Abstract
Severe acute respiratory syndrome (SARS) is a significant emerging infectious disease. Humans infected with the etiological agent, SARS-associated coronavirus (SARS-CoV), primarily present with pneumonitis but may also develop hepatic, gastrointestinal, and renal pathology. We inoculated common marmosets (Callithrix jacchus) with the objective of developing a small nonhuman primate model of SARS. Two groups of C. jacchus were inoculated intratracheally with cell culture supernatant containing SARS-CoV. In a time course pathogenesis study, animals were evaluated at 2, 4, and 7 days after infection for morphological changes and evidence of viral replication. All animals developed a multifocal mononuclear cell interstitial pneumonitis, accompanied by multinucleated syncytial cells, edema, and bronchiolitis in most animals. Viral antigen localized primarily to infected alveolar macrophages and type-1 pneumocytes by immunohistochemistry. Viral RNA was detected in all animals from pulmonary tissue extracts obtained at necropsy. Viral RNA was also detected in tracheobronchial lymph node and myocardium, together with inflammatory changes, in some animals. Hepatic inflammation was observed in most animals, predominantly as a multifocal lymphocytic hepatitis accompanied by necrosis of individual hepatocytes. These findings identify the common marmoset as a promising nonhuman primate to study SARS-CoV pathogenesis.
An outbreak of the emerging infectious disease, severe acute respiratory syndrome (SARS), began in November of 2002 and was eventually contained in July of 2003 by means of an extensive and impressive coordinated worldwide effort to diagnose infected individuals, and to provide care and containment.1 These efforts involved dedicated clinical caretakers, public health agencies, and research laboratories. By the end of the epidemic there were more than 8000 probable cases of SARS and nearly 800 deaths.2
Humans with SARS primarily presented with pneumonitis, and mortality was primarily attributed to respiratory failure or related complications. Many studies have documented that other organs were likely involved in significant ways. Initial clinical findings other than pneumonitis in carefully monitored individuals included diarrhea (up to 25% of patients), lymphocytopenia (up to 90% of patients), and abnormal plasma concentrations of liver enzymes (up to 66% of patients).2 Cardiac failure has been listed as a clinical sign in case reports of unusual presentations.3,4 Abnormal urinalyses have been correlated with viral shedding in the urine.5
The primary functional receptor for SARS-CoV is angiotensin converting enzyme-2 (ACE-2), a metalloproteinase involved in hemodynamic homeostasis.6 ACE-2 is expressed in a tissue distribution (largely pulmonary and intestinal epithelial cells) that is primarily concordant with histopathology obtained at autopsy of individuals who succumbed to the infection.7 Human disease is marked primarily by viral shedding from both the respiratory and the gastrointestinal tracts.5 Viruria, detectable in a significant proportion of infected individuals, suggests that renal tubular epithelial cells expressing ACE-2 are infected.5,8 ACE-2 is also expressed at high levels in vascular endothelial cells, a finding that may explain inflammatory changes in other organ systems and vasculitis that is observed in some individuals.9 Viral RNA has been demonstrated in liver, lymph node, spleen, heart, and skeletal muscle in studies of fatal SARS.10 Viremia has been detected, albeit inconsistently,5,11 and as yet, there has been no report demonstrating infection of vascular endothelial cells.
The SARS-CoV was identified and its genome fully sequenced 1 month after the World Health Organization global alert.2 Ultimately, the demonstration of SARS-CoV infection and disease in primates provided the elements of Koch’s postulates that proved causality.12,13 The reproducibility of SARS-CoV-related pneumonitis in old world primate species has been somewhat problematic. Infected animals rarely show clinical signs of disease, and virus concentrations in tissues tend to be relatively low.14,15 Other small animals that have proven useful in studies of efficacy of certain interventions include the BALBc mouse, Syrian hamsters, and the ferret.16–18 In the murine model, histopathology is minimal despite relatively high viral concentrations in lung tissues, but reliable reductions in viral titers have demonstrated effectiveness of vaccines and neutralizing antibodies.19–21 Syrian hamsters develop a consolidative pneumonitis that resolves within 1 week,16 and ferrets develop a patchy pneumonitis along with hepatitis.18 These latter two animal models have shown promise in measuring effectiveness of vaccines and therapeutics, and also have been used to evaluate the risk of disease enhancement.16,22,23
Reasoning that a small nonhuman primate model for SARS would be a useful research tool to study pathogenesis, vaccines, and therapeutics, we performed pilot studies on the common marmoset (C. jacchus). We selected this animal for study because of its small size (250 to 400 g), the availability of a large breeding colony, its susceptibility to various human respiratory viruses, and the relative wealth of reagents for immunohistochemistry and flow cytometry, and information regarding its immunology.24
Materials and Methods
Virus
The isolate of SARS-CoV used for these studies was the Urbani strain (kindly provided by Larry Anderson, Centers for Disease Control, Atlanta, GA). A low passage stock was generated by growth in Vero E6 cells (medium: Dulbecco’s modified Eagle’s medium, 10% fetal calf serum, Gentamicin). The infectious titer was determined using Vero E6 cells in microtiter plates as described.25
Animals
C. jacchus were bred and housed at the New England Primate Research Center in accordance with the Harvard Medical School’s Standing Committee on Animals and The Guide for the Care and Use of Laboratory Animals (National Academy Press, 1996). Clinical and husbandry techniques have previously been described.24,26 Before experiments animals were moved and acclimatized to an Animal Biosafety Level 3 (ABSL-3) facility for experimental inoculations. For inoculations animals were anesthetized, intubated tracheally with a flexible polyethylene catheter, and 400 μl of cell culture supernatant (infected or uninfected) was infiltrated.
Experimental Design
Two groups of six juvenile or young adult C. jacchus each were intratracheally inoculated with 400 μl of cell culture supernatant containing 106 TCID50 of the Urbani strain of the SARS-CoV. In a time course pathogenesis study animals were evaluated at 2, 4, and 7 days after inoculation for morphological changes and evidence of viral replication. Animals were euthanized at day 2, 4, or 7 after inoculation by intravenous administration of ≫50 mg/kg of Na pentobarbital. Necropsies were performed at the New England Primate Research Center.
Histology
After euthanasia a complete necropsy was performed and tissues collected in 10% neutral buffered formalin. Representative tissues were paraffin-embedded, sectioned at 5 μm, and stained with hematoxylin and eosin for routine evaluation. Immunohistochemistry and quantitative image analysis were used to characterize the immunophenotypic composition of inflammatory cell infiltrates observed in lung and other tissues and to localize viral antigens. Immunohistochemistry was performed using an avidin-biotin-horseradish peroxidase complex technique with 3,3′-diaminobenzidine (DAB) chromogen (DAKO, Carpinteria, CA) as previously described.27 Briefly, sectioned tissues were labeled using monoclonal antibodies for CD3, CD8, CD20, HLA-DR, CD68, or T-cell intracytoplasmic antigen I (TIA-1) and examined with an Olympus Vanox-S AHBS microscope interfaced with a Leica QWin image analysis system (Leica Imaging Systems, Cambridge, UK) via a DEI 750 charge-coupled device camera (Optronics, Goleta, CA).
To localize viral antigen, immunohistochemistry was performed using rabbit anti-SARS surface (S) glycoprotein polyclonal serum. Rabbits were immunized with recombinant S-glycoprotein ectodomain during the development of a human monoclonal antibody.25 The serum of an animal with high SARS-CoV neutralization titers (50% effective concentration >1:1000 dilution) was used for these studies. Sections of formalin-fixed paraffin-embedded tissue were stained with serum at a dilution of 1:400 for 30 minutes at room temperature. The primary antibody was detected with a biotinylated goat anti-rabbit secondary antibody (Vector Laboratories, Burlingame, CA) at a 1:200 dilution followed by ABC immunostaining (Vector Laboratories) and utilization of a DAB chromogen (DAKO). Controls for nonspecific staining included sections from SARS-infected marmosets stained with rabbit preimmune serum (collected before immunization of rabbits with recombinant S-glycoprotein), and sections from mock-infected marmosets stained with rabbit anti-S-glycoprotein serum.
Virology
Quantitative reverse transcriptase (RT)-polymerase chain reaction (PCR) was used to examine viral load in biological specimens and tissues obtained at necropsy. Viral RNA was extracted from specimens using the QIAamp viral RNA or RNeasy kit according to the manufacturer’s protocol (Qiagen Inc., Valencia, CA). RNA quantitation was performed on a Roche LightCycler Instrument (Roche Applied Science, Indianapolis, IN) with the Roche SARS-CoV, and the Artus RealArt HPA-coronavirus quantitation kits according to the manufacturers’ instructions (Roche Diagnostics, Indianapolis, IN; Artus Biotech, Hamburg, Germany). As reported by Hourfar and colleagues,28 we found that the Artus RealArt assay had greater sensitivity than the Roche assay and was more accurate in quantifying lower concentrations of RNA.
Biosafety
Animals were housed in an ABSL-3 facility that is certified by the Harvard Medical School Commission on Microbiological Safety, and exceeds requirements of the Biosafety in the Microbiological and Biomedical Laboratory (Centers for Disease Control/National Institutes of Health, 2000). Personnel handling infected animals used powered air-purifying respirators (PAPRs) and all clinical procedures including necropsies were performed in a biocontainment glove box. At the University of Massachusetts Medical School, personnel used PAPRs in the BSL-3 laboratory during the processing of infected samples.
In an effort to minimize the risk of aerosol formation, two different methods of tissue homogenization that allow tissue disruption in a sealed vessel were tested. We used a Stomacher Lab Blender (Seward, Norfolk, UK) in the first experiment and a mini beadbeater horizontal shaker (BioSpec Products, Bartlesville, OK) in the second experiment. Tissues that had been flash-frozen were thawed and weighed, minced, and placed in a sealable container (bag or tube) with Leibovitz-15 medium (final concentration, 0.5 to 40% w/v) with antibiotics according to Subbarao and colleagues.17 For the beadbeater processing, tubes were half-filled with 1-mm zirconium beads. For each type of tissue disruption, processing was performed at the highest setting for 3 minutes. The RNeasy mini kit was adapted for direct RNA extraction from lung tissues using the mini beadbeater according to the manufacturer’s instructions (Qiagen).
Statistical Analysis
Groups were compared statistically using commercially available software (Sigma Stat, Jandel Scientific) by t-test or the Mann-Whitney rank sum test where appropriate.
Results
Clinical Observations
As with other animal models of SARS-CoV infection, clinical signs were generally mild after inoculation (Table 1). Animals continued to eat and remained active; however there was a statistically significant increase in rectal temperature in animals at 4 and 7 days after inoculation. Although the increased body temperatures were relatively modest, animals were anesthetized during all physical exams and as such commonly have lower body temperatures than active animals. Watery and nonhemorrhagic diarrhea developed in 7 of 12 animals after inoculation and two animals developed dyspnea under anesthesia on day 4 of the first experiment.
Table 1.
Clinical Signs in Common Marmosets Inoculated with SARS-CoV
| Experiment day | Number of observations, experiment 1 | Number of observations, experiment 2 | T > 36.8* | Diarrhea | Dyspnea |
|---|---|---|---|---|---|
| 2 | 6 | 0 | 0/6 | 3/6 | 0/6 |
| 4 | 4 | 6 | 5/10 | 6/10 | 2/10 |
| 7 | 2 | 3 | 2/5 | 4/5 | 0/5 |
T, rectal temperature under anesthesia.
Pulmonary Pathology
There was subtle, but clear evidence of disease in most lung specimens at days 4 and 7. Grossly, the lungs varied from being normal in appearance to having multifocal areas of atelectasis and consolidation. Tracheobronchial lymphadenopathy was observed in two animals. Otherwise no other gross anatomical pathology was observed in heart, spleen, liver, kidney, or gastrointestinal tract.
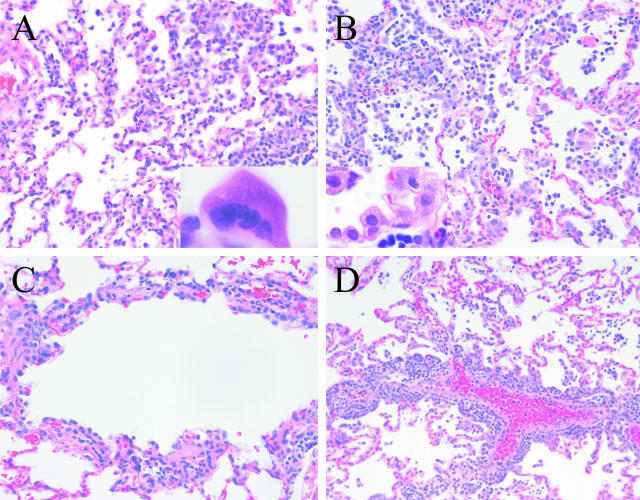
Animals developed a pneumonia characterized by a multifocal to coalescing interstitial pneumonitis (Table 2). At 2 days after infection, infiltrates were mild and characterized by an increased number of neutrophils within septal walls. By 4 days after infection, interstitial disease was predominantly mononuclear and was accompanied variably by increased numbers of alveolar macrophages and exudates (Figure 1, A and B). Hyaline membranes were rarely observed, however, multinucleated syncytia were first apparent at this time (Figure 1A). By day 7 after infection, pneumonitis appeared to be resolving in some animals, but edema was observed in a patchy distribution. In others, pulmonary inflammation persisted and was accompanied by consolidation of alveolar spaces and proliferation of type II pneumocytes (Figure 1B). Microscopically, tracheobronchial lymph node enlargement was associated with parafollicular lymphocytic hyperplasia and histiocytosis within the subcapsular sinuses. Given the small numbers of animals at each time point, it was not possible to reach a conclusion about the peak in pathology. In some animals, it appeared that pneumonitis was resolving by day 7, but the two animals with the greatest amount of disease were euthanized on day 7 (Table 2). As a control to rule out nonspecific reactivity to components of culture medium, two animals were mock infected with Vero E6 culture supernatant and euthanized at 4 days after infection. Both remained healthy and neither developed pneumonitis.
Table 2.
Pulmonary Pathology in Common Marmosets Inoculated with SARS-CoV
| Experiment day | Animal no. | Interstitial pneumonitis | Multinucleated syncytia | Type 2 pneumocyte hyperplasia | Hyaline membrane | Edema | Perivasculitis |
|---|---|---|---|---|---|---|---|
| 2 | 433 | +/− | − | − | − | − | − |
| 145 | + | − | − | − | − | − | |
| 4 | 351 | ++ | + | − | + | + | − |
| 153 | ++ | + | + | + | − | − | |
| 3 | + | − | − | − | − | − | |
| 304 | + | + | − | − | − | ++ | |
| 281 | ++ | + | + | − | − | − | |
| 7 | 478 | + | + | + | + | + | − |
| 114 | + | − | ++ | − | ++ | − | |
| 206 | ++ | + | + | − | + | − | |
| 128 | +++ | + | ++ | − | + | +++ | |
| 110 | ++ | + | + | − | − | − |
−, absent; +, mild; ++, moderate; +++, severe.
Figure 1.
Pulmonary pathology in common marmosets inoculated with SARS-CoV. A: Multifocal interstitial pneumonitis 4 days after inoculation [inset: multinucleated syncytial cell (lung, H&E-stained section)]. B: Multifocal interstitial pneumonitis with type 2 pneumocyte hyperplasia 7 days after inoculation (inset: type 2 pneumocyte hyperplasia). C: Moderate lymphocytic bronchiolitis accompanied by loss of respiratory epithelium integrity. D: Pulmonary perivasculitis 7 days after inoculation.
In addition to this interstitial pneumonitis, airway pathology observed in animals at later time points also involved terminal and respiratory bronchioles (Figure 1C). A mild lymphocytic infiltrate was observed in submucosa and accompanied by loss and irregularity of surface respiratory epithelial cells. In two animals lymphocytic perivascular infiltrates were also apparent. In one of these animals these findings were striking and accompanied by severe coalescing interstitial pneumonia (Figure 1D).
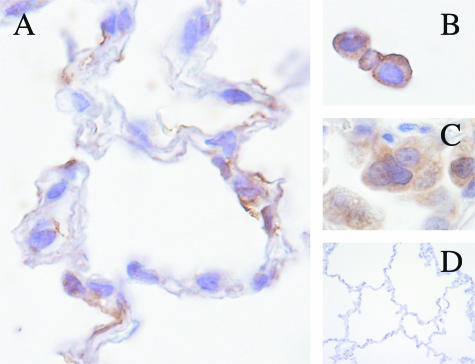
Immunohistochemistry was performed to localize virus and characterize the inflammatory cell infiltrate within pulmonary tissue. Virus antigen (S-glycoprotein) was observed in multinucleated syncytial cells in alveoli and in type I pneumocytes. Macrophages also were positive for viral antigen expression in some animals (Figure 2). Viral antigen was not observed in hepatic, intestinal, renal, or dermal tissues. Immunohistochemistry revealed that the inflammatory infiltrate was composed primarily of CD68-positive macrophages with smaller number of CD3-positive lymphocytes. Increased expression of the MHC II antigen HLA-DR was observed in foci of inflammation at 4 and 7 days.
Figure 2.
Localization of SARS-CoV viral antigen by immunohistochemistry. A–C: Viral antigen within cells morphologically compatible with type 1 pneumocytes (A); mononuclear alveolar exudates (B); and multinucleated syncytial cells (C). D: Mock-infected control lung (rabbit anti-S-glycoprotein, ABC immunostain technique, DAB chromogen).
Extrapulmonary Pathology
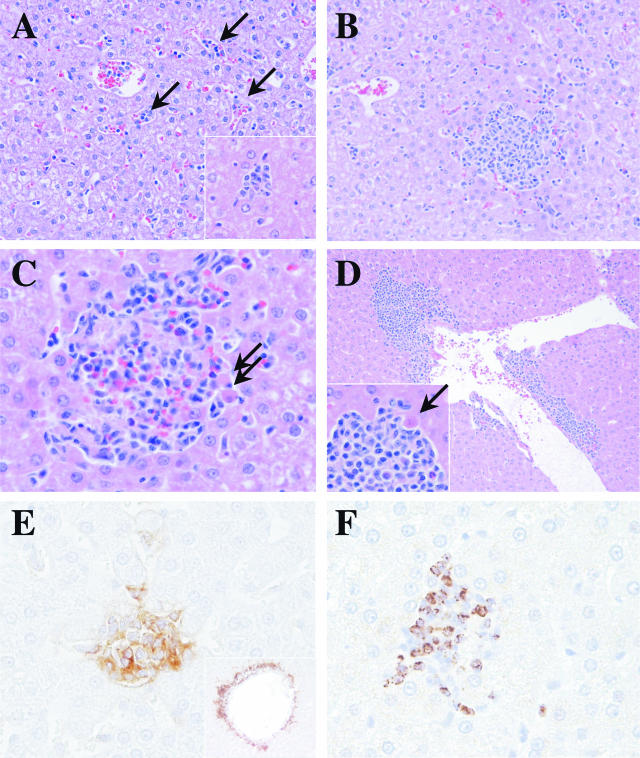
Hepatic inflammatory cell infiltrates were observed in 11 of 12 animals. At the earliest time point, 2 days after inoculation, this was characterized by small aggregates of mononuclear cells within hepatic sinusoids that occasionally coalesced to form a multifocal hepatitis (Figure 3A). By 4 days after infection, hepatitis was readily apparent in four of five animals examined and well-formed lymphocytic inflammatory cell infiltrates were associated with death of individual hepatocytes that appear rounded and densely eosinophilic (Figure 3, B and C). By 7 days after infection, all animals had evidence of multifocal hepatitis. Superimposed on this pattern in 4 of 11 animals with hepatic lesions was a multifocal inflammatory process centered on hepatic veins (Figure 3D). This pattern was characterized by a mononuclear cell cuff of variable thickness that extended into adjacent hepatic parenchyma and was associated with death of individual hepatocytes. Occasionally this process was associated with fewer than expected endothelial cells and fibrin deposition at the vessel surface. Both animals with pulmonary perivasculitis also had this hepatic lesion. Immunostaining demonstrated HLA-DR expression on inflammatory cell infiltrates, and a preponderance of cells staining positive for TIA-1 consistent with cytolytic lymphocytes (NK or T cells) (Figure 3, E and F).
Figure 3.
Hepatic pathology in common marmosets inoculated with SARS-CoV. A: Small aggregates of mononuclear cells within hepatic sinusoids (arrows) and multifocally defined mild lymphocytic hepatitis (higher power inset) 2 days after inoculation (H&E-stained section). B: More pronounced multifocal random lymphocytic hepatitis in four of five animals 4 days after inoculation. C: Hepatitis in five of five animals associated with death of individual hepatocytes (arrows) 7 days after inoculation. D: A second pattern superimposed on hepatic lesions in 4 of 11 animals characterized by inflammation centered on hepatic veins and accompanied by death of hepatocytes (inset, arrow). E: Increased expression of the MHC II antigen HLA-DR at sites of lymphocytic hepatitis and perivascular inflammation (inset) (HLA-DR, ABC immunostain technique, DAB chromogen). F: Foci of inflammation contained T-cell intracytoplasmic antigen (TIA-1)-positive cells compatible with cytotoxic T lymphocytes or NK cells (TIA-1, ABC immunostain technique, DAB chromogen).
A mild diffuse colitis was apparent in 8 of 12 animals and correlated with clinical signs of diarrhea. There was marked colonic crypt epithelial cell hyperplasia characterized by loss of goblet cells, cytoplasmic basophilia, and loss of nuclear polarity. Evidence of enteropathogenic Escherichia coli (EPEC) was observed microscopically in four of the animals with colitis. EPEC are a common enteric pathogen of marmosets and thus these findings (diarrhea and colitis) may have resulted from an infection with a concurrent pathogen. Neither diarrhea nor colitis were observed in the mock-infected animals. A mild multifocal lymphocytic myocarditis was observed in three animals, and was most pronounced in animals examined on day 7. No renal pathology was evident in specimens from any time point.
Virology
In animal experiment 1, we found tissue disruption to be inefficient using the Stomacher. Larger volumes of medium were required, resulting in homogenates of relatively low concentration. It was necessary to concentrate samples using Centricon columns (Millipore, Billerica, MA) to improve yields in PCR assays. Virus isolation and titration were attempted, however, no viral growth was detected in samples that included oral swabs, rectal swabs, and tissue homogenates from the first group of six animals. Despite the sample-processing limitations described above, viral RNA was detected in tissue homogenates from two of the six animals. One animal (no. 433) had high concentrations of SARS-CoV RNA in lung (more than 107 copies/g of tissue), and detectable RNA in tracheobronchial lymph node and heart tissues on day 2 after infection. The other animal (no. 351) had high concentrations of viral RNA in one of two lung samples at day 4.
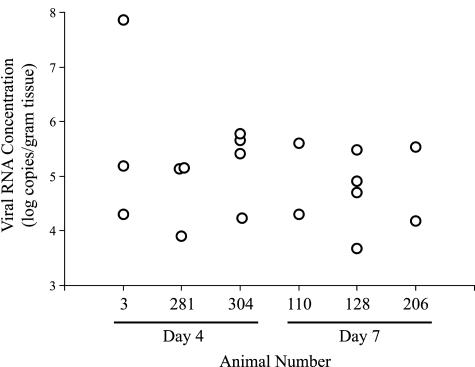
The mini beadbeater enabled the use of smaller volumes in animal experiment 2, resulting in generally higher tissue homogenate concentrations. We also observed that greater tissue disruption was achieved compared with the first experiment. The mini beadbeater protocol was easily adapted to direct RNA extraction from tissues for even greater sensitivity in detection of viral RNA. Given the culture results of the initial experiment, we focused primarily on viral RNA detection and quantification in the second experiment. Viral RNA was detected in lung tissue samples from all six animals. Viral RNA concentrations ranged from undetectable (detection threshold values ranging from 2500 to 17,000 copies/g of tissue) to more than 70 × 106 copies/g of tissue. Multiple lung samples taken from each animal at necropsy indicated wide variations in viral RNA concentrations in different areas of the lungs (Figure 4).
Figure 4.
Viral RNA concentrations in pulmonary tissue (animal experiment 2). Each symbol represents results from a separate lung specimen. Specimens with positive results are shown. Each animal had five separate lung specimens.
Overall, a greater fraction of specimens were positive at day 4 than at day 7, and both average and peak concentrations of viral RNA were higher on day 4 than at day 7 (Table 3), but these differences did not reach statistical significance. Two animals (nos. 3 and 281) had detectable viral RNA in both tracheobronchial lymph node and heart, and one (no. 128) had viral RNA in heart. No viral RNA was detected in hepatic or intestinal homogenates, however all extracts from intestinal homogenates and half from the hepatic homogenates were inhibitory in the PCR assays. A nested PCR reaction to detect viral RNA29 also gave negative results for liver homogenates (data not shown).
Table 3.
Summary of SARS-CoV Pulmonary Viral Load in Common Marmoset by Quantitative PCR (Experiment 2)
| Experiment day | PCR-positive lung samples (%)* | Average concentration† (log10 copies/g tissue) | Peak concentration (log10 copies/g tissue) |
|---|---|---|---|
| Day 4 | 12 of 15 (80%) | 5.0 | 7.9 |
| Day 7 | 8 of 15 (53%) | 4.3 | 5.6 |
| Total | 20 of 29 (69%) | 4.7 | 7.9 |
Combined results from routine processing and direct tissue RNA extractions.
This calculation includes maximum possible negative (threshold) values for samples that were negative by PCR.
Discussion
The common marmoset (C. jacchus) is a small New World nonhuman primate with distinct advantages for infectious disease research. We have shown with these experiments that C. jacchus is susceptible to SARS-CoV infection and develops pulmonary pathology principally characterized by a multifocal interstitial pneumonitis with multinucleated syncytial cells and accompanied by a lymphocytic bronchiolitis at early time points. Viral replication was demonstrated in pulmonary tissue through immunohistochemistry and RT-PCR. It should be noted that the pilot experiments undoubtedly underestimated the concentrations of SARS-CoV in tissues and biological specimens. Our efforts to minimize potential for aerosol formation likely limited the yield of virus from tissues. For this reason, our findings cannot be compared directly with other studies that have used different techniques for tissue disruption and RNA extraction.14,15,17,18,30 More sensitive techniques are currently being used to measure viral concentrations.
Although pulmonic disease predominated, histological evidence of additional organ system involvement existed for cardiac, lymphoid, and hepatic tissues. Data provided direct evidence that virus was present in lymphoid and cardiac tissues. The presence of inflammation in liver, heart, and bowel suggests that this animal model of SARS shares features of extrapulmonary disease commonly found in humans.5,8,10,31,32 In humans, there is generally good concordance between the expression of the primary functional receptor, ACE-2, and SARS-CoV associated pathology or distribution of viral RNA.7,32 The presence of inflammation and/or SARS-CoV RNA in a similar tissue distribution in marmosets suggests that ACE-2 expression levels are similar to humans. The importance of L-SIGN, a less efficient, but functional receptor for SARS-CoV, is yet to be defined, but could be studied in this animal model.33
Hepatic lesions were a conspicuous finding in 11 of 12 animals after inoculation. Elevations of liver enzymes and hepatic lesions have also been described as a clinical entity in human patients infected with the SARS-CoV.34,35 Viral RNA has been demonstrated in hepatic tissues of individuals with SARS.10 Hepatocytes expressing the SARS-CoV N-protein have been demonstrated in fatal cases of SARS.8 The pattern and early appearance of these lesions in the marmoset suggest that direct viral infection of hepatocytes may play a role. We were unable to demonstrate viral antigen or viral RNA in hepatic tissues using the reagents and techniques available at the time. There is direct evidence that sample inhibition limited our ability to amplify gene products in hepatic tissue RNA extracts. The direct infection of hepatocytes, as yet unproven, remains our leading hypothesis for the hepatitis observed in the SARS-CoV-infected marmosets. This hypothesis will be tested in future studies using different techniques for RNA extraction and gene amplification, and different antibodies for immunohistochemistry in an effort to enhance sensitivity. An alternate hypothesis would be a triggering of a cross-reactive autoimmune response. This is considered less likely given the presence of lesions in 11 of 12 animals and the relative lack of similar inflammatory cell infiltrates in other organs (eg, kidneys).
Recently, prominent hepatic lesions were found in a ferret model after vaccination and subsequent SARS-CoV challenge. The greater severity of hepatitis in immunized animals suggested immune enhancement of pathology.22 Enhanced pathology has been described in other coronavirus infections after repeated exposure, and this has implications in the design of vaccine and therapeutic interventions in SARS.36–38 The rapid appearance of a random mononuclear hepatitis in infected marmosets might point toward an innate immune response with infiltrates of NK cells. Such a pattern is observed in mice infected with MCMV with a peak level observed on the second day after infection.39 However, the expanding pattern of the hepatic lesions through day 7 (the latest time point of these pilot studies) and the presence of cytolytic lymphocytes in aggregates of activated cells expressing HLA-DR would also be consistent with virus-specific recognition by T lymphocytes of infected cells that are scattered in the hepatic parenchyma and lining hepatic veins. The enhancement of SARS-CoV-induced hepatitis in vaccinated ferrets would also be consistent with an earlier and more pronounced accumulation of virus-specific T lymphocytes at sites of virus replication. Additional studies will be required to determine whether pre-existing immunity alters hepatic pathology in marmosets.
Although infection of the common marmoset mimics some aspects of primary SARS-CoV infection of man, diffuse alveolar damage, the hallmark morphological feature of SARS, was not observed throughout the 7-day time course evaluated. Epidemiological studies indicate that there may be a range of disease severity in human patients, with advancing age and concurrent disease such as diabetes being clear risk factors for the development of diffuse alveolar damage.2 Our studies focused on the early events of SARS-CoV infection and used juvenile to young adult animals. Studies of longer duration and utilization of animals of different age groups will help determine whether these aspects of SARS-CoV infection may be appropriately studied using this species.
Additional studies are necessary to understand the complete disease course and its manifestations. Variability that exists between infected animals may be problematic, but also potentially valuable in defining determinants of disease course. The larger nonhuman primates cannot be tested in sufficiently large numbers to define host characteristics that influence pathogenesis. This small nonhuman primate model will likely be a useful tool to investigate these aspects of pathogenesis. The common marmoset is particularly well suited for defining immunological correlates of disease.24,40
In conclusion, we have demonstrated that the common marmoset is susceptible to SARS-CoV infection, and develops disease that shares features with SARS in humans. In the absence of a disease outbreak (as is currently the case for SARS), a nonhuman primate model for pathogenesis and treatment becomes increasingly important. Based on promising preliminary data, we believe that the common marmoset will serve as a useful surrogate system for assessing treatment efficacy. As critical host determinants of pathogenesis are defined in this animal model, their relevance to human disease may be better defined. These types of analyses will enhance our understanding of the disease process, and will allow for efficient testing of vaccines and various treatment modalities in the common marmoset.
Acknowledgments
We thank Donna Ambrosino for many helpful discussions and support of the project; and Kanta Subbarao, Anjeanette Roberts, and Leatrice Vogel for technical assistance and advice.
Footnotes
Address reprint requests to Thomas Greenough, M.D., University of Massachusetts Medical School, 373 Plantation St., Biotech 2, Suite 318, Worcester, MA 01605. E-mail: thomas.greenough@umassmed.edu.
Supported by the New England Regional Center of Excellence/Biodefense and Emerging Infectious Diseases (developmental project award AI0571590) and the National Institutes of Health (grant RR00168, NIH contract NO1 AI65315).
References
- Christian MD, Poutanen SM, Loutfy MR, Muller MP, Low DE. Severe acute respiratory syndrome. Clin Infect Dis. 2004;38:1420–1427. doi: 10.1086/420743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Fisher DA, Lim TK, Lim YT, Singh KS, Tambyah PA. Atypical presentations of SARS. Lancet. 2003;361:1740. doi: 10.1016/S0140-6736(03)13336-3. [DOI] [PubMed] [Google Scholar]
- Tee AK, Oh HM, Lien CT, Narendran K, Heng BH, Ling AE. Atypical SARS in geriatric patient. Emerg Infect Dis. 2004;10:261–264. doi: 10.3201/eid1002.030322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IF, Cheng VC, Wu AK, Chan KH, Chu CM, Wong MM, Poon LL, Tse DM, Chan KS, Woo PC, Lau SK, Peiris JS, Yuen KY. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550–1557. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I, Timens W, Bulthuis ML, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, He L, Zhang Q, Huang Z, Che X, Hou J, Wang H, Shen H, Qiu L, Li Z, Geng J, Cai J, Han H, Li X, Kang W, Weng D, Liang P, Jiang S. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang H, Shen H, Li Z, Geng J, Han H, Cai J, Li X, Kang W, Weng D, Lu Y, Wu D, He L, Yao K. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas GA, Poutanen SM, Mazzulli T, Willey BM, Butany J, Asa SL, Faure P, Akhavan P, Low DE, Kain KC. Fatal severe acute respiratory syndrome is associated with multiorgan involvement by coronavirus. J Infect Dis. 2005;191:193–197. doi: 10.1086/426870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Hui DS, Chan KC, Hung EC, Chiu RW, Lee N, Wu A, Chim SS, Tong YK, Sung JJ, Tam JS, Lo YM. Quantitative analysis and prognostic implication of SARS coronavirus RNA in the plasma and serum of patients with severe acute respiratory syndrome. Clin Chem. 2003;49:1976–1980. doi: 10.1373/clinchem.2003.024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stohr K, Osterhaus AD. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, Ling AE, Chan PK, Tam JS, Zambon MC, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra JC, Stohr K, Peiris JS, Osterhaus AD. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe J, Vogel L, Roberts A, Fahle G, Fischer S, Shieh WJ, Butler E, Zaki S, St. Claire M, Murphy B, Subbarao K. Replication of SARS coronavirus administered into the respiratory tract of African green, rhesus and cynomolgus monkeys. Virology. 2004;330:8–15. doi: 10.1016/j.virol.2004.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A, Lamirande EW, Buchholz UJ, Vogel LN, Elkins WR, St. Claire M, Murphy BR, Subbarao K, Collins PL. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363:2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosino DM, Roberts A, Vogel L, Thomas WDJ, Babcock GJ, Greenough TC, Guarner J, Zaki S, Sullivan JL, Subbarao K: Treatment of SARS infected hamsters with human monoclonal antibody (HuMAb 201). Presented at the 42nd Annual Meeting of the Infectious Diseases Society of America, Boston, Oct 2004, 2:2004 [Google Scholar]
- Subbarao K, McAuliffe J, Vogel L, Fahle G, Fischer S, Tatti K, Packard M, Shieh WJ, Zaki S, Murphy B. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina BE, Haagmans BL, Kuiken T, Fouchier RA, Rimmelzwaan GF, Van Amerongen G, Peiris JS, Lim W, Osterhaus AD. Virology: SARS virus infection of cats and ferrets. Nature. 2003;425:915. doi: 10.1038/425915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, Murphy BR, Rappuoli R, Lanzavecchia A. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZY, Kong WP, Huang Y, Roberts A, Murphy BR, Subbarao K, Nabel GJ. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht H, Roberts A, Vogel L, Bukreyev A, Collins PL, Murphy BR, Subbarao K, Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci USA. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H, Czub M, Czub S, Neufeld J, Marszal P, Gren J, Smith G, Jones S, Proulx R, Deschambault Y, Grudeski E, Andonov A, He R, Li Y, Copps J, Grolla A, Dick D, Berry J, Ganske S, Manning L, Cao J. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78:12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J, Bakker AB, van den Brink EN, Weverling GJ, Martina BE, Haagmans BL, Kuiken T, de Kruif J, Preiser W, Spaan W, Gelderblom HR, Goudsmit J, Osterhaus AD. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet. 2004;363:2139–2141. doi: 10.1016/S0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield K. Marmoset models commonly used in biomedical research. Comp Med. 2003;53:383–392. [PubMed] [Google Scholar]
- Greenough TC, Babcock GJ, Roberts A, Hernandez HJ, Thomas WD, Jr, Coccia JA, Graziano RF, Srinivasan M, Lowy I, Finberg RW, Subbarao K, Vogel L, Somasundaran M, Luzuriaga K, Sullivan JL, Ambrosino DM. Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J Infect Dis. 2005;191:507–514. doi: 10.1086/427242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlage E, Mansfield K. Clinical care and diseases of the common marmoset (Callithrix jacchus). Comp Med. 2003;53:369–382. [PubMed] [Google Scholar]
- Mansfield KG, Veazey RS, Hancock A, Carville A, Elliott M, Lin KC, Lackner AA. Induction of disseminated Mycobacterium avium in simian AIDS is dependent upon simian immunodeficiency virus strain and defective granuloma formation. Am J Pathol. 2001;159:693–702. doi: 10.1016/S0002-9440(10)61740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourfar MK, Roth WK, Seifried E, Schmidt M. Comparison of two real-time quantitative assays for detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42:2094–2100. doi: 10.1128/JCM.42.5.2094-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Mazzulli T, Farcas GA, Poutanen SM, Willey BM, Low DE, Butany J, Asa SL, Kain KC. Severe acute respiratory syndrome-associated coronavirus in lung tissue. Emerg Infect Dis. 2004;10:20–24. doi: 10.3201/eid1001.030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Hattermann K, Marzi A, Gramberg T, Geier M, Krumbiegel M, Kuate S, Uberla K, Niedrig M, Pohlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KF, Lo AW. Exploring the pathogenesis of severe acute respiratory syndrome (SARS): the tissue distribution of the coronavirus (SARS-CoV) and its putative receptor, angiotensin-converting enzyme 2 (ACE2). J Pathol. 2004;203:740–743. doi: 10.1002/path.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD, Jr, Thackray LB, Young MD, Mason RJ, Ambrosino DM, Wentworth DE, Demartini JC, Holmes KV. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Lu SN, Changchien CS, Chiu KW, Kuo CH, Chuah SK, Liu JW, Lin MC, Eng HL, Chen SS, Lee CM, Chen CL. Sequential changes of serum aminotransferase levels in patients with severe acute respiratory syndrome. Am J Trop Med Hyg. 2004;71:125–128. [PubMed] [Google Scholar]
- Cui HJ, Tong XL, Li P, Hao YX, Chen XG, Li AG, Zhang ZY, Duan J, Zhen M, Zhang B, Hua CJ, Gong YW. Serum hepatic enzyme manifestations in patients with severe acute respiratory syndrome: retrospective analysis. World J Gastroenterol. 2004;10:1652–1655. doi: 10.3748/wjg.v10.i11.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers WS, Horsburgh BC, Baxendale W, Brown TD. Enhancement of FIP in cats immunised with vaccinia virus recombinants expressing CCV and TGEV spike glycoproteins. Adv Exp Med Biol. 1993;342:359–364. doi: 10.1007/978-1-4615-2996-5_55. [DOI] [PubMed] [Google Scholar]
- Perlman S. Pathogenesis of coronavirus-induced infections. Review of pathological and immunological aspects. Adv Exp Med Biol. 1998;440:503–513. [PubMed] [Google Scholar]
- Olsen CW, Corapi WV, Jacobson RH, Simkins RA, Saif LJ, Scott FW. Identification of antigenic sites mediating antibody-dependent enhancement of feline infectious peritonitis virus infectivity. J Gen Virol. 1993;74:745–749. doi: 10.1099/0022-1317-74-4-745. [DOI] [PubMed] [Google Scholar]
- Salazar-Mather TP, Hokeness KL. Calling in the troops: regulation of inflammatory cell trafficking through innate cytokine/chemokine networks. Viral Immunol. 2003;16:291–306. doi: 10.1089/088282403322396109. [DOI] [PubMed] [Google Scholar]
- Abbott DH, Barnett DK, Colman RJ, Yamamoto ME, Schultz-Darken NJ. Aspects of common marmoset basic biology and life history important for biomedical research. Comp Med. 2003;53:339–350. [PubMed] [Google Scholar]