Abstract
Cyclosporine A, which has been the foremost immunosuppressive agent since the early 1980’s, significantly improves the success of organ transplantation. However, common complications of cyclosporine A therapy, such as severe renal tubulointerstitial fibrosis, limit the drug’s clinical use. Although the exact mechanisms driving cyclosporine A-induced tubulointerstitial fibrosis remain elusive, we hypothesized that epithelial-mesenchymal transition (EMT) may play a major role. We investigated this in vitro by treating human proximal tubular cells with cyclosporine A. Morphological changes were observed after cyclosporine A treatment, including cell elongation (with a large degree of detachment), cytoskeletal rearrangement, and junctional disruption. In addition, expression of the myofibroblast-specific marker α-smooth muscle actin was detected in treated cells. These observations are consistent with events described during EMT. Using Affymetrix gene microarrays, we identified 128 genes that were differentially regulated in renal tubular cells after cyclosporine A treatment, including known profibrotic factors, oncogenes, and transcriptional regulators. Cyclosporine A induced a dose-dependent increase in transforming growth factor-β secretion from proximal tubular cells. Subsequent functional studies revealed that protein kinase C-β isoforms play a key role in cyclosporine A-induced effects. These findings provide novel insights into cyclosporine A-induced renal fibrosis and the molecular mechanisms underlying EMT, events that may be relevant in other disease states.
Since the discovery of its powerful immunosuppressive actions in the 1970’s, cyclosporine A (CsA) has revolutionized the field of whole organ transplantation, with greatly improved survival in kidney, heart, liver, and pancreas graft recipients.1 In more recent times, the use of CsA has expanded to the treatment of autoimmune diseases such as psoriasis and rheumatoid arthritis.2 The immunosuppressive action of CsA is believed to be mediated through preventing T-cell activation through inhibition of calcineurin, thereby preventing the phosphorylation of NFAT family members responsible for the transcriptional activation of the interleukin-2 and interleukin-4 genes.3
The major limiting factors in the use of CsA are the acute and long-term toxic effects in the kidney.4,5 In the short term, CsA induces a reversible reduction in renal blood flow and glomerular filtration rate.6 In the long term, CsA treatment leads to irreversible renal failure characterized by extensive tubulointerstitial fibrosis.7 Tubulointerstitial fibrosis is the final common endpoint in renal disease, involving a number of initiating conditions including diabetes, hypertension, and CsA treatment. Tubulointerstitial fibrosis is marked by tubular atrophy, extracellular matrix (ECM) accumulation, and thickening of the basement membrane resulting in loss of tubular function. It has been proposed that the long-term effects of CsA toxicity can be attributed to sustained restriction of renal blood supply, believed to be caused by decreased nitric oxide production in the renal arterioles.8 However, we have previously reported evidence that the proximal tubular epithelial cells (PTECs) themselves are direct targets for CsA-induced injury.9,10 The essential role that the proximal tubule plays in reabsorption and homeostasis makes it highly susceptible to such injury.
In chronic renal disease, the decline in renal function correlates closely with the degree of tubulointerstitial fibrosis. A striking feature of this fibrosis is interstitial myofibroblast activation, which is thought to be central in the pathogenesis of renal disease.11,12 Previously it was believed that the main cause of the accumulation of ECM in tubulointerstitial fibrosis was infiltrating fibroblasts that became active in the interstitium. Evidence is now accumulating to suggest that a major source of these myofibroblasts may be the epithelium itself. It is now proposed that epithelial-mesenchymal transition (EMT) plays a major role in the progression and maintenance of tubulointerstitial fibrosis.13,14 EMT is a developmental process in which numerous phenotypic changes occur leading to the loss of epithelial markers and function and the acquisition of a more fibroblastic phenotype, termed a myofibroblast.15 The myofibroblast is a morphological intermediate between fibroblasts and smooth muscle cells. Like fibroblasts, they have the ability to produce and secrete ECM components such as collagen I and III and fibronectin, and like smooth muscle cells they express α-smooth muscle actin (α-SMA) and have the ability to contract equipping the myofibroblast with enhanced locomotive ability.15,16 Thus, EMT may contribute significantly to renal failure through the accumulation of ECM and perhaps more significantly, through loss of epithelial integrity and function.
EMT is now recognized as a significant step in the acquisition of an aggressive, invasive phenotype in tumor cells.17 In recent times, data from studies into the effect of long-term CsA use suggest that CsA treatment may lead to an increased susceptibility to certain tumors and tumor metastasis.18 In the past, this was attributed to decreased host vigilance due to CsA immunosuppression, however a number of key studies suggest that CsA has direct cellular effects that may promote tumor cell invasiveness by cell autonomous mechanisms.19,20 Considering these facts, we hypothesized that CsA may induce EMT in human proximal tubular cells. We therefore examined the effects of CsA on the phenotype and the transcriptome of HK-2 renal PTECs and investigated some possible mechanisms of the changes observed.
Materials and Methods
Reagents
CsA was a generous gift from Novartis Pharmaceuticals Ltd (UK). All tissue culture reagents were from Costar. Alamar Blue reagents were obtained from AGB Ltd (Ireland). The lactate dehydrogenase (LDH) assay kit was obtained from Sigma (Ireland). The nature of antibodies used is described in the Western Blot Analysis section. The transforming growth factor (TGF)-β1 enzyme-linked immunosorbent assay (ELISA) kit was obtained from R&D Systems (UK). Primers for reverse transcriptase (RT)-polymerase chain reaction (PCR) were obtained from Sigma Genosys Biotechnologies (Woodlands, TX). The protein kinase C (PKC)-β inhibitor Hispidin was obtained from Calbiochem (La Jolla, CA). All other reagents were of the highest available purity from commercial sources.
Cell Culture
Primary human renal proximal tubular epithelial cells (RPTECs) were purchased from Clonetics (Walkersville, MD). Primary RPTECs were maintained in renal epithelial cell growth medium (REGM) supplemented with REGM SingleQuots and growth factors (also Clonetics). The human renal proximal tubular cell line, HK-2, was purchased from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco’s modified Eagles medium containing 100 U/ml penicillin, 100 g/ml streptomycin, 5 μg/ml insulin, 10 ng/ml epidermal growth factor, and 2 mmol/L l-glutamine. Cell culture medium was changed every 48 hours. Cells were maintained in 75-cm2 Costar flasks at 37°C in a humidified atmosphere containing 95% air and 5% CO2.
Cell Treatment
CsA was prepared as a stock solution (4.2 mmol/L) by dissolving the powder in 100% ethanol. Cells were seeded at 1 × 105 cells/ml 24 hours before treatment resulting in ∼90% confluency at treatment. For viability studies and TGF-β1 ELISA, cells were treated with a range of CsA concentrations as indicated. In all other cases, cells were treated with 4.2 μmol/L CsA and the effects were examined at 12, 24, 48, and 72 hours after treatment. In all experiments with CsA, control cells were exposed to an appropriate concentration of ethanol vehicle. Hispidin was prepared as a stock solution of 10 mmol/L in dimethyl sulfoxide and stored in aliquots at −20°C. Cells were preincubated for 1 hour with 10 μmol/L hispidin before treatment with 4.2 μmol/L CsA. In experiments involving hispidin, control cells were also exposed to an appropriate concentration of dimethyl sulfoxide and ethanol vehicles.
Viability and Cytotoxicity Studies
Viable cell numbers were assessed using an Alamar Blue reduction assay. Briefly, after the indicated treatments, cells were incubated with a solution of 10% Alamar Blue in normal culture medium for 2 hours in the dark. Fluorescence was then measured at 540 nm excitation wavelength and 590 nm emission wavelength using a Wallac 1420 plate reader. Cytotoxicity levels were assessed using an LDH assay kit. After the indicated treatments, 100 μl of cell supernatants were assayed for LDH activity according to the manufacturer’s instructions.
Immunofluorescent Microscopy
Briefly, cells were fixed in 3.7% (v/v) formaldehyde/phosphate-buffered saline (PBS). Cells were permeabilized with Triton X-100/PBS. To stain F-actin, rhodamine phalloidin (no. R415; Molecular Probes) was added as a 1:40 dilution. After blocking, cells being stained for β-catenin, ZO-1, and α-SMA were incubated with a 1:100 dilution of goat anti-β-catenin (no. SC-1496; Santa Cruz Technology), mouse anti-ZO-1 (no. 33-9100; Zymed), or mouse anti-α-SMA (no. A-2547, Sigma). Subsequently the cells were incubated with secondary antibody [1:100 rabbit anti-mouse fluorescein isothiocyanate (no. F02323; DAKO) or 1:100 donkey anti-goat fluorescein isothiocyanate (no. SC 349, Santa Cruz)]. The cells were washed, followed by incubation with 4,6-diamidino-2-phenylindole (no. D1306, Molecular Probes). Cells were incubated with secondary antibodies [1:100 rabbit anti-mouse fluorescein isothiocyanate (DAKO) or 1:100 donkey anti-goat fluorescein isothiocyanate] in the absence of primary antibody to demonstrate background staining. Slides were mounted and viewed using the Zeiss Axioplan 2 imaging fluorescent microscope.
Cell Motility
Analysis of cell motility was performed using a wound-healing assay and a Transwell migration assay. HK-2 cells were plated on uncoated 60-mm Petri dishes and cultured in the presence or absence of 4.2 μmol/L CsA. Light microscopic analysis of the cells was performed immediately after wound formation at 0 hours and subsequently at 48 hours. Cell migration was quantified in Transwell cell culture chambers. Cells were seeded at 1 × 105 cells/ml on the apical side of polyvinylpyrrolidone-free polycarbonate filters with 8.0-μm pore size (no. 3422; Costar, Cambridge, MA). The filters were incubated overnight at 37°C before 4.2 μmol/L CsA was applied to the apical side for 48 and 72 hours. After treatment, medium was aspirated from the wells and the filters were removed. Cell stain containing 0.5% toluidine blue and 0.5% sodium borate was added to the lower chamber of each well. Cells were removed carefully from the apical side of the filters using a cotton tip and the filters were replaced. The migrated cells were incubated for 3 hours at 37°C. Stain was then removed by aspiration and the wells were washed three times using ddH2O. Filters were then placed in extraction buffer (20 mmol/L Tris, 0.2% sodium dodecyl sulfate). Stained solution (150 μl) was removed and absorbance was read at 590 nm.
Western Analysis
Equal total protein amounts of cell lysates or concentrated supernatants were electrophoresed using the procedure of Laemmli.21 For detection of E-cadherin, ZO-1, TGF-β, PKC-β isoforms, and α-SMA, membranes were probed with antibodies specific for E-cadherin (no. 610404, 1:400 dilution; Transduction Laboratories), ZO-1 (no. 33-9100, 1:333 dilution; Zymed), TGF-β (no. AB-100-NA, 1:1000 dilution; R&D Systems), PKC-β isoforms (no. 610127, 1:500 dilution; Transduction Laboratories), α-SMA (no. A2547, 1:1000 dilution; Sigma), and E2A (no. SC 349, 1:1000 dilution; Santa Cruz). Results shown are representative of at least three experiments with similar results.
RNA Isolation
Total RNA was isolated using Trizol reagent (Invitrogen) and quantitated by absorbance at 260 nm. RNA integrity was controlled by electrophoretic analysis on 1.2% agarose gels. RNA was purified for microarray analysis using Qiagen Mini-Spin clean up columns.
Microarray Analysis
Human proximal tubular cells were incubated in the presence of 4.2 μmol/L CsA for 48 hours. RNA was isolated at 0, 12, and 48 hours after treatment. At each time point, RNA from three independent cell treatments of low, medium, and high passage number was pooled to minimize the effects of passage. After RNA clean up, cDNA was synthesized from the total RNA using Superscript choice kit (Invitrogen) with a T7-(dT)24 primer. cRNA was prepared and biotin labeled by in vitro transcription (Enzo Biochemical). Labeled cRNA was fragmented by incubation at 94°C for 35 minutes in the presence of 40 μmol/L Tris acetate, pH 8.1, 100 mmol/L potassium acetate, and 30 mmol/L magnesium acetate. Fifteen mg of fragmented cRNA from each time point was hybridized for 16 hours at 45°C to a HuGene FL array (Affymetrix, Santa Clara, CA). After hybridization, each gene chip was automatically washed and stained with streptavidin-phycoerythrin using a fluidics station. Finally, probe arrays were scanned at 3-mm resolution using GeneChip System confocal scanner made for Affymetrix by Aligent. Affymetrix Microarray suite 5.0 was used to analyze the relative abundance of each gene. Data were analyzed with the GeneSpring version 4.2 (Silicon Genetics, San Carlos, CA) to generate lists of genes with differential expression. The raw data values for genes included in the analysis had to be at least 2 SDs greater than the average absent call value in the particular data set. Additional data and statistical analyses were performed using Microarray Suite version 4.0.1 (Affymetrix). To select differentially expressed genes, we directly compared gene expression of samples taken at the 0-hour treatment time point with that of samples from cells treated for 12 and 48 hours using GeneSpring version 4.2 and restrictions were applied. Genes had to exhibit a fold change of greater than 2 to be regarded as differentially expressed.
RT-PCR
Validation of differentially expressed genes was performed by semiquantitative RT-PCR for a range of selected genes. RT-PCR primer pairs were designed using PRIMER 3 software (available at www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) and using sequence data available at the National Center for Biotechnology Information database. Primers for selected genes are shown in Table 1. Total RNA was extracted as described above. The Gibco-Brl RT kit was used to generate cDNA. Two μl of the cDNA was taken from each sample and inserted into 0.5-ml PCR tubes along with 1 μl of forward primer (100 ng/μl), 1 μl of reverse primer (100 ng/μl), 2 μl of 10 mmol/L dNTPs, 5 μl of 10× buffer, 3 μl of 25 mmol/L MgCl2, 0.25 μl of Taq polymerase (Promega), and 35.75 μl of sterile distilled water to bring the volume up to 50 μl. The number of PCR cycles used were determined to be within the linear range of the reactions. mRNA levels of the glyceraldehyde phosphate dehydrogenase gene (GAPDH), were used as a quantity control. Results shown are representative of at least three experiments with similar results
Table 1.
Genes Up-regulated More Than Two Fold in Response to Treatment with 4.2 μmol/L CsA for 48 Hours
| Enzymes | Fold change |
| Membrane associated | |
| Serine protease hepsin | 8.24 |
| Liver dipeptidyl peptidase IV | 4.56 |
| Autotaxin | 3.21 |
| Prostaglandin D2 synthase | 2.46 |
| Receptor tyrosine kinase (DTK) | 2.31 |
| Receptor tyrosine kinase DDR | 2.04 |
| Cytosolic | |
| Protein kinase C (PKC) type β | 4.79 |
| Long-chain acyl-CoA synthetase | 3.15 |
| Catechol estrogen UDP-glucuronosyl- transferase | 2.87 |
| Transketolase | 2.80 |
| Cysteinyl-tRNA synthetase | 2.76 |
| Long-chain acyl-coenzyme A synthetase | 2.48 |
| Sulfotransferase, Phenol-Preferring | 2.44 |
| Mitochondrial isocitrate dehydrogenase | 2.44 |
| p33/HEH epoxide hydrolase | 2.38 |
| Hydroxymethylglutaryl-CoA lyase | 2.34 |
| Argininosuccinate synthetase | 2.25 |
| Neuron specific (gamma) enolase | 2.23 |
| P1MT isozyme I | 2.11 |
| Red cell-type low MW acid phosphatase | 2.06 |
| Arg protein tyrosine kinase-binding protein | 2.04 |
| Extracellular | |
| Plasma (extracellular) glutathione peroxidase | 2.53 |
| Cytokines & secreted proteins | |
| Extracellular proteinase inhibitor homologue | 6.31 |
| Transforming growth factor-beta 1* | 2.91 |
| Pre-B cell enhancing factor (PBEF) | 2.32 |
| Transcobalamin II | 2.30 |
| Epithelin 1 and 2 | 2.28 |
| TGF-beta superfamily protein | 2.03 |
| Membrane Associated | |
| Cell membrane | |
| Epithelial mucin 1* | 4.58 |
| Cardiac gap junction protein | 3.81 |
| Natriuretic peptide receptor (ANP-A receptor) | 3.51 |
| Lutheran blood group glycoprotein | 3.50 |
| C5a anaphylatoxin receptor | 3.27 |
| NMB | 3.13 |
| Glucose transporter-like protein-III | 2.86 |
| Human B61 | 2.55 |
| Heparan sulfate proteoglycan | 3.55 |
| Glycophorin C* | 2.41 |
| Membrane cofactor protein | 2.36 |
| Pancreatic mucin | 2.29 |
| APXL | 2.23 |
| Amyloid A4 precursor | 2.27 |
| Amyloidβ(A4) Precursor | 2.17 |
| Na,K-ATPase gamma subunit | 2.04 |
| Ryudocan core protein | 2.01 |
| Extracellular barriers | |
| Laminin-related protein | 3.28 |
| Collagen XIV | 2.93 |
| Laminin B1 chain | 2.71 |
| Collagen IV | 2.12 |
| Nuclear | |
| Transcription factors | |
| Apolipoprotein AI regulatory protein | 3.40 |
| Nuclear Factor Nf-IL6 | 2.69 |
| Transcription elongation factor S-II, hS-II-T1 | 2.39 |
| Class I homeoprotein | 2.31 |
| Id1 | 2.17 |
Table 1.
Continued
| Nuclear | Fold change |
| AF-4 | 2.13 |
| Transcription factor E2A | 2.11 |
| Pax8* | 2.11 |
| Co-factors | |
| SWI/SNF complex 60-Kda subunit | 2.72 |
| Repressors | |
| HLH 1R21 helix-loop-helix protein | 3.64 |
| Zinc-finger protein bcl-6 | 3.21 |
| Transcription/translation-associated elements | |
| Eukaryotic initiation factor 4All | 2.65 |
| hnRNPcore protein | 2.51 |
| mRNA for Werner syndrome-1/type 4 | 2.45 |
| Translation initiation factor eIF-2 | 2.02 |
| Oncogenes/Oncogenic markers | |
| Oncogene TIs/Chop, Fusion Activated* | 4.46 |
| Meningioma-expressed antigen | 3.25 |
| HHCPA78[acute promyelocytic leukemia] | 3.02 |
| Human DD96 | 2.72 |
| CD24 signal transducer | 2.19 |
| Metastasis-associated[highly metastatic lung cells] | 2.11 |
| Immune related | |
| hBD-1 protein | 3.73 |
| Human complement component C4A | 3.33 |
| Human interferon-inducible protein | 2.76 |
| Ly-6-related protein | 2.54 |
| Autotaxin-t | 2.30 |
| Interferon-inducible gene family | 2.22 |
| HLA-DMB | 2.06 |
| 4F2 glycosylated heavy chain antigen | 2.04 |
| Intracellular | |
| Human Rad | 2.93 |
| Retinole acid-binding protein 11 | 2.77 |
| Butyrophilin | 2.43 |
| Calcium modulating cyclophilin ligand | 2.31 |
| Apolipoprotein E | 2.14 |
| DNA-PK interaction protein | 2.10 |
| Lysosome-associated membrane protein-2 | 2.07 |
| Gp25L2 protein | 2.03 |
| Undefined | |
| GOS2 gene | 4.48 |
| Hypothetical protein A4 | 2.61 |
| ARSE mRNA | 2.45 |
| mRNA for KIAA0207 | 2.22 |
| mRNA for KIAA0382 | 2.19 |
| mRNA for KIAA0099 | 2.08 |
| mRNA for KIAA0279 | 2.05 |
Denotes genes also significantly up-regulated after 12 hours CsA treatment.
TGF-β1 ELISA
TGF-β1 in HK-2 cell supernatants was quantitatively measured by specific ELISA method according to the manufacturer’s instructions.
Online Supplemental Material
Microarray data files are available for review from the National Center for Biotechnology Information gene expression data repository (GEO, http://www.ncbi.nlm.nih.gov/geo/). The accession numbers for the data sets are; control array, GSM23216; 12 hour array, GSM23217; 48 hour array, GSM23218.
Results
Effect of CsA on the Viability of Proximal Tubular Cells
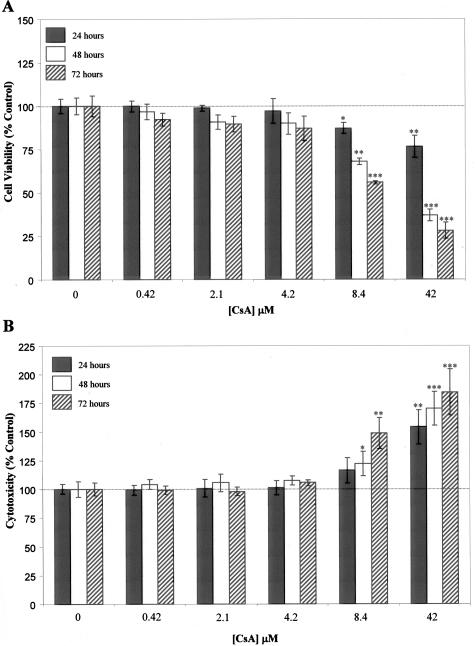
Initially, dose-dependent effects of CsA on PTEC viability were examined. HK-2 cells were cultured in the absence and presence of CsA (0.42, 2.1, 4.2, 8.4, 42 μmol/L) for 24, 48, and 72 hours. Cell viability was assessed by Alamar Blue assay with Alamar Blue reduction used as an index to quantify the amount of viable cells (Figure 1A). CsA was seen to have both time- and dose-dependent effects on HK-2 viability. At 4.2 μmol/L CsA and below, no statistically significant effects were observed on cell viability using the Alamar Blue assay. At 8.4 μmol/L and 42 μmol/L CsA, significant reductions in cell viability were observed at all time points. These results were further confirmed using the LDH assay kit. At 4.2 μmol/L CsA and below, no significant cytotoxic effect was observed. At 8.4 μmol/L CsA, significant cytotoxicity was observed from 48 hours on, whereas at 42 μmol/L CsA significant cytotoxicity was observed at all time points. From these findings, it was decided to further examine the effects of 4.2 μmol/L CsA as the highest noncytotoxic concentration.
Figure 1.
Effects of CsA on the viability of HK-2 cells. HK-2 cells were grown to confluency on 12-well tissue culture plates. Cells were cultured in the absence and presence of CsA for the indicated times with the indicated concentrations. Quantitation of viable cells by Alamar Blue reduction assay: A displays the effect of CsA on cell viability throughout time. Quantitation of cytotoxicity by LDH activity assay: B displays the effect of CsA on cytotoxicity throughout time. In both cases, results are expressed as percentage of time-matched control cells and are shown as mean ± SEM of three independent experiments performed in duplicate for each time point. A one-way analysis of variance was performed, multiple comparisons between control and treatment groups were made using the Bonferroni post test. *Statistically different to control (*P < 0.05, **P < 0.01, ***P < 0.001).
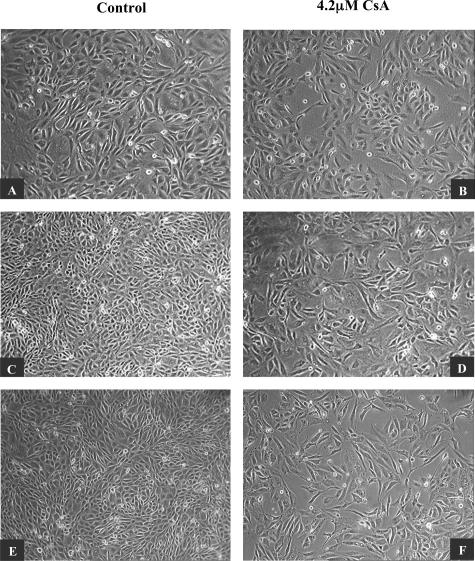
CsA Induced Morphological Changes in Proximal Tubular Cells
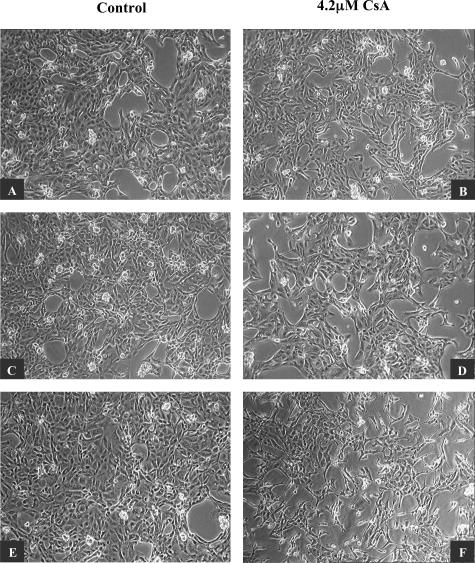
Cell morphology was assessed by phase contrast microscopy. Throughout time, control HK-2 cells formed a confluent monolayer exhibiting normal epithelial characteristics. Individual cells displayed typical polygonal shape with a high degree of attachment to neighboring cells (Figure 2; A, C, and E). Cells exposed to 4.2 μmol/L CsA exhibited striking morphological changes compared to control cells. At 24 hours, both control and treated cells display similar morphology, however by 48 hours morphological differences were detectable. By 72 hours, there were striking differences between the CsA-treated cells and their control counterparts. CsA-treated cells displayed a more elongated shape with a large degree of cell detachment (Figure 2; B, D, and F). These studies were performed in the same manner using human primary RPTECs (Figure 3). The effects of CsA were observed at the same time points. As in the HK-2 cell line, CsA induced morphological effects at 48 hours (Figure 3, C and D) in the primary cells. After 72 hours (Figure 3, E and F), CsA effects were even more pronounced, with treated cells displaying an extremely elongated and fibroblast-like morphology.
Figure 2.
CsA induced phenotypic changes in HK-2 cells. HK-2 cells were grown to confluency on 60-mm tissue culture dishes. Cells were cultured in the absence (A, C, E) and presence (B, D, F) of 4.2 μmol/L CsA for 24, 48, and 72 hours. Morphological changes were examined by phase-contrast microscopy. One representative experiment of at least three individual replicate experiments is shown. Original magnifications, ×100.
Figure 3.
CsA induced phenotypic changes in human primary RPTECs. RPTECs were grown to confluency on 60-mm tissue culture dishes. Cells were cultured in the absence (A, C, E) and presence (B, D, F) of 4.2 μmol/L CsA for 24, 48, and 72 hours. Morphological changes were examined by phase-contrast microscopy. One representative experiment of at least three individual replicate experiments is shown. Original magnifications, ×100.
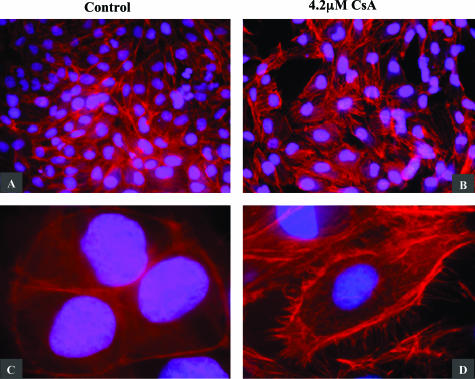
F-actin filaments determine the shape of a cell’s surface and are essential for the maintenance of cell polarity. Filaments are dispersed throughout the cell but in normal epithelial cells, are concentrated in the cytoplasmic cortex beneath the plasma membrane. Immunofluorescent microscopy was used to examine the effect of 4.2 μmol/L CsA on F-actin cytoskeletal arrangement in human proximal tubular cells. HK-2 cells were grown to confluency on Falcon 8-chamber slides and cultured for 48 hours in the absence and presence of 4.2 μmol/L CsA. In control cells, F-actin fibers were densely arranged just inside the cell periphery, where they associate to form a circular bundle. Slim central fibers are also visible (Figure 4, A and C). This organization is characteristic of cells with apical-basolateral polarity, such as normal epithelial cells. In cells exposed to 4.2 μmol/L CsA, the reorganization of the F-actin cytoskeleton can be seen. The dense peripheral arrangement of F-actin has been redistributed into strong central fibers termed stress fibers. The stress fibers are aligned in parallel, and at higher magnification can be seen apparently projecting out of the cell (Figure 4, B and D). F-actin fibers arranged in parallel contribute to the elongated shape of a cell, which is characteristic of cells with front end-back end polarity such as the myofibroblast.
Figure 4.
CsA induced alterations in HK-2 F-actin cytoskeletal arrangement. Cells were grown to confluency on Falcon 8-chamber glass slides. Cells were cultured in the absence (A, C) and presence (B, D) of 4.2 μmol/L CsA for 48 hours. F-actin cytoskeletal arrangement was examined by immunofluorescent microscopy as described in the Materials and Methods section. One representative experiment of at least three individual replicate experiments is shown. Original magnifications: ×200 (A, B); ×630 (C, D).
CsA Disrupted Junctional Structures in Proximal Tubular Cells
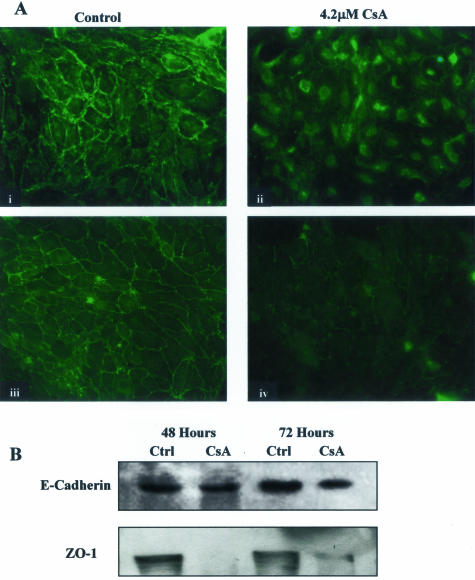
Junctional integrity is essential for normal epithelial barrier function. E-cadherin and β-catenin are key adherens junction components. Cell-cell contact is maintained through the interaction of the extracellular domains of cadherin molecules of neighboring epithelial cells. E-cadherin is the most commonly expressed cadherin in epithelial cells and is a typical epithelial marker. β-Catenin is crucial for two reasons: the anchorage of transmembrane cadherins to the actin cytoskeleton at adherens junctions and the regulation of gene expression through Wnt signaling. Exposure to CsA for 48 hours caused a slight down-regulation of E-cadherin expression levels. After 72 hours of exposure, a significant decrease in E-cadherin expression levels in HK-2 cells was observed, suggesting reduced cell attachment and a loss of epithelial phenotype (Figure 5B). In the normal epithelial monolayer (Figure 5A, i), β-catenin can be seen at the cell periphery where it acts as part of a linking complex between actin filaments and transmembrane cadherins to maintain junctional integrity. When cells were exposed to CsA for 72 hours, the translocation of β-catenin from its normal junctional position to a nuclear or perinuclear position was evident indicating the disruption of the adherens junctions. This translocation can be seen as intense staining around the nucleus (Figure 5A, ii). In terms of cell morphology, β-catenin translocation demonstrates the dissociation of junctional complexes. However, the movement of β-catenin may also suggest the activation of Wnt signaling. In addition, exposure to 4.2 μmol/L CsA resulted in a significant reduction in the expression of the major tight junction protein ZO-1 after both 48 and 72 hours [Figure 5, A (iii, iv) and B]. The effect of CsA on ZO-1 appears to be more severe than the effects on the adherens junction proteins suggesting that the tight junction may be particularly susceptible to CsA-induced changes.
Figure 5.
CsA disrupted junctional structures in HK-2 cells. HK-2 cells were grown to confluency on 60-mm tissue culture dishes. Cells were exposed to 4.2 μmol/L CsA for the indicated time periods and lysed. A: The effect of 4.2 μmol/L CsA on junctional integrity was assessed by immunofluorescent microscopic examination of β-catenin localization (i, ii) and ZO-1 localization (iii, iv). Images shown are representative of at least eight experiments with similar results. B: Equal amounts of whole cell lysate were subjected to Western analysis using an anti-E-cadherin antibody and an anti-ZO-1 antibody, respectively. One representative experiment of at least three individual replicate experiments is shown. Original magnifications, ×200 (i–iv).
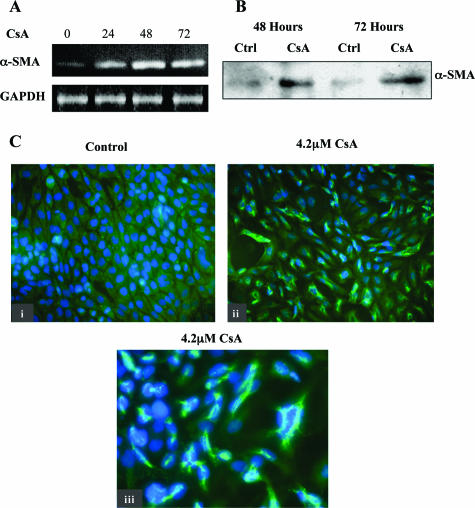
CsA Induced de Novo Synthesis of α-SMA in Proximal Tubular Cells
α-SMA is a phenotypic marker of myofibroblast cells not normally evident in epithelial cells. α-SMA expression is a feature of advanced stages of EMT and a defining characteristic of a true transdifferentiation event. The production of α-SMA and the consequent assembly of α-SMA stress fibers are likely to be critical to the generation of contractile force and hence the motility of a myofibroblast. In human renal proximal tubular cells, incubation with 4.2 μmol/L CsA induced de novo α-SMA mRNA production throughout 48 and 72 hours (Figure 6A). This de novo expression was also evident at the protein level after 48 hours, detected by Western blot analysis (Figure 6B). In addition, the appearance of α-SMA protein was visualized by immunofluorescent microscopy (Figure 6C, ii) as compared with control cells (Figure 6C, i). At a magnification of ×630, the formation of α-SMA fibers is visible as early as 48 hours after treatment (Figure 6C, iii).
Figure 6.
CsA induces de novo α-SMA expression in HK-2 cells. HK-2 cells were grown to confluency on 60-mm tissue culture dishes. Cells were exposed to 4.2 μmol/L CsA for the indicated time periods and lysed or total RNA extracted. A: RT-PCR was performed using primers designed specifically for α-SMA and GAPDH as described in Materials and Methods. The result shown is representative of at least three experiments with similar results. B: Equal amounts of whole cell lysates were subjected to Western analysis using an anti-α-SMA antibody. The blot shown is representative of at least three experiments with similar results. C: Control (i) or CsA-treated (ii, iii) cells were fixed and stained for α-SMA as described in Materials and Methods. One representative experiment of at least three individual replicate experiments is shown. Original magnifications: ×400 (i, ii); ×630 (iii).
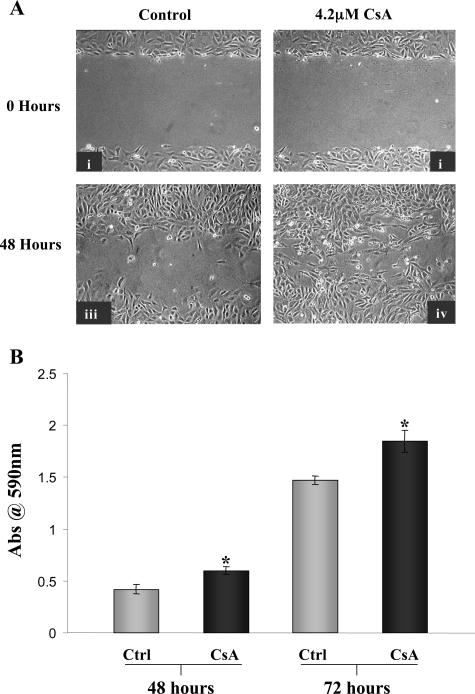
CsA Enhanced the Motility of Proximal Tubular Cells
Epithelial cells grown to confluency in vitro to form a monolayer are relatively static as migratory signals are inhibited by contact with neighboring cells. The wound-healing assay can be used to visualize the migratory ability of cells grown to confluency and then experimentally wounded by scratching a track through the monolayer. Cells at the edge of the wound become migratory and move to repair the damage. Cells exposed to CsA (Figure 7A, ii and iv) were seen to migrate across the wound at a faster rate and in greater numbers than control cells (Figure 7A, i and iii). To assess this in a quantitative manner, a transwell migration assay was performed. HK-2 cells were exposed to 4.2 μmol/L CsA apically for 48 and 72 hours (Figure 7B). At both time points, the migration of cells was significantly higher with CsA treatment as compared with time-matched controls (P < 0.05). This observation is perhaps more significant because the transwell assay assessed the effects of CsA on a normal monolayer of cells as opposed to cells that have been primed for migratory signals in the wound-healing assay by disrupting the confluent monolayer.
Figure 7.
CsA enhanced the migratory potential of HK-2 cells. Analysis of cell motility by wound healing assay. HK-2 cells were grown to confluency on 60-mm tissue culture dishes and experimentally wounded. A: i and ii are phase contrast microscopic analysis of cells immediately after wound formation. Cells were cultured in the absence (i, iii) and presence (ii, iv) of 4.2 μmol/L CsA for 48 hours. B: The effect of 4.2 μmol/L CsA on HK-2 motility as assessed by transwell migration assay. Cells were seeded on polyvinylpyrrolidone-free polycarbonate filters with 8.0-μm pore size. Cells were grown in the absence and presence of 4.2 μmol/L CsA for the indicated time periods. Results shown as mean ± SEM of four independent experiments performed in duplicate for each time point. A one-way analysis of variance was performed, multiple comparisons between control and treatment groups were made using the Bonferroni post test. *Statistically different (P < 0.05) to control.
Differential Gene Expression in Cells Treated with CsA
To identify expression alterations involved in CsA-mediated EMT, large-scale gene expression analysis of the HK-2 cell model was performed using Affymetrix GeneChip technology. Three independent RNA samples were prepared, respectively, in independent experiments at each time point and pooled. Isolated RNA was labeled and hybridized to the Affymetrix HuGene oligonucleotide probe arrays. To minimize the occurrence of false-positives, only genes presenting linear patterns of expression over the three time points were considered, when a signal threshold was reached. Data analysis was performed using the Array suite 4 from Affymetrix and GeneSpring from Silicon Genetics as described in the Materials and Methods.
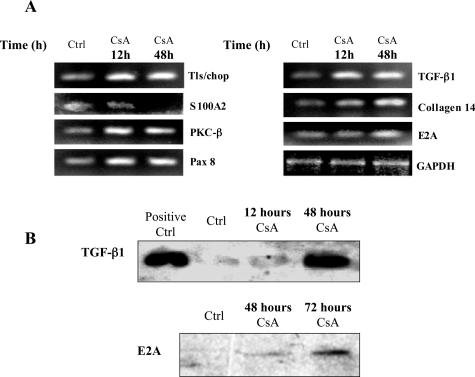
Of the 7070 genes represented on the microarray, 128 genes were significantly differentially regulated after 48 hours of exposure to CsA (93 up, for than twofold; 35 down, more than twofold). These genes are listed in Tables 1 and 2. Differentially expressed genes were from a range of different functional classes. A number of genes from classes of particular interest were assessed by RT-PCR to confirm the accuracy of the array data (Figure 8A). The primer sequences for these genes are given in Table 3. The time course for changes in expression observed by PCR analysis on selected genes paralleled those observed in the array data with some genes exhibiting early responses (12 hours after treatment, denoted by an asterisk in Table 1) while the differential expression of other genes appears to be a later effect (48 hours after treatment).
Table 2.
Genes Down-regulated More Than Two Fold in Response to Treatment with 4.2 μmol/L CsA for 48 Hours
| Enzymes | Fold change |
| Cytosolic acetoacetyl-coenzyme A thiolase | 4.90 |
| Tissue plasminogen activator (PLAT) | 3.52 |
| Squalene epoxidase | 2.33 |
| Ornithine decarboxylase | 2.30 |
| Glutamine PRPP amidotransferase | 2.12 |
| ADE2H1 homology to SAICAR synthetase | 2.10 |
| Dual-specificity protein phosphatase | 2.07 |
| Deoxyguanosine kinase | 2.05 |
| DNA topoisomerase 1 | 2.00 |
| Cytokinase and secreted proteins | |
| Heparin-binding EGF-like growth factor | 2.18 |
| Membrane associated | |
| [Alpha]tubulin | 3.72 |
| Transferrin receptor | 3.61 |
| Neurofilament-66 | 2.17 |
| Annexin II | 2.09 |
| Myosin light chain 2 | 2.06 |
| Nuclear | |
| Transcription factors/Co-factors | |
| HMGI-C protein | 3.58 |
| Nuclear localization sequence receptor | 2.63 |
| Acid finger protein | 2.11 |
| Hepatocyte nuclear factor-3/fork head homolog 11A | 2.05 |
| Centromere protein-A | 2.05 |
| Human C2f | 2.04 |
| Intracellular | |
| Cell cycle | |
| Cyclin B | 2.77 |
| p55CDC | 2.75 |
| Cyclin F | 2.45 |
| Cyclin A2 | 2.35 |
| Other | |
| S100A2 gene | 4.10 |
| Cyr61 gene | 3.01 |
| H105e3 | 2.73 |
| [Mu]crystallin | 2.54 |
| Pre-mRNA splicing factor | 2.44 |
| Heterogeneous Nuclear Ribonucleoprotein | 2.36 |
| Nucleolar protein p40 | 2.32 |
| Nm23 gene | 2.21 |
| Ubiquitin fusion-degradation protein | 2.03 |
| Undefined | |
| mRNA for KIAA0095 | 2.20 |
Figure 8.
Effect of CsA on the expression profile of HK-2 cells. HK-2 cells were grown to confluency on 60-mm tissue culture dishes. Cells were exposed to 4.2 μmol/L CsA for the indicated time periods and total RNA was extracted. A: Semiquantitative RT-PCR analysis of mRNA levels of a range of genes identified by microarray analysis was performed. GAPDH served as an internal control. B: Equal amounts of whole cell lysates were subjected to Western analysis using an anti-TGF-β and E2A antibodies. One representative experiment of at least three individual replicate experiments is shown.
Table 3.
Sequence of Primers for RT-PCR Analysis of Selected Genes
| Gene | Forward primer | Reverse primer |
|---|---|---|
| TLS/FUS | ACGTCATGACTCCGAACAGG | AAAGAGACCGTTGCCTCTCC |
| S100A2 | GGGCTGAAGAAGCTGATGG | CTCAAAGGCATCAACAGTCC |
| PKC-β | CCCCATGCTGTATTTTACGC | GGCGATGTAGTCTGGAGTGC |
| Pax 8 | GAGCAACAGGAGGACTCAGC | CATGGCATGGTTCTCTTTCC |
| TGF-β1 | CCCTGGACACCAACTATTGC | GTCCAGGCTCCAAATGTAGG |
| E2A | CATGGAGCAGAGGTGAACG | GAGTAGATCGAGGCCAGTGC |
| Collagen XIV | ACGCCATGTCTTCTTTGTGG | GCTCCATAGAAACCCCTTCC |
| α-SMA | GCGTGGCTATTCCTTCGTTAC | CATAGTGGTGCCCCCTGATAG |
| GAPDH | ACCACAGTCCATGCCATCAC | TCCACCACCCTGTTGCTGTA |
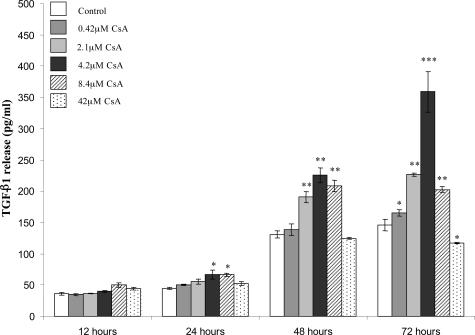
A number of important profibrotic elements were significantly up-regulated. Among these were TGF-β1. CsA treatment caused significant up-regulation of TGF-β1 mRNA levels by 12 hours and by 48 hours this up-regulation had increased by almost threefold. This up-regulation of TGF-β1 was also confirmed at the protein level (Figure 8B). We examined the effect of increasing concentrations of CsA on the levels of TGF-β1 protein secreted by PTECs using a TGF-β1-specific ELISA (Figure 9). TGF-β1 release was seen to increase in a time- and dose-dependent manner at CsA concentrations up to 4.2 μmol/L CsA. At 8.4 μmol/L and 42 μmol/L CsA this trend was not preserved, especially at the later time point, most likely due to CsA cytotoxicity at these concentrations. Collagen XIV also called undulin, was significantly up-regulated by exposure to CsA (Figure 8A). Collagen XIV is a member of the fibronectin-tenascin family of extracellular glycoproteins that account for a large proportion of fibrotic lesion content. PKC-β mRNA was significantly up-regulated by CsA treatment (Figure 8A). PKC-β may play an essential role in this model because its up-regulation is an early event and may be key to the propagation of downstream effects. This up-regulation was also detected at the protein level (Figure 10A). Interestingly, a number of transcriptional regulators were induced, particularly mRNA for the E2A gene that was up-regulated by the 48-hour time point. Changes in E2A protein levels were also detected after 48 and 72 hours of CsA exposure (Figure 8B). The role that E2A gene products may play in our model will be discussed later. The paired box protein, Pax8, which was significantly up-regulated by the 12-hour time point, is a key regulator in the developing kidney (Figure 8A). Pax8 is not believed to play an active role in the adult kidney but, on reactivation, may have a role to play in renal disease development. A number of genes proposed to be involved in the development of tumor cells were significantly up-regulated. The TLS/FUS oncogene was strongly up-regulated early in the course of the changes observed (Figure 8A). TLS/FUS plays a critical role in the pathogenesis of liposarcoma.
Figure 9.
CsA-induced up-regulation of TGF-β1 in HK-2 cells is dose-dependent. HK-2 cells were grown to confluency on 60-mm tissue culture dishes. Cells were exposed to a range of CsA concentrations for the indicated time periods. TGF-β1 protein levels in supernatants were assessed using a TGF-β1-specific ELISA as described in the Materials and Methods. Results are shown as mean ± SEM of three independent experiments performed in duplicate for each time point. A one-way analysis of variance was performed, multiple comparisons between control and treatment groups were made using the Bonferroni post test. *Statistical difference to control (*P < 0.05, **P < 0.01, ***P < 0.001).
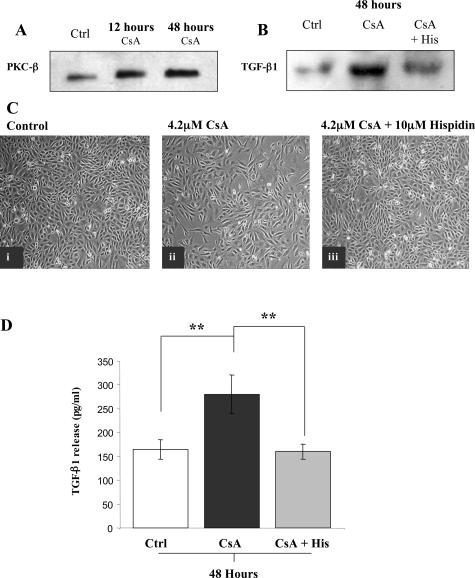
Figure 10.
CsA induced up-regulation of PKC-β and inhibition of PKC-β attenuates CsA-induced TGF-β1 induction and morphological effects on HK-2 cells. HK-2 cells were grown to confluency on 60-mm tissue culture dishes. Cells were exposed to 4.2 μmol/L CsA or a combination of 4.2 μmol/L CsA and 10 μmol/L hispidin for the indicated time periods. Control cells were exposed to an appropriate concentration of vehicle [ethanol (A) or ethanol plus dimethyl sulfoxide (B–D)]. Cell supernatants and lysates were collected. A: Equal amounts of whole cell lysates were subjected to Western analysis using an anti-PKC-β antibody. B: Equal amounts of concentrated supernatants were subjected to Western analysis using an anti-TGF-β antibody. C: Cells were cultured in the absence (i) and presence (ii and iii) of 4.2 μmol/L CsA for 48 hours. In iii, cells were pretreated with 10 μmol/L hispidin for 1 hour before CsA treatment. Cell morphology was assessed by phase contrast microscopy. Cell supernatants were collected and concentrated. D: TGF-β1 protein levels in supernatants were assessed using a TGF-β1-specific ELISA as described in the Materials and Methods. Results are shown as mean ± SEM of three independent experiments performed in duplicate for each time point. A one-way analysis of variance was performed, multiple comparisons between control and treatment groups were made using the Bonferroni post test. *Statistical difference to control (*P < 0.05, **P < 0.01, ***P < 0.001). In A–C, one representative experiment of at least three individual replicate experiments is shown. Original magnifications, ×100 (A–C).
CsA treatment of PTECs also resulted in the down-regulation of 36 genes (Table 2). Down-regulated genes identified included transcriptional regulators, various structural molecules, and genes involved in the regulation of the cell cycle. A number of genes involved in normal cell metabolism were also down-regulated, as were several uncharacterized genes. Of particular interest was the reduced expression of the S100A2 gene. The product of this gene acts as a modulator against excess calcium accumulation in normal human epithelial cells and is thought to also play a role in suppressing tumor cell growth. α-Tubulin, a key component of microtubules was significantly down-regulated as were a number of cyclins. B-cyclins are involved in the apoptotic decision whereas A-cyclins are involved in G0/M phase transitions, perhaps suggesting an increased potential for proliferation in the cells. Hepatocyte growth factor-3, down-regulated by CsA treatment, is a transcriptional regulator of a number of cyclins and its down-regulation may be responsible for decreased cyclin expression. The high mobility group isoform C protein belongs to the nonhistone chromosomal high mobility group protein family. Identification of the deletion and rearrangement of this gene that are associated with myxoid liposarcoma suggests a role in adipogenesis and mesenchymal differentiation.
PKC-β Inhibition Abrogated CsA-Induced TGF-β1 Up-Regulation and Phenotypic Effects in Proximal Tubular Cells
Up-regulation of PKC-β by CsA was detected at the mRNA and protein level (Figures 8 and 10). We therefore examined the effects of inhibition of PKC-β. Figure 10 illustrates the effect of pretreatment with a PKC-β-specific inhibitor, hispidin, on CsA-induced morphological changes to HK-2 proximal tubular cells. Cells exposed to 4.2 μmol/L CsA for 48 hours displayed striking morphological changes, as described above (Figure 10C, ii, compared to vehicle control, Figure 10C, i). In contrast, cells pretreated with 10 μmol/L hispidin for 1 hour and then exposed to 4.2 μmol/L CsA for 48 hours appeared to retain the normal phenotype associated with PTECs (Figure 10C, iii). Cells exposed to hispidin (10 μmol/L) alone, did not show any alterations in morphology (data not shown). To further investigate the mechanism of this protection, supernatants were collected from cells during the experiment and assayed for TGF-β1 levels by Western analysis (Figure 10B) and ELISA (Figure 10D). Cells that were pretreated with hispidin secreted significantly less TGF-β1 than cells treated with CsA alone. Therefore, pretreatment with the PKC-β inhibitor completely blocked the CsA-induced increase in secreted TGF-β1 from treated cells. Cells exposed to hispidin (10 μmol/L) alone, did not show any significant changes in TGF-β1 secretion (data not shown).
Discussion
EMT is now acknowledged to play a significant role in a number of disease states including renal fibrosis and tumor metastasis.13,14,17 The development of renal fibrosis with long-term CsA use is now well established.4,5 With analysis of patient outcomes, the enhancement of tumor formation in long-term CsA users has been recognized.22 In this study, we have demonstrated that exposure to a clinically relevant dose of the immunosuppressant agent CsA, provokes an EMT event in human proximal tubular cells.
PTECs may no longer be regarded as passive victims in renal disease, but as active contributors to renal fibrosis.23 PTECs are capable of secreting various profibrotic cytokines in response to certain stimuli. However, the ability of PTECs to undergo EMT suggests a more significant role. After EMT, the myofibroblast is free to migrate into the interstitium where it can release significant amounts of ECM, factors promoting fibrosis development and indeed, factors inducing other epithelial cells to undergo EMT, forming part of a vicious cycle of fibrosis.24 The development of renal fibrosis associated with CsA treatment has previously been attributed to a number of indirect factors including angiotensin II induction and nitric oxide insufficiency.25 However, this present study demonstrates direct profibrotic effects of CsA on tubular epithelial cells that would also contribute significantly to fibrotic development. Johnson and colleagues26 reported profibrotic effects of CsA on primary PTECs with up-regulation of TGF-β1. This present study is the first demonstration that CsA can induce EMT in PTECs. The findings of this current study may have implications beyond those relating to CsA-induced renal fibrosis. Vaquero and colleagues27 reported that CsA-treated rats displayed significantly impaired pancreatic repair with marked increases in α-SMA-expressing interstitial cells in a model of pancreatitis. It is possible that some of the mediators identified in our model are relevant to other disease states giving rise to transdifferentiation of epithelial cells, including malignant tumor development and metastasis.
EMT is currently viewed as the end result of a number of diverse signals acting on the PTEC. EMT can be induced in vitro by a range of stimuli including fibroblast growth factor, advanced glycation end products, factors released by activated immune cells, and TGF-β1.28–31 The variety of EMT inducers seems to suggest a multitude of potential mechanisms. However, Yang and colleagues15 have succinctly described key events critical to EMT progression, suggesting common downstream mechanisms must be involved regardless of the initiating factors. To gain insight into the mechanisms involved in this novel model of CsA-induced EMT, we performed a time course analysis of the alterations in gene expression due to CsA treatment throughout 12 and 48 hours. The direct effects of CsA are likely reflected in relatively early events at the transcriptional level, detected at the 12-hour time point. Equally, by assessing the later changes in gene expression, perhaps not caused directly by CsA but by CsA-induced effectors, a greater insight can be gained into some of the mechanisms involved in EMT.
As mentioned previously, there is now a large body of evidence linking CsA treatment to increased incidence of certain tumors.18 Compelling evidence is accumulating to support the idea that this increase is not solely reliant on the suppression of the host immune system but may be related to the direct cellular effects of CsA.32 Because EMT is now viewed as the secondary mechanism of tumor development,33 our model may reveal mechanisms that are also relevant in this context.
TGF-β1 is widely proposed to be a major mediator of CsA-induced renal fibrosis.34 The profibrotic profile of TGF-β1 is well described, including its ability to induce EMT in cells of various types.35 Hojo and colleagues32 reported that the use of a TGF-β1-neutralizing antibody abrogated the prometastatic effects of CsA in vitro and in vivo. Because the EMT events observed in our model are a late response (48 hours after treatment and later), we believe that a secondary signaling molecule, induced by CsA is the main stimulus. The present findings of dose- and time-dependent effects of CsA on TGF-β1 secretion provide further evidence that TGF-β1 may be a key mediator of CsA-induced EMT.
The effects of CsA and similar immunosuppressive agents have long been viewed as unique effects on T cells involving the inhibition of genes encoding factors affecting T-cell growth and differentiation via calcineurin inhibition. However, the weight of evidence now suggests that CsA is capable of both inducing and repressing the expression of a wide range of genes in various cell types, in both calcineurin-dependent and -independent manners.36 Mascarell and colleagues37 reported the differential regulation of more than 100 proteins by CsA in activated T cells, a figure in close agreement with this present report. Tomita and colleagues38 demonstrated that CsA blocked chondrocyte differentiation in a calcineurin-dependent manner resulting in the mesenchymal precursor being preserved, an effect that is perhaps related to the mechanisms we now report. Because EMT is a gross change in cell phenotype, it follows that the key events governing the changes occur at the level of gene expression. In the present study, a number of transcriptional regulators were differentially regulated by CsA treatment. The E2A gene was identified as being up-regulated in response to CsA exposure. The E2A gene encodes two basic helix-loop-helix (bHLH) transcription factors, E12 and E47.39 In combination with other bHLH factors, as homodimers or heterodimers, E12/E47 can interact with E-boxes in the promoters of target genes to modulate their transcription.40 E2A products have been identified as being involved in the regulation of a number of genes. Perez-Moreno and colleagues41 demonstrated that transfection of MDCK renal epithelial cells with a construct encoding the E47 transcript caused EMT, resulting from repression of E-cadherin expression. In addition, Kumar and colleagues42 have demonstrated the involvement of E2A in α-SMA induction in smooth muscle cells in vivo. This suggests that up-regulation of E2A may play a key role in CsA-induced EMT, mediating E-cadherin down-regulation and α-SMA induction. This hypothesis is further supported by investigations into the expression pattern of E2A in human tissues. Rutherford and colleagues43 reported that E2A proteins were not detectable in adult human kidney samples. However, E2A expression was detected in the first trimester kidney samples, most abundantly in primitive renal tubules, epithelioid cells, and renal interstitial cells. This would support the notion of the reactivation of developmental pathways in an effort to facilitate repair in the damaged kidney. Given the importance of E2A in lymphocyte development and IgG isotype switching,44,45 this may also identify a novel mechanism of CsA’s immunosuppressive effects. Further investigation will reveal if E2A induction is a direct effect of CsA or TGF-β1-dependent.
Up-regulation of PKC-β was an early event in our model, in response to CsA treatment. CsA-mediated activation of PKC isoforms has been described in a rat model of CsA-induced vascular dysfunction.46 PKC-β plays a role in normal B-cell and T-cell function.19 In addition, PKC-β isoenzymes are believed to play a role in the development of fibrotic disorders associated with diabetes, including diabetic nephropathy.47 PKC-β isoenzymes are now under investigation as potential therapeutic targets in the treatment of a number of such disorders. The effects of PKC-β activation have been reported to be mediated, at least in part through regulation of ERK signaling cascade.48 We have previously demonstrated the involvement of ERK1/2 in CsA-mediated effects in MDCK cells.49 In addition, we have previously reported that CsA induction of TGF-β1 in PTECs was abrogated through use of an ERK1/ERK2 inhibitor.50 We now report that CsA-induced TGF-β1 up-regulation is abrogated by treatment with a PKC-β inhibitor hispidin. These observations suggest that in PTECs, CsA induction of TGF-β1 may occur via a cascade of signals involving PKC-β activation, leading to ERK activation resulting in increased transcription of the TGF-β1 gene, perhaps due to nuclear factor AP-1 activation. AP-1 factor activation by ERK signaling, has been shown to up-regulate TGF-β1 expression,51 although there is evidence that CsA exposure prolongs the presence of AP-1 factors in the nucleus.52 The fact that pretreatment with the PKC-β inhibitor also protected against many of the morphological effects observed with CsA treatment alone, further supports the theory that the EMT events observed are mediated primarily by TGF-β1. PKC-β has been suggested to play a role in colon carcinogenesis as mice overexpressing the gene are highly susceptible to these tumors, whereas in normal mice, PKC-β up-regulation was an early event in carcinogenesis.53 Increased PKC activity has been reported to result in decreased E-cadherin expression in MDCK renal epithelial cells, due to increased E-cadherin endocytosis and decreased trafficking of E-cadherin to the cell membrane.54 Taken in conjunction with the ability of E2A to repress E-cadherin expression at a transcriptional level, it appears that, in this model of EMT, E-cadherin’s role in maintaining cell-cell contact is being severely compromised. Currently, the mechanism by which CsA caused PKC-β up-regulation in our model is under investigation. Elucidation of these mechanisms will identify potential targets for therapeutic intervention.
In conclusion, we provide evidence that CsA induced EMT in these PTECs. Potential mediators and downstream effectors of CsA-induced effects were identified by large-scale expression analysis using Affymetrix microarrays. The findings of this study suggest new potential mediators of CsA nephrotoxicity and new potential mechanisms of CsA immunosuppression. The present findings identified PKC-β as a potentially important mediator, which may be responsible for CsA-induced TGF-β1 up-regulation. In addition, the E2A transcription factors E12/E47, may play a key role in the altered expression profile of CsA-treated cells and thus, cell phenotype. Understanding the mechanisms involved in CsA-induced EMT may lead to the development of novel therapeutic targets and strategies for the treatment of renal fibrosis.
Footnotes
Address reprint requests to Michael P. Ryan, Department of Pharmacology, Conway Institute of Biomolecular and Biomedical Research, University College Dublin, Belfield, Dublin 4, Ireland. E-mail: michael.p.ryan@ucd.ie.
Supported by the Conway Institute of Biomolecular and Biomedical Research; the Dublin Molecular Medicine Centre, under the Higher Education Authority Programe for Research in Third Level Institutions; the Health Research Board of Ireland, Enterprise, Ireland; the Irish Nephrological Society; and Amgen Ltd.
References
- Kronke M, Leonard WJ, Depper JM, Arya SK, Wong-Staal F, Gallo RC, Waldmann TA, Greene WC. Cyclosporin A inhibits T-cell growth factor gene expression at the level of mRNA transcription. Proc Natl Acad Sci USA. 1984;81:5214–5218. doi: 10.1073/pnas.81.16.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren SB, Bosmans JL, Elseviers MM, Verpooten GA, De Broe ME. A meta-analysis and morphological review of cyclosporine-induced nephrotoxicity in auto-immune diseases. Kidney Int. 1998;54:536–545. doi: 10.1046/j.1523-1755.1998.00017.x. [DOI] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Bennett WM. The nephrotoxicity of immunosuppressive drugs. Clin Nephrol. 1995;43(Suppl 1):S3–S7. [PubMed] [Google Scholar]
- Bennett WM, DeMattos A, Meyer MM, Andoh T, Barry JM. Chronic cyclosporine nephropathy: the Achilles’ heel of immunosuppressive therapy. Kidney Int. 1996;50:1089–1100. doi: 10.1038/ki.1996.415. [DOI] [PubMed] [Google Scholar]
- Remuzzi G, Perico N. Cyclosporine-induced renal dysfunction in experimental animals and humans. Kidney Int Suppl. 1995;52:S70–S74. [PubMed] [Google Scholar]
- Myers BD, Ross J, Newton L, Luetscher J, Perlroth M. Cyclosporine-associated chronic nephropathy. N Engl J Med. 1984;311:699–705. doi: 10.1056/NEJM198409133111103. [DOI] [PubMed] [Google Scholar]
- Oriji GK, Keiser HR. Role of nitric oxide in cyclosporine A-induced hypertension. Hypertension. 1998;32:849–855. doi: 10.1161/01.hyp.32.5.849. [DOI] [PubMed] [Google Scholar]
- Healy E, Dempsey M, Lally C, Ryan MP. Apoptosis and necrosis: mechanisms of cell death induced by cyclosporine A in a renal proximal tubular cell line. Kidney Int. 1998;54:1955–1966. doi: 10.1046/j.1523-1755.1998.00202.x. [DOI] [PubMed] [Google Scholar]
- Keane T, Egan D, Ryan MP. Signalling pathways involved in cyclosporine A-induced TGF-β1 secretion from a renal tubular epithelial cell line. J Am Soc Nephrol. 1998;9:A3045. [Google Scholar]
- Hewitson TD, Becker GJ. Interstitial myofibroblasts in IgA glomerulonephritis. Am J Nephrol. 1995;15:111–117. doi: 10.1159/000168813. [DOI] [PubMed] [Google Scholar]
- Essawy M, Soylemezoglu O, Muchaneta-Kubara EC, Shortland J, Brown CB, el Nahas AM. Myofibroblasts and the progression of diabetic nephropathy. Nephrol Dial Transplant. 1997;12:43–50. doi: 10.1093/ndt/12.1.43. [DOI] [PubMed] [Google Scholar]
- Strutz F, Muller GA. Transdifferentiation comes of age. Nephrol Dial Transplant. 2000;15:1729–1731. doi: 10.1093/ndt/15.11.1729. [DOI] [PubMed] [Google Scholar]
- Rastaldi MP, Ferrario F, Giardino L, Dell’Antonio G, Grillo C, Grillo P, Strutz F, Muller GA, Colasanti G, D’Amico G. Epithelial-mesenchymal transition of tubular epithelial cells in human renal biopsies. Kidney Int. 2002;62:137–146. doi: 10.1046/j.1523-1755.2002.00430.x. [DOI] [PubMed] [Google Scholar]
- Yang J, Liu Y. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am J Pathol. 2001;159:1465–1475. doi: 10.1016/S0002-9440(10)62533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer B, Valles AM, Edme N. Induction and regulation of epithelial-mesenchymal transitions. Biochem Pharmacol. 2000;60:1091–1099. doi: 10.1016/s0006-2952(00)00427-5. [DOI] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Haberal M, Karakayali H, Emiroglu R, Basaran O, Moray G, Bilgin N. Malignant tumors after renal transplantation. Artif Organs. 2002;26:778–781. doi: 10.1046/j.1525-1594.2002.07070.x. [DOI] [PubMed] [Google Scholar]
- Long A, Kelleher D, Lynch S, Volkov Y. Cutting edge: protein kinase C beta expression is critical for export of IL-2 from T cells. J Immunol. 2001;167:636–640. doi: 10.4049/jimmunol.167.2.636. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Takahara S, Hatori M, Suzuki K, Wang J, Ichimaru N, Suzuki S, Morozumi K, Okuyama A, Yamanaka H. A novel immunosuppressive drug, FTY720, prevents the cancer progression induced by cyclosporine. Cancer Lett. 2002;181:165–171. doi: 10.1016/s0304-3835(01)00799-6. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Nabel GJ. A transformed view of cyclosporine. Nature. 1999;397:471–472. doi: 10.1038/17207. [DOI] [PubMed] [Google Scholar]
- Becker GJ, Hewitson TD. The role of tubulointerstitial injury in chronic renal failure. Curr Opin Nephrol Hypertens. 2000;9:133–138. doi: 10.1097/00041552-200003000-00006. [DOI] [PubMed] [Google Scholar]
- Lan HY. Tubular epithelial-myofibroblast transdifferentiation mechanisms in proximal tubule cells. Curr Opin Nephrol Hypertens. 2003;12:25–29. doi: 10.1097/00041552-200301000-00005. [DOI] [PubMed] [Google Scholar]
- Shihab FS. Cyclosporine nephropathy: pathophysiology and clinical impact. Semin Nephrol. 1996;16:536–547. [PubMed] [Google Scholar]
- Johnson DW, Saunders HJ, Johnson FJ, Huq SO, Field MJ, Pollock CA. Fibrogenic effects of cyclosporin A on the tubulointerstitium: role of cytokines and growth factors. Exp Nephrol. 1999;7:470–478. doi: 10.1159/000020626. [DOI] [PubMed] [Google Scholar]
- Vaquero E, Molero X, Tian X, Salas A, Malagelada JR. Myofibroblast proliferation, fibrosis, and defective pancreatic repair induced by cyclosporin in rats. Gut. 1999;45:269–277. doi: 10.1136/gut.45.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy E, Leonard M, Madrigal-Estebas L, O’Farrelly C, Watson AJ, Ryan MP. Factors produced by activated leukocytes alter renal epithelial cell differentiation. Kidney Int. 1999;56:1266–1269. doi: 10.1046/j.1523-1755.1999.00694.x. [DOI] [PubMed] [Google Scholar]
- Oldfield MD, Bach LA, Forbes JM, Nikolic-Paterson D, McRobert A, Thallas V, Atkins RC, Osicka T, Jerums G, Cooper ME. Advanced glycation end products cause epithelial-myofibroblast transdifferentiation via the receptor for advanced glycation end products (RAGE). J Clin Invest. 2001;108:1853–1863. doi: 10.1172/JCI11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutz F, Zeisberg M, Ziyadeh FN, Yang CQ, Kalluri R, Muller GA, Neilson EG. Role of basic fibroblast growth factor-2 in epithelial-mesenchymal transformation. Kidney Int. 2002;61:1714–1728. doi: 10.1046/j.1523-1755.2002.00333.x. [DOI] [PubMed] [Google Scholar]
- Masszi A, Di Ciano C, Sirokmany G, Arthur WT, Rotstein OD, Wang J, McCulloch CA, Rosivall L, Mucsi I, Kapus A. Central role for Rho in TGF-beta1-induced alpha-smooth muscle actin expression during epithelial-mesenchymal transition. Am J Physiol. 2003;284:F911–F924. doi: 10.1152/ajprenal.00183.2002. [DOI] [PubMed] [Google Scholar]
- Hojo M, Morimoto T, Maluccio M, Asano T, Morimoto K, Lagman M, Shimbo T, Suthanthiran M. Cyclosporine induces cancer progression by a cell-autonomous mechanism. Nature. 1999;397:530–534. doi: 10.1038/17401. [DOI] [PubMed] [Google Scholar]
- Ruiz P, Gunthert U. The cellular basis of metastasis. World J Urol. 1996;14:141–150. doi: 10.1007/BF00186893. [DOI] [PubMed] [Google Scholar]
- Campistol JM, Inigo P, Larios S, Bescos M, Oppenheimer F. Role of transforming growth factor-beta1 in the progression of chronic allograft nephropathy. Nephrol Dial Transplant. 2001;16(Suppl 1):114–116. doi: 10.1093/ndt/16.suppl_1.114. [DOI] [PubMed] [Google Scholar]
- Bottinger EP, Bitzer M. TGF-beta signaling in renal disease. J Am Soc Nephrol. 2002;13:2600–2610. doi: 10.1097/01.asn.0000033611.79556.ae. [DOI] [PubMed] [Google Scholar]
- Mascarell L, Truffa-Bachi P. New aspects of cyclosporin a mode of action: from gene silencing to gene up-regulation. Mini Rev Med Chem. 2003;3:205–214. doi: 10.2174/1389557033488150. [DOI] [PubMed] [Google Scholar]
- Mascarell L, Frey JR, Michel F, Lefkovits I, Truffa-Bachi P. Increased protein synthesis after T cell activation in presence of cyclosporin A. Transplantation. 2000;70:340–348. doi: 10.1097/00007890-200007270-00019. [DOI] [PubMed] [Google Scholar]
- Tomita M, Reinhold MI, Molkentin JD, Naski MC. Calcineurin and NFAT4 induce chondrogenesis. J Biol Chem. 2002;277:42214–42218. doi: 10.1074/jbc.C200504200. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, Cano A. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem. 2001;276:27424–27431. doi: 10.1074/jbc.M100827200. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Hendrix JA, Johnson AD, Owens GK. Smooth muscle alpha-actin gene requires two E-boxes for proper expression in vivo and is a target of class I basic helix-loop-helix proteins. Circ Res. 2003;92:840–847. doi: 10.1161/01.RES.0000069031.55281.7C. [DOI] [PubMed] [Google Scholar]
- Rutherford MN, LeBrun DP. Restricted expression of E2A protein in primary human tissues correlates with proliferation and differentiation. Am J Pathol. 1998;153:165–173. doi: 10.1016/S0002-9440(10)65557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CP, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- Oriji GK, Keiser HR. Nitric oxide in cyclosporine A-induced hypertension: role of protein kinase C. Am J Hypertens. 1999;12:1091–1097. doi: 10.1016/s0895-7061(99)00089-8. [DOI] [PubMed] [Google Scholar]
- Koya D, King GL. Protein kinase C activation and the development of diabetic complications. Diabetes. 1998;47:859–866. doi: 10.2337/diabetes.47.6.859. [DOI] [PubMed] [Google Scholar]
- Lee SW, Kwak HB, Chung WJ, Cheong H, Kim HH, Lee ZH. Participation of protein kinase C beta in osteoclast differentiation and function. Bone. 2003;32:217–227. doi: 10.1016/s8756-3282(02)00976-6. [DOI] [PubMed] [Google Scholar]
- Kiely B, Feldman G, Ryan MP. Modulation of renal epithelial barrier function by mitogen-activated protein kinases (MAPKs): mechanism of cyclosporine A-induced increase in transepithelial resistance. Kidney Int. 2003;63:908–916. doi: 10.1046/j.1523-1755.2003.00804.x. [DOI] [PubMed] [Google Scholar]
- Keane T, McGlynn H, Egan D, Ryan MP. Potential mechanisms of cyclosporine A-induced renal fibrosis. J Am Soc Nephrol. 1999;10:A3284. [Google Scholar]
- Yue J, Mulder KM. Requirement of Ras/MAPK pathway activation by transforming growth factor beta for transforming growth factor beta 1 production in a Smad-dependent pathway. J Biol Chem. 2000;275:30765–30773. doi: 10.1074/jbc.M000039200. [DOI] [PubMed] [Google Scholar]
- Bemer V, Truffa-Bachi P. T cell activation by concanavalin A in the presence of cyclosporin A: immunosuppressor withdrawal induces NFATp translocation and interleukin-2 gene transcription. Eur J Immunol. 1996;26:1481–1488. doi: 10.1002/eji.1830260712. [DOI] [PubMed] [Google Scholar]
- Gokmen-Polar Y, Murray NR, Velasco MA, Gatalica Z, Fields AP. Elevated protein kinase C betaII is an early promotive event in colon carcinogenesis. Cancer Res. 2001;61:1375–1381. [PubMed] [Google Scholar]
- Le TL, Joseph SR, Yap AS, Stow JL. Protein kinase C regulates endocytosis and recycling of E-cadherin. Am J Physiol. 2002;283:C489–C499. doi: 10.1152/ajpcell.00566.2001. [DOI] [PubMed] [Google Scholar]