Abstract
Prostate cancer is frequent among men over 45 years of age, but it generally only becomes lethal with metastasis. In this study, we identified a gene called cellular stress response 1 (CSR1) that was frequently down-regulated and methylated in prostate cancer samples. Survival analysis indicated that methylation of the CSR1 promoter, and to a lesser extent down-regulation of CSR1 protein expression, was associated with a high rate of prostate cancer metastasis. Forced expression of CSR1 in prostate cancer cell lines DU145 and PC3 resulted in a two- to threefold decrease in colony formation and a 10-fold reduction in anchorage-independent growth. PC3 cells stably expressing CSR1 had an average threefold decrease in their ability to invade in vitro. Expression of CSR1 in PC3 cell xenografts produced a dramatic reduction (>8-fold) in tumor size, rate of invasion (0 versus 31%), and mortality (13 versus 100%). The present findings suggest that CSR1 is a potent tumor suppressor gene.
Prostate cancer is one of the leading causes of death for men in the United States. Since the implementation of the serum prostate-specific antigen (PSA) screening program, the clinical detection rate of prostate cancer has increased substantially as otherwise unnoticed micro-adenocarcinomas of the prostate gland are found.1 Even though many of these prostate cancers are very slow growing and are likely not to be clinically relevant for many patients, nearly 30,000 patients die of prostate cancer annually.2 Despite the tremendous advance in our knowledge about prostate cancer in recent years, the molecular mechanism by which a relatively indolent disease is converted into a highly aggressive and lethal one remains unclear.
Our recent comprehensive gene expression analysis study of prostate cancer tissue revealed that the expression levels of 671 genes and expressed sequence tags were significantly altered in prostate cancer tissues relative to control tissues.3 In this study, we identified one of these genes, called cellular stress response 1 (CSR1), a homolog of macrophage scavenger receptor, to be consistently down-regulated in prostate cancer tissue and to be closely associated with cancer progression. CSR1, located in one of the loci (8p21) frequently linked to prostate cancer progression,4–6 was found to be frequently methylated among prostate cancers that metastasize. CSR1 immunohistochemistry using a prostate tissue array that indicated that the level of CSR1 protein was down-regulated in most of the aggressive prostate cancer cases. These findings strongly suggest that prostate cancer tumor growth and metastasis may be affected by reduction of CSR1 expression and/or methylation. We combined multiple approaches to examine this possibility. First, we conducted a small meta-analysis of three human prostate cancer CSR1 expression data sets. We then transfected PC3 and DU145 prostate cancer cell lines with CSR1-expressing vectors. The effects of CSR1 expression on colony formation, anchorage-independent growth, and invasiveness in a Matrigel transmigration assay were examined. In addition, we examined whether tumorigenesis and metastasis differ between severe combined immunodeficiency mice (SCID) injected with CSR1-expressing or control cells.
Materials and Methods
Cell Lines and Specimens
PC3 and DU145 cell lines, purchased from American Type Culture Collection, Inc. (Manassas, VA), were propagated in monolayer cultures in F-12K medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin at 37°C in 5% CO2. Tissue array blocks were constructed from 343 formalin-fixed and paraffin-embedded tissue blocks that had been stored at the University of Pittsburgh Medical Center between 1981 and 2001. Tissue sections were prepared from formalin-fixed paraffin-embedded tissue blocks. The sections were individually deparaffinized in xylene, submerged in ethanol, rinsed with water, and air-dried. All slides were stained with hematoxylin and eosin and viewed under a light microscope to confirm the histological presence of tumor cells.
Bisulfite Treatment and Methylation-Specific Polymerase Chain Reaction (PCR)
Microdissection was performed using a laser capture microdissection microscope (PixCell II LCM System; Arcturus Engineering, Mountain View, CA). The protocols for tissue collection and experiments were approved by our institutional review board. Each cell sample was considered to be 100% “homogeneously” malignant, as determined by a careful microscopic visualization of the cells captured. DNA was extracted using QIAamp DNA Mini kit (QIAGEN, Valencia, CA). Two micrograms of DNA was denatured in 0.3 mol/L NaOH at 37°C for 30 minutes. The denatured DNA was incubated in 40.5% sodium hydrogen sulfite (Sigma, Columbus, OH) and 10% hydroquinone (Sigma) at 50°C for 16 hours. The modified DNA was then purified by a Wizard DNA Cleanup System (Promega, Madison, WI), eluted in 100 μl of tris-EDTA buffer (pH 8.0), further denatured in 0.3 mol/L NaOH at 37°C for 15 minutes, and precipitated with 7 mol/L ammonium acetate in 80% ethanol. The modified DNA was used as a template for methylation-specific PCR. PCR was performed in a final 50-μl reaction mixture containing 10 mmol/L Tris-HCl, pH 8.3, 1.5 mmol/L MgCl2, 50 mmol/L KCl, 100 μmol/L of each dNTP, 0.5 μmol/L of each primer, and 1.25 units of TaqDNA polymerase (Perkin Elmer, CT). The mixture was heated at 94°C for 5 minutes then subjected to 45 cycles of the following program: 94°C for 30 seconds, 62°C for 1 minute, and 72°C for 2 minutes. The primers used for methylated CSR1 were 5′-GGCGGTTTCGATAGTGAAATGGAT-3′ and 5′-CCCCACAACCCAACCTCGAACC-3′, and for unmethylated CSR1 were 5′-GGTGGTTTTGATAGTGAAATGGAT-3′ and 5′-CCCCACAACCCAACCTCAAACC. The sequence of degenerative primers are 5′-GG(T/C)GGTTT(T/C)GATAGTGAAATGGAT-3′ and 5′-CCCCACAACCCAACCTC(A/G)AACC-3′ for pair 1, and 5′-G(T/C)GGGGTTTAAGAAGTGA(T/C)G-3′ and 5′-ACCCAA-AAC(A/G)C(A/G)CATCCC(A/G)CC-3′ for pair 2.
Construction of Stable CSR1 Expression in PC-3 and DU145 Cells
CSR1 cDNA was generated by reverse transcription PCR on RNA purified from donor blood by standard methods using 5′-GCATTGCCCTTCTTCTAACGGA-3′ as the sense primer and 5′-ATAGCAGATTGACAGTGACAG-C-3′ as the antisense primer. The PCR was performed using the following program: 94°C for 1 minute, 65°C for 1 minute, and 72°C for 2 minutes for 30 cycles. The PCR product was gel-purified, directly cloned into a pCR-XL-TOPO vector (Invitrogen, Carlsbad, CA), and sequenced. The CSR1 cDNA was subsequently digested with NotI and KpnI and ligated into the similarly restricted pCMV-script vector. The pCMV-CSR1 construct was sequenced, and it was confirmed that no mutation was introduced into the insert during the cloning process. The constructs were transfected into PC-3 cells and Du145 cells using the protocols of Superfect (Qiagen, Valencia, CA). Stable CSR1 expression cell lines were generated with G418 (400 μg/ml) selection.
For construction of CSR1-inducible expression vector, the CSR1 full-length cDNA produced above was restricted and ligated into the NotI and KpnI sites of pcDNA4.1 (Invitrogen). This plasmid was then co-transfected into PC3 cells with pcDNA6 encoding tetracycline repressor. Transfected cells were selected using Blasticidin (500 μg/ml) and Zeocin (1 μg/ml) (Invitrogen). Clones were expanded and tested for inducibility by exposure to 1 μg/ml doxycycline and by Western blot analysis with antibody specific for CSR1 and β-actin.
Generation of Antibodies against CSR1
CSR1 antisera were raised in rabbits immunized with peptides corresponding to CSR1 coding sequence regions (AGLDLSLKDLTQECYDVKAAVHQINF). These antisera were peptide affinity-purified using the ami-nolink kit from Pierce (Rockford, IL). The purified antisera were tested for binding specificity for CSR1 in a Western blot with protein extracts from cells overexpressing CSR1 (PDC1 and PDC4) or cells not expressing CSR1 (PC3 and LNCaP cells). The antisera detected a single band of 72-kd protein (the predicted molecular weight of CSR1).
Colony Formation and Soft Agar Colony Formation Assay
Five thousand cells were plated in 60-mm dishes. Triplicate experiments were performed for each cell clone. The medium was changed every 4 days. On the 10th day, the plates were stained with 1% crystal violet, and the colonies were quantified. For soft agar colony formation assay, 5000 cells were plated and grown on a plate containing 2% base agar and 0.43% top agar. Plated cells were incubated at 37°C for 21 days. Plates were stained with 0.005% crystal violet for 1 hour. Colonies were counted under a dissecting microscope.
Invasion Assay
Invasion of tumor cells was assessed by counting the number of cells that migrated through Matrigel inserts with 8-μm pores (Becton Dickinson, Woburn, MA), according to the protocol recommended by the manufacturer. Briefly, cells were plated in the top chamber (4 × 104/chamber). An 8-μm-pore size Matrigel-coated polycarbonated filter separated the top and bottom chamber. The bottom chamber contained 5% fetal bovine serum as a chemoattractant. After a 24-hour incubation period at 37°C in a 5% CO2 atmosphere, the noninvasive cells were removed with a cotton swab. The cells that had migrated through the membrane and stuck to the lower surface of the membrane were fixed with methanol and stained with hematoxylin and eosin. For quantification, cells were counted under a light microscope within fields at magnification ×40. For each membrane, the mean number of cells in each of five randomly selected fields was determined.
Tumor Growth and Spontaneous Metastasis Assay
SCID mice, purchased from Taconic, Inc. (Germantown, NY), were subcutaneously implanted at the abdominal flank with 1 × 107 viable cells and suspended in 0.2 ml of Hanks’ balanced salt solution. The animals were observed daily. Body weight, tumor size, and other special findings, including lymph-node enlargement, were recorded weekly. Six weeks after cell incubation, the mice were sacrificed, and necropsies were performed. Serial sections of lung, brain, liver, kidneys, vertebra, and lymph nodes were performed. These tissues were formalin-fixed and paraffin-embedded. The sections were stained with hematoxylin and eosin and subjected to histological examination. All studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Immunohistochemistry and Tissue Array Analysis
Formalin-fixed and paraffin-embedded human prostate tissues including prostate cancer (n = 210) and normal prostate specimens (n = 133) from 220 individuals were arrayed onto slides in a format of single or duplicate representation for each individual. The ages of patients ranged from 15 to 79. Complete 5-year follow-up data are available for 185 of the prostate cancer samples. For immunohistochemistry, 4-μm-thick sections of tissue array were cut and mounted onto glass slides. The sections were heated at 60°C for 12 hours and deparaffinized in xylene and ethanol. Antigen retrieval was performed using 25 mmol/L sodium citrate buffer (pH 9.0) at 90°C for 15 minutes, followed by treatment of 3% H2O2 to block endogenous peroxidase. The slides were incubated at room temperature for 2 hours with anti-CSR1 antibodies at a 1:400 dilution. The sections were then incubated with horseradish peroxidase-conjugated anti-rabbit IgG for 30 minutes at room temperature. For visualization, horseradish peroxidase was reacted with 3,3′-diaminobenzidine solution (DAKO, Carpinteria, CA). Cells were counterstained with hematoxylin. Immunohistochemical specificity was verified by incubating similar slides with preimmune sera.
Results
CSR1 Expression Is Down-Regulated in Prostate Cancer
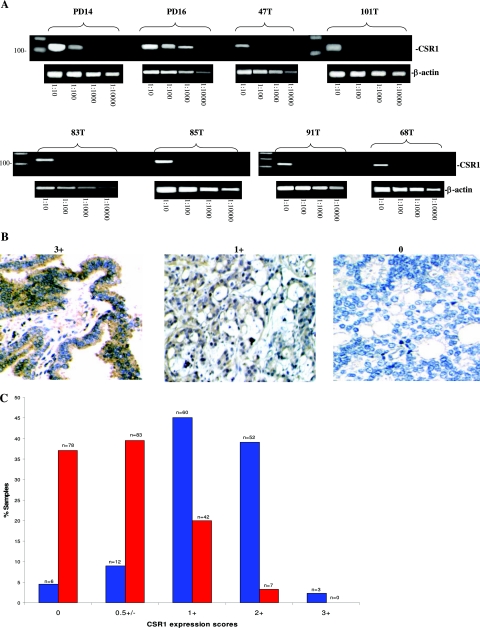
The CSR1 gene was originally identified as a cellular stress response protein. It is widely expressed in most tissues, and it functions to protect cells from the damaging effects of reactive oxygen intermediates.7 Analysis of three data sets of Affymetrix oligo array indicates that CSR1 mRNA expression was down-regulated in the prostate cancer tissues (Table 1). CSR1 was down-regulated in 9 of 15 prostate cancer samples overall in the Luo et al8 data set and in 5 of 7 relapsed prostate cancers. An average 1.93-fold decrease was observed for nonrelapsing prostate cancer, and an average 4.3-fold decrease was observed for relapsing prostate cancer. In the data of Yu et al,3 CSR1 was down-regulated 3.26-fold in prostate cancer that did not metastasize within 5 years of prostatectomy. CSR1 was down-regulated 4.49-fold in prostate cancer samples with PSA relapse. The higher differential data of the most recent analysis is a result of using completely normal prostate tissues as the controls. When using 23 prostate cancer samples from LaTulippe et al9 to compare with donors samples, similar results were obtained. To validate the microarray data, semiquantitative reverse transcriptase-PCR (RT-PCR) on eight selected prostate samples and three prostate cancer cell lines were performed. The results showed consistent and substantial down-regulation of CSR1 mRNA in cancerous prostate tissues (Figures 1A and 2C).
Table 1.
Down-Regulation of CSR1 mRNA Expression in Prostate Cancer
Figure 1.
Down-regulation of CSR1 expression in prostate cancer. A: Semiquantitative RT-PCR. Total RNA (1 μg) of normal prostate tissues (PD14 and PD16) and prostate cancer samples (47T, 101T, 83T, 85T, 91T, and 68T) were reverse-transcribed. PCR was performed on diluted templates using primers specific to CSR1 and β-actin. B: Immunostaining of CSR1 protein on prostate samples. Representative images of CSR1 expression scored as 3+ (normal), 1+ (cancer), and 0 (cancer) are shown. C: Expression of CSR1 protein was down-regulated in prostate cancer samples (red) relative to normal prostate (blue).
Figure 2.
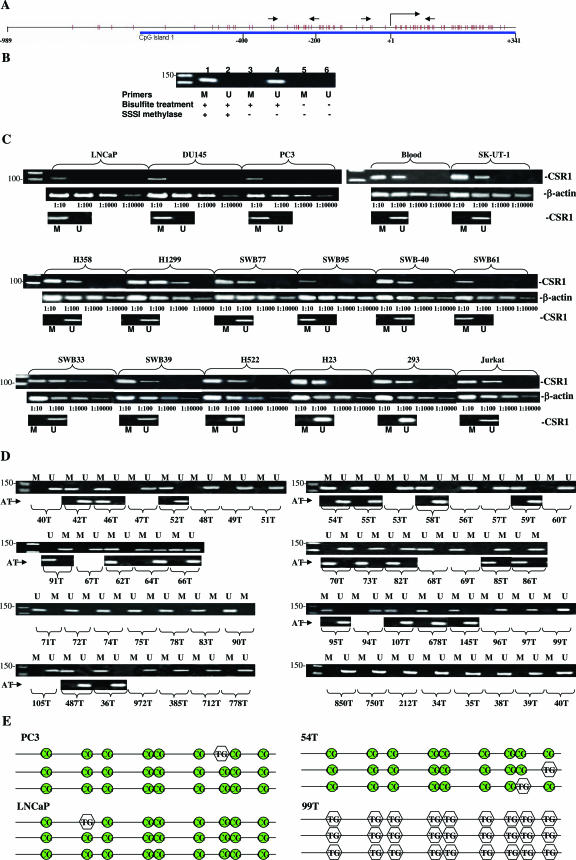
Methylation of CpG island in the promoter region of CSR1. A: Diagram of CpG island in promoter/exon1 region of CSR1. Orange spikes indicate CpG. Straight arrows indicate the position of the PCR primers. Putative mRNA start site is indicated by right angle arrow. B: Primer specificity of CSR1 methylation and nonmethylation primers. Unmethylated DNA templates were treated with (lanes 1 and 2) or without (lanes 3–6) SSSI methylase. The samples were subsequently treated with (lanes 1–4) or without (lanes 5 and 6) sodium bisulfite. PCR was performed with methylation primers (lanes 1, 3, and 5) or unmethylation primers (lanes 2, 4, and 6). C: Correlation of methylation status and suppression of CSR1 expression. Semiquantitative RT-PCR was performed on diluted templates of 16 cell lines and one blood sample using primers specific to CSR1 and β-actin. Methylation status was examined with methylation-specific PCR. M, methylation; U, unmethylation. D: Methylation-specific PCR was performed with 58 prostate cancer samples and 22 benign prostate tissues adjacent to tumor (AT). M, PCR products of methylation-specific primers; U, products of unmethylation-specific primers. Individual tumor samples are labeled. Rows marked with AT represent methylation-specific PCR performed on prostate tissues adjacent to tumor. E: Representation of methylation sequencing of selected samples. The PCR product (−193 to −315) contains nine CpG, excluding those in the primer region and 76-bp nonprimer sequence (122 bp if primers are included). Preservation of cytosine nucleotide after bisulfite treatment in CpG is indicated as CG. Conversion to thymidine after treatment is indicated as TG. Sequencing was performed on templates from clones of TA vector ligated with the PCR products.
To investigate whether CSR1 protein expression was similarly down-regulated in prostate cancer, we performed an immunostaining analysis on 343 samples of prostate tissues using tissue array slides, including 210 prostate cancer and 133 normal prostate tissues. As shown in Figure 1B, CSR1 expression was primarily localized to the cytosol and plasma membrane. To quantify CSR1 expression in prostate tissues, the expression of CSR1 was graded as strong (3 point), moderate (2 point), weak (1 point), focal positive (0.5 point), or negative (0 point) for each sample. The overall score for each sample represented the average of scores by three observers who were blinded to the clinical outcomes. CSR1 protein expression was down-regulated in prostate cancer tissue: 77% (161 of 210) of prostate cancers showed immunostaining ≤0.5 for CSR1 whereas only 23% (49 of 210) showed ≥1 (Figure 1B). In contrast, 84% (112 of 133) of normal prostate samples showed ≥1 CSR staining, and only 21 of 133 had immunostaining ≤0.5. The rank-sum test yielded a P value <0.001, indicating that the difference in expression between the two groups was significant.
Promoter/Leader Sequence of CSR1 Is Hypermethylated in Prostate Cancer
CSR1 is located in 8p21, a genetic locus associated with deletion or hypermethylation in advanced prostate cancer. To examine whether CSR1 is deleted in prostate cancer, PCR was performed on microdissected prostate cancer samples using primers specific to regions including exons 2 and 6. The results indicated that CSR1 sequence was present in 59 of 60 prostate cancer samples, including three prostate cancer cell lines, suggesting that deletion of CSR1 is not a main mechanism of down-regulation of its expression. A CpG island was identified in the promoter/exon1 region of CSR1, spanning from −650 to at least + 341 of the mRNA start site (Figure 2A) using CpG island search tool by Takai et al.10 We designed a set of methylation-specific PCR primers within the CpG island encompassing a region of −193 to −315. To test the specificity of these primers, an unmethylated DNA template was artificially methylated with SSSI methylase and treated with bisulfite. PCR was performed with primers specific for methylated and unmethylated DNA. The results showed a good specificity of these primers (Figure 2B). Subsequently, we examined the methylation status of CSR1 in 16 human cancer cell lines and a blood sample, including three from prostate cancer (PC3, Du145, and LNCaP), four from lung cancer (H358, H1299, H522, and H23), one from leiomyosarcoma (SK-UT-1), one from lymphoma (Jurkat), six from glioblastoma multiforme (SWB95, SWB77, SWB-40, SWB61, SWB33, and SWB39), and one from kidney (293). Five of these cell lines, including three from prostate cancer (PC-3, Du145, and LNCaP) and two from glioblastoma multiforme (SWB95 and SWB61), were found to contain methylated CSR1 gene. To test whether methylation status correlates with mRNA expression of CSR1, semiquantitative RT-PCR of CSR1 was performed on these samples. As shown in Figure 2C, 10- to 100-fold decreases in expression of CSR1 were found when there was methylation in the promoter region, suggesting that methylation in the promoter region of CSR1 suppresses its expression.
Methylation-specific PCR was then performed on samples from 58 prostate cancer tumors. Thirty-eight percent (22 of 58) of primary prostate cancer samples were hypermethylated in the CSR1 promoter region (Figure 2D). To examine whether methylation of CSR1 promoter/exon1 region also occurs in the benign tissue adjacent to prostate cancer, methylation-specific PCR was performed on 22 benign tissues adjacent to tumors (AT) that were shown to contain methylated CSR1 in the tumors. More than 90% of AT were negative (20 of 22) for CSR1 methylation, and less than 10% (2 of 22) were positive. These results suggest that CSR1 methylation is largely a cancer-specific event and probably represents an epigenetic alteration in the early stage of prostate cancer. Twenty-two of these samples, including 12 methylation PCR products and 10 unmethylation ones were ligated into a TA cloning vector, and selected clones were sequenced. Some typical results are shown in Figure 2E. Generally, the results were consistent with the methylation status determined by methylation-specific PCR. In addition, two pairs of degenerative primers capable of amplifying both methylated and unmethylated sequences (Figure 2A) were used to amplify these samples. The results of subsequent sequencing were in complete concordance with those of the initial methylation sequencing.
Down-Regulation of CSR1 Expression and Methylation of the CSR1 Promoter Are Associated with Prostate Cancer Relapse
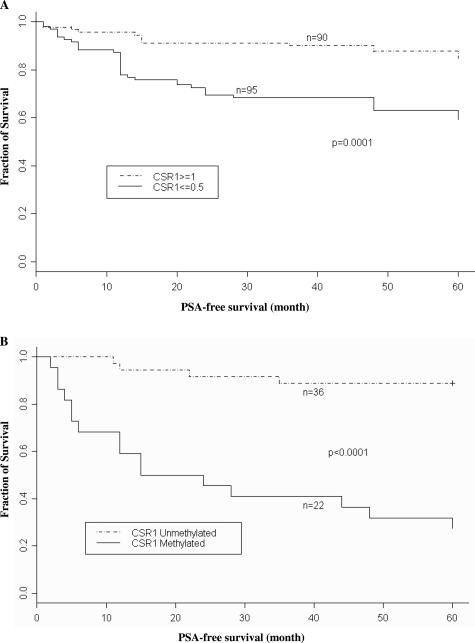
To determine whether down-regulation of CSR1 is associated with poor clinical outcome, tissue samples were divided into two groups: one group with at least weakly expressed CSR1 (≥1) and the other with focal positive (≤0.5) or less expression. These samples were analyzed for PSA relapse or outright physical evidence of metastasis 5 years after prostatectomy. Among the samples with CSR1 scores ≥1, 16.6% (15 of 90) experienced prostate cancer relapse within 5 years of prostatectomy. In contrast, patients with a CSR1 score ≤0.5 had a 41.1% (39 of 95) relapse rate (Figure 3A). Log-rank test results (P = 0.0001) indicated that the absence of CSR1 was associated with a significantly higher risk of prostate cancer recurrence.
Figure 3.
Down-regulation of CSR1 expression and methylation of CSR1 promoter is associated with decrease of PSA-free survival. A: Kaplan-Meier curve of PSA-free survival based on immunostaining of CSR1 in prostate cancer samples. Prostate cancer samples were divided into CSR ≥1 or ≤0.5. B: Kaplan-Meier curve of PSA-free survival based on the methylation status of the CSR1 promoter.
When samples were divided into those with CSR1 methylation and those without methylation, a more striking result was obtained. Among the methylated samples, 72.7% (16 of 22) of the patients experienced clinical relapse within 5 years of prostatectomy (Figure 3B). On the other hand, only 4 of 36 (11%) patients experienced relapse in the same period when CSR1 was not methylated, even though CSR1 RNA levels were similarly down-regulated in 34 (94%) of these samples. These results indicate that even though down-regulation of CSR1 can be induced by various mechanisms, methylation of the CSR1 promoter is the most critical step in inducing prostate cancer progression.
Inactivation of CSR1 Expression in Prostate Cancer Cell Lines Results from CpG Island Methylation in the CSR1 Promoter
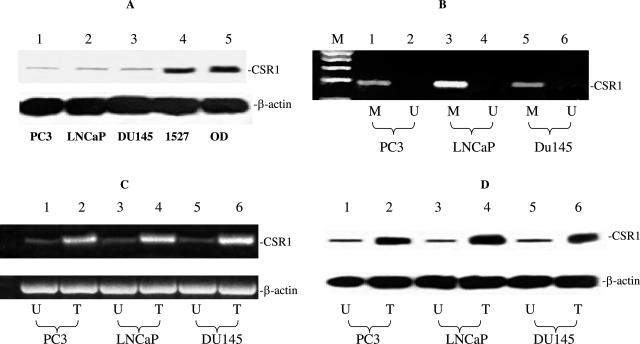
We found down-regulation of CSR1 expression in PC3 (6-fold), DU145 (5.7-fold), and LNCaP (8-fold) cell lines relative to the levels in 23 normal prostate tissues. Western blot analysis revealed small amounts of CSR1 protein in all three cell lines (Figure 4A). PCR specific for methylated CSR1 promoter was performed. The results indicated that the CSR1 promoter was hypermethylated in all three cancer cell lines (Figure 4B). 5-Aza 2′deoxycytidine treatment of cells resulted in dramatic increases of CSR1 mRNA (Figure 4C) and protein expression (Figure 4D). These results support our hypothesis that down-regulation of CSR1 expression in prostate cancer cell lines are caused by methylation of its promoter. Alternatively, these results can be interpreted as consequence of activation of upstream transcription factors due to demethylation.
Figure 4.
CSR1 promoter methylation down-regulates CSR1 expression in prostate cancer cell lines. A: Immunoblotting of CSR1 with antibodies specific to CSR1 and β-actin. Proteins obtained from prostate cancer cell lines (PC3, LNCaP, and DU145), renal cell line 1527, and normal prostate tissue OD were used. B: Methylation-specific PCR of prostate cancer cell lines. M, PCR products of methylation-specific primers. U, unmethylation primers. C: Treatment of 5-aza-2′-deoxycytidine reverses CSR1 expression suppression. T, cells treated with 5-aza-2′-deoxycytidine treatment; U, untreated cells. D: Immunoblotting of CSR1 expression in cell lines treated with 5-aza-2′-deoxycytidine.
CSR1 Contains Tumor Suppressor Activity
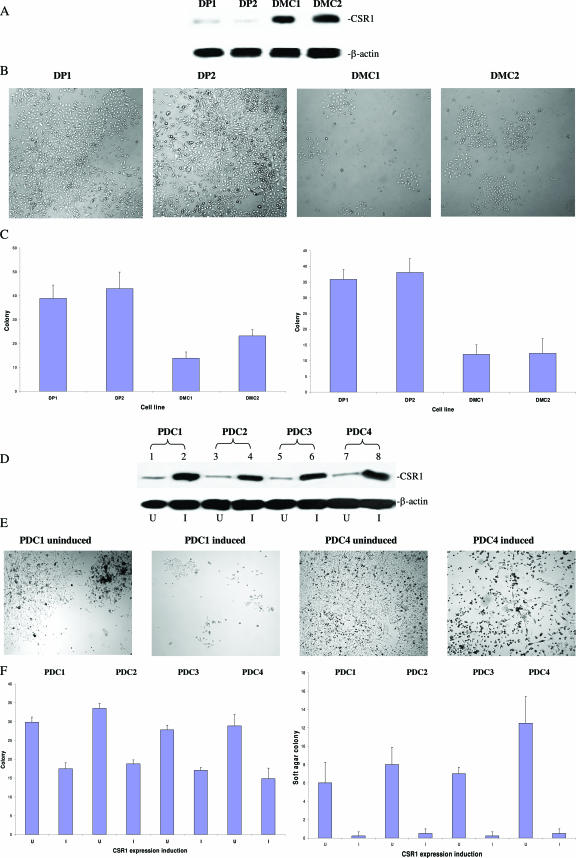
To examine the role of CSR1 in regulating the invasiveness of prostate cancer cells, cDNA of CSR1 was inserted into the eukaryotic expression vector pCMVscript. PC3 cells were transfected with the pCMVscript and pCMV-CSR1 vectors, and 10 colonies were maintained. Two of these colonies were selected for further analysis based on their level of CSR1 overexpression (Figure 5A). These two cell lines (DMC1 and DMC2) and vector-alone controls (DP1 and DP2) were tested for colony formation and their ability for anchorage-independent growth in soft agar. The DMC1 and DMC2 cell lines exhibited threefold and twofold decreases in colony formation, respectively, relative to vector controls. Similar fold changes in the number of colonies formed in soft agar were observed for both DMC1 and DMC2. This observation supports the hypothesis that CSR1 has tumor suppressor activity.
Figure 5.
Suppression of colony formation and anchorage-independent growth of PC3 and DU145 cells expressing CSR1. A: Immunoblots of CSR1 protein from PC3 cells stably expressing CSR1 (DMC1 and DMC2) and vector controls (DP1 and DP2). B: Images of DP1, DP2, DMC1, and DMC2 colony formation. C: Suppression of colony formation and anchorage-independent growth with CSR1 expression. Colonies were allowed to form on a hard surface (left) or on soft agar (right). D: Immunoblotting of CSR1 protein on Du145 cells transformed with pCDNA4-CSR1. I, CSR1 expression induced by tetracycline; U, uninduced. Four clones were selected for the study (PDC1, PDC2, PDC3, and PDC4). E: Representative image of colony formation from PDC1 and PDC4 (uninduced and induced with tetracycline). F: Suppression of colony formation and anchorage-independent growth by CSR1 in PDC1, PDC2, PDC3, and PDC4. Colony formation on hard surface (left) and on soft agar (right) are shown.
To test whether CSR1 tumor suppressor activity is instantaneous and can be induced by transient expression, a tetracyclin-inducible eukaryotic expression vector (pCDNA-4) overexpressing CSR1 mRNA was constructed and transfected into DU145 cell line. Fifteen colonies were chosen and examined for the CSR1 expression levels. Relative to uninduced transformed cells, 14 of these clones were found to have significant increases in CSR1 protein level. Four of these colonies were selected to test for transient tumor suppressor activity. Induction of CSR1 expression suppressed colony formation of DU145 cells by 40 to 60% in four different pCDNA4-CSR1 isolates (Figure 5A). Ten- to 15-fold decreases in colony-forming ability in soft agar were observed in these colonies when CSR1 expression was induced (Figure 5B). This finding provides further support for the hypothesis that CSR1 possesses tumor suppressor activity.
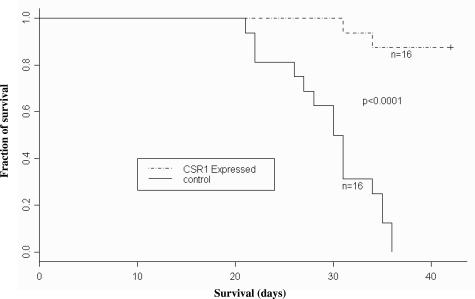
We tested whether CSR1 expression affects prostate cancer cell invasiveness in an in vitro Matrigel transmigration experiment and in an in vivo mouse tumorigenesis experiment. Three-fold fewer CSR1-expressing PC3 cells (pCMV-CSR1 transformants) traversed the Matrigel than did control cells (Table 2). Moreover, the propensity for PC3 cells injected into mice to develop into tumors was suppressed by CSR1 expression (Table 3). Relative to the pCMVscript controls, mice injected with CSR1-expressing cells developed tumors that were >8-fold smaller (mean, 3.9 versus 0.46 cm3), displayed a lower incidence of metastasis (0 of 16 versus 5 of 16 mice), and had a markedly reduced 6-week mortality (2 of 16 versus 16 of 16 mice) (Figure 6). These experiments clearly indicate that CSR1 suppresses tumor growth, decreases metastasis, and reduces cancer-related mortality.
Table 2.
CSR1 Inhibits PC3 Cell Invasiveness
| Cells | Cells invading through Matrigel* | Cells in control membrane* | Invasion/migration |
|---|---|---|---|
| PC3 + pCMVscript | |||
| Clone DP1 | 46.6 ± 3.5 | 63.4 ± 3.8 | 73.5 |
| Clone DP2 | 54.4 ± 4.6 | 66.6 ± 4.5 | 82.4 |
| Clone DP3 | 24.6 ± 1.4 | 38.9 ± 1.4 | 64.7 |
| Clone DP4 | 29.8 ± 2.5 | 41.8 ± 1.8 | 71.3 |
| PC3 + pCMV-CSR1 | |||
| Clone DMC1 | 2.6 ± 1.2 | 15.8 ± 2.1 | 16.5 |
| Clone DMC2 | 7.4 ± 0.7 | 30.4 ± 2.6 | 24.3 |
| Clone DMC3 | 4.2 ± 0.5 | 16.4 ± 1.7 | 25.6 |
| Clone DMC4 | 5.6 ± 1.4 | 18.4 ± 3.5 | 30.4 |
Mean of five experiments with SEM. Five high-power fields were counted for each experiment.
Table 3.
CSR1 Inhibits Growth and Metastasis of Xenografted PC-3 Cell Tumors in SCID Mice
| PC3 + pCMVscript
|
PC3 + pCMV-CSR1
|
||||||
|---|---|---|---|---|---|---|---|
| Mouse | Tumor size (cm3) | Metastasis | PD | Mouse | Tumor size (cm3) | Metastasis | PD |
| DP1 tumor | DMC1 tumor | ||||||
| 1 | 3.41 | None | Yes | 1 | 0.84 | None | No |
| 2 | 1.63 | None | Yes | 2 | 0.37 | None | No |
| 3 | 2.32 | None | Yes | 3 | <0.1 | None | No |
| 4 | 2.84 | Abdominal wall and liver | Yes | 4 | 0.92 | None | Yes |
| 5 | 3.11 | Liver | Yes | 5 | 0.32 | None | No |
| 6 | 1.77 | Bladder | Yes | 6 | 0.44 | None | No |
| 7 | 4.12 | None | Yes | 7 | 0.11 | None | No |
| 8 | 3.62 | None | Yes | 8 | <0.1 | None | No |
| DP2 tumor | DMC2 tumor | ||||||
| 1 | 1.72 | Bladder | Yes | 1 | 0.70 | None | No |
| 2 | 2.34 | None | Yes | 2 | 0.44 | None | No |
| 3 | 4.65 | Liver | Yes | 3 | 0.61 | None | No |
| 4 | 2.74 | None | Yes | 4 | 0.58 | None | No |
| 5 | 3.31 | None | Yes | 5 | 1.12 | None | Yes |
| 6 | 1.76 | None | Yes | 6 | 0.27 | None | No |
| 7 | 2.18 | Lung | Yes | 7 | 0.94 | None | No |
| 8 | 4.28 | None | Yes | 8 | 0.22 | None | No |
PD, died within 6 weeks of inoculation.
Figure 6.
CSR1 expression in PC3 cell xenograft reduces mortality. Mice inoculated with 107 cells from control (DP1 and DP2) or experimental (DMC1 and DMC2) clones were observed, and the percentages of mice in each group that died within a 6 week period were recorded. Note that mice inoculated with an experimental clone exhibited dramatically reduced mortality relative to controls (data pooled).
Discussion
CSR1 was initially identified as a macrophage scavenger receptor homolog. A study suggested that expression of CSR1 was induced under stress condition, and thus, the name of cellular stress response (CSR) gene was coined.7 CSR1 contains three possible functional domains: a membrane-spanning region, an α-helix coiled-coil domain, and a collagen-like domain. Because CSR1 has an extremely short putative extracellular domain, it is unlikely that it functions independently as a receptor. We speculate that CSR1 may combine with other molecules to form heterodimeric receptors. Alternatively, CSR1 may function as an intracellular signaling molecule. Findings that CSR1 overexpression confers resistance to cytotoxicity from both UV irradiation and hydrogen peroxide are consistent with the view that the function of CSR is stress related.7 CSR1 expression has been shown to reduce the level of reactive oxygen species and to protect cells from injury. Thus, CSR1 may function as a protector of genetic material and prevent DNA damage from chemical or physical assaults. Our findings indicate that CSR1 also possesses tumor suppressor activity. Thus, the membrane-bound protein CSR1 may perform dual signaling functions: one to activate enzymes that reduce oxidative free-radicals under stress and the other to suppress cell growth and modulate cell-cell interactions and thereby reduce tumor cell invasiveness.
Abnormalities of 8p21-23 are frequently associated with advanced prostate cancer. Several putative tumor suppressor genes have been identified in this region. The most noted ones are DBC-2, DLC-1, and NKX3.1. DBC-2 deletion has been linked with breast cancer,11 and DLC-1 deletion has been linked with human liver cancer, breast cancer, and non-small cell carcinoma in lung.12–15 NKX3.1 was found to be deleted in a subset of prostate cancers.16 However, none of these genes were found to be related to prostate cancer survival. DBC-2, DLC-1, and NKX3.1 are located within 300 kb of one another, whereas CSR1 is 4 megabases away from these genes. Deletion of CSR1 in prostate cancer is unusual at best, suggesting that the frequently observed down-regulation of CSR1 in prostate cancer is mediated by a distinctive inactivation mechanism rather than a genetic deletion. There are several putative mechanisms that may suppress CSR1 protein expression. These may include expression of transcription repressors, inactivation of transcription factors, modification of histones, genome mutations, and DNA methylation. Our study indicates that suppression of CSR1 expression through methylation, but not through other means, is linked with a poor prognosis of prostate cancer, suggesting that the mechanism leading to suppression of expression of CSR1 is important in determining the behavior of the cancer. One explanation for this novel observation is that histone deacetylation and DNA methylation suppress CSR1 expression in such a consistent and permanent manner that there is little chance for leaky expression. Meanwhile, down-regulation of CSR1 expression resulting from inactivation of transcription factors is less tight, and leaky expression might occur because of redundancy of transcription factors. We should also note that the percentage (59% or 13 of 22) of the tumor samples with CSR1 methylation in invasive (T3a or more) pathological stages is similar (P = 0.79) to that (58.3 or 21 of 36) of those without methylation, suggesting that CSR1 methylation is independent of tumor stages.
To our knowledge, this is the first report that identifies and describes the relationship between CSR1 and malignancy. Because this gene appears both to reduce oxygen free radicals and to modulate tumor behavior, it is possible that CSR1 plays an important role in suppressing other malignancies besides prostate cancer. CSR1 down-regulation and methylation is associated with a poor prognosis for prostate cancer. Forced expression of CSR1 in highly aggressive prostate cancer cell lines retards xenograft tumor growth, decreases metastasis, and extends life span. These findings suggest a potential application of CSR1 as a clinical prognosticator predicting prostate cancer clinical outcomes and as a therapeutic target for treating aggressive types of prostate cancer.
Footnotes
Address reprint requests to Jian-Hua Luo, Department of Pathology, University of Pittsburgh School of Medicine, 3550 Terrace Street, Pittsburgh, PA 15261. E-mail: luoj@msx.upmc.edu.
Supported by the National Cancer Institute (grants 1UO1CA88110-01 to G.M. and R01 CA098249 to J.H.L.) and by the John Rangos Foundation for Enhancement of Research in Pathology.
References
- Isaacs JT. Molecular markers for prostate cancer metastasis: developing diagnostic methods for predicting the aggressiveness of prostate cancer. Am J Pathol. 1997;150:1511–1521. [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Emi M, Komiya A, Fujiwara Y, Yatani R, Nakamura Y, Shimazaki J. Localization of a tumor suppressor gene associated with progression of human prostate cancer within a 1.2 Mb region of 8p22–p21.3. Genes Chromosomes Cancer. 1995;13:168–174. doi: 10.1002/gcc.2870130306. [DOI] [PubMed] [Google Scholar]
- Bova GS, Carter BS, Bussemakers MJ, Emi M, Fujiwara Y, Kyprianou N, Jacobs SC, Robinson JC, Epstein JI, Walsh PC. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Res. 1993;53:3869–3873. [PubMed] [Google Scholar]
- Luo JH, Yu YP. Genetic factors underlying prostate cancer. Expert Rev Mol Med. 2003;2003:1–26. doi: 10.1017/S1462399403006057. [DOI] [PubMed] [Google Scholar]
- Han HJ, Tokino T, Nakamura Y. CSR, a scavenger receptor-like protein with a protective role against cellular damage caused by UV irradiation and oxidative stress. Hum Mol Genet. 1998;7:1039–1046. doi: 10.1093/hmg/7.6.1039. [DOI] [PubMed] [Google Scholar]
- Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, Finkelstein S, Michalopoulos G, Becich M. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- Hamaguchi M, Meth JL, von Klitzing C, Wei W, Esposito D, Rodgers L, Walsh T, Welcsh P, King MC, Wigler MH. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci USA. 2002;99:13647–13652. doi: 10.1073/pnas.212516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- Yuan BZ, Zhou X, Durkin ME, Zimonjic DB, Gumundsdottir K, Eyfjord JE, Thorgeirsson SS, Popescu NC. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 2003;22:445–450. doi: 10.1038/sj.onc.1206064. [DOI] [PubMed] [Google Scholar]
- Wong CM, Lee JM, Ching YP, Jin DY, Ng IO. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63:7646–7651. [PubMed] [Google Scholar]
- Yuan BZ, Jefferson AM, Baldwin KT, Thorgeirsson SS, Popescu NC, Reynolds SH. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- He WW, Sciavolino PJ, Wing J, Augustus M, Hudson P, Meissner PS, Curtis RT, Shell BK, Bostwick DG, Tindall DJ, Gelmann EP, Abate-Shen C, Carter KC. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]