Abstract
Platelet-derived growth factor (PDGF) has been implicated in the pathogenesis of vascular occlusive disorders such as atherosclerosis and restenosis in part due to its regulation of smooth muscle cell phenotype. The molecular mechanisms regulating the expression of PDGF-Rα, which binds all known dimeric forms of PDGF except PDGF-DD, are poorly understood. Here we demonstrate that the winged helix-turn-helix proto-oncogene Ets-1 controls PDGF-Rα transcription and mRNA expression in smooth muscle cells. Mutational analysis, electrophoretic mobility shift assay, and chromatin immunoprecipitation revealed the existence of a reverse Ets binding motif (−45TTCC−42) in the proximal region of the PDGF-Rα promoter, which bound both recombinant and endogenous Ets-1. Ets-1-inducible PDGF-Rα expression depended on the integrity of both the −45TTCC−42 motif and the −61G10−52 element, which resides upstream of −45TTCC−42 and mediates Sp1 induction. Hydrogen peroxide (H2O2) at nanomolar concentrations stimulated levels of Ets-1 and increased PDGF-Rα transcription and mRNA expression without affecting Sp1 expression. H2O2 activation of the PDGF-Rα promoter was abolished by disrupting −45TTCC−42 or −61G10−52. These studies identify a functional Ets motif in the PDGF-Rα promoter that plays a pivotal role in agonist-inducible PDGF-Rα transcription.
Platelet-derived growth factor (PDGF) is a family of potent mitogenic and chemoattractant proteins for cells of mesenchymal origin such as vascular smooth muscle cells (SMCs) and fibroblasts.1,2 PDGF proteins consist of four ligands, PDGF-A, -B, -C, and -D that form disulfide-linked homodimers or heterodimers and bind to the transmembrane receptor tyrosine kinases, PDGF receptor-α and -β (PDGF-Rα and PDGF-Rβ).3 PDGF-Rα can bind PDGF-A, -B, or -C,4,5 whereas PDGF-Rβ binds PDGF-B and -D.3,6 The PDGF-Rs have distinct and overlapping biological functions. For example, PDGF-Rα is involved in cell hyperplasia and hypertrophy whereas PDGF-Rβ is involved in both mitogenesis and migration.7 Cells that express both PDGF-Rs can exploit receptor differences by activating PDGF-Rα that inhibits β-receptor-induced migration.2,8 PDGF-Rs activate signaling cascades including extracellular signal-regulated kinases (ERKs) and stress-activated protein kinase-1/c-Jun NH2-terminal kinase-1 (JNK-1).9
Numerous lines of evidence indicate that the PDGF/PDGF-R system plays an important role in SMC growth and the development of vascular occlusive disorders. For example, PDGF-Rα is expressed in advanced atherosclerotic lesions.10 Cardiac allograft arteriosclerosis in rats is inhibited by CGP 53716, a protein tyrosine kinase inhibitor selective for PDGF-R.11 PDGF-Rα levels increase after balloon injury to rat carotid arteries.12,13 Balloon injury also increases PDGF-Rα tyrosyl phosphorylation.14 Anti-sense oligonucleotides targeting PDGF-Rβ inhibit neointima formation in balloon-injured rat carotid arteries.15 Chimeric antibodies raised against PDGF-Rα significantly reduced SMC density in the newly formed neointima in a baboon model.7 Finally, blockade of PDGF signaling with antibodies against PDGF-Rα and PDGF-Rβ in a baboon vascular graft model caused intimal atrophy.16
The Ets family of transcription factors regulate gene expression via physical interactions with DNA (by virtue of their evolutionarily conserved DNA-binding domain and their core recognition motif, 5′GGAA/T3′) and protein-protein interactions with co-factors and other transcription factors. Posttranslational modifications (such as phosphorylation) can also alter the transcriptional activity of Ets factors.17 We have recently shown that Ets-1 physically interacts with Sp1 and regulates Fas ligand gene transcription in SMCs.18 Ets-1 has also been shown to interact with nuclear factors such as nuclear factor-κB,19 AP-1,20 and GHF-1.21 Ets-1, like PDGF-Rα, may play a permissive role in vascular remodeling. For example, Ets-1 expression is induced after arterial injury and exposure to PDGF-BB.22 Moreover, Ets-1 regulates the expression of many genes involved in degradation and remodeling of the extracellular matrix.23
Here we demonstrate that PDGF-Rα expression is under the transcriptional control of Ets-1. A functional Ets motif lies in the proximal region of the PDGF-Rα promoter 5′ of a recently identified Sp1-binding element.24 Ets-1-inducible PDGF-Rα expression is critically dependent on the Sp1 site. Surprisingly, Sp1 transactivation of the PDGF-Rα promoter does not require the Ets motif. The integrity of the Ets site is critical for agonist-inducible PDGF-Rα transcription in vascular SMCs. Peroxide stimulates PDGF-Rα and Ets-1 expression without affecting Sp1 expression. Induction of the PDGF-Rα promoter by H2O2 is abolished by disruption of either −45TTCC−42 or −61G10−52. These studies show that PDGF-Rα transcription is controlled by Ets-1, which mediates peroxide-inducible PDGF-Rα transcription.
Materials and Methods
Immunohistochemical Detection of PDGF-Rα, Ets-1, Sp1, and α-Smooth Muscle Actin (α-SMA) in Human Carotid Arteries
Immunohistochemical analysis was performed with antibodies to PDGF-Rα, Ets-1, Sp1 (all from Santa Cruz Biotechnology, Inc., Santa Cruz, CA), or α-SMA (Novocastra, New Castle upon Tyne, UK) on sister sections of formalin-fixed, paraffin-embedded atherosclerotic carotid artery specimens obtained by endarterectomy at St. Vincent’s Hospital, Sydney, Australia. Immunohistochemical analysis was performed essentially as previously described.25 Briefly, before staining, deparaffinized sections were treated with 3% hydrogen peroxide and boiled in citrate buffer, pH 6.0, to retrieve antigenicity. The standard avidin-biotin complex (ABC) immunoperoxidase technique was used.26 After washing in Tris-buffered saline, pH 7.6, sections were incubated in primary antibody for 60 minutes, followed by incubation with the appropriate secondary antibody for 20 minutes, and finally with ABC (Elite Vector PK-6100) for 30 minutes. Immunogenicity was visualized by treatment in 3,3′-diaminobenzidine solution for 2 minutes, which produced brown coloration. Sections were counterstained with Mayer’s hematoxylin. As negative control, the primary antibody was omitted, or the sections were treated with the immunoglobulin fraction of appropriate nonimmune serum as a substitute for the primary antibody. No positive staining was observed in any of the negative control sections.
Cell Culture
Rat aortic (pup) WKY12-22 SMCs (obtained as a generous gift from Dr. Stephen M. Schwartz, University of Washington, Seattle, WA) were cultured in Waymouth’s MB752/1 medium (Life Technologies, Inc.), pH 7.4, supplemented with 10% fetal calf serum, 10 U/ml penicillin, and 10 μg/ml streptomycin at 37°C and 5% CO2. The cells were passaged every 3 to 4 days in 75-cm2 flasks. Primary bovine aortic endothelial cells (Cell Applications, Inc., CA) were maintained in 10% fetal bovine serum/Dulbecco’s modified Eagle’s medium, pH 7.4, with antibiotics and not used beyond passage 8.
Cell Proliferation Assay
SMCs (2000 cells/well) and endothelial cells (3000 cells/well) were seeded into 96-well titer plates and rendered growth-quiescent by incubation in medium containing 0.05% fetal bovine serum for 24 hours and transfected with 0.1 or 0.3 μmol/L PDGF-Rα antisense (AS) oligonucleotide (ODN) or its size-matched scrambled (SCR) counterpart, PDGF-Rα SCR ODN, using FuGENE6 (3 μl/μg). After 72 hours, the cells were trypsinized, resuspended in Isoton II, and quantitated using a Coulter Z1 counter (Beckman). PDGF-Rα AS and PDGF-Rα SCR ODN sequences were 5′-GTTCTCCTCGGTTCT-3′ and 5′-GTGTCTCCTCCTGTC-3′, respectively, based on analysis of rat and bovine PDGF-Rα in MFOLD.
Plasmid Constructs
pLuc-a227 containing the promoter region of PDGF-Rα was a generous gift from Dr. Yutaka Kitami (Ehime University School of Medicine, Ehime, Japan). pLuc-a2m.Sp1 and pLuc-a2m.Ets were produced using the QuikChange XL site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer’s instructions. The primers were pLuc-a2m.Sp124 forward 5′-TTTATTTTGAAGAGACCATTTTTTTTTTCTTCATTTCCTGACAGCT-3′ and pLuc-a2m.Ets forward 5′-GGGGGGGGCTTCATGGAAATGCAGCTATTTAC-3′. CMV-Sp1 was received from Dr. Robert Tjian (Howard Hughes Medical Institute, University of California, Seattle, WA). CMV-gutless was generated by excising Sp1 cDNA from CMV-Sp1. Ets-1 cDNA was excised from pKCR3-Ets-1 (received from Dr. Ian Cassidy, Department of Biochemistry and Molecular Biology, University of Queensland, Australia) and cloned into pcDNA3. pcDNA3-Fli-1, used in co-transfection experiments, was a gift of Dr. Michael Eisbacher (Centre for Vascular Research, University of New South Wales, Sydney, Australia).28
Transient Transfection Analysis
For reporter gene analysis, SMCs were transfected with 10 μg of pLuc-a2, pLuc-a2.mSp1, or pLuc-a2.mEts. Cells were also transfected with 1 μg of pRL-null (Promega) to correct for transfection efficiency. Firefly luciferase activity was normalized to Renilla. Transient transfections were performed using FuGENE6 (Roche). After incubation at 22°C for 10 minutes, the DNA/FuGENE6 mixture was added to cells containing 10 ml complete medium. Twenty-four hours after transfection cell lysates were prepared for assessment of luciferase activity in which the dual luciferase assay reporter system (DLR) was performed on a manual luminometer, (model TD-20/20; Turner Designs, Quantum Science). Hydrogen peroxide (Sigma) was incubated for 24 hours after transfection for a further 24 hours and cell lysates were prepared as above.
mRNA Expression Analysis
Cells were washed two times with ice-cold phosphate-buffered saline (PBS) and harvested with 4.5 ml of TRIzol reagent (Gibco) in accordance with the manufacturer’s instructions. cDNA synthesis from total RNA was performed using Superscript II RT (Promega) in accordance with the manufacturer’s instructions. Thermal cycling conditions were as follows: PDGF-Rα: 94°C for 10 seconds, 62°C for 30 seconds, 72°C for 1.5 minutes for 40 cycles; GAPDH: 94°C for 30 seconds, 60°C for 30 seconds, 68°C for 2 minutes for 18 cycles; and u-PA: 94°C for 30 seconds, 64°C for 30 seconds, 72°C for 1 minute for 33 cycles. PDGF-Rα primers were: forward 5′-AGATAGCTTCATGAGCCGAC-3′ and reverse 5′-GGAACAGGGTCAATGTCTGG-3′, GAPDH primers: forward 5′-ACCACAGTCCATGCCATCAC-3′ and reverse 5′-TCCACCACCCTGTTGCTGTA-3′; u-PA primers: forward 5′-GCCCAAAGAAATTCAAAGGGGAG-3′ and reverse 5′-CGTATCTTCAGCAAGGCTATG-3′. Densitometric analyses were performed using Quantity One V4.1.0 (Bio-Rad).
Preparation of Nuclear Extracts
SMC monolayers were washed two times with ice-cold PBS, pH 7.4, then scraped into 10 ml of cold PBS. The cells were pelleted by centrifugation at 250 × g for 10 minutes at 4°C. Cells were lysed by the addition of ice-cold hypotonic solution (buffer A) consisting of 10 mmol/L HEPES, pH 8.0, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L dithiothreitol, 200 mmol/L sucrose, 0.5% Nonidet P-40, 0.5 mmol/L phenylmethyl sulfonyl fluoride (PMSF), and 1 μg/ml aprotinin. The suspension was recentrifuged and the nuclei were lysed in an ice-cold solution (buffer C) consisting of 20 mmol/L HEPES, pH 8.0, 100 mmol/L KCl, 0.2 mmol/L ethylenediamine tetraacetic acid (EDTA), 20% glycerol, 1 mmol/L dithiothreitol, 0.5 mmol/L PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin. The nuclear fraction was combined with an equal volume of buffer D (20 mmol/L HEPES, pH 8.0, 100 mmol/L KCl, 0.2 mmol/L EDTA, 20% glycerol, 1 mmol/L dithiothreitol, 0.5 mmol/L PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin) and stored at −80°C.
Electrophoretic Mobility Shift Assays (EMSAs)
Binding reactions for gel shift assays were performed in 20 μl of 10 mmol/L Tris-HCl, 50 mmol/L NaCl2, 1 mmol/L EDTA, 2 mmol/L dithiothreitol, 5% glycerol, 0.5% Nonidet P-40, 1 mg/ml bovine serum albumin, 32P-labeled oligonucleotide probe (150,000 cpm), and 1 μl of relevant recombinant protein or 2 μl of nuclear extract. The recombinant protein reaction was incubated for 20 minutes at 22°C. In supershift studies, the appropriate affinity-purified anti-peptide polyclonal antibody (Santa Cruz Biotechnology) was incubated with binding mix for 20 minutes at 37°C. Bound complexes were separated from free probe by loading samples onto a 6% nondenaturing polyacrylamide gel and electrophoresing at 120 V for 2.5 hours. The gels were vacuum-dried at 80°C and subjected to autoradiography overnight at −80°C. Ets consensus oligonucleotide sequence: Ets cons: 5′-ATC AGA AAA TTG TGG GCG GAA ACT TCC AGG-3′.18 Generation of recombinant Ets-1 protein was previously described.18 Recombinant Sp1 was purchased from Promega (Madison, WI). Sp1 consensus oligonucleotide sequence: Oligo A represents proximal PDGF-A promoter and interacts with Sp1.29 Oligo A: 5′-GGG GGG GGC GGG GGC GGG GGC GGG GGA GG-3′.
Chromatin Immunoprecipitation Analysis
SMCs were washed in PBS, pH 7.4, incubated with 1% formaldehyde for 10 minutes, and quenched with glycine (final concentration, 0.1 mol/L). The cells were washed with PBS, trypsinized, then resuspended and incubated in buffer A (100 mmol/L Tris-HCl, pH 9.4, 10 mmol/L dithiothreitol) for 15 minutes at 30°C. The cells were precipitated by spinning at 14,000 × g for 1 minute, washed sequentially in cold PBS, buffer I (0.25% Triton X-100, 10 mmol/L EDTA, 0.5 mmol/L EGTA, 10 mmol/L HEPES, pH 6.5), buffer II (200 mmol/L NaCl, 1 mmol/L EDTA, 0.5 mmol/L EGTA, 50 mmol/L HEPES, pH 6.5), lysed in lysis buffer [1% sodium dodecyl sulfate (SDS), 10 mmol/L EDTA, 50 mmol/L Tris-HCl, pH 8.1, 0.5 mmol/L PMSF, 0.5 μg/ml leupeptin, 2 μg/ml aprotinin] and sonicated. After spinning, the supernatant was mixed with dilution buffer (1% Triton X-100, 2 mmol/L EDTA, 150 mmol/L NaCl, 20 mmol/L Tris-HCl, pH 8.1). Protein A- and G-Sepharose (previously incubated with salmon sperm DNA and preimmune serum) was added to the supernatant and gently mixed at 4°C for 2 hours. The suspension was evenly divided and 5 μg of rabbit polyclonal antibodies to Sp1 and Ets-1 (Santa Cruz Biotechnology) were added or no antibody was added, and incubated overnight at 4°C. The suspensions were washed sequentially in TSEI (0.1% SDS, 1% Triton X-100, 2 mmol/L EDTA, 20 mmol/L Tris-HCl, pH 8.1, 150 mmol/L NaCl), TSEII (0.1% SDS, 1% Triton X-100, 2 mmol/L EDTA, 20 mmol/L Tris-HCl, pH 8.1, 500 mmol/L NaCl), buffer III (0.25 mol/L LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mmol/L EDTA, 10 mmol/L Tris-HCl, pH 8.1), 1× TE, and elution was performed in elution buffer (1% SDS, 0.1 mol/L NaHCO3) at 65°C for 8 hours. The suspension was treated with proteinase K at 37°C overnight, before phenol-chloroform extraction and ethanol precipitation in the presence of tRNA as a carrier. Primer sequences for PDGF-Rα were: forward 5′-GCACACCATCTCACAATCCA-3′ and reverse 5′-AGTCATCCTCCGAAAACTAATCA-3′. The fragment of PDGF-R-α promoter was amplified by polymerase chain reaction. Thermal cycling conditions were as follows: 94°C for 30 seconds, 56°C for 10 seconds, 72°C for 45 seconds for 46 cycles, generating a 628-bp product.
Western Immunoblot Analysis
Cells treated with 10 nmol/L H2O2 for designated times were washed two times with ice-cold PBS before being lysed on ice in 1× RIPA buffer (50 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% SDS) containing protease inhibitors (2 mmol/L PMSF, 5 mmol/L EDTA, 10 μg/ml leupeptin, and 1% aprotinin). Cell lysates were collected after centrifugation at 14,000 rpm for 20 minutes at 4°C and the protein concentration was determined by BCA protein assay (Pierce). Lysates containing 10 μg or 20 μg of protein were prepared in SDS sample buffer (50 mmol/L Tris, pH 6.8, 10% glycerol, 2% SDS, 0.01% bromophenol blue, and 30 mmol/L dithiothreitol) and boiled for 5 minutes with 0.5 mol/L iodoacetamide added before loading on a 10% SDS-polyacrylamide gel. Gels were blotted onto Immobilon-P (polyvinylidene difluoride) membranes (Millipore) and blocked overnight at 4°C in 5% skim milk powder, 0.05% Tween 20, and PBS. Ets-1, Sp1, and PDGF-Rα protein were detected using rabbit polyclonal antibodies (Santa Cruz Biotechnology) and chemiluminescence detection (NEN Life Sciences Products) according to the manufacturer’s instructions. Densitometric analyses were performed using Quantity One V4.1.0 (Bio-Rad).
Results
PDGF-Rα Is Required for SMC Growth in Vitro
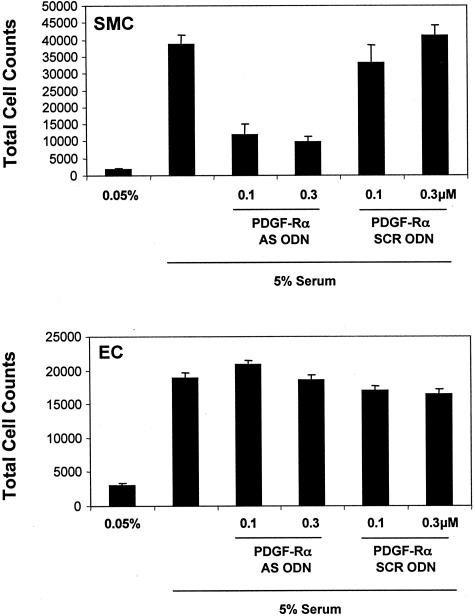
To demonstrate the reliance of cultured SMCs on PDGF-Rα for cell growth, we exposed WKY12-22 cells (SMCs bearing remarkable similarity to SMCs of the synthetic phenotype30,31) to antisense oligonucleotides targeting PDGF-Rα. These agents blocked SMC growth in both a dose-dependent and sequence-specific manner (Figure 1). In contrast, these agents had no effect on endothelial cell replication (Figure 1), consistent with the poor expression of PDGF-Rα in the latter cell type.32,33
Figure 1.
Anti-sense oligonucleotides targeting PDGF-Rα block SMC growth in vitro. WKY12-22 SMCs were incubated overnight in 0.05% fetal bovine serum/Waymouth’s medium and transfected with 0.1 or 0.3 μmol/L PDGF-Rα AS ODN or its size-matched scrambled counterpart PDGF-Rα SCR ODN. The medium was changed to 0.05% fetal bovine serum/Waymouth’s and after 2 days the cells were trypsinized, resuspended in Isoton II, and quantitated using a Coulter counter. Endothelial cells (EC) grown in Dulbecco’s modified Eagle’s medium with the equivalent amounts of serum were treated identically. The data are representative of two experiments.
PDGF-Rα, Ets-1, and Sp1 Are Expressed by SMCs in Human Atherosclerotic Plaque
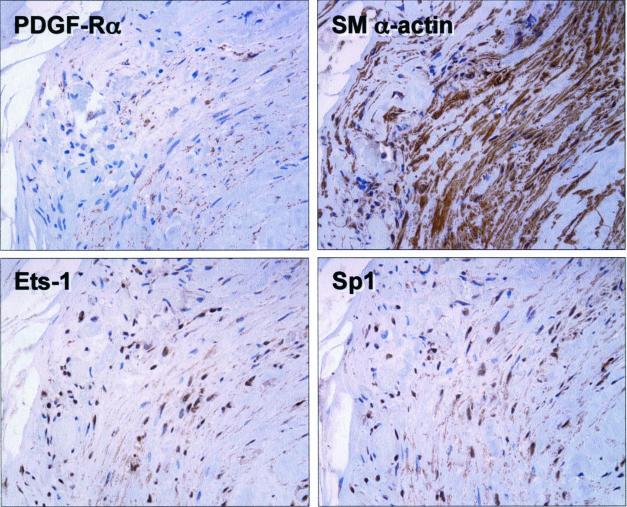
Immunohistochemical analysis on sister sections of formalin-fixed, paraffin-embedded human atherosclerotic carotid artery specimens obtained by endarterectomy revealed that PDGF-Rα, and the transcription factors Ets-1 and Sp1 are expressed in SMCs in these lesions (Figure 2). Unlike the spatial pattern of PDGF-Rα and α-SMA staining, Ets-1 and Sp1 expression was confined mainly to cell nuclei (Figure 2), consistent with their roles as transcription factors.
Figure 2.
PDGF-Rα, Ets-1, and Sp1 are expressed in SMCs of human carotid atheroma. Paraffin sections of formalin-fixed human carotid atherosclerotic plaques were stained with polyclonal antibodies to PDGF-Rα, Ets-1, Sp1, and α-SMA. No staining was observed in the absence of primary antibody.
Ets-1 Induces PDGF-Rα mRNA Expression and Transcription
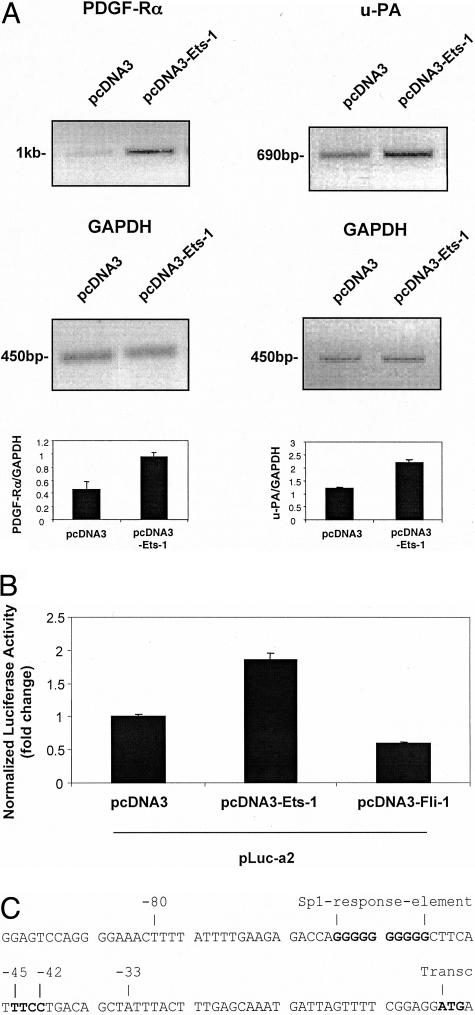
Because PDGF-Rα and Ets-1 are expressed in SMC after balloon injury12,14,34 and in atheroma (Figure 2), we explored whether Ets-1 regulates PDGF-Rα in this cell type. PDGF-Rα mRNA expression was assessed 24 hours after transfection of SMCs with pcDNA3-Ets-1 or its backbone control pcDNA3. Semiquantitative reverse transcriptase-polymerase chain reaction analysis showed that Ets-1 up-regulated endogenous steady-state PDGF-Rα mRNA (Figure 3A, top left). Urokinase-type plasminogen activator (u-PA) is a known transcriptional target of Ets-1.35 Levels of endogenous u-PA, like those of PDGF-Rα, increased twofold 24 hours after the forced expression of Ets-1 (Figure 3A, top right) confirming therefore, that PDGF-Rα expression is positively influenced by Ets-1. Corresponding GAPDH transcript levels demonstrated unbiased sample loading (Figure 3A, middle). Additionally, we assessed the effect of overexpressing the cell-restricted Ets family member Fli-136 on PDGF-Rα transcription. Figure 3B demonstrates that although Ets-1 activates the PDGF-Rα promoter, Fli-1 was inactive.
Figure 3.
Ets-1 induces PDGF-Rα mRNA expression and transcription. A: Cells were transfected with 20 μg of pcDNA3-Ets-1 or pcDNA3 and PDGF-Rα or u-PA mRNA expressions were assessed by reverse transcriptase-polymerase chain reaction after 24 hours. The expected amplification product of PDGF-Rα is 1017 bp, GAPDH is 450 bp, and u-PA is 690 bp. Densitometric analysis was performed on PDGF-Rα, u-PA, and GAPDH mRNA bands. The result is representative of at least two separate determinations performed in duplicate. B: Ets-1 activates the PDGF-Rα promoter, whereas Fli-1 does not. SMCs were co-transfected with the PDGF-Rα promoter-reporter construct pLuc-a2, pRL-Null, and either pcDNA3-Ets-1 or pcDNA3-Fli-1. Luciferase activity was assessed by luminometry after 24 hours. C: Nucleotide sequence of the PDGF-Rα proximal promoter. The transcriptional (transc) start site (ATG)27 is indicated. The reverse Ets (−45TTCC−42) binding site, that is the subject of this study, and the upstream Sp1 element (−61/−52)24 are indicated. Oligo −80/−33 contains both putative Ets and Sp1 sites. The sequence was obtained from EMBL RN 13172 Rattus norvegicus.
We also assessed the effect of overexpressing Ets-1 on PDGF-Rα transcription. SMCs were co-transfected with pcDNA3-Ets-1 and pLuc-a2, a Firefly luciferase-based reporter construct driven by 1.3 kb of PDGF-Rα promoter.27 Luciferase activity 24 hours after transfection revealed that Ets-1 activates PDGF-Rα transcription (Figure 3B). The proximal region of the PDGF-Rα promoter contains a putative reverse Ets-binding motif (5′TTCC3′) located at −45/−42 bp relative to the transcriptional start site (Figure 3C).
Abrogation of Ets-1 Induction of PDGF-Rα Transcription on Mutation of a Putative Reverse Ets Motif (−45TTCC−42)
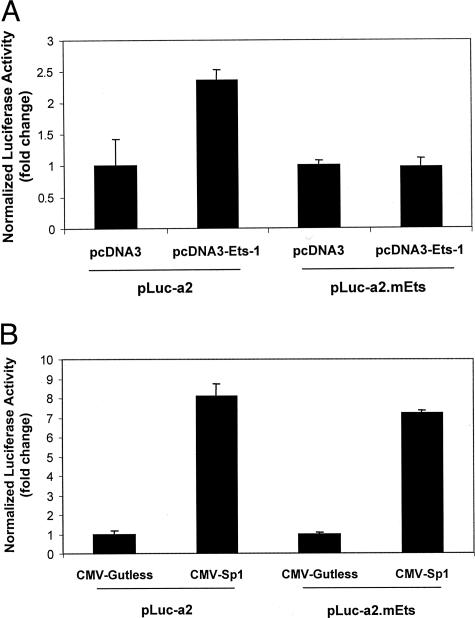
To determine the functional relevance of the −45TTCC−42 element in the PDGF-Rα promoter we mutated −45TTCC−42 to −45AAGG−42 in pLuc-a2 plasmid to generate pLuc-a2.mEts. We assessed the effect of the mutation on PDGF-Rα transcription by co-transfecting SMCs with pLuc-a2 or pLuc-a2.mEts and pcDNA3-Ets-1. Luciferase activity 24 hours after transfection revealed that Ets-1 no longer up-regulated PDGF-Rα promoter-dependent expression when the reverse Ets binding element was disrupted (Figure 4A).
Figure 4.
Ets-1 induction of PDGF-Rα is abolished on mutation of the Ets (−45TTCC−42) motif whereas Sp1 induction is unaffected. A: Cells were transfected with 10 μg of pLuc-a2 or pLuc-a2.mEts and 1 μg of pcDNA3-Ets-1 or pcDNA3. Firefly luciferase was determined in cell lysates after 24 hours. The y axis indicates the ratio of Firefly luciferase activity over Renilla to normalize for transfection efficiency. B: Sp1 does not require the Ets site to up-regulate PDGF-Rα transcription. The cells were transfected with 10 μg of pLuc-a2 or pLuc-a2.mEts and 3 μg of CMV-Sp1 or CMV-gutless. Firefly luciferase was determined in cell lysates after 24 hours. The y axis represents the ratio of Firefly luciferase activity over the Renilla to normalize for transfection efficiency. The result is representative of at least two independent observations.
The Ets Recognition Element (−45TTCC−42) in the PDGF-Rα Promoter Is Not Required for Sp1-Inducible PDGF-Rα Transcription
We have previously shown that Sp1 regulates PDGF-Rα transcription in SMCs via an atypical Sp1 binding element located at −61G10−52.24 Our previous studies in SMCs reveal that endogenous Ets-1 and Sp1 physically interact.18 Because co-operative regulation of PDGF-Rα transcription has not been described, we hypothesized that Ets-1 and Sp1 together mediate expression of this gene.
To determine whether Sp1 required the −45TTCC−42 element for Sp1-inducible PDGF-Rα promoter-dependent transcription, we co-transfected SMCs with either pLuc-a2 or pLuc-a2.mEts, and CMV-Sp1 or its backbone control CMV-gutless. Luciferase activity, determined 24 hours after transfection, demonstrated that integrity of the Ets binding motif (−45/−42 bp) is not essential for Sp1-stimulated PDGF-Rα transcription (Figure 4B).
Ets-1 Induction of PDGF-Rα Transcription Is Critically Dependent on an Intact Sp1-Response Element
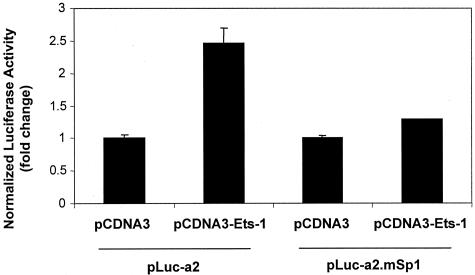
When we addressed the question of Sp1/Ets-1 interplay in the opposite manner we were surprised by the outcome. Ets-1 failed to transactivate the PDGF-Rα promoter if the Sp1 response element (−61G10−52) was disrupted (−61G10−52 changed to −61T10−52 in construct pLuc-a2.mSp1).24 PDGF-Rα transcription in this experiment was assessed in cells co-transfected with pLuc-a2 or pLuc-a2.mSp1 and pcDNA3-Ets-1 or pcDNA3. Luciferase activity 24 hours after transfection revealed Ets-1 no longer altered PDGF-Rα promoter-dependent transcription (Figure 5).
Figure 5.
Ets-1 induction of PDGF-Rα is abolished by mutation of the proximal Sp1 (−61G10−52) element in PDGF-Rα promoter. This site (−61G10−52) mediates Sp1-inducible PDGF-Rα expression.24 Cells were transfected with 10 μg of pLuc-a2 or pLuc-a2.mSp1 and 1 μg of pcDNA3-Ets-1 or pcDNA3. Firefly luciferase was determined in cell lysates after 24 hours. The y axis represents the ratio of Firefly luciferase activity over Renilla to normalize for transfection efficiency. The result is representative of at least two independent observations.
Interaction of Ets-1 with the Oligo −80/−33 Is Critically Dependent on Both Proximal Sp1 and Ets Sites
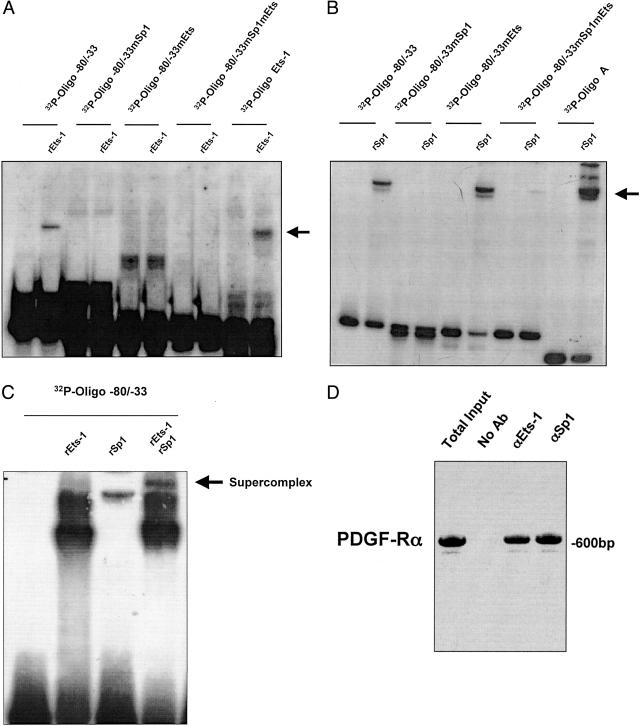
To provide a mechanistic explanation for these observations we performed EMSA studies using Ets-1 and Sp1 and this proximal region of the PDGF-Rα promoter (Figure 3C). Recombinant Ets-1 (rEts-1) formed a nucleoprotein complex with 32P-Oligo −80/−33 (Figure 6A, lane 2). rEts-1 no longer bound the promoter fragment when 32P-Oligo −80/−33 bore a mutation (mEts) in the Ets motif (Figure 6A, lane 6). rEts-1 also failed to bind the oligonucleotide when 32P-Oligo −80/−33 carried a mutation (mSp1) in the Sp1 motif (Figure 6A, lane 4). These findings support our transient transfection studies in which Ets-1 failed to up-regulate PDGF-Rα transcription when either the Ets or Sp1 elements were mutated (Figures 4A and 5). Mutation of both elements (mSp1mEts) in 32P-Oligo −80/−33 failed to support formation of a rEts-1 nucleoprotein complex (Figure 6A, lane 8). The binding of rEts-1 protein to a previously used Ets-binding oligonucleotide (32P-Oligo Ets-1)29 served as a positive control (Figure 6A, lane 10) in this analysis.
Figure 6.
Interaction of Ets-1 and Sp1 with −80/−33 region of the PDGF-Rα promoter requires −45TTCC−42 and −61G10−52. Oligonucleotides spanning the −80/−33 region of the PDGF-Rα proximal promoter were incubated with indicated recombinant (r) protein. EMSA using rEts-1 (A), rSp1 (B), or rEts-1 and rSp1 (C). The arrow indicates the nucleoprotein complex. Wild-type 32P-Oligo −80/−33, or 32P-Oligo −80/−33mEts (where −45TTCC−42 was changed to −45TACA−42), an Sp1 mutant, 32P-Oligo −80/−33mSp1 (where −61G10−52 was changed to −61T10−52) and a double mutant, 32P-Oligo −80/−33mSp1mEts (where both sites were mutated). D: Chromatin immunoprecipitation analysis demonstrating amplification of the PDGF-Rα promoter in the total input, without antibody pull-down (no Ab), or pulled down with Sp1 and Ets-1 antibodies. The data are representative of at least two independent determinations.
Sp1 Requires the Sp1 Element but Not the Ets Site to Bind the PDGF-Rα Promoter
Our PDGF-Rα promoter-dependent reporter studies revealed that the Ets binding motif (−45/−42 bp) is not required for Sp1-inducible PDGF-Rα transcription (Figure 4B). This, together with binding analyses using rEts-1 (Figure 6A), led us to investigate the capacity of recombinant Sp1 (rSp1) to bind Oligo −80/−33. 32P-Oligo A,29 which bears multiple consensus Sp1 recognition elements served as a positive control for rSp1 binding (Figure 6B, lane 10). rSp1 formed a nucleoprotein complex with 32P-Oligo −80/−33 (Figure 6A, lane 2). 32P-Oligo −80/−33 bearing a mutation in the Sp1 element (mSp1), as expected, did not bind rSp1 (Figure 6B, lane 4). When the Ets site in 32P-Oligo −80/−33 was mutated (mEts), rSp1 binding was unaffected (Figure 6B, lane 6) indicating therefore, that Sp1 does not require the Ets site to bind the PDGF-Rα promoter. In contrast, both sites are required for Ets-1 binding and PDGF-Rα transactivation (Figures 4A, 5, and 6A). rSp1 failed to bind 32P-Oligo −80/−33 when both the Ets and Sp1 sites were disrupted (Figure 6B, lane 8). A supercomplex formed when Ets-1 and Sp1 were incubated simultaneously with 32P-Oligo −80/−33 bearing both respective sites (Figure 6C). Chromatin immunoprecipitation analysis shows that the authentic PDGF-Rα promoter is indeed occupied by Ets-1 and Sp1 (Figure 6D). Ets-1/Sp1 binding and transactivation of the PDGF-Rα promoter are summarized in tabular form in Table 1. Cooperative interactions between Ets-1 and Sp1 mediate the expression of genes such as Fas ligand,18 CD53,37 SERA3,38 and fgl2,39 and more recently PDGF-A.40
Table 1.
PDGF-Rα Promoter Transactivation and Binding Analyses to Ets-1 and Sp1
| WT and mutant EMSA probes or reporter constructs | Ets-1 activation | Sp1 activation | Ets-1 binding | Sp1 binding |
|---|---|---|---|---|
| WT | + | + | + | + |
| mEts | − | + | − | + |
| mSp1 | − | − | − | − |
The table summarizes the results of transient transfection and binding studies with wild-type (WT) and mutant PDGF-Rα promoter oligonucleotides (Oligo −33/−80) and reporter constructs (pLuc-a2). PDGF-Rα promoter interactions and transactivation by Ets-1or Sp1 are indicated by + or −.
Hydrogen Peroxide Stimulates Ets-1 and Activates PDGF-Rα Expression via the −45TTCC−42 and −61G10−52 Elements in the Proximal Promoter
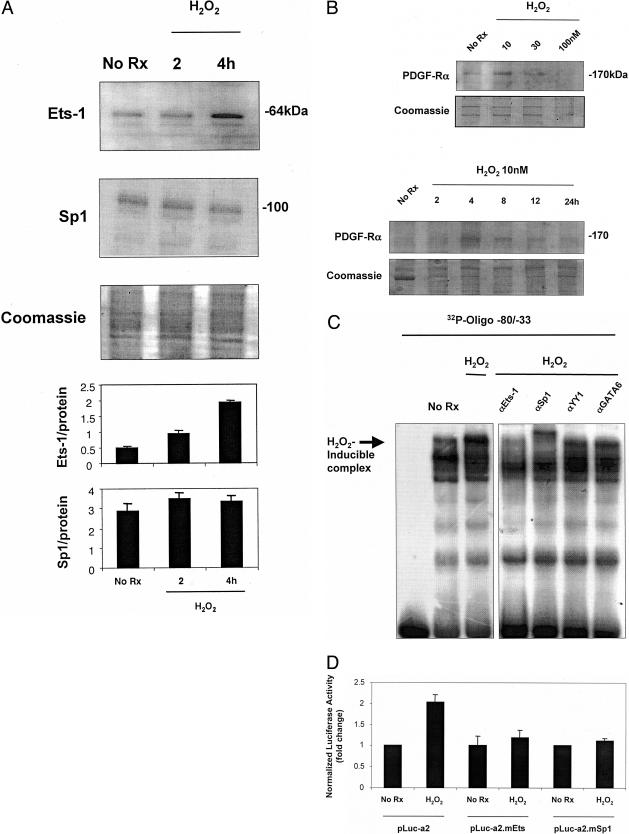
The preceding studies demonstrate that Ets-1 binds to and activates the PDGF-Rα promoter in an Sp1-dependent manner. These findings alone, however, do not establish whether the Ets site plays an important role in the context of a pathophysiologically relevant agonist. Reactive oxygen species are produced in human atheroma41 and in the balloon-injured artery wall.42 Western immunoblot analysis revealed that low concentrations of H2O2 (10 nmol/L) stimulate Ets-1 expression in SMCs within 4 hours (Figure 7A) without affecting Sp1 levels (Figure 7A), consistent with previously demonstrated H2O2 up-regulation of Ets-1 but not Sp1, albeit in endothelial cells.43 PDGF-Rα protein levels increased in SMCs exposed to the same concentration of H2O2 after 4 hours (Figure 7B).
Figure 7.
H2O2 induces Ets-1 protein expression and PDGF-Rα transcription via the −45TTCC−42 motif in the proximal promoter. A: Growth quiescent cells were incubated with 10 nmol/L H2O2 for 2 and 4 hours. Ets-1 and Sp1 protein levels were assessed by Western immunoblot analysis (20 μg total protein for Ets-1 analysis and 10 μg for Sp1). Ets-1 protein levels increased compared to basal levels with addition of 10 nmol/L H2O2 compared to untreated cells. Sp1 protein levels were unchanged compared to basal levels with addition of 10 nmol/L H2O2 compared to untreated cells. Coomassie-stained gel demonstrates unbiased loading. Densitometric analysis was performed on bands of interest and relative to Coomassie-stained protein. B: SMCs were incubated with various concentrations of H2O2 for different times and PDGF-Rα protein levels assessed by Western immunoblot analysis (10 μg of total protein). Coomassie-stained gels demonstrate unbiased loading. C: EMSA using nuclear extracts of SMCs that had been incubated with 10 nmol/L H2O2 for 4 hours. The H2O2-inducible nucleoprotein complex was eliminated by antibodies to Ets-1 and Sp1, but not YY1 or GATA6. D: SMCs were transfected with 10 μg of either pLuc-a2, pLuc-a2.mEts, or pLuc-a2.mSp1 for 24 hours followed by the addition of 10 nmol/L H2O2 for 24 hours. Firefly luciferase was determined in cell lysates after 24 hours. The y axis represents the fold change in PDGF-Rα promoter activity. The result is representative of at least two independent observations.
EMSA performed using nuclear extracts of SMCs incubated with H2O2 for 4 hours revealed a single peroxide-inducible nucleoprotein complex that was eliminated in supershift analysis using antibodies to Ets-1 and Sp1 (Figure 7C). The Ets-1 elimination was achieved under conditions different from those previously reported.24 This complex was unaffected by antibodies to YY1, GATA6, or several other Ets family members including Ets-2 (Figure 7C and data not shown). Transient transfection analysis revealed that H2O2 activated PDGF-Rα promoter-dependent expression within 24 hours (Figure 7D). Mutation of the −45TTCC−42 element in construct pLuc-a2.mEts completely blocked H2O2 activation of the PDGF-Rα promoter (Figure 7D). Moreover, H2O2 activation of PDGF-Rα transcription was absolutely dependent on element −61G10−52 (Figure 7D). This represents the first demonstration of the capacity of H2O2 to modulate PDGF-Rα transcription in any cell type.
Discussion
The capacity of H2O2 to induce PDGF-Rα expression in vascular SMCs has implications to PDGF-Rα-dependent hyperplasia in the injured artery wall. That synthetic WKY12-22 SMCs were used in the present study may account for the nanomolar concentrations of H2O2 that stimulated Ets-1 and PDGF-Rα expression. Ets-1, PDGF-Rα, and reactive oxygen species are found in arterial lesions (present study).12,23,34,44 Superoxide dismutase, glutathione peroxidase, and catalase are intensely expressed by migrating SMCs in human atherosclerotic lesions.41 There is evidence that reactive oxygen species can influence SMC accumulation in the artery wall as a consequence of increased migration and proliferation.41,45 Accordingly, a reactive oxygen species/Ets-1/PDGF-Rα axis may influence the formation of the SMC-rich neointima, just as Ets-1 serves as a transcriptional mediator of peroxide-inducible Ets-1-dependent changes in the endothelial cell phenotype.43 This is particularly relevant to vascular complications of diabetes wherein reactive oxygen species are generated under hyperglycemic conditions.45
The functional interplay of transcription factors like Ets-1 in the promoter regions of key genes exerts a profound phenotypic influence in the vessel wall. This study provides greater insight into the transcriptional regulation of PDGF-Rα expression by demonstrating Ets-1 and Sp1 co-occupancy and the critical requirement of Sp1 in Ets-1-inducible PDGF-Rα transcription. That Ets-1 and Sp1 also modulate transcription of one of the ligands of PDGF-Rα in vascular SMCs,40 which is also inducibly expressed in vascular lesions,34,46 suggests a more general theme in the transcriptional control of the PDGF ligand-receptor system.
Acknowledgments
We thank Mr. Fernando S. Santiago for expert technical assistance.
Footnotes
Address reprint requests to Professor Levon M. Khachigian, Ph.D., D.Sc., Centre for Vascular Research, Department of Pathology, The University of New South Wales, Sydney NSW 2052 Australia. E-mail: l.khachigian@unsw.edu.au.
Supported by the National Health and Medical Research Council, a New South Wales State Department of Health Research infrastructure grant, and an Australian postgraduate research award (to M.R.B.).
L.M.K. is a Senior Principal Research Fellow of the National Health and Medical Research Council.
References
- Ross R, Raines EW, Bowen-Pope DF. The biology of platelet-derived growth factor. Cell. 1986;46:155–169. doi: 10.1016/0092-8674(86)90733-6. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Westermark B. Mechanisms of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1300. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- Uutela M, Lauren J, Bergsten E, Li X, Horelli-Kuitunen N, Eriksson U, Alitalo K. Chromosomal location, exon structure, and vascular expression patterns of the human PDGFC and PDGFD genes. Circulation. 2001;103:2242–2247. doi: 10.1161/01.cir.103.18.2242. [DOI] [PubMed] [Google Scholar]
- Fang L, Yan Y, Komuves LG, Yonkovich S, Sullivan CM, Stringer B, Galbraith S, Lokker NA, Hwang S, Nurden P, Phillips DR, Giese NA. PDGF C is a selective alpha platelet-derived growth factor receptor agonist that is highly expressed in platelet alpha granules and vascular smooth muscle. Arterioscler Thromb Vasc Biol. 2004;24:787–792. doi: 10.1161/01.atv.0000120785.82268.8b. [DOI] [PubMed] [Google Scholar]
- Li X, Ponten A, Aase K, Karlsson L, Abramsson A, Uutela M, Backstrom G, Bostrom H, Li H, Soriano P, Betsholtz C, Heldin C, Alitalo K, Ostman A, Eriksson U. PDGF-C is a new protease-activated ligand for the PDGF α-receptor. Nat Cell Biol. 2000;2:302–309. doi: 10.1038/35010579. [DOI] [PubMed] [Google Scholar]
- Seifert R, van Koppen A, Bowen-Pope DF. PDGF-AB requires PDGF receptor alpha-subunits for high-affinity, but not for low-affinity, binding and signal transduction. J Biol Chem. 1993;268:4473–4480. [PubMed] [Google Scholar]
- Davies MG, Owens EL, Mason DP, Lea H, Tran K, Vergel S, Hawkins SA, Hart CE, Clowes W. Effect of platelet-derived growth factor receptor-alpha and -beta blockade on flow-induced neointimal formation in endothelialized baboon vascular grafts. Circ Res. 2000;86:779–786. doi: 10.1161/01.res.86.7.779. [DOI] [PubMed] [Google Scholar]
- Eriksson A, Siegbahn A, Westermark B, Heldin CH, Claesson-Welsh L. PDGF α- and β-receptors activate unique and common signal transduction pathways. EMBO J. 1992;11:543–550. doi: 10.1002/j.1460-2075.1992.tb05085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Deuel TF, Kim HC. Platelet-derived growth factor (PDGF) receptor-alpha activates c-Jun NH2-terminal kinase-1 and antagonized PDGF receptor-beta-induced phenotypic transformation. J Biol Chem. 2000;275:19076–19082. doi: 10.1074/jbc.M910329199. [DOI] [PubMed] [Google Scholar]
- Irvine CD, George SJ, Sheffield E, Johnson JL, Davies AH, Lamont PM. The association of platelet-derived growth factor receptor expression, plaque morphology and histological features with symptoms in carotid atherosclerosis. Cardiovasc Surgery. 2000;8:121–129. doi: 10.1016/s0967-2109(99)00090-3. [DOI] [PubMed] [Google Scholar]
- Sihvola R, Koskinen P, Myllarniemi M, Loubtchenkov M, Hayry P, Buchdunger E, Lemstrom K. Prevention of cardiac allograft arteriosclerosis by protein tyrosine kinase inhibitor selective for platelet-derived growth factor receptor. Circulation. 1999;99:2295–2301. doi: 10.1161/01.cir.99.17.2295. [DOI] [PubMed] [Google Scholar]
- Panek RL, Dahring TK, Olszewski BJ, Keiser JA. PDGF receptor protein tyrosine kinase expression in the balloon-injured rat carotid artery. Arterioscler Thromb Vasc Biol. 1997;17:1283–1288. doi: 10.1161/01.atv.17.7.1283. [DOI] [PubMed] [Google Scholar]
- Majesky MW, Giachelli CM, Reidy MA, Schwartz SM. Rat carotid neointimal smooth muscle cells reexpress a developmentally regulated mRNA phenotype during repair of arterial injury. Circ Res. 1992;71:759–768. doi: 10.1161/01.res.71.4.759. [DOI] [PubMed] [Google Scholar]
- Abe J, Deguchi J, Mastsumoto T, Takuwa N, Noda M, Ohno M, Makuchi M, Kurokawa K, Takuwa Y. Stimulated activation of platelet-derived growth factor receptor in vivo in balloon-injured arteries: a link between angiotensin II and intimal thickening. Circulation. 1997;96:1906–1913. doi: 10.1161/01.cir.96.6.1906. [DOI] [PubMed] [Google Scholar]
- Sirois MG, Simons M, Edelman ER. Antisense oligonucleotide inhibition of PDGFR-beta receptor subunit expression directs suppression of intimal thickening. Circulation. 1997;95:669–676. doi: 10.1161/01.cir.95.3.669. [DOI] [PubMed] [Google Scholar]
- Englesbe MJ, Hawkins SM, Hsieh PC, Daum G, Kenagy RD, Clowes AW. Concomitant blockade of platelet-derived growth factor receptors alpha and beta induces intimal atrophy in baboon PTFE grafts. J Vasc Surg. 2004;39:440–446. doi: 10.1016/j.jvs.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2002;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- Kavurma MM, Bobryshev Y, Khachigian LM. Ets-1 positively regulates Fas ligand transcription via cooperative interactions with Sp1. J Biol Chem. 2002;277:36244–36252. doi: 10.1074/jbc.M200463200. [DOI] [PubMed] [Google Scholar]
- Bassuk AG, Anandappa RT, Leiden JM. Physical interactions between Ets and NF-kappaB/NFAT proteins play an important role in their cooperative activation of the human immunodeficiency virus enhancer in T cells. J Virol. 1997;71:3563–3573. doi: 10.1128/jvi.71.5.3563-3573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, Leiden JM. A direct physical association between ETS and AP-1 transcription factors in normal T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- Bradford AP, Wasylyk C, Wasylyk B, Gutierrez-Hartmann A. Interaction of Ets-1 and the POU-homeodomain protein GHF-1/Pit-1 reconstitutes pituitary-specific gene expression. Mol Cell Biol. 1997;17:1065–1074. doi: 10.1128/mcb.17.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito S, Shimizu S, Maeda S, Wang J, Paul R, Fagin JA. Ets-1 is an early response gene activated by ET-1 and PDGF-BB in vascular smooth muscle cells. Am J Physiol. 1998;274:C472–C480. doi: 10.1152/ajpcell.1998.274.2.C472. [DOI] [PubMed] [Google Scholar]
- Hultgardh-Nilsson A, Cercek B, Wang J, Naito S, Lovdahl C, Sharifi B, Forrester JS, Fagin JA. Regulated expression of the Ets-1 transcription factor in vascular smooth muscle cells in vivo and in vitro. Circ Res. 1996;78:589–595. doi: 10.1161/01.res.78.4.589. [DOI] [PubMed] [Google Scholar]
- Bonello MR, Khachigian LM. Fibroblast growth factor-2 represses platelet-derived growth factor receptor-alpha (PDGFR-alpha) transcription via ERK1/2-dependent Sp1 phosphorylation and an atypical cis-acting element in the proximal PDGFR-alpha promoter. J Biol Chem. 2004;279:2377–2382. doi: 10.1074/jbc.M308254200. [DOI] [PubMed] [Google Scholar]
- Kavurma MM, Bobryshev Y, Khachigian LM. Ets-1 positively regulates Fas ligand transcription via cooperative interactions with Sp1. J Biol Chem. 2002;277:36244–36252. doi: 10.1074/jbc.M200463200. [DOI] [PubMed] [Google Scholar]
- Hsu S-M, Raine L, Fanger H. Use of avidin-biotin-peroxidase complex (ABC) in immunoperoxidase techniques: a comparison between ABC and unlabeled antibody (PAP) procedures. J Histochem Cytochem. 1981;29:577–580. doi: 10.1177/29.4.6166661. [DOI] [PubMed] [Google Scholar]
- Kitami Y, Inui H, Uno S, Inagami T. Molecular structure and transcriptional regulation of the gene for platelet-derived growth factor alpha receptor in cultured vascular smooth muscle cells. J Clin Invest. 1995;96:558–567. doi: 10.1172/JCI118068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisbacher M, Holmes ML, Newton A, Hogg PJ, Khachigian LM, Crossley M, Chong BH. Protein-protein interaction between Fli-1 and GATA-1 mediates synergistic expression of megakaryocyte-specific genes through cooperative DNA binding. Mol Cell Biol. 2003;23:3427–3441. doi: 10.1128/MCB.23.10.3427-3441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafty LA, Santiago FS, Khachigian LM. NF1/X represses PDGF A-chain transcription by interacting with Sp1 and antagonizing Sp1 occupancy of the promoter. EMBO J. 2002;21:334–343. doi: 10.1093/emboj/21.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire JM, Covin CW, White S, Giachelli CM, Schwartz SM. Characterization of cloned aortic smooth muscle cells from young rats. Am J Pathol. 1994;144:1068–1081. [PMC free article] [PubMed] [Google Scholar]
- Rafty LA, Khachigian LM. Zinc finger transcription factors mediate high constitutive PDGF-B expression in smooth muscle cells derived from aortae of newborn rats. J Biol Chem. 1998;273:5758–5764. doi: 10.1074/jbc.273.10.5758. [DOI] [PubMed] [Google Scholar]
- Lindner V, Reidy MA. Platelet-derived growth factor ligand and receptor expression by large vessel endothelium. Am J Pathol. 1995;146:1488–1497. [PMC free article] [PubMed] [Google Scholar]
- Mondy JS, Lindner V, Miyashiro JK, Berk BC, Dean RH, Geary RL. Platelet-derived growth factor ligand and receptor expression in response to altered blood flow in vivo. Circ Res. 1997;81:320–327. doi: 10.1161/01.res.81.3.320. [DOI] [PubMed] [Google Scholar]
- Majesky MW, Reidy MA, Bowen-Pope DF, Hart CE, Wilcox JN, Schwartz SM. PDGF ligand and receptor gene expression during repair of arterial injury. Cell Biology. 1990;111:2149–2158. doi: 10.1083/jcb.111.5.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaka C, Tanaka K, Abe M, Sato Y. Ets-1 regulates angiogenesis by inducing the expression of urokinase-type plasminogen activator and matrix metalloproteinase-1 and the migration of vascular endothelial cells. J Cell Physiol. 1996;169:522–531. doi: 10.1002/(SICI)1097-4652(199612)169:3<522::AID-JCP12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Hewett PW, Nishi K, Daft EL, Clifford Murray J. Selective expression of erg isoforms in human endothelial cells. Int J Biochem Cell Biol. 2001;33:347–355. doi: 10.1016/s1357-2725(01)00022-x. [DOI] [PubMed] [Google Scholar]
- Hernandez-Torres J, Yunta M, Lazo PA. Differential cooperation between regulatory sequences required for human CD53 gene expression. J Biol Chem. 2001;276:35405–35413. doi: 10.1074/jbc.M104723200. [DOI] [PubMed] [Google Scholar]
- Hadri L, Ozog A, Soncin F, Lompre A. Basal transcription of the mouse sarco(endo)plasmic reticulum Ca2+-ATPase type 3 gene in endothelial cells is controlled by Ets-1 and Sp1. J Biol Chem. 2002;277:36471–36478. doi: 10.1074/jbc.M204731200. [DOI] [PubMed] [Google Scholar]
- Liu M, Leibowitz JL, Clark DA, Mendicino M, Ning Q, Ding JW, D’Abreo C, Fung L, Marsden PA, Levy GA. Gene transcription of fgl2 in endothelial cells is controlled by Ets-1 and Oct-1 and requires the presence of both Sp1 and Sp3. Eur J Biochem. 2003;270:2274–2286. doi: 10.1046/j.1432-1033.2003.03595.x. [DOI] [PubMed] [Google Scholar]
- Santiago FS, Khachigian LM. Ets-1 stimulates platelet-derived growth factor A-chain gene transcription and vascular smooth muscle cell growth via cooperative interactions with Sp1. Circ Res. 2004;95:479–487. doi: 10.1161/01.RES.0000141135.36279.67. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Inoue N, Azumi H, Seno T, Hirata K, Kawashima S, Hayashi Y, Itoh H, Yokozaki H, Yokoyama M. Expressional changes of the vascular antioxidant system in atherosclerotic coronary arteries. J Atheroscler Thromb. 2002;9:184–190. doi: 10.5551/jat.9.184. [DOI] [PubMed] [Google Scholar]
- Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin CH, Alitalo K, Eriksson U. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nat Cell Biol. 2001;3:512–516. doi: 10.1038/35074588. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Ohzeki Y, Shimizu S, Naito S, Ohtsuru A, Yamamoto T, Kuroiwa Y. Stimulation of in vitro angiogenesis by hydrogen peroxide and the relation with ETS-1 in endothelial cells. Life Sci. 1999;64:249–258. doi: 10.1016/s0024-3205(98)00560-8. [DOI] [PubMed] [Google Scholar]
- Azumi H, Inoue N, Ohashi Y, Terashima M, Mori T, Fujita H, Awano K, Kobayashi K, Maeda K, Hata K, Shinke T, Kobayashi S, Hirata K, Kawashima S, Itabe H, Hayashi Y, Imajoh-Ohmi S, Itoh H, Yokoyama M. Superoxide generation in directional coronary atherectomy specimens of patients with angina pectoris: important role of NAD(P)H oxidase. Arterioscler Thromb Vasc Biol. 2002;22:1838–1844. doi: 10.1161/01.atv.0000037101.40667.62. [DOI] [PubMed] [Google Scholar]
- Srivastava AK. High glucose-induced activation of protein kinase signaling pathways in vascular smooth muscle cells: a potential role in the pathogenesis of vascular dysfunction in diabetes. Int J Mol Med. 2002;9:85–89. [PubMed] [Google Scholar]
- Barrett TB, Benditt EP. Platelet-derived growth factor gene expression in human atherosclerotic plaques and normal artery wall. Proc Natl Acad Sci USA. 1988;85:2810–2814. doi: 10.1073/pnas.85.8.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]