Abstract
The 26S proteasome system is involved in eliminating various proteins, including ubiquitinated misfolded/unfolded proteins, and its inhibition results in cellular accumulation of protein aggregates. Intramuscle-fiber ubiquitinated multiprotein-aggregates are char-acteristic of sporadic inclusion-body myositis (s-IBM) muscle fibers. Two major types of aggregates exist, containing either amyloid-β (Aβ) or phosphorylated tau (p-tau). We have now asked whether abnormalities of the 26S proteasome contribute to s-IBM pathogenesis and whether the multiprotein aggregates have features of aggresomes. Using cultured human muscle fibers we also studied the effect of amyloid-β precursor protein (AβPP) overexpression on proteasome function and the influence of proteasome inhibition on aggresome formation. We report that in s-IBM muscle biopsies 26S proteasome subunits were immunodetected in the γ-tubulin-associated aggresomes, which also contained Aβ, p-tau, ubiquitin, and HSP70. In addition, a) expression of proteasome subunits was greatly increased, b) the 20Sα proteasome subunit co-immunoprecipitated with AβPP/Aβ, and c) the three major proteasomal proteolytic activities were reduced. In cultured muscle fibers, AβPP-overexpressing fibers displayed diminished proteasomal proteolytic activities, and addition of proteasome inhibitor strikingly increased aggresome formation. Accordingly, proteasome dysfunction in s-IBM muscle fibers may play a role in accumulation of misfolded, potentially cytotoxic proteins and may be induced by increased intracellular AβPP/Aβ.
Sporadic inclusion body myositis (s-IBM), the most common degenerative muscle disease of patients age 50 years and older, is of unknown etiology and pathogenesis, and it lacks definitive treatment.1,2 The light-microscopic features of s-IBM muscle biopsies include: 1) vacuolated muscle fibers; 2) accumulation of intramuscle fiber multiprotein aggregates; and 3) various degrees of lymphocytic inflammation.1,2 An intriguing feature of the s-IBM muscle-fiber phenotype is its similarity to the Alzheimer’s disease (AD) brain, including accumulation of amyloid-β (Aβ), phosphorylated tau (p-tau), and several other Alzheimer-characteristic proteins.1,2 Two major types of intracellular aggregates contain either Aβ or p-tau,1–4 and both contain ubiquitin.1,2 Both types of aggregates are positive with crystal-violet, thioflavin S, and Congo Red, indicating that they contain proteins in alternate conformation (unfolded or misfolded) that are assembled in the β-pleated-sheet configuration of amyloid.1,2 Both types of aggregates contain several other accumulated proteins,1,2 including mutated ubiquitin (UBB+1)5. Recently the unfolded protein response was demonstrated in s-IBM muscle fibers,6 further suggesting a role for misfolded/unfolded proteins in the s-IBM pathogenesis. Some of the proteins in those aggregates have been shown experimentally to inhibit the 26S proteasome.7–13
The 26S proteasome, composed of a catalytic 20S core and a 19S regulatory complex, is an ∼700-kd multisubunit protease complex present in the cytoplasm and nucleus of eukaryotic cells. It plays a major role in the degradation of normal and abnormal proteins, through a ubiquitin-mediated ATP-independent process.14,15 19S mediates the recognition of polyubiquitinated proteins, permitting their access into the 20S component core, which is comprised of α- and β-subunits. β-Subunits contain trypsin-like (TL), chymotrypsin-like (CTL), and peptidyl-glutamyl-peptide hydrolytic (PGPH) activities.14,15 Three β-subunits, β1, β2 and β5, have γ-interferon-inducible counterparts,14 which increase CTL and TL proteasome activities that are optimal for major histocompatibility complex-I (MHC-I) epitope processing.16,17 The 20S proteasome is involved also in ubiquitin-independent degradation of several proteins,18,19 and in degradation of oxidized proteins in an ATP-independent manner.20
Aggresomes, microtubule-dependent pericentriolar cytoplasmic structures, form when a cell’s capacity to degrade misfolded proteins is diminished.21,22 Their formation requires an intact microtubule system,21,22 and the presence of γ-tubulin is their distinctive feature.21–23 Aggresomes contain multiubiquitinated misfolded proteins, and various other proteins, including heat-shock proteins (HSPs) and 20/26S proteasome components.22–24 In various mononucleated cells, aggresomes have been induced by overexpression of both normal and mutated proteins combined with proteasome inhibition.21–23,25,26 Recently, it has been suggested that Lewy bodies in Parkinson’s disease are related to aggresomes.27 Whether aggresomes contribute to cellular death or protect cells from toxic effects of misfolded proteins remains uncertain. We have now asked whether s-IBM muscle fiber multiprotein-aggregates have features of aggresomes and if proteasome inhibition may contribute to the s-IBM pathogenesis. These questions were further explored in our experimentally induced IBM model, which is based on genetic overexpression of amyloid-β precursor protein (AβPP) in cultured human muscle fibers.
Materials and Methods
Muscle Biopsies
Immunocytochemical studies were performed on 10-μm-thick transverse sections of fresh-frozen diagnostic muscle biopsies obtained with informed consent from 25 patients with these diagnoses: 10 s-IBM, 3 polymyositis, 1 dermatomyositis, 2 morphologically nonspecific myopathy, 4 peripheral neuropathy, 2 amyotrophic lateral sclerosis, and 3 normal muscle. All IBM biopsies had muscle fibers with vacuoles on Engel trichrome staining,28 and 15- to 21-nm paired helical filaments (PHFs) by SMI-31 immunoreactivity4 and by electron microscopy, and Congo Red positivity using fluorescence enhancement.29
Light-Microscopic Immunocytochemistry
Immunocytochemistry was as described.3,4,6,30,31 We used 26 well-characterized monoclonal and polyclonal antibodies against 20S and 19S proteasome (Table 1). Double immunofluorescence used selected antibodies against 20S and 19S proteasome in combination with one of the following: 1) mouse monoclonal antibody 6E10 (Signet, Dedham, MA), diluted 1:100, which morphologically recognizes Aβ in both AD brain32 and s-IBM muscle,31 and on immunoblots it recognizes the Aβ region within the parent AβPP molecule; 2) mouse monoclonal antibody SMI-31 (Sternberger Monoclonals, Inc., Baltimore, MD), diluted 1:1000, which recognizes p-tau of s-IBM4 and AD PHFs;33 3) AT100 antibody (Pierce Biotechnology, Rockford, IL), diluted 1:1000, which recognizes p-tau of AD PHFs.34 Aggresomes were identified by well-characterized mouse monoclonal and rabbit polyclonal antibodies against γ-tubulin (Santa Cruz Biotechnology, Santa Cruz, CA). This was combined with one of the following: 1) mouse monoclonal antibody against ubiquitin (Chemicon, Temecula, CA), diluted 1:100; 2) mouse monoclonal antibody 6E10; 3) mouse monoclonal antibody SMI-31; 4) rabbit polyclonal or mouse monoclonal antibodies against 20S and 19S proteasome subunits; and 5) rabbit polyclonal antibody against HSP70 (StressGene, Victoria, British Columbia) diluted 1:200. To block nonspecific binding of antibody to Fc receptors, sections were preincubated with normal goat or rabbit serum diluted 1:10, as described.3,34,35 Controls for staining specificity were omission of the primary antibody, or its replacement with nonimmune sera or irrelevant antibody.
Table 1.
Intensities of Immunohistochemical Reactions Using Various Antibodies against 20S and 19S Proteasome Subunits in s-IBM Muscle Fibers
| Subunit | Type (clone) | Dilution | Intensity-IHC |
|---|---|---|---|
| Antibodies against 20S proteasome subunits α1,2,3,5,6,7 | mAb (MCP231) | 1:200 | ++ |
| α2 | mAb (MCP21) | 1:50 | − |
| α3 | mAb (MCP257) | 1:200 | + |
| α4 | mAb (MCP34) | 1:200 | + |
| α5 | mAb (MCP196) | 1:200 | ++ |
| α6 | mAb (MCP20) | 1:50 | − |
| α7 | mAb (MCP72) | 1:200 | + |
| α5,α7,β1,β5,β5i,β7 | pAb (MCP231) | 1:200 | + |
| β1 | mAb (MCP421) | 1:200 | ++ |
| 1i (LMP2) | pAb | 1:200 | ++ |
| β1i (LMP2) | mAb | 1:50 | − |
| β2 (Z) | mAb (MCP168) | 1:200 | ++ |
| β2 (Z) | pAb* | 1:200 | ++ |
| β2i (MECL1) | pAb | 1:200 | ++ |
| β3 | mAb (MCP102) | 1:50 | − |
| β4 | pAb | 1:200 | + |
| β5 | pAb | 1:200 | + |
| 5i (LMP7) | mAb | 1:200 | ++ |
| β5i (LMP7) | pAb | 1:50 | + |
| β6 | pAb | 1:50 | − |
| β7 | mAb (MCP205) | 1:50 | + |
| Antibodies against 19S proteasome subunits | |||
| ATPase | |||
| S6a-Rpt5 | mAb (TBP1-19) | 1:300 | ++ |
| S6b-Rpt3 | pAb | 1:500 | ++ |
| S7 | pAb* | 1:100 | + |
| Non-ATPase | |||
| S5a-Rpn10 | mAb (S5a-18) | 1:50 | − |
| S10a-Rpn7 | pAb | 1:50 | − |
All antibodies are from Affiniti Research Products Ltd. except those marked with an asterisk, which are from Affinity Bioreagents. Final staining intensity graded as 0 to ++.
Immunoelectron Microscopy
Single- and double-label gold immunoelectron microscopy was performed on 10-μm unfixed frozen sections adhered to the bottom of 35-mm Petri dishes, as described.3,4,6,30,31 In brief, a primary antibody against the 20S or 19S proteasome subunit was used separately or in combination with each other, and an antibody against 20S proteasome was also combined with antibodies against Aβ or p-tau. After incubation with the appropriate secondary antibodies conjugated to 5-nm and 10-nm gold particles, sections were processed for electron microscopy as described.4,6,30,31 The same procedure ultrastructurally immunolocalized γ-tubulin.
Immunoblotting
Muscle biopsies of six s-IBM and six age-matched control patients were immunoblotted, as recently detailed.6,30 In brief, 10 μg of protein were loaded onto 10% NuPAGE gels (Invitrogen, Grand Island, NY), electrophoresed, transferred to nitrocellulose membranes, and immunoprobed with antibodies against 19S and 20S proteasome. After incubation in the appropriate secondary antibodies, blots were developed using an enhanced chemiluminescence system (Amersham Biosciences, Piscataway, NJ). Protein loading was evaluated by the Coomassie blue-stained myosin band. Quantification of immunoreactivity was performed by densitometric analysis using NIH Image J 1.310. One sample, designated as standard, was used on every gel so that band densities on different blots could be normalized to that standard,35 allowing comparison of protein samples on multiple blots.
Combined Immunoprecipitation/Immunoblot Procedure
To evaluate whether in s-IBM muscle fibers 20S proteasome physically associates with Aβ/AβPP, a combined immunoprecipitation/immunoblot technique was performed, as we recently described in detail.6,30 In brief, 100 μg of total muscle protein from s-IBM and control biopsies were immunoprecipitated with 5 μg of 6E10 antibody, as described.6,30 Immunoprecipitates were separated by electrophoresis, transferred to a nitrocellulose membrane, and immunoblotted with 20Sα4 subunit proteasome antibody, diluted 1:2000. Subsequently, an appropriate secondary antibody and Western Breeze anti-mouse chemiluminescence kit (Invitrogen) were used. To confirm the specificity of those experiments, the following controls were performed: the 6E10 antibody against AβPP/Aβ, which was used for immunoprecipitation, was omitted, and the 20Sα4 primary antibody used for immunoblotting of immunoprecipitates was omitted.
Measurement of Proteasome Activity
Three main proteasome activities were determined by evaluating the cleavage of specific fluorogenic substrates.20 Four s-IBM and four age-matched control muscle biopsies were homogenized in 20 mmol/L Tris-HCl, pH 7.2, containing 0.1 mmol/L ethylenediamine tetraacetic acid, 1 mmol/L 2-mercaptoethanol, 5 mmol/L ATP, 20% glycerol, and 0.04% (v/v) Nonidet P-40,36,37 centrifuged, the supernatant collected, and protein concentration determined using the Bradford method. Subsequently, 200 μg of biopsied muscle protein or 30 μg of protein from cultured muscle (see below), were incubated in 100 μmol/L fluorogenic substrates for the three different protease activities: Z-Leu-Leu-Glu-AMC (substrate II) for PGPH; Suc-Leu-Leu-Val-Tyr-AMC (substrate III) for CTL; and Z-Ala-Arg-Arg-AMC (substrate VI) for TL activity. Fluorescence emission was excited at 360 nm and recorded at 430 nm.37
Statistical Analysis
Statistical analyses were performed by a one-tailed t-test for both the proteasome activity and proteasome protein levels. Significance level was set at P < 0.05. Data are reported as means ± SEM for all groups.
Cultured Human Muscle Fibers
Tissue cultures of normal human muscle were established from satellite cells of portions of diagnostic muscle biopsies from patients who, after all tests were performed, were considered free of muscle disease.38 In five experimental culture sets, each established from a different muscle biopsy, a 3-kb 751 AβPP-cDNA, in sense or in anti-sense orientation, was transferred into 3-week-old cultured muscle fibers using an adenovirus vector, doses as described.39,40 Three days after the transfer, experimental and control (non-AβPP-overexpressing) cultures were treated with 1 μmol/L epoxomicin (Biomol Research Laboratories, Plymouth Meeting, PA), an irreversible proteasome inhibitor,36 and after 24 hours they were processed for light- and electron-microscopic immunocytochemistry, as described.40 At the same time, cultures were harvested for measurements of proteasome activity. Selected cultures were also treated, concurrently with epoxomicin, with 1 μmol/L of microtubule-disruptor nocodazole (Sigma, St. Louis, MO), which has been shown to disrupt aggresome formation.21
Results
s-IBM and Control Muscle Biopsies
Light-Microscopic Immunocytochemistry
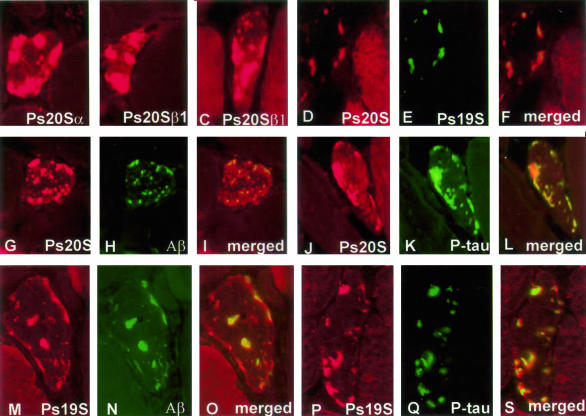
In all s-IBM muscle biopsies, 70 to 80% of the vacuolated muscle fibers contained, mainly in their nonvacuolated cytoplasm, numerous well-defined, plaque-like, dotty or elongated aggregates immunoreactive with antibodies against 20S and 19S proteasome subunits. In addition, in all biopsies, 20 to 30% of the nonvacuolated, otherwise normal-appearing fibers (on a given cross-section) contained similar aggregates (Figure 1; A to F, J, M, P). By double-label immunofluorescence, in all abnormal muscle fibers, aggregates immunoreactive for 20S and 19S proteasome co-localized with each other (Figure 1; D to F) and with Aβ or p-tau immunoreactivity (Figure 1; G to R). Both anti-p-tau antibodies used gave the same results. Immunoreactivity with the 26 antibodies directed against various proteasome subunits is presented in Table 1.
Figure 1.
Immunofluorescence of 26S proteasome subunits in s-IBM muscle. A–C: Single-label immunofluorescence illustrates strongly immunoreactive, various-sized aggregates of Ps20Sα (A), Ps20Sβ1 constitutive (B), and Ps20Sβ1I (inducible LMP2 subunit) (C) in s-IBM abnormal muscle fibers. D–S: Double-label immunofluorescence illustrates that in s-IBM muscle fibers the aggregates immunoreactive for Ps20S (D) are also immunoreactive for Ps19S (D–F merged), and both Ps20S (G, J)- and Ps19S (M, P)-immunoreactive aggregates are immunoreactive for amyloid-β (H and I merged, N and O merged) and for p-tau (K, Q, and S merged). Original magnifications, ×1100.
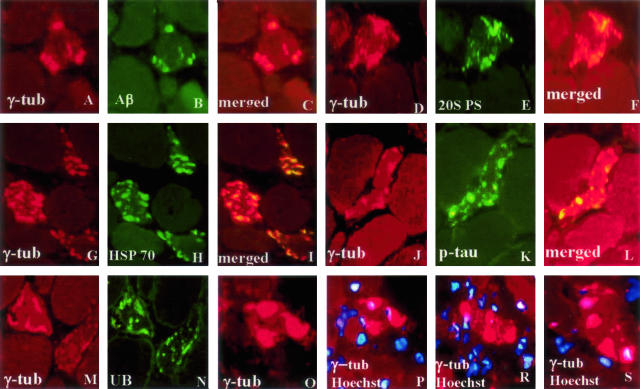
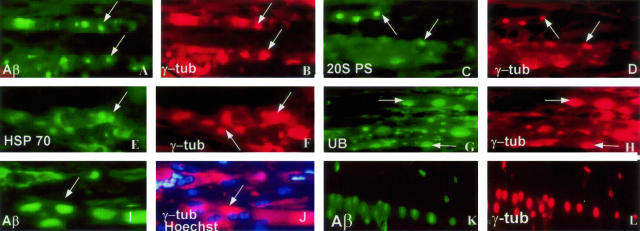
Accumulated aggregates had several features of aggresomes, because 70 to 80% of them were associated with γ-tubulin and they contained ubiquitin and HSP70, in addition to Aβ and p-tau (Figure 2). In contrast to classical aggresomes originally described in mononucleated cells,21,22 aggresomes in s-IBM muscle fibers did not have perinuclear localization (Figure 2; P to S). None of the control non-s-IBM, diseased and normal, human muscle biopsies had muscle fibers containing aggregates immunoreactive with antibodies against 20S, 19S, or γ-tubulin. Eliminating primary antibodies or replacing them with nonrelevant antibodies resulted in nonstaining.
Figure 2.
Double-label immunofluorescence of γ-tubulin with Aβ, 20S PS, HSP70, p-tau, and ubiquitin (UB), indicating aggresomes in s-IBM muscle fibers. P to S illustrate that γ-tubulin-immunoreactive aggregates are not associated with the nuclei marked by the blue nuclei marker Hoechst. Original magnifications, ×1100.
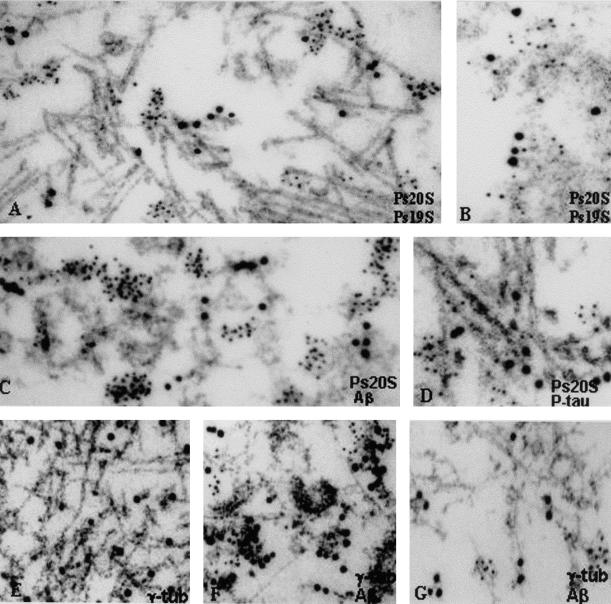
Immunoelectron Microscopy
In s-IBM abnormal muscle fibers, double immunolabeling of both 20S and 19S proteasome revealed that both subunits were associated with the same structures, namely PHFs, 6- to 10-nm amyloid-like fibrils, and floccular material (Figure 3, A and B). Because both the 20S and 19S proteasome subunits localized to the same structures, electron microscopic double immunolabeling with Aβ and p-tau was performed only with the antibody against 20S. This revealed that 20S was associated with both Aβ on 6- to 10-nm amyloid-like fibrils and floccular material (Figure 3C) and p-tau on PHFs (Figure 3D). In contrast to p-tau, which was localized on PHFs themselves, the proteasome subunits were in small clusters adjacent to PHFs but not directly on them (Figure 3D). γ-Tubulin was associated with PHFs (Figure 3E), and together with Aβ it was associated with 6- to 10-nm fibrils (Figure 3F).
Figure 3.
Double-label gold immunoelectron microscopy in s-IBM (A–F) and AβPP-overexpressing cultured muscle fibers (G). Ps20S (5-nm gold particles) and Ps19S (10-nm gold particles) are both associated with PHFs (A), and 6- to 10-nm amyloid-like fibrils and floccular amorphous material (B). Ps20S (5-nm gold particles) and amyloid-β (10-nm gold particles) are both associated on 6- to 10-nm amyloid-like fibrils and floccular amorphous material (C). Ps20S (5-nm gold particles) and p-tau (10-nm gold particles) are both associated with PHFs (D), but in contrast to p-tau, which is localized on the filaments themselves, Ps20S is in small clusters adjacent to PHFs but not directly on them (D). E and F: γ-Tubulin. E: Single label showing association of γ-tubulin with PHFs. F: Double immunolocalization of γ-tubulin (5-nm gold particles) with Aβ (10-nm gold particles) shows that both are associated with 6- to 10-nm fibrils and amorphous material. G: Double immunolocalization of γ-tubulin (5-nm gold particles) with Aβ (10-nm gold particles) in an AβPP-overexpressing plus epoxomicin-treated cultured human muscle fiber. Similar to biopsied s-IBM muscle fibers, both γ-tubulin (5-nm gold particles) and Aβ (10-nm gold particles) are associated with 6- to 10-nm fibrils. Original magnifications: ×48,500 (A–C, E–G); ×61,400 (D).
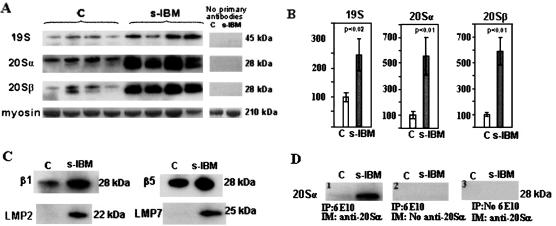
Immunoblots of Proteasome Subunits
In s-IBM and normal muscle biopsies, the 20S α- and β-subunits each migrated as either single or double bands of ∼28 kd, whereas the 19S Rpn10 subunit migrated as a 45-kd band, in the typical positions for each. Expression of subunits 20Sα4, 20Sβ1, β2 and β5, and of 19S Rpn10 was increased in s-IBM muscle biopsies as compared to control biopsies (Figure 4, A and C). Omitting the primary antibody resulted in nonstaining (Figure 4A).
Figure 4.
Immunoblots of 26S proteasome subunits and immunoprecipitation in control and s-IBM muscle biopsies. A: Immunoblots of muscle homogenates of normal control and s-IBM muscle biopsies demonstrate in s-IBM a much stronger expression of Ps 19S, Ps 20Sα4, and Ps 20Sβ2 subunits as compared to control muscle biopsies. Omission of primary antibodies resulted in negative reactions. B: Densitometric analysis of the blots in A, performed using NIH Image J 1.310. Protein loading was evaluated by the Coomassie blue-stained myosin band. One sample, designated as standard, was used on every gel so that band densities on different blots could be normalized to this standard. Data are indicated as mean ± SE. Significance was determined by one-tailed unpaired t-test. The level of significance was set at P < 0.05. C: Immunoblots of the constitutive β1 and β5, and the inducible LMP2 and LMP7, subunits in control and s-IBM muscle biopsies illustrate increased expression of both β1 and β5 in s-IBM as compared to controls: LMP2 and LMP7 subunits are present in s-IBM but are not detectable in controls. D: Immunoprecipitations of control and s-IBM biopsies with 6E10 antibody against AβPP/Aβ, followed by immunoprobing with an antibody against 20Sα4 proteasome subunit. A strong band of the appropriate molecular weight corresponding to the 20Sα proteasome subunit is revealed in s-IBM muscle, whereas the control has only a very weak band (1). Omission of the antibody against 20Sα (2) or omission of the antibody against AβPP/Aβ (3) resulted in negative reactions.
In six s-IBM muscle biopsies, densitometry of 20S proteasome subunits α4 and β2, and of 19S subunit Rpn10, evaluated after normalization to the corresponding myosin band, revealed, as compared to the six age-matched control muscle biopsies, that α4 was increased 5.6 times (P < 0.01), β2 was increased 5.9 times (P < 0.01), and 19S was increased 2.4 times (P < 0.02) (Figure 4B). In addition, LMP2 and LMP7, inducible counterpart subunits, respectively, of β1 and β5, were strongly expressed in s-IBM muscle but were virtually undetectable in control muscle biopsies (Figure 5C).
Figure 5.
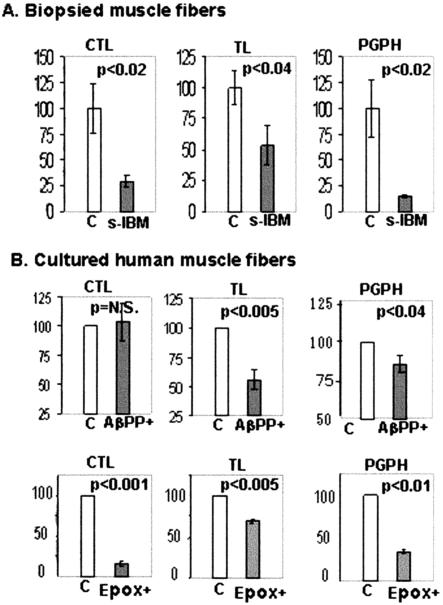
Proteasome enzymatic activities in control and s-IBM muscle biopsies (A), and in cultured control and AβPP-overexpressing (AβPP+), and in not-AβPP-overexpressing but epoxomicin-treated human muscle fibers (B). A: Proteasome TL, CTL, and PGPH activities were measured in four s-IBM and four normal control muscle biopsies and results were normalized to proteasome quantity in the same muscle biopsies. All three proteasome activities are significantly reduced in s-IBM muscle biopsies, CTL to 29.4% (P < 0.02), TL to 53.5% (P < 0.04), and PGPH to 14.5% (P < 0.02) of control muscle biopsies. Data are indicated as mean ± SE. Significance was determined by the one-tailed unpaired t-test. The level of significance was set at P < 0.05. B: TL, CTL, and PGPH activities were measured in five sets of normal cultured human muscle fibers having AβPP-overexpression (AβPP+), in their not-AβPP-overexpressing cultured sister controls, and in not-AβPP+ epoxomicin-treated cultures. The results were normalized to proteasome quantity. In AβPP+ cultures, TL and PGPH activities are reduced to 55.8% and to 85.4% of controls, respectively, with no significant change in CTL. Data are indicated as mean ± SE. Epoxomicin treatment reduced CTL, TL, and PGPH to 16.0%, 69.1%, and 33.4% of controls, respectively.
Immunoprecipitation
Immunoprecipitation of s-IBM and control muscle biopsies with mouse monoclonal 6E10 antibody, which recognizes either Aβ or AβPP, followed by immunoprobing with a mouse monoclonal antibody against 20S, revealed a very strong 20S proteasome band of 28 kd in s-IBM muscle, but only an extremely weak band in control muscle (Figure 4D). Omitting of either the 6E10 antibody used for immunoprecipitation or the primary antibody used for immunoprobing gave negative results.
Proteasome Activity
20S proteasome activities were measured in the same s-IBM and normal control muscle biopsies in which immunoblots were performed, and their activities were normalized to the 20Sβ2 proteasome protein value measured in each patient.35 In s-IBM biopsies, CTL, TL, and PGPH protease activities were reduced to 29.4% (P < 0.02), 53.5% (P < 0.04), and 14.5% (P < 0.02), respectively, as compared to the control biopsies (Figure 5A).
Cultured Muscle Fibers
Light-Microscopic Immunocytochemistry
In all five experimental sets of human cultured muscle fibers, overexpression of AβPP, combined with epoxomicin treatment, induced in the cytoplasm of 85 to 95% of cultured muscle fibers numerous well-defined, various-sized, round, or dotty γ-tubulin-immunoreactive aggregates, which by double-label immunofluorescence were also immunoreactive for Aβ, ubiquitin, HSP70, and proteasome subunits (Figure 6; A to J). Approximately 20% of the control, non-AβPP-overexpressing but epoxomicin-treated cultured muscle fibers had only very small γ-tubulin-immunoreactive aggregates (not shown). AβPP-overexpressing, but not epoxomicin-treated, cultured fibers had well-defined, various-sized γ-tubulin- and Aβ-immunoreactive aggregates in ∼40% of fibers (Figure 6, K and L). As in a previous study,21 nocodazole prevented γ-tubulin-immunoreactive aggregates in the majority of the fibers.
Figure 6.
Double-label immunofluorescence of γ-tubulin with Aβ (A, B), 20S PS (C, D), HSP70 (E, F), and ubiquitin (UB) (G, H), indicating various-sized aggresomes in cultured human muscle fibers, that were AβPP-overexpressing plus epoxomicin-treated (A–J), and in ones AβPP-overexpressing but not treated with epoxomicin (K, L). J illustrates that most of the γ-tubulin-immunoreactive aggregates are not associated with nuclei, which are indicated with the blue nuclei marker Hoechst. Original magnifications, ×1400.
Immunoelectron Microscopy of Cultured Human Muscle Fibers
In AβPP-overexpressing plus epoxomicin-treated cultured human muscle fibers, both γ-tubulin and Aβ were associated with 6- to 10-nm amyloid-like fibrils (Figure 3F).
Proteasome Activity
In AβPP-overexpressing cultured muscle fibers, after normalization to the 20Sβ2 proteasome level, TL activity was reduced to 55.8% (P < 0.005), and PGPH to 85.4% (P < 0.04), as compared to control cultured muscle fibers, while there was no significant change in CTL activity (Figure 5). Cultured muscle overexpressing anti-sense AβPP had no reduction of any proteolytic activity (data not shown). A 24-hour treatment with epoxomicin of non-Aβ-overexpressing cultures reduced the CTL activity to 16%, TL to 69.1%, and PGPH to 33.4% (P < 0.01) of control (Figure 5B).
Discussion
In this study we found significant proteasomal abnormalities in s-IBM muscle fibers as evidenced by: 1) abnormal accumulation of 26S proteasome subunits by immunofluorescence and immunoelectron microscopy; 2) increased expression of 26S proteasome subunits by immunoblots; and 3) reduced activities of the three major proteasomal proteolytic enzymes. Previously, in s-IBM muscle biopsies, abnormal proteasomal accumulations were reported, but their relationship to Aβ/AβPP was not studied.41 Our present study strongly suggests that the AβPP/Aβ-proteasome interrelationship may be important in inducing proteasome abnormalities and aggresome formation in s-IBM muscle fibers because: 1) Aβ and proteasome co-localized at the light-microscopy level and were associated ultrastructurally with the same structures; 2) there was a physical association of Aβ/AβPP and proteasome by immunoprecipitation studies; and 3) proteasome activity was inhibited in cultured human muscle fibers overexpressing Aβ/AβPP.
In s-IBM muscle fibers, increased synthesis of AβPP1,2 is associated with its abnormal processing1,2 and with accumulation of its cytotoxic fragment Aβ.1,2 Experimental overexpression of AβPP/Aβ in cultured human muscle fibers was demonstrated to induce in them aspects of the IBM phenotype.1,2,39,40 Accordingly, it was proposed that overproduction of AβPP/Aβ plays an important role in s-IBM pathogenesis.1,2 The inhibition of two major proteasome activities, TL and PGPH, by the overexpressed AβPP/Aβ that we have demonstrated in the cultured muscle fibers suggests that AβPP/Aβ may be causatively involved in proteasome inhibition in s-IBM fibers. Other factors present in s-IBM muscle fibers that might contribute to inhibition of proteasome function include: an aging muscle environment (because s-IBM patients are usually age 50 or older), protein over-crowding, oxidative stress,1,2 and accumulated p-tau,1,2,4 α-synuclein,31 and UBB+1 5, all shown to be capable of inhibiting proteasome activity.8–13 Abnormal proteasome distribution or its inhibition were suggested to contribute to the pathogenesis of several neurodegenerative diseases, including AD, Parkinson’s disease, Lewy-body dementia, and Huntington disease.9,15,42–44
Our demonstrated increase of proteasome-protein subunits may represent the muscle fiber’s attempt to deal with multiprotein overcrowding, but, in the presence of the severely impaired proteasomal enzymatic function those attempts are futile and the unfolded/misfolded proteins accumulate in the form of aggregates. Our study also demonstrated that most s-IBM multiprotein aggregates have some features of aggresomes, because they are associated with γ-tubulin, and contain proteasome subunits, ubiquitin and HSP70. We also induced aggresome-like structures in cultured human muscle fibers by overexpressing AβPP, +/− epoxomicin treatment. Previously, aggresomes have been produced only in cultured mononucleated cells, where they appeared as perinuclear inclusions.21–24 However, in our studies, both in the s-IBM biopsied muscle fibers and in the AβPP-overexpressing cultured muscle fibers, aggresome-like structures were not associated with the nuclei. This might be related to the fact that a muscle fiber is a multinucleated cell and mechanisms responsible for the location of aggresomes might be different from in a mononucleated cell. Our study was not able to demonstrate a protective influence of aggresomes, which was reported in other cell systems.26 Even though the exact role of aggresome-like structures in s-IBM muscle fibers still needs to be investigated, their presence there, as well as their induction in cultured human muscle fibers by AβPP overexpression and proteasome inhibition, further support the prominent role of proteasome inhibition, and protein aggregation and misfolding, in the complex cascade of the s-IBM pathogenesis.
Proteasome is responsible for degradation of most cellular proteins.14,15 A failure to degrade/remove surplus proteins, including abnormal damaged proteins, would be detrimental to the muscle fiber. Furthermore, accumulated ubiquitinated, misfolded, and oxidized protein aggregates themselves can cause inhibition of proteasome.25,45 Also, the still soluble, early intermediates of protein aggregates, in the form of dimers and trimers, are highly toxic to cells,46,47 and they can also induce proteasome inhibition.11 Because proteasome inhibition might, in turn, contribute to the formation of soluble toxic protein oligomers, in a vicious circle, proteasome inhibition may be detrimental to the cells even before protein aggregates can be identified morphologically.
In addition to the formation of multiprotein aggregates, proteasome abnormalities might play another role in the s-IBM pathogenesis, namely the cytotoxic T-cell inflammation. Some s-IBM muscle fibers express MHC-I,48 which probably plays a role in the CD8+ cytotoxic T-cell response49 by enabling a muscle fiber to become antigen-presenting. Proteasomal inducible β-subunits, LMP2 and LMP7, which we have shown increased in s-IBM muscle fibers, are involved in the intracellular processing of peptides for MHC-I expression.16,17 Possibly, in s-IBM muscle fibers LMP2 and LMP7 are involved in antigen presentation development of the T-cell inflammatory response.
In summary, our present study demonstrates proteasome dysfunction in s-IBM muscle fibers, and it suggests that Aβ/AβPP may promote that dysfunction. Our studies raise the possibility of novel therapeutic manipulations to correct that proteasomal malfunction.
Acknowledgments
We thank Maggie Baburyan for providing excellent technical assistance in electron microscopy.
Footnotes
Address reprint requests to Valerie Askanas, M.D., Ph.D., USC Neuromuscular Center, Good Samaritan Hospital, 637 S. Lucas Ave., Los Angeles, CA 90017-1912. E-mail: askanas@hsc.usc.edu.
Supported by grants (to V.A.) from the National Institutes of Health (AG16768 merit award), the Muscular Dystrophy Association, the Myositis Association, and the Helen Lewis Research Fund.
References
- Askanas V, Engel WK. Proposed pathogenetic cascade of inclusion-body myositis: importance of amyloid-beta, misfolded proteins, predisposing genes, and aging. Curr Opin Rheumatol. 2003;15:737–744. doi: 10.1097/00002281-200311000-00009. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK. Inclusion-body myositis: newest concepts of pathogenesis and relation to aging and Alzheimer disease. J Neuropathol Exp Neurol. 2001;60:1–14. doi: 10.1093/jnen/60.1.1. [DOI] [PubMed] [Google Scholar]
- Askanas V, Alvarez RB, Engel WK. Beta-amyloid precursor epitopes in muscle fibers of inclusion body myositis. Ann Neurol. 1993;34:551–560. doi: 10.1002/ana.410340408. [DOI] [PubMed] [Google Scholar]
- Askanas V, Alvarez RB, Mirabella M, Engel WK. Use of anti-neurofilament antibody to identify paired-helical filaments in inclusion-body myositis. Ann Neurol. 1996;39:389–391. doi: 10.1002/ana.410390318. [DOI] [PubMed] [Google Scholar]
- Fratta P, Engel W, Van Leeuwen F, Hol E, Vattemi G, Askanas V. Mutant ubiquitin UBB+1 is accumulated in sporadic inclusion-body myositis muscle fibers. Neurology. 2004;63:1114–1117. doi: 10.1212/01.wnl.0000138574.56908.5d. [DOI] [PubMed] [Google Scholar]
- Vattemi G, Engel WK, McFerrin J, Askanas V. Endoplasmic reticulum stress and unfolded protein response in inclusion body myositis muscle. Am J Pathol. 2004;164:1–7. doi: 10.1016/S0002-9440(10)63089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL. Structure/function in neuroprotection and apoptosis. Ann Neurol. 1998;44(Suppl 1):S65–S71. doi: 10.1002/ana.410440711. [DOI] [PubMed] [Google Scholar]
- Gregori L, Fuchs C, Figueiredo-Pereira ME, Van Nostrand WE, Goldgaber D. Amyloid beta-protein inhibits ubiquitin-dependent protein degradation in vitro. J Biol Chem. 1995;270:19702–19708. doi: 10.1074/jbc.270.34.19702. [DOI] [PubMed] [Google Scholar]
- Keck S, Nitsch R, Grune T, Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer’s disease. J Neurochem. 2003;85:115–122. doi: 10.1046/j.1471-4159.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R, Mayer RJ, Layfield R. Inhibition of the ubiquitin-proteasome system in Alzheimer’s disease. Proc Natl Acad Sci USA. 2000;97:9902–9906. doi: 10.1073/pnas.170173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindersson E, Beedholm R, Hojrup P, Moos T, Gai W, Hendil KB, Jensen PH. Proteasomal inhibition by alpha-synuclein filaments and oligomers. J Biol Chem. 2004;279:12924–12934. doi: 10.1074/jbc.M306390200. [DOI] [PubMed] [Google Scholar]
- Lindsten K, de Vrij FM, Verhoef LG, Fischer DF, van Leeuwen FW, Hol EM, Masucci MG, Dantuma NP. Mutant ubiquitin found in neurodegenerative disorders is a ubiquitin fusion degradation substrate that blocks proteasomal degradation. J Cell Biol. 2002;157:417–427. doi: 10.1083/jcb.200111034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Salon M, Pasquini L, Besio Moreno M, Pasquini JM, Soto E. Relationship between beta-amyloid degradation and the 26S proteasome in neural cells. Exp Neurol. 2003;180:131–143. doi: 10.1016/s0014-4886(02)00060-2. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2:179–187. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- Rock KL, York IA, Saric T, Goldberg AL. Protein degradation and the generation of MHC class I-presented peptides. Adv Immunol. 2002;80:1–70. doi: 10.1016/s0065-2776(02)80012-8. [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Tarcsa E, Cascio P, Goldberg AL. The unfolding of substrates and ubiquitin-independent protein degradation by proteasomes. Biochimie. 2001;83:311–318. doi: 10.1016/s0300-9084(01)01244-5. [DOI] [PubMed] [Google Scholar]
- Jariel-Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Piechaczyk M. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J Biol Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- Shringarpure R, Grune T, Mehlhase J, Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem. 2003;278:311–318. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- Junn E, Lee SS, Suhr UT, Mouradian MM. Parkin accumulation in aggresomes due to proteasome impairment. J Biol Chem. 2002;277:47870–47877. doi: 10.1074/jbc.M203159200. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kim YM, Lee G, Junn E, Iwatsubo T, Mouradian MM. Aggresomes formed by alpha-synuclein and synphilin-1 are cytoprotective. J Biol Chem. 2004;279:4625–4631. doi: 10.1074/jbc.M310994200. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Shashidharan P, Perl DP, Jenner P, Olanow CW. Aggresome-related biogenesis of Lewy bodies. Eur J Neurosci. 2002;16:2136–2148. doi: 10.1046/j.1460-9568.2002.02301.x. [DOI] [PubMed] [Google Scholar]
- Engel WK, Cunningham GG. Rapid examination of muscle tissue and improved trichrome method for fresh-frozen biopsy sections. Neurology. 1963;13:919–923. doi: 10.1212/wnl.13.11.919. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK, Alvarez RB. Enhanced detection of Congo-red-positive amyloid deposits in muscle fibers of inclusion body myositis and brain of Alzheimer’s disease using fluorescence technique. Neurology. 1993;43:1265–1267. doi: 10.1212/wnl.43.6.1265-a. [DOI] [PubMed] [Google Scholar]
- Vattemi G, Engel WK, McFerrin J, Askanas V. Cystatin C colocalizes with amyloid-beta and coimmunoprecipitates with amyloid-beta precursor protein in sporadic inclusion-body myositis muscles. J Neurochem. 2003;85:1539–1546. doi: 10.1046/j.1471-4159.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK, Alvarez RB, McFerrin J, Broccolini A. Novel immunolocalization of alpha-synuclein in human muscle of inclusion-body myositis, regenerating and necrotic muscle fibers, and at neuromuscular junctions. J Neuropathol Exp Neurol. 2000;59:592–598. doi: 10.1093/jnen/59.7.592. [DOI] [PubMed] [Google Scholar]
- Kim KS, Wen GY, Bancher C, Chen J, Sapienza V, Hong H, Wisniewski HM. Detection and quantitation of amyloid B-peptide with 2 monoclonal antibodies. Neurosci Res Commun. 1990;7:113–122. [Google Scholar]
- Nukina N, Kosik KS, Selkoe DJ. Recognition of Alzheimer paired helical filaments by monoclonal neurofilament antibodies is due to crossreaction with tau protein. Proc Natl Acad Sci USA. 1987;84:3415–3419. doi: 10.1073/pnas.84.10.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng-Fischhèofer Q, Biernat J, Mandelkow EM, Illenberger S, Godemann R, Mandelkow E. Sequential phosphorylation of Tau by glycogen synthase kinase-3beta and protein kinase A at Thr212 and Ser214 generates the Alzheimer-specific epitope of antibody AT100 and requires a paired-helical-filament-like conformation. Eur J Biochem. 1998;252:542–552. doi: 10.1046/j.1432-1327.1998.2520542.x. [DOI] [PubMed] [Google Scholar]
- Husom AD, Peters EA, Kolling EA, Fugere NA, Thompson LV, Ferrington DA. Altered proteasome function and subunit composition in aged muscle. Arch Biochem Biophys. 2004;421:67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok B, Elofsson M, Sin N, Crews C. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demasi M, Shringarpure R, Davies KJ. Glutathiolation of the proteasome is enhanced by proteolytic inhibitors. Arch Biochem Biophys. 2001;389:254–263. doi: 10.1006/abbi.2001.2332. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK: Cultured normal and genetically abnormal human muscle. The Handbook of Clinical Neurology, Myopathies, Vol 18. Edited by Rowland LP, Di Mauro S. Amsterdam, North Holland, 1992, pp 85–116 [Google Scholar]
- Askanas V, McFerrin J, Baque S, Alvarez RB, Sarkozi E, Engel WK. Transfer of beta-amyloid precursor protein gene using adenovirus vector causes mitochondrial abnormalities in cultured normal human muscle. Proc Natl Acad Sci USA. 1996;93:1314–1319. doi: 10.1073/pnas.93.3.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askanas V, McFerrin J, Alvarez RB, Baque S, Engel WK. Beta APP gene transfer into cultured human muscle induces inclusion-body myositis aspects. Neuroreport. 1997;8:2155–2158. doi: 10.1097/00001756-199707070-00012. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Martin B, Castano JG, Lucas JJ, Moreno D, Olive M. Proteasomal expression, induction of immunoproteasome subunits, and local MHC class I presentation in myofibrillar myopathy and inclusion body myositis. J Neuropathol Exp Neurol. 2004;63:484–498. doi: 10.1093/jnen/63.5.484. [DOI] [PubMed] [Google Scholar]
- McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW. Altered proteasomal function in sporadic Parkinson’s disease. Exp Neurol. 2003;179:38–46. doi: 10.1006/exnr.2002.8050. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Lindenberg KS, Krebs A, Schèols L, Laccone F, Herms J, Rechsteiner M, Riess O, Landwehrmeyer GB. Protein surveillance machinery in brains with spinocerebellar ataxia type 3: redistribution and differential recruitment of 26S proteasome subunits and chaperones to neuronal intranuclear inclusions. Ann Neurol. 2002;51:302–310. doi: 10.1002/ana.10101. [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- Bence NF, Roopal MS, Kopito RS. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Klein WL. ADDLs & protofibrils—the missing links? Neurobiol Aging. 2002;23:231–235. doi: 10.1016/s0197-4580(01)00312-8. [DOI] [PubMed] [Google Scholar]
- Ellis RJ, Pinheiro TJ. Medicine: danger—misfolding proteins. Nature. 2002;416:483–484. doi: 10.1038/416483a. [DOI] [PubMed] [Google Scholar]
- Dalakas MC. The molecular and cellular pathology of inflammatory muscle diseases. Curr Opin Pharmacol. 2001;1:300–306. doi: 10.1016/s1471-4892(01)00053-4. [DOI] [PubMed] [Google Scholar]
- Murata K, Dalakas MC. Expression of the costimulatory molecule BB-1, the ligands CTLA-4 and CD28, and their mRNA in inflammatory myopathies. Am J Pathol. 1999;155:453–460. doi: 10.1016/s0002-9440(10)65141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]