal (SPA-400), mouse anti-Hsp70 monoclonal (SPA810) (Stressgene, Victoria, CA), rabbit anti-20S proteasome polyclonal (Affiniti), mouse antiubiquitin monoclonal (Chemicon) at 1:100 or 1:200 for 1 h at RT and washed three times. Then cells were incubated with either goat anti-mouse IgG or goat anti-rabbit IgG conjugated to Texas red (Molecular Probes) for 1 h in the dark at RT (1:200 or 1:400; 1:1,000 for HA-tagged constructs), washed three times, dried, and mounted (see above). All wash steps were performed in 1× PBS.
For Western blotting, cell lysates were electrophoresed on 15% resolving gels, and primary antibodies were used at 1:2,000 (HSP70) and 1:3,000 (actin, Sigma). Blots were probed with peroxidase-labeled anti-mouse (HSP70) and anti-rabbit (actin) antibodies at 1:3,000 (Amersham). Bands were detected with the ECC detection reagent (Amersham). Protein loading was controlled by actin.
Statistical Analysis.
We counted between 250 and 600 EGFP- or HA-expressing cells per slide (blinded) in multiple random visual fields. The proportion of HD exon 1-expressing cells with one or more inclusions was used as a measure for inclusion formation, following the precedent of Cummings et al. (19). Pooled estimates for the changes in inclusion formation resulting from perturbations assessed in multiple experiments were calculated as odds ratios (OR) with 95% confidence intervals [(% cells expressing construct with inclusions in perturbation conditions/% cells expressing construct without inclusions in perturbation conditions)/(% cells expressing construct with inclusions in control conditions/% cells expressing construct without inclusions in control conditions)]. OR and P-values were determined by unconditional logistical regression analysis, by using the general loglinear analysis option of SPSS Ver. 6.1 software (SPSS, Chicago). Similarly, we determined the proportion of cells with multiple inclusions by counting 50–100 cells per slide. By focusing through each single cell, the number of inclusions was determined. To simplify statistical analysis, we determined the proportion of treated vs. untreated cells with more than eight inclusions in all experiments, except for proteasome inhibition experiments (>9). These arbitrary cutoff points were chosen in favor of qualitative analyses.
Results
HD Exon 1 Aggregation Is polyQ Length Dependent. Inclusions Colocalize with Ubiquitin, 20S Proteasome, HSP40, and HSP70.
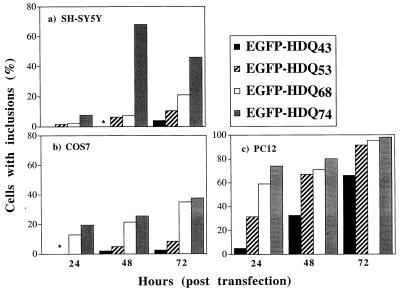
Human neuroblastoma (SH-SY5Y), rat phaeochromocytoma (PC12), and African green monkey (COS-7) cells were transiently transfected with constructs comprising EGFP at the N terminus, fused to an exon 1 fragment of huntingtin containing different polyQ repeat lengths (see Materials and Methods). No aggregates were seen in any of the three cell lines expressing this construct with 23 glutamines (EGFP-HDQ23) at 24, 48, and 72 h after transfection (data not shown). However, aggregates were present in these cells transfected with EGFP-HDQ43, -Q53, -Q68, and -Q74 (Fig. 1). The proportion of EGFP-positive cells containing aggregates increased with time after transfection and was correlated with the length of the polyQ expansion in all cell lines (Fig. 1). The high rate of inclusion formation in PC12 cells, compared with the COS-7 and SH-SY5Y lines, was not simply a function of expression level, because the mean fluorescence of cells 48 h after a typical transfection with empty pEGFP vector was determined by quantitative flow cytometry as 43.7 arbitrary units (A.U.) in COS-7, 6.2 A.U. in SH-SY5Y, and 4.7 A.U. in PC12 cells.
Figure 1.
Time and polyQ repeat dependency of inclusion formation. (a) SH-SY5Y (human neuroblastoma cells; *, inclusions are occasionally present (<1% of EGFP-positive cells). The decrease of FP-positive cells showing inclusions is possibly because of cell death for EGFP-HDQ74 at 72 h. (b) COS-7 (African green monkey cells). (c) PC12 (rat pheochromocytoma cells).
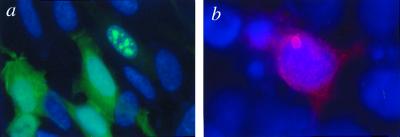
Inclusion formation was associated with weaker EGFP staining in the cytoplasm, compared with cells without inclusions (Fig. 2a). Inclusions generated by pEGFP constructs were star like and either cytoplasmic, perinuclear, or intranuclear in all cell lines (data not shown). To ensure that our interpretations were not confounded by the EGFP tag, we made a second set of vectors (pHM6) comprising the same HD exon 1 fragment tagged with an influenza HA epitope at the N terminus and a 6× His tag at the C terminus. We observed ring-like aggregates with constructs containing 74 glutamines (HA-HDQ74) in COS-7 cells (Fig. 2b) and all other cell types, whereas HA-HDQ23 did not form aggregates (data not shown).
Figure 2.
HD exon 1 protein aggregates in SH-SY5Y and COS-7 cells. (a) SH-SY5Y cells expressing EGFP-HDQ74 (green). Note that the cell with multiple inclusions shows weak EGFP staining in the cytoplasm compared with the other transfected cells. Nuclei were stained with 4′,6-diamidino-2-phenylindole. (b) COS-7 cell transfected with HA-HDQ74 showing a single inclusion.
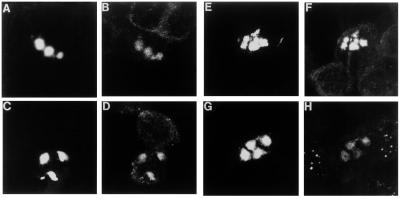
Aggregates in SH-SY5Y (Fig. 3 a–h), COS-7, and PC12 cells (data not shown) were ubiquitinated and sequestered components of the 20S proteasome, HSP40 and HSP70. Cells with 20S proteasome-containing inclusions showed a deficiency in 20S proteasome staining in the rest of the cell compared with cells without inclusions (Fig. 3h), as described by Cummings et al. (19).
Figure 3.
Ubiquitin, HSP40, HSP70, and the 20S proteasome colocalize with inclusions. Colocalization was determined by using confocal image analysis. SH-SY5Y cells were transfected with EGFP-HDQ74. In a, c, e, and g, the cells were microphotographed by using the FITC filter to show the inclusions formed by HD exon 1 constructs. In b, d, f, and h, the same cells are shown by using the Texas red filter to detect the other proteins sequestered into the inclusions. Cells were analyzed with specific antibodies: (a and b) anti-ubiquitin; (c and d) anti-HSP40 (HDJ-2/HSDJ); (e and f) anti-HSP70; (g and h) anti-20S proteasome.
Effects of HDJ-2/HSDJ Overexpression.
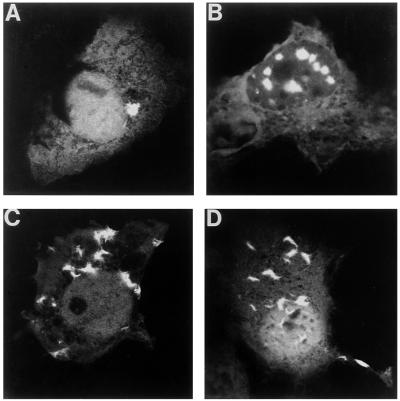
We coexpressed EGFP-HDQ74 with wild-type (wt) HDJ-2/HSDJ, two J-domain mutants of HDJ-2/HSDJ with impaired activity (19) (Δ250: deletion of amino acids 9–46 and Δ450: deletion of amino acids 9–107) or empty vector control (pFlag-CMV2). To ensure that cells expressing the HD exon 1 construct also overexpressed HDJ-2/HSDJ or its mutants, we used a 3:1 ratio of HDJ-2/HSDJ to EGFP-HDQ74. Data in Table 1 are expressed as OR, which summarize a series of experiments comparing the proportions of EGFP-expressing cells with inclusions when cotransfected with wt HDJ-2/HSDJ, to the proportions observed when cotransfected with control vectors. OR were considered to be the most appropriate summary statistic, because the percentage of cells with inclusions under specified conditions varies between experiments on different days, whereas the relative change in the proportion of cells with inclusions induced by an experimental perturbation is expected to be more constant. For example, in line 1 of Table 1, cotransfection of wt HDJ-2/HSDJ with EGFP-HDQ74 resulted in a 1.59-fold increase in the proportion of cells with inclusions vs. HD exon 1-expressing cells without inclusions, compared with cells cotransfected with EGFP-HDQ74 and the empty pFLAG vector (OR = 1.59; P < 0.001). This increase in the proportion of cells with inclusions was confirmed at 24, 48, and 72 h after transfection (Table 1) and was also seen in most experiments when the wt HDJ-2/HSDJ was compared with HDJ-2/HSDJΔ450 and HDJ-2/HSDJΔ250. Increased aggregation was also observed in COS-7 cells coexpressing wt HDJ-2/HSDJ and HA-HDQ74, suggesting that the observed effects were not a function of the EGFP tag. (Table 1). Coexpression of wt HDJ-2/HSDJ and EGFP-HDQ74 in COS-7 cells was generally associated with a higher proportion of cells showing multiple (>8) inclusions per cell, compared with mutant and empty vector controls (Table 1, Fig. 4 a and b). Coexpression of wt HDJ-2/HSDJ and EGFP-HDQ23 did not result in aggregation formation.
Table 1.
Comparison of the number of cells with intracellular inclusions after cotransfection experiments with HDJ-2/HSDJ (HSP40)
| Test constructs | Control constructs | Time, h | OR | 95% CI for OR | MI |
|---|---|---|---|---|---|
| COS-7 | |||||
| HDJ-2 | pFLAG | 24 | 1.59 | 1.23–2.05*** | * |
| 48 | 1.57 | 1.43–3.49*** | *** | ||
| 72 | 1.58 | 1.35–1.85*** | *** | ||
| HDJ-2† | PFLAG† | 48 | 2.27 | 1.86–2.77*** | NS |
| HDJ-2‡ | pFLAG | 48 | 1.92 | 1.55–2.37*** | ND |
| HDJ-2 | Δ250 | 24 | 1.20 | 0.94–1.53 | * |
| 48 | 1.38 | 1.24–1.54*** | * | ||
| 72 | 1.33 | 1.14–1.55*** | ** | ||
| HDJ-2† | Δ250† | 48 | 1.05 | 0.88–1.26 | * |
| HDJ-2‡ | Δ250 | 48 | 1.75 | 1.41–2.15*** | ND |
| HDJ-2 | Δ450 | 24 | 1.36 | 1.06–1.74* | * |
| 48 | 1.56 | 1.42–1.74*** | NS | ||
| 72 | 1.25 | 1.07–1.46** | *** | ||
| HDJ-2† | Δ450† | 48 | 2.24 | 1.82–2.77*** | *** |
| HDJ-2‡ | Δ450 | 48 | 2.80 | 2.13–3.68*** | ND |
| SH-SY5Y | |||||
| HDJ-2 | pFLAG | 48 | 0.82 | 0.60–1.12 | NS |
| 72 | 1.51 | 1.14–2.01** | NS | ||
| Δ250 | 48 | 1.04 | 0.83–1.32 | NS | |
| 72 | 0.94 | 0.72–1.22 | NS | ||
| Δ450 | 48 | 1.09 | 0.79–1.51 | NS | |
| 72 | 1.20 | 0.91–1.57 | NS | ||
| PC-12 | |||||
| HDJ-2 | pFLAG | 24 | 1.21 | 0.96–1.52 | * |
| 48 | 1.31 | 1.05–1.63* | ND | ||
| Δ250 | 24 | 1.04 | 0.83–1.32 | NS | |
| 48 | 1.17 | 0.94–1.46 | ND | ||
| Δ450 | 24 | 1.10 | 0.88–1.39 | NS | |
| 48 | 1.27 | 1.02–1.58 | ND |
We used a 3:1 ratio of HDJ-2/HSDJ to huntingtin (EGFP-HDQ74) construct in all experiments except where marked with †, where we used a 7:1 ratio. An HA-tagged huntingtin construct (HA-HDQ74) was used where indicated by ‡. pFLAG, empty vector; HDJ-2/HSDJ-Δ250 (Δ250) and HDJ-2/HSDJ-Δ450 (Δ450) are deletion mutants of HDJ-2/HSDJ (see Results). Time, length of experiment, hours. OR with 95% confidence intervals (CI) are presented (see Materials and Methods); MI, multiple inclusion phenotype; P-values are represented as follows: *, P < 0.05, **, P < 0.01, ***, P < 0.001; P-values for MI reflect whether there are significantly more cells with >8 inclusions in perturbation vs. control conditions. NS, not significant; ND, not done. All experiments were performed at least three times.
Figure 4.
Cells expressing pEGFP-HDQ74 develop multiple inclusions when they coexpress HDJ-2/HSDJ, when treated with heat shock or lactacystin. Comparisons of typical inclusion phenotypes of COS-7 cells expressing pEGFP-HD74 when (a) cotransfected with pFLAG-CMV-2 (empty vector); (b) cotransfected with wt HDJ-2/HSDJ; (c) treated with heat shock; (d) treated with lactacystin (10 μM).
Because a large molar excess of DnaJ proteins is required to suppress protein aggregation in vitro (in yeast) (27), we tested whether a larger molar ratio of HDJ-2/HSDJ could decrease aggregate formation. However, we still observed increased aggregate formation when we coexpressed wt HDJ-2/HSDJ with EGFP-HDQ74 in a 7:1 ratio (Table 1).
The aggregation-enhancing effects of wt HDJ-2/HSDJ were not observed in SH-SY5Y and PC12 cells (Table 1). This could not be explained by saturation of inclusion formation, because in these cotransfection experiments <30% of EGFP-expressing SH-SY5Y cells showed aggregates up to 72 h after transfection, and <55% of PC12 showed aggregates up to 72 h after transfection. In contrast to COS-7 cells, the multiple-inclusion phenotype was not a typical response to wt HDJ-2/HSDJ overexpression in SH-SY5Y and PC12 cells (Table 1). Because wt HDJ-2/HSDJ reduced aggregation formation in SCA1 and SBMA in HeLa cells (19, 21), we transfected HeLa cells with EGFP-HDQ74. Despite a high transfection efficiency (≈40%), the number of EGFP-HDQ74-positive HeLa cells containing inclusions was very low (<1%), and we did not use HeLa cells for further experiments. Analysis of COS-7, PC12 and SH-SY5Y cells coexpressing HSP70 with EGFP-HDQ74 (3:1 ratio) for 24 and 48 h suggested that this HSP did not modulate inclusion formation (at least four experiments in each case; data not shown).
Heat-Shock and Proteasome Inhibition Enhance Aggregation Formation.
To explore further the effects of HSPs on inclusion formation, we exposed COS-7 and SH-SY5Y cells to elevated temperatures to induce HSP expression (28). Heat shock (see Materials and Methods) resulted in an induction of HSP70 protein levels: in all cell lines, endogenous HSP70 was not detected in untreated cells by Western blot analysis but gave a strong signal in heat-shocked cells (data not shown). Heat shock increased the proportion of EGFP-HDQ74-expressing cells with inclusions (SH-SY5Y, 24 and 48 h; COS-7, 24 h) and also increased the proportions of cells containing multiple inclusions (Table 2 and Fig. 4c), similar to COS-7 cells overexpressing wt HDJ-2/HSDJ (Table 1; Fig. 4b). These results cannot be explained simply by increased pEGFP-HD exon 1 synthesis, because heat shock did not increase the mean fluorescence per cell in COS-7 cells transfected with the empty pEGFP vector (as determined by quantitative flow cytometry) (data not shown). Heat shock did not induce inclusion formation by EGFP-HDQ23 (data not shown).
Table 2.
Effect of heat shock or lactacystin treatment on the number of cells expressing EGFP-HDQ74 containing intracellular inclusions
| Treatment | Time, h | OR | 95% CI for OR | n | MI |
|---|---|---|---|---|---|
| COS-7 | |||||
| Heat shock | 24 | 2.63 | 2.03–3.40*** | 5 | *** |
| 48 | 0.94 | 0.78–1.13 | 5 | *** | |
| Lactacystin | 48 | 2.27 | 1.11–1.92*** | 4 | ** |
| SH-SY5Y | |||||
| Heat shock | 24 | 2.95 | 2.18–3.97*** | 4 | NS |
| 48 | 2.58 | 2.03–3.25*** | 4 | ** | |
| Lactacystin | 48 | 2.33 | 1.73–3.13*** | 3 | * |
Time, length of experiment. OR with 95% confidence intervals (CI) (see Materials and Methods). n, no. of experiments. MI, multiple inclusion phenotype. P-values are represented as follows: *, P < 0.05, **, P < 0.01, ***, P < 0.001. P-values for MI reflect whether there are significantly more cells with >8 or 9 inclusions in heat shock or proteasome inhibition vs. control conditions. NS, not significant.
Lactacystin, a specific, irreversible proteasome inhibitor, caused a significant increase in the proportion of HD exon 1-expressing cells with inclusions, and these cells had a multiple inclusion phenotype (Table 2; Fig. 4d). The 10 μM lactacystin dose, which effectively inhibits the proteasome (21), resulted in an induction of HSP70 protein levels consistent with previous data (29–31): by using Western blotting, a strong signal was detected in lactacystin-treated cells, compared with no signal for untreated cells of all cell types (data not shown). By contrast, HDJ-2/HSDJ gave a strong signal on Western blots in untreated cells and was not obviously induced by heat shock and lactacystin treatment (data not shown). Lactacystin did not induce inclusion formation by EGFP-HDQ23 (data not shown).
COS-7 Cells with Inclusions Are Susceptible to Death.
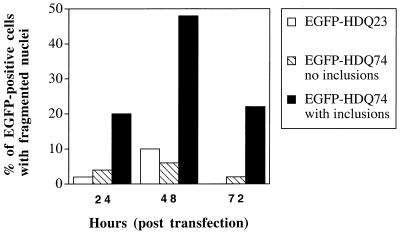
If polyQs are less toxic in an inclusion than when free (15), then inclusion-containing cells may preferentially survive heat shock, HDJ-2/HSDJ transfection, and lactacystin treatment, compared with cells without inclusions. This preferential survival could account for the increased proportion of EGFP-HDQ74-expressing cells with inclusions after these treatments. Because all three manipulations caused an increase in inclusion formation in COS-7 cells (see above), we analyzed nuclear fragmentation in cells expressing EGFP-HDQ23 and EGFP-HDQ74 at 24, 48, and 72 h after transfection (Fig. 5). EGFP-HDQ74-expressing cells with inclusions showed significantly more nuclear fragmentation at all time points compared with either EGFP-HDQ23-expressing cells or EGFP-HDQ74-expressing cells without inclusions (P < 0.02 for all comparisons by using χ2 tests). Treatment of cells with 100 μM Z-VAD-FMK (inhibitor of caspase-1-like proteases) reduced nuclear fragmentation in inclusion-containing cells to levels similar to those in noninclusion-containing cells (data not shown). Thus, inclusion formation is associated with cell death and not with preferential survival.
Figure 5.
Proportion of COS-7 cells showing nuclear fragmentation in EGFP-HDQ23-expressing cells and EGFP-HDQ74-expressing cells with or without inclusions (n = 50). These data are from one experiment that was representative of three independent experiments.
We also dismissed the possibility that our findings were because of the heat shock, HDJ-2/HSDJ transfection, and lactacystin treatments increasing survival of EGFP-HDQ74-expressing cells. Triplicate experiments, which showed increased inclusion formation with these treatments, showed no significant decrease in nuclear fragmentation in test vs. control conditions for all EGFP-containing cells or for inclusion-containing cells (data not shown).
Discussion
We have developed an in vitro model to study inclusion formation in polyQ diseases by using a fragment of exon 1 of huntingtin, because large fragments of the HD, SCA3, and DRPLA gene products do not show inclusion formation/cell death in cell lines (5, 7, 11, 12, 18, 32). Only a small N-terminal polyQ-containing part(s) of huntingtin is found in inclusions in vivo (3), and the exact nature of this fragment(s) is unclear. Although exon 1 models may not be specific for HD, they are powerful tools to study inclusion formation in relation to cell death/dysfunction in polyQ diseases and have been extensively studied in transgenic mice (10, 33) and cell models (e.g., ref. 12). In our model, inclusion formation was CAG-length- and time-dependent in the three cell lines studied (Fig. 1). Our data provide preliminary evidence suggesting that different cell types may show varying susceptibilities to inclusion formation, which cannot be accounted for by differential HD exon 1 expression levels. For instance, PC12 cells showed a higher rate of inclusion formation than COS-7 cells (Fig. 1), whereas the latter cells appear to have about nine times higher mean levels of pEGFP expression per cell (see Results). It is likely that the higher expression in COS-7 cells arises because they contain the SV40 large T antigen that allows for episomal replication of all the expression vectors we used (as they contain the SV40 ori). However, we cannot exclude the possibility that the lower levels of inclusions in COS-7 cells are because of greater toxicity in this cell type vs. PC12 and SH-SY5Y cells.
HDJ-2/HSDJ (HSP40) and HSP70 colocalized with HD exon 1 inclusions in neuronal (SH-SY5Y, PC12) and nonneuronal cells (COS-7), consistent with previous reports for ataxin-1 and androgen receptor in HeLa cells (19, 21). HSPs acting as molecular chaperones mediate appropriate folding of target proteins by controlled binding and release (25). The failure of release of HSPs from their substrate in polyQ diseases is abnormal. As Cummings et al. pointed out, HSP40 and HSP70 are inducible forms, and their presence in aggregates suggests that polyQ proteins may induce a heat-shock response (19).
We have made the observation that overexpression of a wt HSP can increase the aggregation of an abnormally folded protein in mammalian cells, because wt HDJ-2/HSDJ increased the proportion of HD exon 1-expressing COS-7 cells with inclusions. However, no increase or decrease in inclusion formation was seen in PC12 and SH-SY5Y cells. Previous studies with ataxin-1 and the androgen receptor have reported that HDJ-2/HSDJ reduced the proportion of cells with inclusions and suggested that molecular chaperones may have a protective role in polyQ diseases (19, 21). There are several possible explanations for the discrepancies between our data and previous reports. First, the protein sequences outside the polyQ tract may mediate different responses for huntingtin exon 1, the androgen receptor and ataxin-1. Second, both previous studies used HeLa cells (19, 21), whereas we analyzed three different cell lines; the process of inclusion formation may vary in different cell lines. Third, HD exon 1 and HDJ-2/HSDJ constructs may be expressed at higher levels in COS-7 cells. The expression levels of other HSPs in the relevant cell lines may also be important, because HDJ-2/HSDJ acts as a cochaperone with other HSPs, like HSP70, and some HSP70 family members have comparatively low expression in brain and “neuronal” cell lines (34). Recent findings of antagonistic interactions between the yeast chaperones HSP104 and HSP70 in prion curing (35) suggest that the net effect of HSPs on polyQ aggregation in humans may depend on the overall balance of different HSPs. Thus, the apparent contradiction between our HDJ-2/HSDJ data and previous reports may reflect a dual role for HSPs in polyQ aggregation formation.
HDJ-2/HSDJ cooperates with HSP70 by stimulating its low intrinsic rate of ATP hydrolysis, which is thought to be regulated by the J-domain (36). J-domain mutants of HDJ-2/HSDJ were unable to suppress ataxin-1 (19) and androgen receptor aggregation (21). wt HDJ-2/HSDJ resulted in significantly more HD exon 1 aggregation in COS-7 cells compared with J-domain mutants. Thus, HDJ-2/HSDJ may modulate aggregation formation by stimulating ATPase activity of endogenous HSP70.
Ubiquitin and the 20S component of the proteasome are sequestered by aggregates in all three cell lines (Fig. 3), consistent with in vivo and in vitro data from other polyQ diseases (19–21). Cells with 20S proteasome-containing inclusions showed a deficiency in 20S proteasome staining in the rest of the cell compared with cells without inclusions (Fig. 3h), as observed by Cummings et al. (19), who suggested that the ubiquitin–proteasome pathway may be disrupted in cells with aggregates. Our data are consistent with those for ataxin-3, where proteasome inhibitors caused an increase in aggregate formation. As Chai et al. (20) suggested, the simplest model to explain these results is that the proteasome reduces the concentration of polyQ aggregation-prone polypeptides under normal conditions. When this function is blocked, the level of misfolded polyQ proteins increases favoring nucleation and aggregation.
Alternatively, proteasome inhibition could increase polyQ aggregation indirectly. We have confirmed previous data showing that proteasome inhibition induces HSPs (refs. 29–31; this study). Lactacystin and heat-shock treatment resulted in similar cellular phenotypes: an increased proportion of HD exon 1-expressing cells with inclusions and more inclusions per inclusion-containing cell. Thus, we cannot rule out the possibility that these perturbations may be because of induction of HSPs.
In summary, our data suggest that proteasome inhibitors and heat shock increase the proportion of HD exon 1-expressing cells with inclusions. Thus, inclusion formation may be enhanced in polyQ diseases, if the pathological process results in inhibition of the proteasome or a heat-shock response. In COS-7 cells, which had the highest expression of our constructs, we unexpectedly observed an increased proportion of HD exon 1-expressing cells with inclusions as a consequence of overexpression of wt HDJ-2/HSDJ. To our knowledge, this is the first report of an HSP acting to increase aggregation of an abnormally folded protein in mammalian cells. This finding may be important, irrespective of the cell type or polyQ-containing construct used: it suggests additional roles for HDJ-2/HSDJ in protein folding/aggregation, which may be relevant to our general understanding of these fundamental processes.
Acknowledgments
We thank Drs. C. Cummings and H. Zoghbi for HSP40/70 constructs, Dr. A. Carmichael for technical help, and the Rehabilitation and Medical Research Trust, Friends of Peterhouse, the Cambridge University Department of Pathology, the Swiss Academy of Medical Sciences, and the Isaac Newton Trust for financial support. A.W. is grateful to the Swiss National Science Foundation and the Hereditary Disease Foundation for research fellowships; R.A.F. is a Peterhouse Senior Research Fellow in Neuroscience; J.S. is a Merck Sharp and Dohme Research Fellow; J.C. is an Action Research Training Fellow; J.S. and J.C. are grateful for Sackler Studentships; and D.C.R. is a Glaxo Wellcome Research Fellow.
Abbreviations
- HD
Huntington's disease
- HSP
heat shock protein
- polyQ
polyglutamine
- HA
hemagglutinin
- EGFP
enhanced green fluorescent protein
- wt
wild type
- OR
odds ratios
References
- 1.Perutz M F. Trend Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein D C, Wyttenbach A, Rankin J. J Med Genet. 1999;36:265–270. [PMC free article] [PubMed] [Google Scholar]
- 3.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 4.Li M, Miwa S, Kobayashi Y, Merry D E, Yamamoto M, Tanaka F, Doyu M, Hashizume Y, Fischbeck K H, Sobue G. Ann Neurol. 1998;44:249–254. doi: 10.1002/ana.410440216. [DOI] [PubMed] [Google Scholar]
- 5.Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- 6.Skinner P J, Koshy B T, Cummings C J, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Nature (London) 1997;389:971–978. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 7.Paulson H L, Perez M K, Trottier Y, Trojanowski J Q, Subramony S H, Das S S, Vig P, Mandel J L, Fischbeck K H, Pittman R N. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 8.Holmberg M, Duyckaerts C, Durr A, Cancel G, Gourfinkel-An I, Damier P, Faucheux B, Trottier Y, Hirsch E C, Agid Y, et al. Hum Mol Gen. 1998;7:913–918. doi: 10.1093/hmg/7.5.913. [DOI] [PubMed] [Google Scholar]
- 9.Becher M W, Koztuk J A, Sharp A H, Davies S W, Bates G B, Price D L, Ross C A. Neurobiol Dis. 1997;4:1–11. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 10.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 11.Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, McCutcheon K, Singaraja R, Kazemi-Esfarjani P, Devon R, Kim S U, et al. Nat Genet. 1998;18:150–154. doi: 10.1038/ng0298-150. [DOI] [PubMed] [Google Scholar]
- 12.Cooper J K, Schilling G, Peters M F, Herring W J, Sharp A H, Kaminsky Z, Masone J, Khan F A, Delanoy M, Borchelt D R, et al. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- 13.Sato A, Shimohata T, Koide R, Takano H, Sato T, Oyake M, Igarashi S, Tanaka K, Inuzuka T, Nawa H, et al. Hum Mol Genet. 1999;8:997–1006. doi: 10.1093/hmg/8.6.997. [DOI] [PubMed] [Google Scholar]
- 14.Ellerby L M, Hackam A S, Proop S S, Ellerby H M, Rabizadeh S, Cashman N R, Trifiro M A, Pinsky L, Wellington C L, Salvesen G S, et al. J Neurochem. 1999;72:185–195. doi: 10.1046/j.1471-4159.1999.0720185.x. [DOI] [PubMed] [Google Scholar]
- 15.Sadou F, Finkbeiner S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 16.Klement I A, Skinner P J, Kayfor M D, Yi H, Hersch S M, Clark H B, Zoghbi H Y, Orr H T. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 17.Orr H T, Skinner P A, Klement C J, Cummings C J, Zoghbi H J. Am. J. Hum. Genet. Suppl. 1998. A8. [Google Scholar]
- 18.Hackam A S, Singaraja R, Wellington C L, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden M R. J Cell Biol. 1998;141:1097–1105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cummings C J, Mancini M A, Antalffy B, DeFranco D B, Orr H T, Zoghbi H Y. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 20.Chai Y, Koppenhafer S L, Shoesmith S J, Perez M K, Paulson H L. Hum Mol Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 21.Stenoinen D L, Cummings C J, Adams H P, Mancini M G, Patel K, DeMartino G N, Marcelli M, Weigel N L, Mancini M A. Hum Mol Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 22.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 23.Coux D, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 24.Glover J R, Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 25.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 26.Hendricks J P, Hartl F U. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 27.Lu Z, Cyr D M. J Biol Chem. 1998;273:1970–5978. doi: 10.1074/jbc.273.10.5970. [DOI] [PubMed] [Google Scholar]
- 28.Morimoto R I, Kline M P, Bimston D N, Cotto J J. Essays Biochem. 1997;32:17–29. [PubMed] [Google Scholar]
- 29.Bush K T, Goldberg A L, Nigam S K. J Biol Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 30.Kim D, Kim S H, Li G C. Biochem Biophys Res Commun. 1999;254:264–268. doi: 10.1006/bbrc.1998.9840. [DOI] [PubMed] [Google Scholar]
- 31.Kawazoe Y, Nakai A, Tanabe M, Nagata K. Eur J Biochem. 1998;255:356–362. doi: 10.1046/j.1432-1327.1998.2550356.x. [DOI] [PubMed] [Google Scholar]
- 32.Lunkes A, Mandel J L. Hum Mol Genet. 1998;7:1355–1361. doi: 10.1093/hmg/7.9.1355. [DOI] [PubMed] [Google Scholar]
- 33.Schilling G, Becher M W, Sharp A H, Jinnah H A, Duan K, Kotzuk J A, Slunt H H, Ratovitski T, Cooper J K, Jenkins N A, et al. Hum Mol Genet. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- 34.Beck S C, Paidas C N, Tan H, Yang J, De Maio A. Am J Physiol. 1995;269:R608–R613. doi: 10.1152/ajpregu.1995.269.3.R608. [DOI] [PubMed] [Google Scholar]
- 35.Newnam G P, Wegrzyn R D, Lindquist S L, Chernoff Y O. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cyr D M, Langer T, Douglas M G. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]