Abstract
Rheumatoid arthritis (RA) and osteoarthritis (OA) are joint disorders that cause major public health problems. Previous studies of the etiology of RA and OA have implicated Wnt genes, although the exact nature of their involvement remains unclear. To further clarify the relationship between RA, OA, and the Wnt gene family, gene expression analyses were performed on articular cartilage, bone, and synovial tissues in knee joints taken from RA, OA, and normal/control patients. Cytokine assays were also performed in cells transfected with Wnt-7b, a member of the gene family most closely linked to RA and OA. Of the human Wnt genes, real-time PCR analysis revealed significant up-regulation of Wnt-7b in OA cartilage and RA synovium. In situ hybridization and immunohistochemistry also revealed that Wnt-7b was present in articular cartilage, bone, and synovium of RA samples and in osteophytes, articular cartilage, bone marrow, and synovium of OA samples. The levels of the cytokines tumor necrosis factor-α, interleukin-1β, and interleukin-6 were significantly increased in RA synovium and Wnt-7b-transfected normal synovial cells when compared with normal samples. These results point to the potential involvement of Wnt signaling in the pathobiology of both RA and OA.
Rheumatoid arthritis (RA) and osteoarthritis (OA) are two of the most common joint disorders in humans. RA is characterized by synovial inflammation, osteoporosis, generalized loss of cartilage, and bony erosions in joints that ultimately lead to significant disability of joint movement.1 OA also can be defined as an organ-level failure of a diarthrodial joint, which is characterized radiographically by loss of joint space, subchondral sclerosis, bony contour remodeling, and the presence of osteophytes. Osteophytes represent areas of new cartilage and bone formation.2,3 The prevalence of RA or OA is still increasing, and it is estimated that approximately 30 million people aged ≥35 years have RA or OA according to the Third National Health and Nutrition Examination Survey conducted by the National Center for Health Statistics from 1988 to 1994 in the United States.4 However, the etiology of these destructive joint disorders has not been defined. Here, we analyzed molecular events in the joints using Japanese patients with destructive joint disorders.
Over the past few decades, the mechanisms involved in skeletal pattern formation have been clarified at the molecular level, due to the discovery and identification of diffusible factors that regulate skeletal morphogenesis. Of these factors, bone morphogenetic proteins (BMPs), fibroblast growth factors (FGFs). Hedgehog, and Wnts have received considerable attention in the pathomorphology research area. The involvement of these proteins in the formation, growth, maintenance, and turnover of the skeleton has been confirmed by a number of genetic studies.5,6
The Wnt gene was first defined as a proto-oncogene (int-1).7,8 To date, 19 molecular types in the human Wnt family have been reported. Wnt signaling is transduced via the β-catenin-TCF pathway, the Ca2+-releasing pathway, or the Jun-N-terminal kinase pathway.9,10 The Wnt signal transduction system is involved in the determination of polarity, differentiation, and proliferation of zooblasts, and it regulates axial formation, organogenesis, carcinogenesis, embryogenesis, and morphogenesis.11–13 Wnt genes also play a key role in the highly regulated processes of vertebrate skeletogenesis, endochondral bone formation, and fracture repair.12,14–17 Wnt-1, -5a, -7a, and 9a (-14) are thought to inhibit the differentiation of undifferentiated mesenchymal cells into chondrocytes. Wnt-1, -3a, -4, -7a, and -7b also in-hibit early differentiation of chondrocytes in the chick limb.13–20 In addition, these Wnt genes are associated with BMP signaling or synovial joint formation and the maintenance of joint integrity.17,20 Recently, the importance of the Wnt pathway in normal bone accrual and in fibroblast-like cells in RA patients was reported.16,19,21,22 However, the role of the Wnt signaling pathway in association with the onset and/or progress of destructive joint disease has not been fully established compared with other factors, such as BMPs, FGFs, and hepatocyte growth factors. This is due in part to the fact that increasing numbers of subtypes have been identified and added to the Wnt family. Therefore, a comprehensive analysis of Wnt gene family expression in the joint might provide useful insight into the role of Wnt family members in the pathophysiology of joint destruction.
In this setting, we compared quantitatively the expression level of all human Wnt genes reported so far in joint tissues from RA, OA, and normal/control patients, and we investigated the distribution of Wnt proteins in the joint tissues of these patients. We also examined the potential role of Wnt genes by performing human cytokine assays after transfection of Wnt-7b, the most significant gene in RA or OA in this study, into human joint tissues.
Materials and Methods
Patients
We used knees of RA or OA patients based on the criteria of American College of Rheumatology who underwent total knee arthroplasty (TKA). The condition of all samples was in the end stage of the diseases. As control tissues, knee joint tissues of knee injuries that underwent arthroscopies or hip tissues of femoral neck fracture that underwent bipolar hip replacement, were used. Articular cartilage, bone, and synovial tissue from three groups (OA group; RA group; control: non-RA, non-OA, but traumatic group) were removed surgically and processed rapidly to extract RNA. Patients experiencing complications were excluded from the study. Each subject involved in the study had signed informed consent before the operation. This study was carried out in accordance with the World Medical Association Declaration of Helsinki.
Tissue Collection and Processing for Reverse Transcribed-Polymerase Chain Reaction (RT-PCR)
RT-PCR was performed to analyze all human Wnt genes reported so far using samples from the following patient groups: 10 RA patients who underwent TKA varying in age from 27 to 72 years (male, 1; female, 9; averaged age, 53.1 years), 9 OA patients who underwent TKA varying in age from 43 to 81 years (male, 3; female, 6; averaged age, 66.6 years), and 7 trauma patients who underwent bipolar hip replacement varying in age from 32 to 82 years (male, 1; female, 6; averaged age, 65.1). PCR primers were checked by Blast, and their sequences were confirmed to be specific to the designated target.
Total RNA from fresh, rapidly frozen tissue samples was extracted using Isogen (Nippon Gene Co., Tokyo, Japan) according to the manufacturer’s instructions. A portion of the cDNA mixture was used as a template for PCR using primer pairs specific for Human Wnts-1, -2, -3, -3a, -4, -5a, -5b, -6, -7a, -7b, -8a, -8b, -9a, -14, -9b, -15, -10a, -10b, -11, -13, -16, and FRZB. PCR products were separated on a 2% agarose gel. Each PCR product was subcloned and sequenced using a DNA sequencing kit (Applied Biosystems, Warrington, UK). The nucleotide sequences of the cloned PCR products were compared with those in the GenBank databases (GenBank Wnt-3:NM030753, Wnt-3a:XM047539, Wnt-4:NM030761, Wnt-5a:XM016181, Wnt-5b:XM053451, Wnt-7b:NM004625, Wnt-8b:XM005702, Wnt-10a:XM050840, Wnt-10b:XM053707, Wnt-11:XM006222, Wnt-13:AB070218, Wnt-14:NM003395, Wnt-15:NM003396, and Frzb:NM001463). Five or more RT-PCR reactions were run for a single gene in each joint tissue sample.
Tissue Collection and Processing for Real-Time PCR
Real-time PCR was performed to analyze human Wnts -3, -3a, -4, -5a, -5b, -7b, -8b, -10a, -10b, -11, -13, -14, -15 p-catenin, FRZB, and low-density lipoprotein receptor-related protein5 (LRP5). The following tissues were completely different from the sample used by RT-PCR. Eight RA patients varying in age from 52 to 79 years (male, 1; female, 7; averaged age, 70.2 years), 11 OA patients varying in age from 72 to 94 years (male, 1; female, 10; averaged age, 84.4 years) who underwent TKA, and 11 trauma patients varying in age from 17 to 72 years (male, 5; female, 6; averaged age, 34.4 years) who underwent joint arthroscopies were selected for real-time PCR.
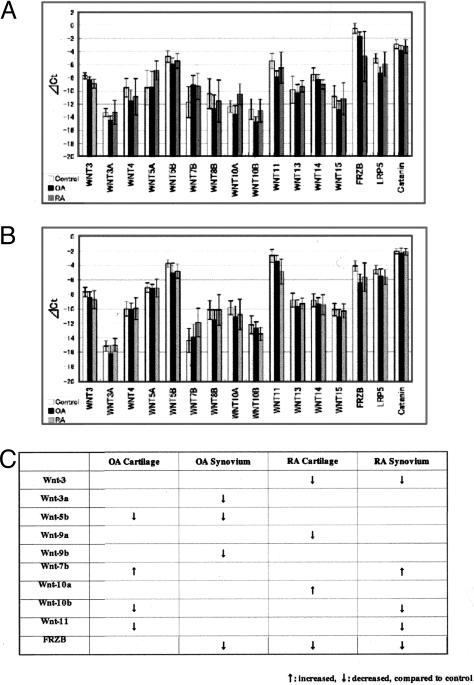
Tissue samples collected during the surgery were frozen immediately after removal and stored at −80°C in a freezer. For RNA preparations, the frozen tissues were crushed in the presence of liquid nitrogen and solubilized with TRIzol (Invitrogen, Tokyo, Japan) at room temperature for 30 minutes. After mixing with CHCl3, the aqueous phase was separated by centrifugation and precipitated with isopropanol. The precipitated nucleic acids were dissolved in RNase-free water and then treated with DNase I (Qiagen, Tokyo, Japan). The concentration of the RNA obtained was determined by using the 2100 BioAnalyzer (Agilent Technologies, Tokyo, Japan). cDNA was synthesized from 1 μg of total RNA with dT primer (Advantage RT for PCR kit; Clontech, Tokyo, Japan). TaqMan reactions were run using the TaqMan Universal PCR Master Mixture (PE Biosystems, Tokyo, Japan). The reaction conditions were as follows: 50°C for 2 minutes, 95°C for 10 minutes, followed by 94°C (15 seconds) and 60°C (1 minute) for 40 cycles. Using the real-time PCR method, the template concentration is determined by the Ct value. Ct or threshold cycle, is the PCR cycle at which an increase in reporter fluorescence above a baseline signal can first be detected. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene is used as an internal control. The abundance of each Wnt transcript is expressed as ΔCt, which is the difference between Ct (GAPDH) and Ct (target gene). Because GAPDH is expressed abundantly, ΔCt has a negative value for all Wnt genes. In Figures 1 and 2, the baseline of ΔCt is set at −20, so more mRNA expressions correspond to longer bars. Sequences of primers and probes for the real-time PCR primers are described in Table 1.
Figure 1.
Comprehensive analyses of Wnt genes, FRZB, LRP5, and β-catenin in human control samples, cartilage, and synovium in RA (A) and cartilage and synovium in OA (B) by real-time PCR. The template concentration is determined by the Ct value. GAPDH gene is used as an internal control. The abundance of each Wnt transcript is expressed as ΔCt, which is the difference between Ct (GAPDH) and Ct (target gene). The baseline of ΔCt is set at −20, so that more mRNA expressions correspond to longer bars. Expression pattern of Wnt genes by real-time PCR in RA, OA cartilage and synovium. C: Summary of the results in A and B. Wnt-7b in OA cartilage, and RA synovium, Wnt-10a in RA cartilage were significantly up-regulated, compared with control samples. Wnt-3 in RA cartilage and synovium, Wnt-3a in OA synovium, Wnt-5b in OA cartilage and synovium, Wnt-9a in RA cartilage, Wnt-9b in OA synovium, Wnt-10b in OA cartilage and RA synovium, Wnt-11 in OA cartilage and RA synovium, FRZB in OA synovium, and RA cartilage and synovium were significantly down-regulated compared with control samples.
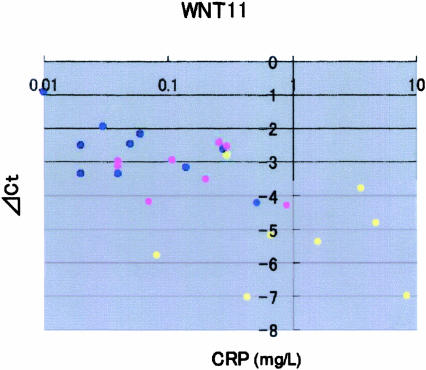
Figure 2.
Clinical data of WBC count, CRP (mg/L), and Hct (%). Blue dots represent the data from control, pink dots from OA, and yellow dots from RA synovium. Wnt-11 expression significantly decreased as CRP increased in RA and OA patients, compared with control patients.
Table 1.
Real-Time PCR Primers
| Gene | Primer sequence | Probe |
|---|---|---|
| WNT3 | GGAGAAGCGGAAGGAAAAATG | TTCCACTGGTGCTGCTACGTCA |
| GCACGTCGTAGATGCGAATACA | ||
| WNT3A | CCTGCACTCCATCCAGCTACA | AGAAGCCTCTCGTCCCGTCCCTCC |
| GACCTCTCTTCCTACCTTTCCCTTA | ||
| WNT4 | GATGTGCGGGAGAGAAGCAA | AACCTCCACAACAATGAGGCCGGC |
| ATTCCACCCGCATGTGTGT | ||
| WNT5A | GAAATGCGTGTTGGGTTGAA | TTCTGCCTCACCCCTTTGTCT |
| ATGCCCTCTCCACAAAGTGAA | ||
| WNT5B | CTGCCTTTCCAGCGAGAATT | TCCACGGTTCACTAGCTCCTACCT |
| AGGTCAAATGGCCCCCTTT | ||
| WNT7B | CCCGGCAAGTTCTCTTTCTTC | AGCAAAGTGATGAGGAGACTGAGCGT |
| GGCGTAGCTTTTCTGTGTCCAT | ||
| WNT8B | TCCCAGAAAAACTGAGGAAACTG | CCCCGGAAAAGCATGTCTTTGGG |
| AACCTCTGCCTCTAGGAACCAA | ||
| WNT10A | GGCAACCCGTCAGTCTGTCT | CATCCTTTCACCCCTTCCCTG |
| CATTCCCCACCTCCCATCT | ||
| WNT10B | CTTTTCAGCCCTTTGCTCTGAT | TCTTGGTCCCTGGAAGCTTAAAGT |
| CCCCTAAAGCTGTTTCCAGGTA | ||
| WNT11 | GGCTTGTGCTTTGCCTTCA | TTGGAAGCCACCAGGAACAGAAGG |
| TTTGATGTCCTGCCCTCCTT | ||
| WNT13 | TGCCAAGGAGAAGAGGCTTAAG | CCCGGGCCCTCATGAACTTACA |
| GTGCGACCACAGCGGTTATT | ||
| WNT14 | CTTAAGTACAGCAGCAAGTTCGTCAA | AGCAAGGATCTGCGAGCCCGTGT |
| CCACGAGGTTGTTGTGGAAGT | ||
| WNT15 | CAGGTGCTGAAACTGCGCTAT | CTCGGCTGTCAAGGTGTCCAGT |
| GCCCAAGGCCTCATTGGT | ||
| β-Catenin | ||
| F | 5′-CTGCTGTTTTGTTCCGAATGTC | |
| R | 5′-CCATTGGCTCTGTTCTGAAGAGA | |
| Probe | 5′-AAACGGCTTTCAGTTGAGCTGACC | |
| FRZB | CCTGCCCTGGAACATGACTAA | CAACCACCTGCACCACAGCACTCA |
| CAGACCTTCGAACTGCTCGAT | ||
| LRP5 | ||
| F | 5′-GCTGTACCCGCCGATCCT | |
| R | 5′-GGCGCCATTCCTCGAAT | |
| Probe | 5′-CAAACATTCCGGCCACTGCGAGAC |
All primer sequences are written from 5′ to 3′. For each primer pair, the top sequence is sense, and the bottom sequence is antisense.
We used the so-called 2ΔΔCt method to analyze relative changes of Wnt-7b or -10a as described elsewhere by Livak et al.23 The means ± SD of each Wnt gene, β-catenin, FRZB, and LRP5 expression were obtained from each RA, OA, or control group. Differences in the mean of each Wnt gene, β-catenin, FRZB, and LRP5 among experimental groups were evaluated with Dunnett’s test.
RNA Probes for in Situ Hybridization
A 354-base fragment of human Wnt-7b cDNA and a 402-base fragment of human Wnt-10a cDNA were subcloned into pBlueScript SK(−) vector (Stratagene, Tokyo, Japan) and used for generation of sense and antisense RNA probes. The sequences of Wnt-7b and Wnt-10a probes correspond to GenBank no. AB062766, position 1105 to 1458, and no. AB059569, position 1199 to 1600, respectively.
In Situ Hybridization
Three severe RA patients and three severe OA patients after TKA were used for histological assessment. A 49-year-old female RA patient after TKA (most severe case) and a 94 year-old male OA patient after TKA (most severe case), are shown here. After the operations, articular cartilage, bone, and synovial tissues were taken out en bloc, cut at 2 mm thickness, and fixed in fresh 10% neutral buffered formalin for 24 hours. The cartilage and bone tissues were decalcified with 20% EDTA after washing with 0.1 mol/L phosphate-buffered saline (PBS). They were then dehydrated through a graded ethanol series and embedded in paraffin. Sections of 4 μm thickness were cut and mounted on α-aminopropyl triethoxysilane (APS)-coated slides. They were stored at 4°C until use.
Sections were blow-dried, deparaffinized, rehydrated, and fixed with 4% paraformaldehyde for 10 minutes at room temperature. They were then successively treated with PBS for 5 minutes, 15 μg/ml proteinase (DAKO Cytomation, Cambridgeshire, UK) in PBS for 10 minutes at 37°C, 4% paraformaldehyde for 10 minutes, 0.2 N HCl for 10 minutes, 0.1 mol/L triethanolamine for 5 minutes, and 1% acetic acid/0.1 mol/L triethanolamine for 10 minutes. Digoxigenin (DIG)-labeled single-strand antisense RNA probes for Wnt-7b and -10a were prepared with a DIG-labeling Mix (×10 concentration). The histological sections were covered with siliconized coverslips and incubated at 60°C for 16 hours in a humid chamber. The hybridization solution contained 50% deionized formamide, 5× standard saline citrate (SSC), 1% sodium dodecyl sulfate (SDS), 50 μg/ml of heparin, and approximately 75 ng/slide of RNA probe. After hybridization, the slides were washed two times for 10 minutes with 2× SSC at 50°C, and 0.2× SSC at 50°C. Hybridized DIG-labeled probes were detected by Tris-buffered saline (TBS) (0.3% Tween 20), blocking solution, anti DIG-alkaline phosphatase-labeled antibody (1:2000), TBS (0.3% Tween 20) twice, APB (0.1 mol/L Tris-HCl (pH 9.5), 0.1 mol/L NaCl, and 50 mmol/L MgCl2) and then 4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) (Boehringer, Tokyo, Japan). Thereafter, the slides were mounted with Kernechtrot stain solution (Muto Chemical Co., Tokyo, Japan). The control procedures consisted of hybridization with the sense probes and omission of either antisense RNA probe or the anti-DIG antibody.
Hematoxylin and Eosin and Immunohistochemical Staining
Hematoxylin and eosin and immunohistochemical staining were performed with the same block used for in situ hybridization, and sections of 4 μm thickness were prepared using a microtome. Anti-human Wnt-7b and -10a antibodies were purchased from Sigma Genosys (Tokyo, Japan). Deparaffinized sections were treated with 0.3% hydrogen peroxide in methanol at room temperature for 30 minutes to quench endogenous peroxidase and then washed in TBS. The sections were treated with 0.05% proteinase K in TBS and then washed in TBS. They were incubated overnight at 4°C with rabbit anti-Wnt-7b or -10a antibody diluted to 2 μg/ml, after they were treated with normal goat serum for 30 minutes at 37°C to avoid nonspecific binding. After washing in TBS, they were incubated at room temperature for 30 minutes with a 1:600 Biotinylated goat anti-rabbit immunoglobulins (DAKO Cytomation). They were treated with peroxidase-conjugated streptavidin (Nichirei Co., Tokyo, Japan) at room temperature for 5 minutes and then washed in TBS. Color was developed by the DAB substrate kit (Nichirei Co., Tokyo, Japan). Finally, the sections were counterstained with hematoxylin. Control sections were treated in the same manner with 1% bovine serum used in place of the primary antibodies.
Examination of Clinical Data
Blood was taken from the patients before operations to obtain clinical data including white blood cell (WBC) count, C-reactive protein (CRP) (mg/L), and Hematocrit (Hct) (%). Samples from the same patients described under “Tissue Collection and Processing for Real-Time PCR” were used here. The latest data from the patients before the surgeries are reported in the present study.
The correlation between expression level of Wnt gene and clinical data were examined. Every possible combination between the order of ΔCt value of Wnt and the order of clinical data value of tissue donor (WBC, CRP, Hct, and age) was tested with Spearman’s Rank Correlation Method.
Cell Cultures, Plasmids, and Transfection Procedures
Considering that the expression of Wnt-7b was up-regulated in OA cartilage and RA synoviums at the mRNA and protein levels, compared with the controls, we next tried to investigate the function of Wnt-7b in those tissues.
Primary synovial cells and chondrocytes in control patients were generated for the following transfection as described previously.24 Primary synovial cells and chondrocytes in RA and OA patients were also generated for the following Western blot analysis and cytokine assay. Cells were plated at 2 × 106 cells/100-mm plastic dish (Falcon #3003; Becton Dickinson Labware, Tokyo, Japan) in 10 ml of Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL, Grand Island, NY) with 10% fetal calf serum (Sigma Chemical Co., St. Louis, MO) at 37°C and incubated for 1 day before transfection. Transfection grade eukaryotic expression vector containing hemagglutinin-tagged mouse Wnt-7b under the control of cytomegalovirus promoter was purified using standard procedures (Upstate Biotechnology, Lake Placid, NY). The amino acid homology of Wnt-7b between mouse and human is approximately 98% (GenBank human Wnt-7b, NP478679; mouse Wnt-7b, NP033554).
For each plate to be transfected, 4 μg of Wnt-7b plasmid with serum-free DMEM was combined with LipofectAmine plus reagent (Invitrogen, Carlsbad, CA) and then incubated at room temperature for 15 minutes. LipofectAmine reagent was also combined with serum-free DMEM and then added dropwise to a small Eppendorf tube containing DNA. After tapping to mix, the tubes were incubated at room temperature for 15 minutes. Without removing the overlying media from cells, the entire LipofectAmine reagent/DNA mixture was added dropwise to the plates. The plates were gently rocked to mix, and then incubated at 37°C, 5% CO2. The following day, medium was replaced with 5 ml of serum-free DMEM. After an incubation of 24 hours, conditioned medium was collected and subject to Western blot analysis.
Western Blot Analysis
Because Wnt-7b is a secreted protein, 0.5 ml of supernatant from each plate was added to 0.5 ml of sample buffer (0.05 mol/L Tris-HCl (pH6.8), 2% SDS, 6% β-mercaptoethanol, and 10% glycerol) and centrifuged at 12,000 × g for 5 minutes at 4°C. Then the protein content of each sample was measured by a UV assay at an OD of 280 nm. Anti- Wnt-7b human antibody was used at a dilution of 1 μg/ml. Aliquots of protein solution (5 μl) were adjusted to 1 μg/μl, mixed with 1% bromophenol blue (1 μl; Sigma, St. Louis, MO), and then boiled for 2 minutes. They were then loaded on SDS (10% ∼ 20%) acrylamide gradient gels (35 mA, low voltage, 90 minutes). The protein bands were transferred to a polyvinylidene difluoride membrane (Immunobilon-P Transfermembrane; Millipore, Bedford, MA) according to the manufacturer’s instructions. After treatment with blocking reagent (Nippon Roche Co., Tokyo, Japan) for 1 hour at room temperature, the membranes were washed with PBS for 5 minutes and then incubated for 1 hour with primary antibody (1:200). After two 5-minute washes with PBS, the membranes were incubated with horseradish peroxidase (HRP)-conjugated rabbit anti-goat antibodies (1:500, Histofine; Nichirei Co.) for 1 hour. After two further 5-minute washes with PBS, the immunoblot was developed using an ImmunoStar kit for Rabbit (Wako Pure Chemical Industries Ltd., Tokyo, Japan) in the process of the detection of biotin and chemiluminescence. We used β-tubulin as an internal control.
Cytokine Assay
Because the expression of Wnt-7b was up-regulated in transfected RA synovial cells at the protein level, we next sought to examine how Wnt-7b would affect inflammatory cytokines tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, and IL-6.
Human cytokine antibody arrays with TNF-α, IL-1β, and IL-6 were purchased from RayBiotech, Inc. (Tokyo, Japan). The supernatant from human primary synovial cells (RA, OA, control patients, and Wnt-7b transfected normal synovial cells) were placed into the array membrane. The array membranes were then washed with wash buffer after treatment with blocking buffer (5% BSA/TBS, 0.01 mol/L Tris-HCl, and 0.15 mol/L NaCl) and then incubated for 2 hours with biotin-conjugated anticytokine antibodies to remove unbounded cytokines. The membranes were incubated with HRP-conjugated streptavidin (2.5 pg/ml) for 60 minutes at room temperature. After further washes with wash buffer to remove any unbound materials, the signals were detected and quantitated by a chemiluminescence imaging device and enhanced chemiluminescence system (Amersham Pharmacia Biotech, Aylesbury, UK). The changes of the signals were compared with the control patients (fold change = 1). The mean of fold change cytokines with SD were obtained from performing the assay five times for each group. Differences in the mean of fold change cytokines between experimental groups were evaluated with Dunnett’s test.
Results
RT-PCR
Wnt-3, -3a, -4, -5a, -5b, -7b, -8b, -9a,14, -9b,15, -10a, -10b, -11, -13, and FRZB were detected among Wnt-1, -2, -3, -3a, -4, -5a, -5b, -6, -7a, -7b, -8a, -8b, -9a, -9b, -10a, -10b, -11, -13, -16, and FRZB in all RA, OA cartilage and synoviums by RT-PCR. Each Wnt gene and FRZB were detected in every RA, OA cartilage and synovium tissue by RT-PCR (data not shown). Then, real-time PCR was performed for quantitative assessment.
Expression Profiles of Some Wnts, FRZB, LRP5, and β-Catenin mRNA in RA, OA, and Normal Tissue by Real-Time PCR
Real-time PCR revealed the overall level of each Wnt family member in human joint tissues. The overall expression patterns were not very different in RA, OA, and control groups. (Figure 1, A and B). In RA patients, Wnt-7b was significantly up-regulated (5.62-fold, P < 0.01) in synovium, and Wnt-10a was significantly up-regulated (3.73-fold, P < 0.05) in cartilage, when compared with control tissues. Wnt-3, -10b, -11, and FRZB were down-regulated in synovium. In OA patients, Wnt-7b was significantly up-regulated (6.25-fold, P < 0.01) in cartilage. Wnt-3a, -5b, -15, and FRZB were down-regulated in synovium, and Wnt-3, -14, and FRZB were down-regulated in cartilage (Figure 1C). LRP5 was down-regulated in RA, OA synovium and cartilage. FRZB was also down-regulated in RA cartilage, and OA synovium and cartilage. β-Catenin mRNA level did not significantly change in RA or OA samples, compared with control tissues.
Examination of Clinical Data
Among every possible combination between the expression level of Wnt genes and clinical data, the most significant relationship was found between Wnt-11 and CRP (correlation, −0.64; P < 0.001). As shown in Figure 2, as CRP increased, the expression of Wnt-11 decreased.
In Situ Hybridization Study
As the expression of Wnt-7b and -10a increased in RA and/or OA groups, compared with the control groups, we next tried to find the localizations of those genes by in situ hybridization. In situ hybridization revealed that in RA patients, Wnt-7b was strongly expressed in the macrophage-like cells, fibroblastic cells, and vessel walls in the synovium, particularly in the region of inflammation. Wnt-10a was weakly expressed in chondrocytes of articular cartilage, bone marrow stromal cells, or hematopoietic cells in subchondral bone but strongly expressed in macrophage-like cells, fibroblastic cells, and vessel walls in the synovium. In OA patients, Wnt-7b was strongly expressed in chondrocytes, osteoclast-like cells in the osteophyte and articular cartilage, and bone marrow stromal cells or hematopoietic cells in subchondral bone. It was also strongly expressed in macrophage-like cells, fibroblastic cells, and vessel walls in the synovium, particularly in the region of inflammation. Wnt-10a was weakly expressed in chondrocytes, osteoclast-like cells in the osteophyte and articular cartilage, and bone marrow stromal cells or hematopoietic cells in subchondral bone. It was also expressed in macrophage-like cells, fibroblastic cells, and vessel walls in the synovium (Figure 3, A to F; Wnt-10a: data not shown).
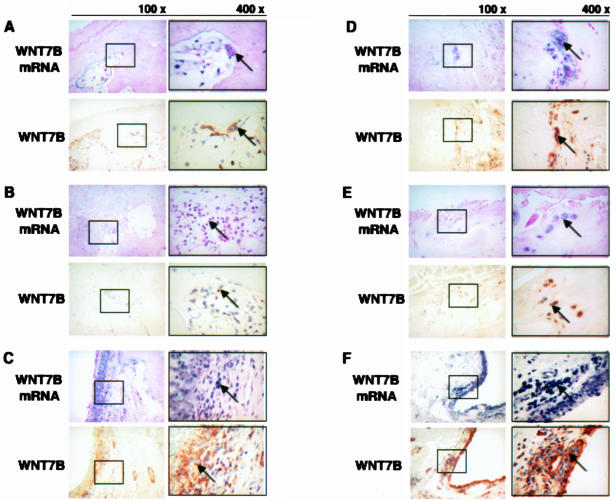
Figure 3.
Histological appearance of Wnt-7b by in situ hybridization (ISH) and immunohistochemistry (IHC). Cross-section at 100× magnification is in the left panel and 400× magnification section is in the right panel. Higher-magnification images corresponding to the box areas in each figure. In RA samples, ISH and IHC show that Wnt-7b is present at osteoclast-like cells of articular cartilage (A, arrows), bone marrow stromal cells or hematopoietic cells of bone marrow (B, arrows), and fibroblastic cells and vessel walls of the synoviums (C, arrows). In OA samples, ISH and IHC reveal that Wnt-7b is expressed in multinucleated osteoclast-like cells in osteophytes (D, arrows), osteoclast-like cells in the articular cartilage (E, arrows), and fibroblastic cells and vessel walls of synovium (F, arrows).
Immunohistochemical Study and Western Blotting
An immunohistochemical study was performed on Wnt-7b and -10a, because of the significant up-regulation of these genes by real-time PCR and the expression of those genes by in situ hybridization in RA and/or OA joint tissues. In RA and OA joint tissues, Wnt-7b was heavily localized in macrophage-like cells, fibroblastic cells, and vessel walls in the synovium, weak and sparse chondrocytes in articular cartilage, bone marrow stromal cells, or hematopoietic cells in subchondral bone. Wnt-10a was not detected in joint tissue (Figure 3, A to F; Wnt-10a: data not shown). Western blotting analysis also confirmed that the expression of Wnt-10a was not detected at the protein level in RA and OA joint tissues (data not shown).
Transfection of Wnt-7b, Western Blotting Analysis, and Cytokine Assay
As the expression of Wnt-7b was enhanced in RA, OA joint tissues at mRNA and the protein levels, we further investigated the secretion of inflammatory cytokines after transfection of Wnt-7b in control chondrocytes and synovial cells. LipofectAmine reagent transfected approximately 10% of the undifferentiated mesenchymal cells confirmed by Western blotting and immunohistochemistry (data not shown).
Western blotting analysis revealed that the secretion of Wnt-7b in the transfected synovial cells in control patients was similar to that of synovial cells in RA patients (Figure 4). The secretion of Wnt-7b in the transfected cartilage cells in control patients was not detected (data not shown).
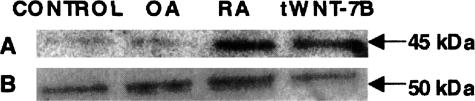
Figure 4.
Western blot analysis of Wnt-7b using primary synovial cells in control, RA, OA patients, and normal synovial cells after transfection of Wnt-7b (tWNT-7B). The expression of Wnt-7b was significantly increased in RA and tWNT-7B, compared with the control and OA samples (A). β-Tubulin protein levels obtained by Western blotting was used as a positive control (B).
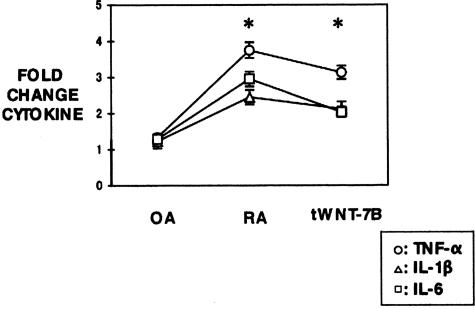
Compared with the empty vector transfected controls, the secretion of TNF-α, IL-1β, and IL-6 in primary synovial cells of OA patients were not significantly changed. However, the secretion of TNF-α, IL-1β and IL-6 in primary synovial cells of RA patients and the Wnt-7b transfected normal synovial cells, especially TNF-α, significantly increased the expression level twofold to fourfold (P < 0.05) (Figure 5).
Figure 5.
The expression changes of TNF-α, IL-1β, and IL-6 in primary synovial cells from RA, OA patients, and normal synovial cells after transfection of Wnt-7b (tWNT-7B). Quantitation of cytokine levels in primary synovial cells was measured by chemiluminescence imaging device in the OA, RA, and tWNT-7B. The score in the controls (the expression in empty vector transfected cells) was used as a standard (fold change of controls = 1). The expression of TNF-α, IL-1β, and IL-6 in RA, and tWNT-7B was twofold to fourfold changes than that of OA. The expression of TNF-α was significantly higher than that of IL-1β and IL-6 in each group (P < 0.05).
Discussion
The etiopathogenesis of RA or OA is still the subject of intense debate and research. This is the first comprehensive study of all known human Wnt genes and the Wnt-related genes, β-catenin, FRZB, and LRP5 in joint tissues from human RA and OA patients. Compared with normal tissues, real-time PCR revealed that Wnt-7b in OA cartilage and RA synoviums and Wnt-10a in RA cartilage were significantly up-regulated at mRNA level. An in situ hybridization study revealed that in OA patients, Wnt-7b and -10a were expressed in chondrocytes or osteoclast-like cells in osteophytes, chondrocytes in articular cartilage, bone marrow stromal cells or hematopoietic cells in subchondral bone, and fibroblastic cells and vessel walls in the synovium. In RA patients, Wnt-7b was expressed in chondrocytes in articular cartilage, bone marrow stromal cells or hematopoietic cells in subchondral bone, and fibroblastic cells and vessel walls in synovium. Wnt-10a was expressed in chondrocytes of articular cartilage and bone marrow stromal cells or hematopoietic stem cells in subchondral bone. Wnt-10a was also expressed in fibroblastic cells and vessel walls in synovium. However, Wnt-10a was not detected at the protein level. Therefore, the increased expression of Wnt-7b may be a relationship with joint disease in RA and OA patients. Wnt-7b was transfected into primary chondrocytes and synovial cells in control patients to elucidate the role of RA or OA. We then investigated the secreted levels of TNF-α, IL-1β, and IL-6 in those cells. As a result, the secretion of TNF-α, IL-1β, and IL-6 in primary synovial cells of RA patients and the Wnt-7b transfected normal synovial cells were significantly enhanced compared with the control tissues. These results suggest that the expression of Wnt-7b in RA synovium might play a few roles in the process of RA inflammation. The reason why Wnt-7b was expressed in vessel walls in RA and OA synoviums is still unclear. However, the results might indicate that Wnt-7b is associated with not only with joint destruction, but also perhaps within the vessel walls. Wnt-7b was expressed in bone marrow stromal cells or hematopoietic cells in RA and OA bone marrow tissues, however, the cells were not specified. Further studies would be necessary to clarify the localization of Wnt-7b in joint tissues.
Osteophytes, which are most notably present at the joint margins, represent areas of new cartilage and bone formation in humans and have been used to characterize OA. The presence of osteophytes, which can distinguish OA from RA, has commonly been used in the definition of OA.2,3,24,25 It seems likely that mechanical, humoral, and other factors are involved in stimulating the formation of osteophytes, although the cause of growth and development of osteophytes remains unclear. In this study, Wnt-7b in OA cartilage was significantly up-regulated by real-time PCR. At mRNA and protein levels, Wnt-7b was expressed in cartilage cells or osteoclast-like cells in osteophytes and bone marrow stromal cells or hematopoietic cells in subchondral bone. Therefore, Wnt-7b might affect osteophyte formation.
There are three pathways in Wnt signaling. The Wnt-β-catenin cascade is thought to be the main pathway.26–28 β-Catenin is the key mediator of the Wnt signal and is usually stored in the cytoplasm. Wnt-1, -8a, and -8b are involved in the Wnt-β-catenin cascade as described previously.29–30 Here, Wnt-8b was significantly down-regulated in OA synovium, compared with normal synovium, although the expression of β-catenin did not significantly change in OA or RA tissue, compared with normal tissues, by real-time PCR. These results suggest that Wnt-β-catenin signaling might not be involved in human joint destruction. Functional investigations would be necessary to support these results. LRP5 is capable of functioning in the Wnt/β-catenin signaling pathway.29 Wnt/LRP5 signaling is necessary for the induction of alkaline phosphatase, and the Wnt/β-catenin signaling pathway can act in a Smad-independent manner in ST2 cells and C3H10T1/2 cells.30 The down-regulation of LRP5 mRNA in OA or RA tissue in this study, compared with normal tissues, might suggest that LRP5 is not activated during human joint destructive processes.
FRZB, which antagonizes Wnt signaling, appears to indirectly promote chondrogenesis by removing the blocking activity of Wnt on chondral differentiation. Cartilage extracts containing FRZB have in vivo chondrogenic activity.31–33 In our study, FRZB was down-regulated in OA synovium and cartilage and in RA cartilage. Therefore, FRZB might be also involved in human joint cartilage destruction. FRZB is also a member of Wnt family, so it would be important to look at the expression of FRZB-related genes in these tissues.
WBC count, CRP, and Hct are useful parameters to judge systemic joint inflammation. In our study, as CRP increased, only Wnt-11 significantly decreased in RA and OA patients. This result suggests that there might be a relationship between Wnt-11 and CRP in the process of inflammation in the destructive joint disease.
In conclusion, our results suggest that Wnt signaling mechanisms are involved in the joint pathology associated with RA and OA disease progression. The secretion of Wnt-7b might be a key factor in inflammation of RA. Therefore, the regulation of Wnt expression may provide opportunities to arrest the joint destruction and pain linked with RA and OA. Future studies will be directed toward understanding the relationship between Wnt genes, Wnt receptors, and the effectors of Wnt signaling in joint tissues.
Acknowledgments
We thank Dr. Hiroyuki Nakaya and Dr. Hiroshi Ota at Department of Orthopaedic Surgery, Shinshu University School of Medicine, and Dr. Takahiro Nomura at Iida Municipal Hospital for providing us with various human tissue samples. We are grateful to Dr. Jun Ishizaki and Dr. Etsuo Nakamura at Shionogi & Co. Ltd, for their technical assistance and valuable discussion. We also thank Dave C. Morris of Discovery Research Biology and Simone Wilson at Case Western Reserve University for their writing assistance. All research was performed at Shinshu University School of Medicine.
Footnotes
Address reprint requests to Shigeyuki Wakitani, M.D., Department of Orthopaedic Surgery, Shinshu University School of Medicine, Asahi 3-1-1, Matsumoto, 390-8621, Japan. E-mail: wakitani@hsp.md.shinshu-u.ac.jp.
A related commentary by Y. Ishikawa appears in this issue (Am J Pathol 2005, 167: 1–3).
References
- Haugeberg G, Orstavik RE, Kvein TK. Effects of rheumatoid arthritis on bone. Curr Opin Rheumatol. 2003;15:469–475. doi: 10.1097/00002281-200307000-00016. [DOI] [PubMed] [Google Scholar]
- Moskowitz RW, Howell DS, Altman RD, Buckwalter JA, Goldberg VM. Osteoarthritis, ed 3. Philadelphia: W.B. Saunders,; Diagnosis and Medical/Surgical Management, 2003 [Google Scholar]
- Hashimoto S, Creghton-Achermann, Takahashi K, Amiel D, Coutts RD. Development and regulation of osteophyte formation during experimental osteoarthritis. Osteoarthritis Cartilage. 2002;10:180–187. doi: 10.1053/joca.2001.0505. [DOI] [PubMed] [Google Scholar]
- National Center for Health Statistics, Centers for Disease Control and Prevention, US Department of Health and Human Services Hyattsville, MD: Division of Data Services, National Center for Health Statistics, US Centers for Disease Control and Prevention,; National Health and Nutrition Examination Survey, III 1988–1994. 1996 [Google Scholar]
- Grotewold L, Ruther U. Bmp, Fgf and Wnt signalling in programmed cell death and chondrogenesis during vertebrate limb development: the role of Dickkopf-1. Int J Dev Biol. 2002;46:943–947. [PubMed] [Google Scholar]
- Tuan RS. Cellular signaling in developmental chondrogenesis: n-cadherin, Wnts, and BMP-2. J Bone Joint Surg Am. 2003;85:137–141. doi: 10.2106/00004623-200300002-00019. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Kitagaki J, Iwamoto M. Regulation of chondrocyte differentiation by a soluble Wnt receptor, Frzb-1. Jikken Igaku. 2001;19:1210–1214. [Google Scholar]
- Nusse R, Brown A, Papkoff J, Scambler P, Shackleford G, McMahon A, Moon R, Vamus H. A new nomenclature for int-1 and related genes: the Wnt gene family. Cell. 1991;64:231. doi: 10.1016/0092-8674(91)90633-a. [DOI] [PubMed] [Google Scholar]
- Miller JR, Hocking AM, Brown JD, Moon RT. Mechanism and function of signal transduction by the Wnt/beta-catenin and Wnt/Ca2+ pathways. Oncogene. 1999;18:7860–7872. doi: 10.1038/sj.onc.1203245. [DOI] [PubMed] [Google Scholar]
- Seidensticker MJ, Behrens J. Biochemical interactions in the wnt pathway. Biochim Biophys Acta. 2000;1495:168–182. doi: 10.1016/s0167-4889(99)00158-5. [DOI] [PubMed] [Google Scholar]
- Sohn P, Crowley M, Slattery E, Serra R. Developmental and TGF-beta-mediated regulation of Ank mRNA expression in cartilage and bone. Osteoarthritis Cartilage. 2002;10:482–490. doi: 10.1053/joca.2002.0810. [DOI] [PubMed] [Google Scholar]
- Moon RT, Brown JD, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Hsieh JC, Rattner A, Smallwood PM, Nathans J. Biochemical characterization of Wnt-frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc Natl Acad Sci USA. 1999;96:3546–3551. doi: 10.1073/pnas.96.7.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;27:3141–3159. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- Sen M, Chamorro M, Reifert J, Corr M, Carson DA. Blockade of Wnt-5A/frizzled 5 signaling inhibits rheumatoid synoviocyte activation. Arthritis Rheum. 2001;44:772–781. doi: 10.1002/1529-0131(200104)44:4<772::AID-ANR133>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/s0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- Rudnicki JA, Brown AM. Inhibition of chondrogenesis by Wnt gene expression in vivo and in vitro. Dev Biol. 1997;185:104–118. doi: 10.1006/dbio.1997.8536. [DOI] [PubMed] [Google Scholar]
- Sen M, Lauterbach K, EI-Gabalawy H, Firestein GS, Corr M, Carson DA. Expression and function of wingless and frizzled homologs in rheumatoid arthritis. Proc Natl Acad Sci U S A. 2000;97:2791–2796. doi: 10.1073/pnas.050574297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letamendia A, Labbe E, Attisano L. Transcriptional regulation by Smads: crosstalk between the TGF-beta and Wnt pathways. J Bone Joint Surg Am. 2001;83:31–39. [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- Ijiri K, Nagayoshi R, Matsushita N, Tsuruga H, Taniguchi N, Gushi A, Sakakima H, Komiya S, Matsuyama T. Differential expression patterns of secreted frizzled related protein genes in synovial cells from patients with arthritis. J Rheumatol. 2002;29:2266–2270. [PubMed] [Google Scholar]
- Barchowsky A, Frleta D, Vincenti MP. Integration of the NF-kappaB and mitogen-activated protein kinase/AP-1 pathways at the collagenase-1 promoter: divergence of IL-1 and TNF-dependent signal transduction in rabbit primary synovial fibroblasts. Cytokine. 2000;12:1469–1479. doi: 10.1006/cyto.2000.0743. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Shepstone L, Rogers J, Kirwan J, Silverman B. Distribution of distal femoral osteophytes in a human skeletal population. Ann Rheum Dis. 2000;59:513–520. doi: 10.1136/ard.59.7.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;9:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Kuhl M, Sheldahl LC, Park M, Miller JR, Moon RT. The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet. 2000;16:279–283. doi: 10.1016/s0168-9525(00)02028-x. [DOI] [PubMed] [Google Scholar]
- Boutros M, Paricio N, Strutt DI, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML. Osteoporosis-Pseudoglioma Syndrome Collaborative Group: LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Mine T, Katoh M. Expression and regulation of WNT8A and WNT8B mRNAs in human tumor cell lines: up-regulation of WNT8B mRNA by beta-estradiol in MCF-7 cells, and down-regulation of WNT8A and WNT8B mRNAs by retinoic acid in NT2 cells. Int J Oncol. 2002;20:999–1003. [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang B, Moos M, Jr, Vukicevic S, Luyten FP. Primary structure and tissue distribution of FRZB, a novel protein related to Drosophila frizzled, suggest a role in skeletal morphogenesis. J Biol Chem. 1996;271:26131–26137. doi: 10.1074/jbc.271.42.26131. [DOI] [PubMed] [Google Scholar]
- Wang S, Krinks M, Moos M., Jr Frzb-1, an antagonist of Wnt-1 and Wnt-8, does not block signaling by Wnts -3A, -5A, or -11. Biochem Biophys Res Commun. 1997;236:502–504. doi: 10.1006/bbrc.1997.6995. [DOI] [PubMed] [Google Scholar]