Abstract
Using a purpose-designed experimental model, we have defined new, statistically significant, differences in gene expression between heavily and weakly metastatic human breast cancer cell populations, in vivo and in vitro. The differences increased under selection pressures designed to increase metastatic proficiency. Conversely, the expression signatures of primary tumors generated by more aggressive variants, and their matched metastases in the lungs and lymph nodes, all tended to converge. However, the few persisting differences among these selectively enriched malignant growths in the breast, lungs, and lymph nodes were highly statistically significant, implying potential mechanistic involvement of the corresponding genes. The evidence that has emerged from the current work indicates that selective enhancement of metastatic proficiency by serial transplantation co-purifies a subliminal gene expression pattern within the tumor cell population. This signature most likely includes genes participating in metastasis pathogenesis, and we document manageable numbers of candidates for this role. The findings also suggest that metastasis to at least two different organs occurs through closely similar genetic mechanisms.
Metastasis, the spread of cancer cells from the primary tumor to distant organs and their treatment-resistant proliferation in multiple locations, remains a major clinical and biological challenge. It is known from previous work that tumor cells that make metastases can be propagated as cell lines that conserve their capabilities to produce secondary cancers in other organs.1,2 The heritable nature of this escalating problem, confirmed by the work of many investigators (reviewed by Fidler3) and by our own work on spontaneous murine and human neoplasms of various histogenetic origins,4,5 demonstrates that it is caused by a genetic disorder governing the behavior of the cancer cells. Also, this inherited behavior pattern, although disruptive of tissues and organs, is a highly coordinated process requiring the completion of several complicated steps in the correct order in time and space. Successful achievement of the metastatic event therefore implies the sequential and orderly mobilization of relevant gene pathways. Knowledge of the genes involved would be of considerable diagnostic, prognostic, and therapeutic value both in patients who have not yet developed clinically detectable metastases and in more advanced cases in which the limitation of further spread would be beneficial.
We therefore used oligonucleotide microarrays to perform high throughput screening of global gene expression in tumors and metastases produced by a unique matched pair of human clonal cell lines of opposite metastatic capabilities, which we have derived from the same breast cancer line, MDA-MB-435,6 and confirmed to be isogenic by several methods, including chromosomal analysis7 and genetic fingerprinting. There are already some reports of high-density microarray profiling of gene expression patterns in metastatic human primary cancers and metastases,8–12 but no consensus has yet emerged on any groups of genes that are consistently involved. This may be due to the masking effects resulting from comparisons between samples from individuals of different genetic backgrounds.
The work presented below makes progress from previous approaches by using a tightly controlled, well characterized, xenograft model of breast cancer metastasis.7,13 This investigative system facilitated direct examination of differences between primary tumors and matched metastases in the lungs and lymph nodes from the same animal and thus eliminated the noise from biological variations between different individuals. To our knowledge, this is the first study to systematically investigate gene expression patterns in matched metastases from both of these organs in the same host. In addition, this investigation provides new data on dynamic gene expression patterns in vivo and in vitro of metastasis-competent and incompetent human cell populations within the same parent tumor, opening a window on the effects of tumor-host interactions on behavior. Such comparison is not possible in samples excised from clinical tumor specimens. Technical advances are also incorporated in this work: tumor cell lineages were labeled with green fluorescent protein (GFP) to enhance accuracy of selection of primary and secondary tumor tissue for analysis. Also, to evaluate the initial screening results, we conducted extensive laboratory studies and computational (training and test) procedures and validated the expression levels of a number of genes of interest.
Our findings indicated that the genes expressed in primary tumors generated by metastasis-competent cell populations differed clearly from those in their metastasis-incompetent counterparts. In contrast, the patterns in metastases were similar to the primary tumors from which they originated, and metastases in the lungs displayed remarkably similar gene expression patterns to those in the lymph nodes of the same animal. Additionally, the patterns observed in tumors and metastases differed from those seen in the parental cell lines in vitro, indicating that the host microenvironment is an active participant in tumor progression and metastasis. These findings have significant implications for defining mechanisms of metastasis and for designing novel effective therapy. They also contribute a manageable list of candidate genes from which to choose targets for interventional studies on mechanisms involved in the process.
Materials and Methods
Cell Lines
The NM-2C5 and M-4A4 lines were isolated in our laboratory from the MDA-MD-435 breast cancer cell line as described by Bao et al6 and subsequently transduced with an enhanced green fluorescent protein-expressing vector.13 These monoclonal cell lines were routinely cultured in RPMI 1640 medium supplemented with 10% newborn calf serum (Invitrogen, Carlsbad, CA), penicillin, and streptomycin at 37°C in a humidified atmosphere of 5% CO2-95% air. Cell line LM3 was derived from a lung metastasis produced by M-4A4, and cell line CL16 was obtained from a lung metastasis made by descendants of LM3 after two more similar selection cycles in nude mice. Both are progressively more metastatic variants of the parent line (see below) grown under the same conditions in vitro.
Murine Xenograft Metastasis Model
One million cells in 50 μl of a mixture of RPMI 1640 medium and ECM gel (Sigma Chemical Co., St. Louis, MO) were inoculated into the mammary fat pad of anesthetized mice. Animals were euthanized and autopsied at 3 to 4 months postinoculation when the primary tumors reached ∼20 mm in diameter. Metastasis formation was assessed by macroscopic observation of all major organs for secondary tumors and confirmed by histological examination. Metastasis was also confirmed by looking for fluorescence of incorporated GFP under blue light (λ = 490 nm), which is sufficiently sensitive to detect single cells. Only cell clusters (>1 mm) are regarded as true metastases. Tissues from primary tumors and metastases were snap-frozen and stored at −80°C until used for RNA or protein extraction.
Protein Analyses
Protein detection and quantification were performed either on primary tumors or sera from tumor-bearing mice depending on the protein localization. Tumor homogenates were prepared in 20 mmol/L Tris-HCl, pH 8.0, 5 mmol/L CaCl2, 1 mmol/L phenylmethylsulfonyl fluoride, 15 μmol/L pepstatin A, and 0.05% (w/v) Brij 35. The Complete EDTA-free protease inhibitor cocktail (Boehringer Ingelheim Chemicals, Petersburg, VA) was added to the extraction buffer. The tissues were homogenized on ice, and the homogenates were centrifuged at 14,000 × g for 40 minutes (4°C). Protein quantitation of the supernatants was performed using the Coomassie Plus Protein Assay kit (Pierce, Rockford, IL). Twenty micrograms of denatured protein samples was separated on a 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis glycine gel, and proteins were transferred onto polyvinylidene difluoride membranes using a semi-dry apparatus (Bio-Rad, Life Science, Hercules, CA) according to the manufacturer’s instructions. Immunodetection was performed as described previously7 using the following specific antibodies: monoclonal anti-silver homolog (Pmel-17) (Neomarkers, Inc., Fremont, CA); monoclonal anti-MITF (Neomarkers, Inc.); polyclonal anti-α1-antichymotrypsin (DAKO, Carpinteria, CA); polyclonal anti-TRP1 (Santa Cruz Biotechnology, Santa Cruz, CA); monoclonal anti-osteopontin (Chemicon International, Inc., Temecula, CA); monoclonal anti-thrombospondin (BD Transduction Laboratories, San Jose, CA); and polyclonal anti-MMP-8 (Chemicon International, Inc). MMP-8 and OPN protein expressions were quantified using commercially available enzyme-linked immunosorbent assay (ELISA) kits from Amersham Biosciences (San Francisco, CA) and Assay Designs (Ann Arbor, MI), respectively.
Frozen sections of xenograft primary tumors were fixed in 4% buffered formaldehyde. Antigen retrieval was performed using the target retrieval solution at 1:10 dilution (DAKO). Endogenous peroxidase activity was blocked by incubation in 3% H2O2. Nonspecific binding of the antibodies to irrelevant proteins was blocked by incubation in 10% goat serum. Proteins of interest were targeted with the same antibodies used in the Western-blot experiments (see above). When monoclonal mouse antibodies were applied, a prior incubation of the section with goat anti-mouse Ig Fab fragments (Jackson Immunoresearch Laboratories, West Grove, PA), which neutralized the Fc domain reactivity of the endogenous host Ig, was performed. The horseradish peroxidase-conjugated secondary antibody was visualized by diaminobenzidine substrate-chromogen 3,3′-diaminobenzidine (DAKO). Sections were counterstained with Hematoxylin-Gills No. 2 solution, dehydrated in alcohol, cleared in xylene, and mounted in Permount (Fisher Chemicals, Lake Forest, CA).
cRNA Preparation and GeneChip Hybridization
Total RNA was extracted from cultured cells and frozen tissue samples with TRIzol reagent (Invitrogen) and cleaned with the DNA-free kit (Ambion, Austin, TX). RNA quality was assessed by running the samples on a native 1% agarose gel and on a Biogem analyzer (Agilent, Palo Alto, CA). cRNA was prepared in the University of California-San Diego Cancer Center Microarray facility as described by the standard Affymetrix microarray protocols. The cRNA was then hybridized to human HG-U133A GeneChip oligonucleotide arrays (Affymetrix, Santa Clara, CA), which interrogate approximately 22,000 transcripts. The arrays were scanned at 560 nm using an argon-ion confocal laser as the excitation source.
Microarray Data Analysis
The DAT files containing the scanned images of each microarray were individually inspected for quality control and digitized by Microarray Analysis Suite 5.0 (Affymetrix). The resultant CEL files containing the raw numerical data for signal intensity at probe level were collectively read and analyzed in dChip software.14,15 Briefly, each microarray was normalized against a common baseline array using the “invariant probe set” method. After normalization, the model-based expression index of each gene was then calculated according to the PM-MM model. To identify candidate genes that were differentially expressed between any two group of arrays, a screening filter consisting of the following criteria was applied: 1) a fold change (fc) larger than 1.5 or 3; 2) two-tailed P values (paired t-test if applicable) smaller than 0.05; and 3) a minimal difference of 100 between the group mean of model-based expression index. The resultant lists of candidate genes were then sorted according to their corresponding P values with a cut-off of 0.005 to ensure high stringency of the analysis (Tables 1–6). For subsequent high-level analysis, candidate gene lists from six group-wise comparisons were combined and subjected to hierarchical clustering (centroid-linkage) or classification by linear discriminant analysis (LDA) within the R environment (http://www.R-project.org).
Table 1.
Lists of Differentially Expressed Genes in the Horizontal and Vertical Comparisons: Nonmetastatic Tumor versus Metastatic Tumor (P < 0.005, fc > 1.5)
| P value | fc | Gene description | Accession no. | Probe set |
|---|---|---|---|---|
| Top 20 genes up-regulated in the nonmetastatic tumor (versus the metastatic tumor) | ||||
| 0.00002 | 3.48 | Cancer/testis antigen 2 | AJ012833.1 | 215733_x_at |
| 0.000029 | 1.98 | Cancer/testis antigen 1 | AF038567.1 | 211674_x_at |
| 0.00003 | 2.17 | Cancer/testis antigen 1 | AJ275978.1 | 217339_x_at |
| 0.000039 | 2.86 | Nucleotide binding protein 2 (MinD homolog, E. coli) | NM_012225.1 | 218227_at |
| 0.000048 | 2.72 | Sulfide quinone reductase-like (yeast) | NM_021199.1 | 217995_at |
| 0.000063 | 2.32 | Membrane-spanning 4-domains, subfamily A, member 3 | L35848.1 | 210254_at |
| 0.000082 | 1.88 | HN1-like | AK023154.1 | 212115_at |
| 0.000087 | 2.29 | Thrombospondin 1 | NM_003246.1 | 201110_s_at |
| 0.000098 | 2.31 | Serologically defined colon cancer antigen 16 | BC001149.1 | 221514_at |
| 0.000099 | 2.43 | Influenza virus NS1A binding protein | AF205218.1 | 201362_at |
| 0.000101 | 2.13 | Guanine nucleotide binding protein (G protein), γ 11 | NM_004126.1 | 204115_at |
| 0.000102 | 2.45 | Splicing factor, arginine/serine-rich 7, 35 kd | NM_006276.2 | 201129_at |
| 0.000106 | 2.33 | DnaJ (Hsp40) homolog, subfamily A, member 3 | NM_005147.1 | 205963_s_at |
| 0.000112 | 1.65 | Kinesin family member 4A | NM_012310.2 | 218355_at |
| 0.000121 | 2.34 | Influenza virus NS1A binding protein | AB020657.1 | 201363_s_at |
| 0.000124 | 3281.94 | Chondroitin sulfate proteoglycan 4 (melanoma-associated) | NM_001897.1 | 204736_s_at |
| 0.000132 | 2.36 | Regulator of G-protein signalling 10 | NM_002925.2 | 204316_at |
| 0.000138 | 1.65 | Uracil-DNA glycosylase | NM_003362.1 | 202330_s_at |
| 0.000144 | 21.5 | Melanoma antigen, family A, 1 (directs expression of antigen MZ2-E) | NM_004988.1 | 207325_x_at |
| 0.000145 | 1.86 | Heat shock protein 75 | NM_016292.1 | 201391_at |
| Top 20 genes up-regulated in the metastatic tumor (versus the nonmetastatic tumor) | ||||
| 0.000001 | 3.44 | Protein kinase C-like 1 | NM_002741.1 | 202161_at |
| 0.000002 | 10.81 | Serine (or cysteine) proteinase inhibitor, clade A, member 3 | NM_001085.2 | 202376_at |
| 0.000003 | 10.34 | Collagen, type IX, α 1 | NM_001851.1 | 222008_at |
| 0.000007 | 4.52 | Serine (or cysteine) proteinase inhibitor, clade F, member 1 | NM_002615.1 | 202283_at |
| 0.00001 | 6.29 | Aldehyde dehydrogenase 1 family, member A1 | NM_000689.1 | 212224_at |
| 0.00001 | 3.92 | Preferentially expressed antigen in melanoma | NM_006115.1 | 204086_at |
| 0.00001 | 2.61 | Retinol dehydrogenase 11 (all-trans and 9-cis) | NM_016026.1 | 217775_s_at |
| 0.000013 | 2.32 | SH3-domain binding protein 4 | AF015043.1 | 222258_s_at |
| 0.000015 | 3.57 | Dynein, cytoplasmic, intermediate polypeptide 1 | NM_004411.1 | 205348_s_at |
| 0.000023 | 6.64 | Ribonuclease, RNase A family, 1 (pancreatic) | NM_002933.1 | 201785_at |
| 0.000028 | 2.14 | Proteasome (prosome, macropain) 28S subunit, non-ATPase, 8 | NM_002812.1 | 200820_at |
| 0.000032 | 3.1 | Dudulin 2 | NM_018234.1 | 218424_s_at |
| 0.00004 | 3.23 | Likely ortholog of mouse semaF cytoplasmic domain-associated protein 3 | AL569804 | 212915_at |
| 0.000048 | 4.11 | LIM domain protein | BE043700 | 214175_x_at |
| 0.000051 | 1.63 | ADP-ribosylation factor interacting protein 2 (arfaptin 2) | NM_012402.1 | 202109_at |
| 0.000052 | 47.46 | G antigen 4 | NM_021123.1 | 208235_x_at |
| 0.000052 | 2.2 | Protein tyrosine phosphatase type IVA, member 1 | BF576710 | 200732_s_at |
| 0.000061 | 79.78 | G antigen 4 | NM_001476.1 | 208155_x_at |
| 0.000061 | 2.41 | Eukaryotic translation initiation factor 2, subunit 1 α, 35 kd | BC002513.1 | 201143_s_at |
| 0.000062 | 3.55 | Retinoblastoma-associated factor 600 | AB007931.1 | 211950_at |
Table 2.
Lists of Differentially Expressed Genes in the Horizontal and Vertical Comparisons: Nonmetastatic Tumor versus Lung Metastases (P < 0.005, fc > 1.5)
| P value | fc | Gene description | Accession no. | Probe set |
|---|---|---|---|---|
| Top 20 genes up-regulated in the nonmetastatic tumor (versus the lung metastases) | ||||
| 0.000027 | 3.49 | Guanine nucleotide binding protein (G protein), gamma 11 | NM_004126.1 | 204115_at |
| 0.000027 | 1.92 | Non-POU domain containing, octamer-binding | NM_007363.2 | 200057_s_at |
| 0.000031 | 2.03 | Chromosome 11 hypothetical protein ORF3 | NM_020154.1 | 217898_at |
| 0.000037 | 2.4 | Mahogunin, ring finger 1 | AB011116.1 | 212576_at |
| 0.000041 | 8.62 | Similar to X-linked ribosomal protein 4 (RPS4X) | AL137162 | 217019_at |
| 0.000052 | 2.22 | Cytoplasmic FMR1-interacting protein 1 | BC005097.1 | 208923_at |
| 0.000056 | 1.63 | Dual specificity phosphatase 4 | BC002671.1 | 204015_s_at |
| 0.000058 | 2.43 | RNA binding protein S1, serine-rich domain | NM_006711.1 | 207939_x_at |
| 0.00007 | 1.55 | Superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) | NM_000454.1 | 200642_at |
| 0.000072 | 2.6 | Synaptosomal-associated protein, 23 kd | BC003686.1 | 209130_at |
| 0.000081 | 1.91 | Succinate-CoA ligase, GDP-forming, β subunit | AL050226.1 | 215772_x_at |
| 0.000088 | 1.68 | PVVP2 periodic tryptophan protein homolog (yeast) | U56085.1 | 209336_at |
| 0.000089 | 2.01 | Similar to RPS3A (ribosomal protein S3A) | AL356115 | 216823_at |
| 0.000093 | 1.7 | Sialyltransferase 4C (β-galactoside α-2,3-sialyltransferase) | NM_006278.1 | 203759_at |
| 0.000096 | 477.5 | Cancer/testis antigen 1 | AF038567.1 | 211674_x_at |
| 0.000096 | 2.74 | Regulator of G-protein signaling 10 | NM_002925.2 | 204316_at |
| 0.000097 | 2.58 | TYRO3 protein tyrosine kinase | U05682.1 | 211432_s_at |
| 0.000097 | 1.91 | ALEX3 protein | NM_016607.1 | 217858_s_at |
| 0.000111 | 2.09 | Hypothetical gene supported by AK000185 | AK000185.1 | 216644_at |
| 0.000115 | 2.12 | MAX interacting protein 1 | NM_005962.1 | 202364_at |
| Top 20 genes up-regulated in the lung metastases (versus the nonmetastatic tumor) | ||||
| 0.000002 | 2.12 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 | NM_005804.1 | 201584_s_at |
| 0.000002 | 5.17* | Neuroblastoma, suppression of tumorigenicity 1 | NM_005380.1 | 201621_at |
| 0.000003 | 5.28 | Serine (or cysteine) proteinase inhibitor, clade F, member 1 | NM_002615.1 | 202283_at |
| 0.000003 | 7.75 | Ribonuclease, RNase A family, 1 (pancreatic) | NM_002933.1 | 201785_at |
| 0.000004 | 3.72 | Neuroblastoma, suppression of tumorigenicity 1 | D28124 | 37005_at |
| 0.000004 | 6.18 | Osteopontin | M83248.1 | 209875_s_at |
| 0.000005 | 2.03 | Adaptor-related protein complex 2, σ 1 subunit | BC006337.1 | 211047_x_at |
| 0.000006 | 8.71 | LIM domain protein | BC003096.1 | 211564_s_at |
| 0.000009 | 13.92 | Baculoviral IAP repeat-containing 7 (livin) | NM_022161.1 | 220451_s_at |
| 0.00001 | 3.06 | KIAA0930 protein | AK025608.1 | 217118_s_at |
| 0.000011 | 1.65 | Cytochrome c oxidase subunit Vlb | NM_001863.2 | 201441_at |
| 0.000013 | 13.79 | Ocular albinism 1 (Nettleship-Falls) | NM_000273.1 | 206696_at |
| 0.000017 | 2.69 | Six transmembrane epithelial antigen of the prostate | NM_012449.1 | 205542_at |
| 0.000017 | 3.4 | Retinoblastoma-associated factor 600 | AB007931.1 | 211950_at |
| 0.00002 | 82.72 | G antigen 4 | NM_001476.1 | 208155_x_at |
| 0.000024 | 6.76 | Slalyltransferase | NM_006456.1 | 204542_at |
| 0.000027 | 2.36 | N-Acylsphingosine amidohydrolase (acid ceramidase) 1 | AI934569 | 213702_x_at |
| 0.000028 | 3.62 | Arginase, type II | U75667.1 | 203946_s_at |
| 0.000037 | 1.68 | Heme binding protein 1 | NM_015987.1 | 218450_at |
| 0.000047 | 7.32 | Proteoglycan 1, secretory granule | J03223.1 | 201858_s_at |
Refuted by Q-PCR.
Table 3.
Lists of Differentially Expressed Genes in the Horizontal and Vertical Comparisons: Nonmetastatic Tumor (versus LN Metastases (P < 0.005, fc > 1.5)
| P value | fc | Gene description | Accession no. | Probe set |
|---|---|---|---|---|
| Top 20 genes up-regulated in the nonmetastatic tumor (versus the LN metastases) | ||||
| 0.000024 | 1.56 | Dual specificity phosphatase 4 | BC002671.1 | 204015_s_at |
| 0.000028 | 3.08 | Transforming growth factor, β-induced, 68 kd | NM_000358.1 | 201506_at |
| 0.00003 | 1.99 | Ras-GTPase-activating protein SH3-domain-binding protein | BG500067 | 201503_at |
| 0.000035 | 1.85 | Non-POU domain containing, octamer-binding | NM_007363.2 | 200057_s_at |
| 0.00004 | 1.56 | Superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult) | NM_000454.1 | 200642_at |
| 0.000048 | 2.54 | KIAA0843 protein | NM_014945.1 | 205730_s_at |
| 0.000052 | 80.77 | Cancer/testis antigen 1 | AF038567.1 | 211674_x_at |
| 0.000056 | 1.85 | Nuclear pore complex interacting protein | NM_006985.1 | 204538_x_at |
| 0.000068 | 1.67 | Integrin, α 6 | NM_000210.1 | 201656_at |
| 0.000074 | 3.11 | Plectin 1, intermediate filament binding protein 500 kd | Z54367 | 216971_s_at |
| 0.00008 | 2.15 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 | NM_016222.1 | 217840_at |
| 0.000085 | 1.6 | ATPase, H+ transporting, lysosomal 16 kd, V0 subunit c | NM_001694.1 | 200954_at |
| 0.000088 | 1.96 | Cytoplasmic FMR1 interacting protein 1 | BC005097.1 | 208923_at |
| 0.000089 | 1.91 | Parvulin | BE674061 | 214224_s_at |
| 0.000094 | 2.19 | Sialyltransferase 4C (β-galactoside α-2,3-sialyltransferase) | NM_006278.1 | 203759_at |
| 0.000095 | 2.36 | STIP1 homology and U-Box containing protein 1 | NM_005861.1 | 217934_x_at |
| 0.000096 | 2.49 | Mahogunin, ring finger 1 | AB011116.1 | 212576_at |
| 0.000099 | 2.23 | Nonmetastatic cells 4, protein expressed in | AL523860 | 212739_s_at |
| 0.000104 | 1.97 | MAX interacting protein 1 | NM_005962.1 | 202364_at |
| 0.000121 | 2.54 | Regulator of G-protein signaling 10 | NM_002925.2 | 204316_at |
| Top 20 genes up-regulated in the LN metastases (versus the nonmetastatic tumor) | ||||
| 0 | 6.63 | Osteopontin | M83248.1 | 209875_s_at |
| 0.000007 | 3.4 | Ornithine decarboxylase 1 | NM_002539.1 | 200790_at |
| 0.000007 | 2.2 | RAB27A, member RAS oncogene family | BE502030 | 209514_s_at |
| 0.00001 | 13 | Baculoviral IAP repeat-containing 7 (livin) | NM_022161.1 | 220451_s_at |
| 0.00001 | 6.23 | Sulfotransferase family, cytosolic, 1C, member 1 | AF026303.1 | 205342_s_at |
| 0.00001 | 3.94 | Preferentially expressed antigen in melanoma | NM_006115.1 | 204086_at |
| 0.00001 | 3.67 | KIAA0930 protein | AK025608.1 | 217118_s_at |
| 0.00001 | 2.4 | Mitochondrial ribosomal protein L35 | NM_016622.1 | 218890_x_at |
| 0.000013 | 39.88 | Chromosome 1 open reading frame 34 | BC004399.1 | 210652_s_at |
| 0.000021 | 3.73 | GREB1 protein | NM_014668.1 | 205862_at |
| 0.000023 | 2.45 | Malate dehydrogenase 1, NAD (soluble) | NM_005917.1 | 200978_at |
| 0.000023 | 2.24 | Adaptor-related protein complex 2, σ 1 subunit | NM_021575.1 | 208074_s_at |
| 0.000024 | 2.42 | N-Acylsphingosine amidohydrolase (acid ceramidase) 1 | U47674.1 | 210980_s_at |
| 0.000025 | 2.07 | Adaptor-related protein complex 2, σ 1 subunit | BC006337.1 | 211047_x_at |
| 0.000027 | 1.9 | Sorting nexin 10 | NM_013322.1 | 218404_at |
| 0.000028 | 11.05 | Aldehyde dehydrogenase 1 family, member A1 | NM_000689.1 | 212224_at |
| 0.00003 | 6.06 | Serine (or cysteine) proteinase inhibitor, clade F, member 1 | NM_002615.1 | 202283_at |
| 0.00003 | 2.92 | Neutral sphingomyelinase (N-SMase) activation associated factor | NM_003580.1 | 203269_at |
| 0.000032 | 4519.89 | G antigen 4 | NM_001473.1 | 207663_x_at |
| 0.000032 | 2.36 | N-acylsphingosine amidohydrolase (acid ceramidase) 1 | AI934569 | 213702_x_at |
Table 4.
Lists of Differentially Expressed Genes in the Horizontal and Vertical Comparisons: Metastatic Tumor versus Lung Metastases (P < 0.005, fc > 1.5)
| P value | fc | Gene description | Accession no. | Probe set |
|---|---|---|---|---|
| Genes up-regulated in the metastatic tumor (versus the lung metastases) | ||||
| 0.000728 | 1.72* | GalNAc-T1 | NM_020474.2 | 201724_s_at |
| 0.001816 | 1.61 | Oxysterol binding protein-like 10 | NM_017784.1 | 219073_s_at |
| 0.001868 | 1.87 | Solute carrier family 23 (nucleobase transporters), member 2 | AL389886 | 209236_at |
| 0.002554 | 2.09 | Transmembrane, prostate androgen-induced RNA | NM_020182.1 | 217875_s_at |
| 0.00297 | 1.66 | Plasminogen activator, tissue | NM_000930.1 | 201860_s_at |
| 0.00342 | 1.63 | Likely ortholog of rat GRP78-binding protein | NM_017870.1 | 218834_s_at |
| 0.003664 | 1.66 | SRY (sex determining region Y)-box 4 | NM_003107.1 | 201417_at |
| 0.004619 | 1.57 | Scavenger receptor class B, member 1 | NM_005505.1 | 201819_at |
| 0.004706 | 1.6 | Hypothetical protein LOC283687 | AF249277.1 | 210242_x_at |
| Genes up-regulated in the lung metastases (versus the metastatic tumor) | ||||
| 0.00052 | 6.83 | Surfactant, pulmonary-associated protein C | J03553 | 38691_s_at |
| 0.000576 | 2† | Neuroblastoma, suppression of tumorigenicity 1 | NM_005380.1 | 201621_at |
| 0.001028 | 1.75 | Tight junction protein 1 (zona occludens 1) | NM_003257.1 | 202011_at |
| 0.001141 | 2.77 | Human HL14 gene encoding β-galactoside-binding lectin, 3 end, clone 2 | M14087.1 | 216405_at |
| 0.001547 | 1.53 | Phosphoenolpyruvate carboxykinase 2 (mitochondrial) | NM_004563.1 | 202847_at |
| 0.001648 | 1.8† | Neuroblastoma, suppression of tumorigenicity 1 | D28124 | 37005_at |
| 0.001781 | 751.48 | Surfactant, pulmonary-associated protein C | BC005913.1 | 211735_x_at |
| 0.002132 | 1.68 | VAMP (vesicle-associated membrane protein)-associated protein A, 33 kd | AF154847.1 | 208780_x_at |
| 0.002596 | 2.57 | New member of the thymosininterferon-inducible multigene family | AL133228 | 216438_s_at |
| 0.003235 | 1.93 | Pilin-like transcription factor | NM_012228.1 | 218773_s_at |
| 0.003408 | 1.53 | Ets variant gene 5 (ets-related molecule) | X76184.1 | 216375_s_at |
| 0.003577 | 2.02 | Ubiquitin specific protease 1 | AW499935 | 202412_s_at |
| 0.003774 | 1.68 | Apolipoprotein C-I | NM_001645.2 | 204416_x_at |
| 0.003932 | 1.82 | Serine/arginine repetitive matrix 2 | AI655799 | 208610_s_at |
| 0.004159 | 1.94 | Serotonin-7 receptor pseudogene | U86813.1 | 216098_s_at |
| 0.004508 | 1.68 | Endothelin receptor type B | M74921.1 | 204271_s_at |
| 0.004889 | 2.37 | Transmembrane 4 superfamily member 2 | NM_004615.1 | 202242_at |
Refuted by Q-PCR.
Validated by Q-PCR.
Table 5.
Lists of Differentially Expressed Genes in the Horizontal and Vertical Comparisons: Metastatic Tumor versus LN Metastases (P < 0.005, fc > 1.5)
| P value | fc | Gene description | Accession no. | Probe set |
|---|---|---|---|---|
| Top 20 genes up-regulated in the metastatic tumor (versus the LN metastases) | ||||
| 0.000098 | 3.18† | Matrix metalloproteinase 14 (membrane-inserted) | NM_004995.2 | 202828_s_at |
| 0.000295 | 1.61 | Protocadherin gamma subfamily C, 3 | AK026188.1 | 215836_s_at |
| 0.000365 | 1.55 | Cold-inducible RNA binding protein | NM_001280.1 | 200810_s_at |
| 0.000378 | 1.62 | Transforming growth factor β-stimulated protein TSC-22 | AK027071.1 | 215111_s_at |
| 0.000401 | 2.47 | Scavenger receptor class B, member 1 | NM_005505.1 | 201819_at |
| 0.000432 | 1.76 | Plasminogen activator, tissue | NM_000930.1 | 201860_s_at |
| 0.000475 | 1.95 | KIAA0121 gene product | D50911.2 | 212399_s_at |
| 0.000483 | 1.67 | Protocadherin γ subfamily C, 3 | NM_002588.1 | 205717_x_at |
| 0.00055 | 5.28 | Transmembrane, prostate androgen induced RNA | NM_020182.1 | 217875_s_at |
| 0.000641 | 1.63 | Protocadherin gamma subfamily C, 3 | AF152318.1 | 209079_x_at |
| 0.000793 | 1.77 | Chondroitin polymerizing factor | NM_024536.1 | 202175_at |
| 0.000814 | 1.57 | Protocadherin gamma subfamily C, 3 | BC006439.1 | 211066_x_at |
| 0.000823 | 2.33† | Matrix metalloproteinase 14 (membrane-inserted) | AU149305 | 202827_s_at |
| 0.000835 | 1.77 | T-box 2 | AW173045 | 213417_at |
| 0.001002 | 2.1 | Solute carrier family 23 (nucleobase transporters), member 2 | AL389886 | 209236_at |
| 0.001067 | 1.7 | Centaurin, γ 2 | NM_014914.1 | 204066_s_at |
| 0.001507 | 1.71 | Unc-84 homolog B (C. elegans) | AL021707 | 212144_at |
| 0.001677 | 1.55 | Tumor differentially expressed 1 | U49188.1 | 221473_x_at |
| 0.001701 | 1.8 | Plexin B2 | BC004542.1 | 208890_s_at |
| 0.001844 | 1.85 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 | NM_016222.1 | 217840_at |
| Top 20 genes up-regulated in the LN metastases (versus the metastatic tumor) | ||||
| 0.000114 | 2.35 | 3-hydroxylsobutyryl-Coenzyme A hydrolase | AW000964 | 213374_x_at |
| 0.000338 | 1.54 | ATP synthase, H+ transporting, mitochondrial F1 cpx, γ polypeptide 1 | AV711183 | 213366_x_at |
| 0.000378 | 1.64 | Isocitrate dehydrogenase 3 (NAD+) α | AI826060 | 202069_s_at |
| 0.00043 | 3.36 | Sulfotransferase family, cytosolic, 1C, member 1 | AF026303.1 | 205342_s_at |
| 0.000464 | 2.28 | Endothelin receptor type B | NM_003991.1 | 206701_x_at |
| 0.000491 | 1.89 | Endothelin receptor type B | NM_000115.1 | 204273_at |
| 0.000585 | 4.76 | Silver homolog (mouse) | U01874.1 | 209848_s_at |
| 0.000625 | 1.97* | Neuroblastoma, suppression of tumorigenicity 1 | D28124 | 37005_at |
| 0.000665 | 1.87 | RAB38, member RAS oncogene family | NM_022337.1 | 219412_at |
| 0.000666 | 2.06 | Endothelin receptor type B | M74921.1 | 204271_s_at |
| 0.000743 | 1.52 | Malate dehydrogenase 1, NAD (soluble) | NM_005917.1 | 200978_at |
| 0.000786 | 1.7 | Colony stimulating factor 2 receptor, α, low-affinity | M64445.1 | 211287_x_at |
| 0.000791 | 3.05 | IQ motif containing GTPase-activating protein 2 | NM_006633.1 | 203474_at |
| 0.000917 | 1.67 | RAB27A, member RAS oncogene family | BE502030 | 209514_s_at |
| 0.001092 | 5.28 | Cell adhesion molecule with homology to L1CAM (close homolog of L1) | NM_006614.1 | 204591_at |
| 0.001149 | 1.57 | ATP synthase, H+ transporting, mitochondrial F1 complex, polypeptide 1 | BC000931.2 | 208870_x_at |
| 0.001212 | 1.74 | Cell division cycle 2, G1 to S and G2 to M | NM_001786.1 | 203214_x_at |
| 0.001251 | 1.61 | ATP synthase, H+ transporting, mitochondrial F1 complex, polypeptide 1 | NM_005174.1 | 205711_x_at |
| 0.001343 | 1.57 | Syntaxin 7 | NM_003569.1 | 203457_at |
| 0.001352 | 1.89 | Heat shock 70 kd protein 4 | AA043348 | 208814_at |
Refuted by Q-PCR.
Validated by Q-PCR.
Table 6.
Lists of Differentially Expressed Genes in the Horizontal and Vertical Comparisons: Lung Metastases versus LN metastases (P < 0.005, fc > 1.5)
| P value | fc | Gene description | Accession no. | Probe set |
|---|---|---|---|---|
| Genes up-regulated in the lung metastases (versus the LN metastases) | ||||
| 0.000266 | 6.63 | Surfactant, pulmonary-associated protein C | J03553 | 38691_s_at |
| 0.000335 | 1.5† | S100 calcium binding protein A9 (calgranulin B) | NM_002965.2 | 203535_at |
| 0.000925 | 1.62† | Homeo box B13 | U57052.1 | 209844_at |
| 0.00099 | 1.99† | Matrix metalloproteinase 14 (membrane-inserted) | NM_004995.2 | 202828_s_at |
| 0.001212 | 2.42 | Sine oculis homeobox homolog 3 (Drosophila) | NM_005413.1 | 206634_at |
| 0.001642 | 1.63 | Discs, large (Drosophila) homolog-associated protein 1 | NM_004746.1 | 206490_at |
| 0.001999 | 1.56 | Interleukin 1 receptor antagonist | BE563442 | 216245_at |
| 0.002368 | 1.62 | Transition protein 2 (during histone to protamine replacement) | NM_005425.1 | 207736_s_at |
| 0.00255 | 1.55 | Suppression of tumorigenicity 7 like | NM_017744.1 | 219964_at |
| 0.003281 | 1.64 | Homo sapiens cDNA: FLJ21911 fls, clone HEP03855 | AK025564.1 | 216780_at |
| 0.003664 | 1.54 | H. sapiens cDNA: FLJ21198 fls, clone COL00220. | AK024851.1 | 216740_at |
| 0.004192 | 1.67 | KIAA0570 gene product | AK023845.1 | 215013_s_at |
| 0.004537 | 1.85 | SPARC-like 1 (mast9, hevin) | NM_004884.1 | 200795_at |
| 0.004661 | 2.05 | Transmembrane 4 superfamily member 1 | M90657.1 | 209387_s_at |
| Genes up-regulated in the LN metastases (versus the lung metastases) | ||||
| 0.00249 | 1.51 | Syndecan 2 | J04621.1 | 212154_at |
Validated by Q-PCR.
Quantitative PCR
mRNA from the same total RNA samples used for the microarray analyses was reverse transcribed using M-MLV reverse transcriptase and oligo(dT) from the Retroscript cDNA synthesis system (Ambion). The amplification reactions were conducted in 96-well plates in 25-μl reaction volumes containing 12.5 μl of 2X SYBR Green Master Mix (PE Applied Biosystems, Foster City, CA), 50 nmol/L each of forward and reverse primers, and 1 μl of the cDNA and monitored in an ABI Prism 7700 Sequence Detector System (PE Applied Biosystems). The thermal profile for the PCR was 50°C for 2 minutes and 95°C for 10 minutes followed by 40 cycles of 95°C for 10 seconds (denaturation step) and 60°C for 1 minute (annealing and elongation steps). Measurements on each sample were performed in triplicate, and the expression of the tested gene was normalized to a GAPDH standard curve run in duplicates on the same plate.
Results
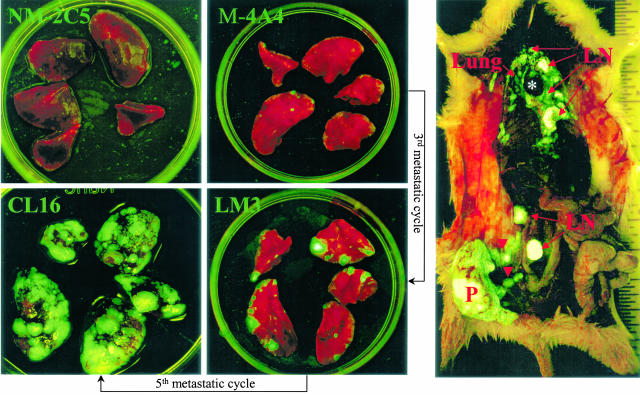
Our investigative strategy compared the gene expression patterns associated with the metastatic behaviors of three isogenic human breast tumor cell lines and the primary tumors that they generated after orthotopic inoculation in nude or SCID mice. It also examined and compared gene expression profiles of metastases in various organs with each other and with the primary tumors. One of these lines generates tumors that are essentially nonmetastatic (NM-2C5), whereas the other two produce ones that are moderately (LM3) or highly (CL16) metastatic to the lungs and lymph nodes (Figure 1). The differences are great in magnitude and obvious but not absolute. In some batches of mice inoculated with NM-2C5, occasional metastases are seen, whereas in other batches, there are none. For convenience and brevity, we shall refer to metastatic or nonmetastatic primary tumors. LM3 and CL16 are third and fifth generation descendents of the M-4A4 metastatic line originally isolated by Bao et al6 from MDA-MB-435 and were obtained by cyclically culturing and orthotopically re-inoculating the cells of successive generations of metastases as originally described by Fidler.16 The degree of metastatic aggressiveness attained by the fifth cycle compared with ancestral lines is readily seen from the overwhelming burden of fluorescent cancer cells colonizing the lungs and lymph nodes (Figure 1).
Figure 1.
Visualization of GFP-labeled tumors and metastases in our xenogeneic breast cancer metastasis model. Left: Increasing metastatic loads in the five pulmonary lobes of mice bearing 2-cm primary tumors developed after orthotopic inoculation of isogenic clonal cell lines with progressively enriched metastatic capabilities, as indicated. Right: A survey view of the heavy metastatic spread of a CL16 primary tumor in the whole animal. Numerous tumor cells leaving the primary tumor (P) distended the afferent lymphatic vessels (arrowheads). Pulmonary and lymphatic metastases are also indicated. An asterisk indicates the heart.
Screening Studies with Oligonucleotide Microarrays
The tissue samples were obtained from two animal experiments; one in severe combined immunodeficient (SCID) mice and a second in nude mice. In SCIDs, the metastatic line CL16 is overwhelmingly more metastatic than in nude mice, although the metastatic capability of NM-2C5 remains low in both strains. The SCID mouse study was, therefore, the primary experiment, and the one in nude mice was a backup for testing conclusions in another strain. In tissues from SCID mice, the expression levels of 22,000 genes were screened in three NM-2C5 and five CL16 primary tumors as well as dissected and cleaned lung and thoracic lymph node metastases from the same five animals bearing the CL16 tumors. RNA from each primary tumor or metastasis sample was hybridized to a separate individual GeneChip, comprising a total of 18 microarrays on SCID samples (see supplemental material at http://ajp.amjpathol.org). The studies on samples from nude mice were conducted on 4 microarrays hybridized with primary NM-2C5 tumors, 7 arrays with primary LM3 or CL16 tumors, and 10 with lung metastases, totaling 21 arrays. In addition, we screened gene expression patterns in the parent NM-2C5 and M-4A4 cell lines and in CL16 in vitro using RNA from three distinct cultures of each line pooled and hybridized to a separate individual array. The grand total of arrays used in the work described in this communication, therefore, amounts to 42.
To evaluate the degree of consistency of the global gene expression profiles among samples in each biological category, chip-to-chip comparisons of the signal intensities from the complete set of genes present on the array were performed for the data from the SCID animals, which we later used to train the algorithms we used to test the data from the nude animals. The scatter plots obtained by comparing data from each SCID microarray with the others from the same tumor category (ie Mor NM primaries or metastases) showed tight grouping of the points, and the correlation coefficients ranged between 0.89 and 0.98, with the majority between 0.97 and 0.98. These analyses, using thousands of data points from each chip, established that the biological samples in each category showed good consistency and were suitable for more detailed studies.
Quantitative PCR Validation of a Subset of Differentially Expressed Genes Selected from the Microarray Data Analyses
To evaluate the microarray results using a different method, we selected 41 genes of interest to us, from the 22,000 present on the HG-U133A chip, and quantified their expression in vitro (cell lines) and in vivo (primary tumor/metastasis samples) by real-time PCR using human-specific primers. The genes chosen for validation by this independent technology covered the low- and high-signal-intensity range and included 37 differentially expressed genes in one or the other of the pathologically relevant comparisons (nonmetastatic versus metastatic primary tumor or metastases versus primary), as well as four genes that were not differentially expressed (fc <1.5). Numerical data and gene identities are available online on the Web site (http://ajp.amjpathol.org).
The real-time PCR data for the cultured cell lines showed that 76% of the microarray fc were confirmed to be in the same trend and that only 9% had a trend in the opposite direction. The remaining 15% were distributed between 12% false positive (fc >1.5 detected only by microarray quantification) or 3% false negative (fc >1.5 detected only by quantitative PCR (Q-PCR)) categories. It was also found that, because Q-PCR had much greater sensitivity and dynamic range of quantitation, the magnitudes of fold changes of differentially expressed genes were underestimated by microarray analysis. Similar analysis of the datasets for the tumor tissue samples in vivo showed that 49% of the fold changes identified as differentially regulated by microarrays were validated by Q-PCR and that 15% displayed a reverse trend. The frequency of false-positive results was approximately similar to that seen in the pure cell lines, but the false-negative results increased from 3 to 22%. Because the chips used in all our experiments were of the same type, the difference in validated fold changes identified by them in cells versus tissues is attributed to a confounding effect from host (mouse) transcripts in the latter. The value of 76% fc validated on pure human cells with an additional 12% false positive by microarray indicates that the technique is not missing much important information.
Collectively, these results indicate that oligonucleotide microarray technology is a valuable screening tool for selecting potentially interesting candidate genes for further study, but real-time PCR validation is an essential requirement, especially in a xenogeneic system in which human mRNA is co-extracted with mouse (host) transcripts from the intermingled cell populations in the tumors and the metastases. Our validation Q-PCR studies, using confirmed human-specific primers, established that many of the differences in gene expression revealed by HG-U133A microarray analysis between nonmetastatic and metastatic primary tumors and distant metastases were attributable to the human tumor cells within them. It should be noted that, to be very conservative in interpretation, any differential expression of less than fc 1.5 measured by Q-PCR in our validation experiments was recorded as not validated. Therefore, although differential changes seen by microarray were indicated as correct by the Q-PCR readings for many genes, we chose to exclude them in case they were within the margin of experimental error.
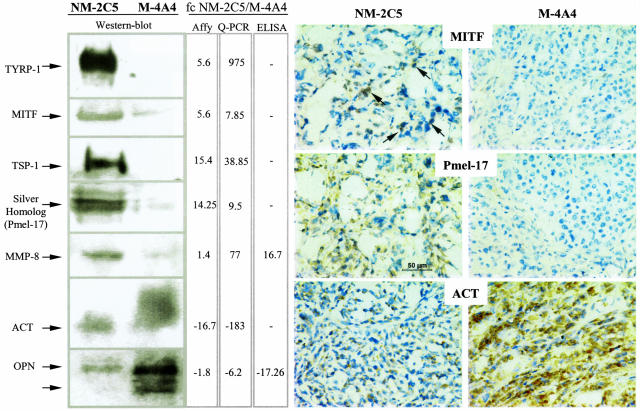
Confirmation of differential gene expression at the protein level is also desirable, and we did this by Western blot, ELISA, or immunohistochemistry (Figure 2) for seven of these genes (TYRP-1, MITF, TSP-1, Pmel-17, MMP-8, ACT, and OPN) using antibodies raised against human antigens. Several of these proteins (OPN, ACT, MMP-8, and TSP-1) were differentially detectable in the serum of the tumor-bearing animals, indicating the possibility of detecting potential biomarkers by this approach.
Figure 2.
Protein validations of candidates selected on microarray-based differential expression between the nonmetastatic and the metastatic tumors in BalbC mice. Changes in mRNA expressions estimated by microarray analyses and quantified by real-time PCR (middle columns) were validated at the protein level by quantitative (ELISA, third column) or semiquantitative methods (Western blotting, left; immunohistochemistry, right) as described in Materials and Methods. Both the “Affy” and the “Q-PCR” fold changes came from the Q-PCR validation table provided in the supplemental material. This explains why, for some genes, the microarray data indicated in this figure may not exactly match those in Table 1, which are based on SCID comparisons. All of the stained tumor sections were observed at 400× magnification, and a scale bar was inserted in the NM-2C5 tumor section stained with the anti-Pmel-17 antibody. Arrows in the upper left show examples of positive signals of MITF localization in the nuclei of nonmetastatic tumor cells.
Candidate Genes Putatively Involved in the Metastatic Process
Microarray Experiment Design and Comparative Analysis
To enhance discrimination of the metastasis-related signals from nonspecific background expression due to the biological variations of an in vivo system, a “training” data set derived from CL16 tumors and their matched metastases to lymph nodes or lungs from the SCID experiments was first used for preliminary comparisons. The results were then validated with the “test” data set consisting of biological samples from a different strain of host, namely nude mice. This policy enabled us to perform paired t-tests with more statistical power to eliminate gene expression due to individual variation that is irrelevant to metastasis.
All biologically relevant comparisons illustrated in Figure 3 were performed using dChip software as described in Materials and Methods. The comparison of expression levels for each gene between two biological sample groups generated fold changes with attached P values. Instead of focusing on high fold change based on group means, we opted to rank-order the candidate gene lists by high consistency of differential expression (low P values) in specimens from a given category (Tables 1–6). This approach of emphasizing statistical measures of data consistency over fold change is a defining feature of this work, a direct benefit of the tightly controlled experiment design. In addition, it was chosen because of the limited dynamic range and accuracy of the microarray results, revealed by the Q-PCR validation experiment, and our intention to have a high level of confidence in the candidate genes selected for further study.
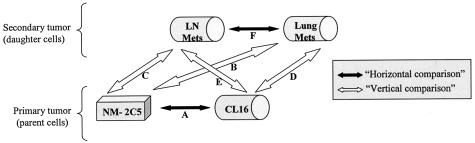
Figure 3.
Schematic diagram of the “horizontal” (black arrows) and “vertical” (white arrows) comparisons performed in the microarray data analyses. dChip software14,15 was used to compare global expression datasets obtained with the HG-U133A array hybridized with the nonmetastatic (NM-2C5) and metastatic (CL16) primary tumors and the spontaneous metastatic deposits developing in the lung (Lung Mets) or in the thoracic lymph nodes (LN Mets) in SCID mice.
Six categories of in vivo comparisons made in the horizontal (nonmetastatic versus metastatic primary tumors) or vertical dimensions (metastatic or nonmetastatic primary tumors versus metastases) (Figure 3) and the distribution of differentially expressed genes are displayed according to their P values in Table 7. This results in 12 groups of differentially expressed genes (ie, each either up- or down-regulated in the partners of a given comparison). Although values of P < 0.05 are adopted in most published microarray studies, we were able, because of our matched biological samples, to use more stringent P values (in the range P < 0.001 to P < 0.005) in pair-wise comparisons. This had the additional benefit of reducing our lists of differentially expressed genes to manageable numbers, even when the threshold fc was lowered to 1.5, which still represents a change of expression of 50% for any given gene. The combination of parameters that we used (low P value and low fold change) was designed to detect reliable minor changes, which could be biologically significant, depending on the function of the gene.
Table 7.
Distribution of the Differentially Expressed Genes in Metastasis-Related Groups Generated from the Horizontal and Vertical Comparisons in Vivo and from the Cell Line Comparisons in Vitro
| In vivo | Genes up-regulated
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nonmetastatic tumor
|
Metastatic tumor
|
Lung metastases
|
LN metastases
|
|||||||||||
| Compared samples | fc | P < 0.001 | P < 0.005 | P < 0.05 | P < 0.001 | P < 0.005 | P < 0.05 | P < 0.001 | P < 0.005 | P < 0.05 | P < 0.001 | P < 0.005 | P < 0.05 | |
| NM-2C5T | M-4A4T | fc > 1.5 | 142 | 386 | 1068 | 219 | 416 | 697 | ||||||
| fc > 3 | 17 | 43 | 92 | 55 | 77 | 101 | ||||||||
| NM-2C5T | M-4Lu | fc > 1.5 | 117 | 323 | 969 | 193 | 403 | 660 | ||||||
| fc > 3 | 12 | 31 | 82 | 51 | 74 | 90 | ||||||||
| NM-2C5T | M-4LN | fc > 1.5 | 114 | 329 | 886 | 190 | 379 | 680 | ||||||
| fc > 3 | 15 | 35 | 87 | 48 | 67 | 95 | ||||||||
| M-4A4T | M-4Lu | fc > 1.5 | 1 | 9 | 69 | 2 | 17 | 88 | ||||||
| fc > 3 | 0 | 0 | 6 | 1 | 2 | 8 | ||||||||
| M-4A4T | M-4LN | fc > 1.5 | 14 | 56 | 279 | 14 | 58 | 184 | ||||||
| fc > 3 | 2 | 8 | 27 | 3 | 9 | 17 | ||||||||
| M-4Lu | M-4LN | fc > 1.5 | 4 | 14 | 50 | 0 | 1 | 20 | ||||||
| fc > 3 | 1 | 1 | 4 | 0 | 0 | 0 | ||||||||
| In vitro | Nonmetastatic line | Metastatic line | Highly metastatic line | ||
|---|---|---|---|---|---|
| NM-2C5 | M-4A4-GFP | fc > 1.5 | 363 | 361 | |
| fc > 3 | 40 | 28 | |||
| NM-2C5 | LM3 clone 16 | fc > 1.5 | 1708 | 930 | |
| fc > 3 | 198 | 113 | |||
| M-4A4-GFP | LM3 clone 16 | fc > 1.5 | 1473 | 336 | |
| fc > 3 | 155 | 70 |
Horizontal Comparisons among Primary Tumors
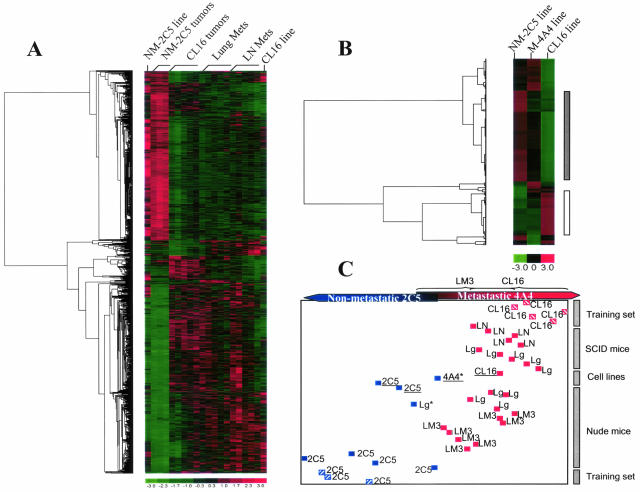
Screening among the 22,000 features on the human HG-U133A chip for differentially expressed genes between the NM-2C5 and CL16 tumors in the SCID mice resulted in a list of candidate genes ranging from 72 (55+17) to 1765 (1068+697) genes when different filtering criteria were applied (Table 2). To evaluate the biological significance of the resultant gene lists, we performed linear discriminant analysis (LDA) to evaluate whether each list would classify additional test samples (from the experiment in nude mice) into the nonmetastatic group or the metastatic group (Figure 4C) correctly. The results suggested that at P < 0.005 and fc >1.5, the classification of the 17 samples from a separate (duplicate) experiment in a different mouse strain was close to optimal, with only two arrays being borderline misclassified (93.5% accuracy). In the corresponding gene list (P < 0.005), 386 candidates were found to be up-regulated more than 1.5 times in the nonmetastatic primary tumor compared with its metastatic counterpart, and 416 were up-regulated in the metastatic primary tumor (Table 2). Thus, of a total of 802 genes that are consistently differentially expressed in the CL16 tumors, approximately one-half of them have to be down-regulated, and the other one-half, up-regulated. It follows that some genes must be “turned on,” but also that others need to be “turned off,” for the metastatic phenotype to be triggered. This observation highlights the potential importance of negative regulators (metastasis suppressor) in metastasis.
Figure 4.
Signature gene clustering and sample classification. Hierarchical cluster analyses of the differentially expressed genes from in vivo and in vitro comparisons. A: Candidate genes differentially expressed (with P < 0.005 and fc >1.5) at least in one of the six in vivo comparisons (illustrated in Figure 3) were clustered according to their expression levels in the NM-2C5 and CL16 lines in vitro and in the primary and secondary tumors they generated in SCID mice. B: Candidate genes differentially expressed (with fc >3) at least in one of the three in vitro comparisons of the cell lines (Table 7) were clustered according to their expression levels in NM-2C5, LM3, and CL16 lines. The vertical bars on right of this panel indicate the candidate genes gradually overexpressed (white bar) or down-regulated (gray bar) in regard with the progressive acquirement of high metastatic capabilities. As shown in the color bar, red indicates high expression; green, low expression; and black, intermediate expression. The dendogram on the left indicates the pairing of genes, and the branch length is proportional to the distances between the clusters. C: Linear discriminant analysis using NM-2C5 (blue striped squares) and CL16 (red striped squares) primary tumors in SCID mice as the training set to classify unknown samples (test set) according to the combined candidate gene list used in A for the clustering analysis. Plain blue squares: Samples classified into the nonmetastatic group; plain red squares: samples classified into the metastatic group. The unknown samples include primary tumors (NM-2C5 and LM3), metastases in the lung (Lg) or lymph nodes (LN), and cell lines (underlined). The asterisk shows misclassified samples.
Vertical Comparisons among Primary Tumors and Metastases
An identical statistical strategy was used in a vertical comparison (Figure 3) between the primary (CL16) tumors and their metastatic deposits in the lungs and lymph nodes. The term “vertical” is used to describe a spatio-temporal comparison between a sample and its biological progeny located in a different organ. The tumor cells constituting the secondary deposits are the direct descendants of the experimentally selected clone of neoplastic cells forming the primary tumor and are, therefore, genetically almost identical to them. Theoretically, one might speculate that the changes that generate metastasis might be evident in both populations and that their expression profiles might be very similar. Indeed, we found (Table 2) that their expression profiles showed little differences in contrast to when the metastatic and nonmetastatic primary tumors were compared (ie, in the horizontal comparison): only 26 (17+49) genes were more than 1.5-fold differentially expressed between lung metastases and their corresponding CL16 primaries at the high level of significance (P < 0.005). For lymph node metastases, the corresponding figure was 114 (56+58) genes.
Because the original breast tumor from which MDA-MB-435 was derived was composed of a mixture of metastatic, less metastatic, and nonmetastatic tumor cell populations, we also compared the expression profiles of metastases with that of sister (isogenic) cells in nonmetastatic NM-2C5 primary tumors, growing in a different set of mice of the same strain (Table 3). This comparison is clinically and pathogenetically relevant, because it represents the extreme ends of the phenotype but cannot be performed in “wild-type” tumors and is, uniquely, only possible in this experimental system. It would be expected to reveal metastasis-relevant genes that are expressed in both primary and secondary tumors of the metastatic phenotype (CL16) and therefore are not seen as differentially expressed when these two categories are compared. Surprisingly, both lung and lymph node metastases harbor slightly less differentially expressed genes than the parental metastatic primary tumors (CL16) when compared with the NM-2C5 tumors. This result suggests that the CL16 cells residing in a primary tumor have already acquired an enriched metastatic gene expression profile as a result of the cycling and selection procedures and that this profile is inherited by the cells in the metastases.
Once again, samples of metastases in the lungs and primary tumors in the breast from the duplicate experiment in nude mice were correctly classified by the LDA procedure (Figure 4C).
Combination of Comparisons
Each of the pair-wise comparisons performed above only represents a segment of the “metastatic spectrum.” Therefore, we combined all six candidate gene lists to create a working database comprising 1127 differentially expressed genes putatively involved in progression from a basal to an enhanced metastatic potential in the primary tumor, extending to a consummated metastatic profile in lymph node and lung metastases. To visualize changing expression patterns of these genes accompanying transitions between tumors of low, higher, and highest metastatic potency, we performed supervised hierarchical clustering of the biological samples using the “pooled” candidate gene list (Figure 4A). This procedure graphically demonstrated the similarities between prevailing expression patterns in the cells in metastatic lesions in different organs and in their parent tumors as well as the differences from their phenotypically opposite counterparts. Additionally, we included in this analysis the expression data from cultures of NM-2C5 and CL16 cell lines in vitro. This permitted side-by-side comparison with the in vivo data and indicated that several genes were regulated by the host microenvironment while many others were expressed at similar levels both in vitro and in vivo. Within the NM-2C5 category, the expression levels of these 1127 pooled candidate genes were more stable and conserved between in vivo and in vitro samples. In contrast, the CL16 samples displayed greater magnitudes of changes in expression levels for these genes, and approximately one-half of them were highly inducible by the host microenvironment. Therefore, our in silico reconstruction of tumor progression toward metastasis indicates that the M-4A4 family of cells (ie, M-4A4, LM-3, and CL16) responded more dynamically to the environment in vivo and developed a characteristic signature that segregated with the metastatic phenotype during accentuation by cyclical selection and reinoculation.
Comparisons of Cell Lines in Vitro
To ascertain whether the changing pattern of expression seen in primary tumors and metastases, as the phenotype became enriched, reflected inherited intrinsic changes in the tumor cells as they were cycled, we compared data from cell lines NM-2C5, M-4A4, and CL16 cells growing in vitro. In this situation, there are no intermingled mouse cells, and the results reflect gene expression patterns in “purified” human cells of these tumor lines growing under essentially identical conditions. It was found that 724 genes (Table 2) were differentially expressed by 1.5-fold or more between the first generation M-4A4 and NM-2C5 cell lines. This number increased to 2638 when the fifth generation CL16 was similarly compared with the NM-2C5 line. More importantly, 377 of the total 724 genes generated by the first comparison were also found in the second comparison. In addition, very few of these 377 genes were regulated in opposite directions in M-4A4 and CL16, consistent with the notion that specific gene expression was segregated along with the metastatic phenotype. Hierarchical clustering of the lines demonstrated the inheritance of the changing gene expression pattern as the cell lines evolved more metastatic phenotype (Figure 4B). Therefore, in the absence of host influence, comparisons of autonomous gene expression profiles reveal a group of human genes that likely represent an essential requirement for metastasis.
Discussion
Although comprising a powerful investigation tool, microarray experiments provide, by design, a summary analysis of (biologically) averaged gene expression profiles. As a result, microarray studies conducted on clinical cancers are often obscured by the uneven amount of cancerous tissues within samples that were harvested from patients of unmatched genetic backgrounds. For the study of metastasis-related gene expression, the difficulties are further compounded by tumor heterogeneity, which translates into uncertainty about the ratio of cell populations in the primary tumor that inherited variable genetic potential for metastasis.
In this study, we therefore designed the biological system to control, if not overcome, the above problems. By using clonal isogenic tumor cell lines of divergent metastatic performance, we focused the enquiry on metastasis-related differential expression and enhanced the potency of the metastatic line by recycling it several times. Also, the use of GFP-labeled human cell lines in immunocompromised unlabeled mice improved the identification of metastases while minimizing contamination by irrelevant tissues during sample dissection, hence enriching gene expression truly associated with the tumor or metastases. On the other hand, the host genetic backgrounds were carefully controlled by performing independent animal experiments in two different stains of mice.
The main findings emerging from this work are 1) that gene expression in clonal metastatic primary breast tumors differed clearly from that seen in isogenic nonmetastatic tumors generated by a different clonal cell population isolated from the same patient; and 2) that the expression patterns in matched pulmonary and lymph nodal metastases closely resembled those in the primary tumor within the same animal. Hierarchical clustering and linear discriminant analysis of the differentially expressed genes from all of the comparisons in two successive experiments in different mouse strains cross-corroborated each other: that metastatic primary tumors and their metastases were very distinct from nonmetastatic primary tumors. Thus the evidence that emerges from this specially designed study of human cancer cells favors the view that cells with a distinct gene expression signature associated with metastasis can be isolated from human neoplasms and that this profile faithfully segregates with the phenotype during cyclical selection procedures designed to enrich metastatic capability.4,17–22 The tumor cells within the primary tumors and metastases in SCIDs were in the fifth cycle of re-derivation and orthotopic inoculation, and the relatively small numbers of differences between lung and lymph node metastasis signatures suggest that many similar biological mechanisms are being executed in reaching and growing in these two different favored sites. Expression profiles of matched bone, brain, or liver metastases would inform us whether the results obtained in this study apply to other preferred sites for human breast cancer metastasis. Unfortunately, these samples are rarely available in this model. Collectively, this is potentially valuable information for further investigations aiming to find prognostic markers and therapeutic targets.
Although the identities of the candidate genes that may be “driving” the process are of substantial interest, one cannot be sure that they can be extrapolated to naturally occurring “wild-type” human cancers. We believe that, at present, the observations are informative only about the system under study, but they do provide potentially valuable leads about genes and pathways that merit functional validation in the experimental model and further investigation in pure human tumors. Also, the candidates, which we provide in Table 2 and on the study Web site, are composite collections of genes that are differentially expressed in the tumor cells and in the host tissues intermingled with them. Thus, some of the results obtained will be reflecting differences between the mouse stroma in the tumors in different organs. The investigative power of this xenogeneic system lies in the opportunity now available to identify which components of this joint signature are contributed by the tumor cells and which by the host stroma, by the use of species-specific primers and antibodies. Earlier studies23–25 involving orthotopic versus ectopic inoculation of carcinoma cells have demonstrated that local host tissues influence metastatic behavior by metastasis-competent cells, and opportunities to analyze host gene expression in the process are therefore valuable. In this way, the present work contributes a foundation for the further study of tumor-stromal interactions in metastasis and for the identification of intrinsic tumor cell genes driving the process.
It is appropriate to compare these findings with previous microarray studies describing gene expression profiles in metastases and primary human tumors. In general, such work has been hampered by the limited availability of suitable human tissue samples. However, lately, two major studies investigated gene expression patterns in metastases from carcinomas of the prostate8 and from primary adenocarcinomas of different organs9 and compared the results with those in corresponding primary tumors from other patients (ie, unmatched primaries). Both concluded that secondary tumors display similar transcriptional profiles to the primary neoplasm, in agreement with Adib et al,26 who described observations on primary ovarian cancers compared with matched omental metastases (although metastasis was hematogenous in the first two studies cited but transcoelomic in the third), and with Weigelt et al,27, who compared individual hematogenous metastases excised from various organs of eight patients with their primary breast cancers. Conversely, two earlier studies on prostate cancer metastasis reported that expression in metastatic samples differed from that in the primary tumor,11,12 although, once again, the primaries and metastases were not matched. The discordance of these observations might result either from primary tumor cell heterogeneity, resulting from differences in the time of emergence of the metastatic cell phenotype, or from comparison of metastases and primary tumors obtained from patients with different genotypes. Both issues were bypassed in our model.
Recently Lee and coworkers28 described a study of xenografted tumors profiled with cDNA spotted microarrays comprising 5800 known genes on glass slides. They had derived two MDA-MB-435 variant cell lines, LN435 and Tho435, which preferentially metastasize to the lymph nodes or to the thoracic cavity, respectively, when orthotopically injected in SCID mice. Gene expression profiles of the primary tumors that they generated after inoculation were compared with the profiles of primary tumors generated by the parent MDA-MB-435 line to derive candidate genes potentially involved in organ specificity of breast cancer metastasis. Although the experimental strategy of our work differed from that of Lee et al,28 by profiling metastatic deposits taken directly from the lungs and lymph nodes and comparing their expression patterns with their isogenic primary tumors in the mammary gland, using 22,000 gene Affymetrix arrays, there are several overlaps between the gene lists provided in the Lee et al28 study and in our own. Despite many conceptual differences between these two studies based on the MDA-MB-435 breast cancer line, the cross-corroboration of a number of interesting candidates by separate laboratories strongly supports their involvement in the metastatic process.
The pioneering work of Fidler1,2 and of many other subsequent investigators4,22,29–34 has established beyond any reasonable doubt that tumors are composed of heterogeneous populations that differ from each other in many qualities including metastatic capability. This has been confirmed by pathologists who have shown that human tumors show zonal heterogeneity with regard to pigmentation, fibrosis, vascular supply, and many other properties. Our own work, reported here and previously,6,7 shows conclusively that the parent (MDA-MB-435) cell line from which we obtained NM-2C5 and M-4A4 clones was heterogeneous and that it contained nonmetastatic cell populations. Thus, the deconstruction exercise by which we derived these clones with polar opposite behavior and the subsequent screening process by which we chose the most suitable tumors for study facilitated and clarified our gene expression analysis. This helps to illuminate how investigations on unmatched and unselected fresh wild-type human tumors may give variable results.
In conclusion, this work, based on oligonucleotide microarray gene profiling, adopted a purpose-designed, new strategy to define patterns of gene expression relevant to the dynamic process of tumor metastasis, using a unique isogenic breast cancer model. The gene-sorting strategy was designed to provide very robust candidates (P < 0.005), even with low fold change values (fc >1.5). We recognize that this statistical stringency could potentially exclude some genes of interest, but we concluded that combining it with pair-wise analysis of signatures from other neoplastic lesions in the same animal offered the best chance of serving our intention to identify genes worthy of further work. Moreover, if this strategy was to exclude an important gene related to metastasis, the global microarray screening technique, by nature, would detect related changes in its relevant gene network and thereby could lead to the identification of the missing candidate. Statistical “training and test” validation of the observations made on repeat experiments in different mouse strains together with biochemical validation of expression of 41 genes constitute further novel aspects of this study building upon and extending previous investigations by other laboratories. These results show the power of microarray analysis for initial high-throughput screening while demonstrating the necessity of corroboration with other techniques. The combination of the methods described enables one to have reasonable confidence that some of the genes driving metastasis lie within these lists, although further sifting is needed. The study design permitted previously impossible comparisons to be made (such as between metastases and isogenic nonmetastatic breast cancers) and revealed molecular information pertinent to the emergence and to the ongoing cyclical continuation of the metastatic phenotype. The main purpose of the work has been to design and use a new tool for the identification of genes that are of biological or clinical interest. The candidate genes so obtained can be screened for new information on possible pathways involved and tested in knock-down and overexpression assays, using this model as well as assayed in fresh human tumor samples.
The new information provided here, showing the importance of the inductive effects of the stroma of the host breast on gene expression by mammary carcinoma cells that are about to metastasize and the preservation of this distinctive expression pattern when their descendants make deposits in the lungs and lymph nodes, opens new insights into the mechanisms of the biology of the phenomenon and fresh possibilities for finding prognostic markers and therapeutic targets.
Supplementary Material
Acknowledgments
We thank Dr. Luminita Castillos, who kindly shared her personal data. We also acknowledge the administrative help of Ms. Diane Sweet and Ms. Linda Mellor, all from University of California-San Diego, in the conduct of this study. We thank the staff of the Moores/University of California-San Diego Cancer Center Microarray facility for providing high-quality work on the oligonucleotide microarrays.
Footnotes
Address reprint requests to David Tarin, Rebecca and John Moores Comprehensive Cancer Center and Department of Pathology, University of California, San Diego, 9500 Gilman Drive, M/C 0912 La Jolla, CA 92093-0912. E-mail: dtarin@ucsd.edu.
Supported by a grant from the Loppieda Foundation.
V.M. and T.-Y.H. contributed equally to this work.
References
- Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978;38:2651–2660. [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Price JE, Carr D, Tarin D. Spontaneous and induced metastasis of naturally occurring tumors in mice: analysis of cell shedding into the blood. J Natl Cancer Inst. 1984;73:1319–1326. [PubMed] [Google Scholar]
- Tarin D, Price JE, Kettlewell MG, Souter RG, Vass AC, Crossley B. Mechanisms of human tumor metastasis studied in patients with peritoneovenous shunts. Cancer Res. 1984;44:3584–3592. [PubMed] [Google Scholar]
- Bao L, Pigott R, Matsumura Y, Baban D, Tarin D. Correlation of VLA-4 integrin expression with metastatic potential in various human tumour cell lines. Differentiation. 1993;52:239–246. doi: 10.1111/j.1432-0436.1993.tb00636.x. [DOI] [PubMed] [Google Scholar]
- Urquidi V, Sloan D, Kawai K, Agarwal D, Woodman AC, Tarin D, Goodison S. Contrasting expression of thrombospondin-1 and osteopontin correlates with absence or presence of metastatic phenotype in an isogenic model of spontaneous human breast cancer metastasis. Clin Cancer Res. 2002;8:61–74. [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, Ekman P, DeMarzo AM, Tibshirani R, Botstein D, Brown PO, Brooks JD, Pollack JR. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Goodison S, Kawai K, Hihara J, Jiang P, Yang M, Urquidi V, Hoffman RM, Tarin D. Prolonged dormancy and site-specific growth potential of cancer cells spontaneously disseminated from nonmetastatic breast tumors as revealed by labeling with green fluorescent protein. Clin Cancer Res. 2003;9:3808–3814. [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH: Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol 2001, 2:RESEARCH0032. Epub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ. Selection of successive tumour lines for metastasis. Nat New Biol. 1973;242:148–149. doi: 10.1038/newbio242148a0. [DOI] [PubMed] [Google Scholar]
- Brunson KW, Beattie G, Nicolsin GL. Selection and altered properties of brain-colonising metastatic melanoma. Nature. 1978;272:543–545. doi: 10.1038/272543a0. [DOI] [PubMed] [Google Scholar]
- Tao T, Matter A, Vogel K, Burger MM. Liver-colonizing melanoma cells selected from B-16 melanoma. Int J Cancer. 1979;23:854–857. doi: 10.1002/ijc.2910230618. [DOI] [PubMed] [Google Scholar]
- Edel G, Grundmann E. Selection of liver-colonizing tumor cells from a murine fibrosarcoma induced by methylcholanthrene. J Cancer Res Clin Oncol. 1984;108:274–280. doi: 10.1007/BF00390457. [DOI] [PubMed] [Google Scholar]
- Pettaway CA, Pathak S, Greene G, Ramirez E, Wilson MR, Killion JJ, Fidler IJ. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin Cancer Res. 1996;2:1627–1636. [PubMed] [Google Scholar]
- Barroga EF, Kadosawa T, Okumura M, Fujinaga T. Establishment and characterization of the growth and pulmonary metastasis of a highly lung metastasizing cell line from canine osteosarcoma in nude mice. J Vet Med Sci. 1999;61:361–367. doi: 10.1292/jvms.61.361. [DOI] [PubMed] [Google Scholar]
- Price JE, Carr D, Jones LD, Messer P, Tarin D. Experimental analysis of factors affecting metastatic spread using naturally occurring tumours. Invasion Metastasis. 1982;2:77–112. [PubMed] [Google Scholar]
- Price JE, Polyzos A, Zhang RD, Daniels LM. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res. 1990;50:717–721. [PubMed] [Google Scholar]
- Morikawa K, Walker SM, Nakajima M, Pathak S, Jessup JM, Fidler IJ. Influence of organ environment on the growth, selection, and metastasis of human colon carcinoma cells in nude mice. Cancer Res. 1988;48:6863–6871. [PubMed] [Google Scholar]
- Bao L, Matsumura Y, Baban D, Sun Y, Tarin D. Effects of inoculation site and Matrigel on growth and metastasis of human breast cancer cells. Br J Cancer. 1994;70:228–232. doi: 10.1038/bjc.1994.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adib TR, Henderson S, Perrett C, Hewitt D, Bourmpoulia D, Ledermann J, Boshoff C. Predicting biomarkers for ovarian cancer using gene-expression microarrays. Br J Cancer. 2004;90:686–692. doi: 10.1038/sj.bjc.6601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Glas AM, Wessels LF, Witteveen AT, Peterse JL, van’t Veer LJ. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc Natl Acad Sci USA. 2003;100:15901–15905. doi: 10.1073/pnas.2634067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Lin ECK, Liu L, Smith JW. Gene expression profiling of tumor xenografts: in vivo analysis of organ-specific metastasis. Int J Cancer. 2003;107:528–534. doi: 10.1002/ijc.11428. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Withers HR, Koehler MW. Heterogeneity and variability of artificial lung colony-forming ability among clones from mouse fibrosarcoma. Cancer Res. 1978;38:3349–3351. [PubMed] [Google Scholar]
- Tarin D, Price JE. Metastatic colonization potential of primary tumour cells in mice. Br J Cancer. 1979;39:740–754. doi: 10.1038/bjc.1979.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri A, Nicolson GL. Phenotypic drift of metastatic and cell-surface properties of mammary adenocarcinoma cell clones during growth in vitro. Int J Cancer. 1981;28:731–738. doi: 10.1002/ijc.2910280612. [DOI] [PubMed] [Google Scholar]
- Poste G, Doll J, Fidler IJ. Interactions among clonal subpopulations affect stability of the metastatic phenotype in polyclonal populations of B16 melanoma cells. Proc Natl Acad Sci USA. 1981;78:6226–6230. doi: 10.1073/pnas.78.10.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poste G. Cellular heterogeneity in malignant neoplasms and the therapy of metastases. Ann NY Acad Sci. 1982;397:34–48. doi: 10.1111/j.1749-6632.1982.tb43415.x. [DOI] [PubMed] [Google Scholar]
- Wang N, Yu SH, Liener IE, Hebbel RP, Eaton JW, McKhann CF. Characterization of high- and low-metastatic clones derived from a methylcholanthrene-induced murine fibrosarcoma. Cancer Res. 1982;42:1046–1051. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.