Abstract
Hepatitis C virus (HCV) infection is frequently complicated by glomerulonephritis with immune complexes containing viral RNA. We examined the potential influence of Toll-like receptors (TLRs), specifically TLR3 recognition of viral dsRNA exemplified by polyriboinosinic:polyribocytidylic acid [poly(I:C) RNA]. Normal human kidney stained positive for TLR3 on mesangial cells (MCs), vascular smooth muscle cells, and collecting duct epithelium. Cultured MCs have low TLR3 mRNA levels with predominant intracellular protein localization, which was increased by tumor necrosis factor-α, interleukin (IL)-1β, interferon (IFN)-γ, and the TLR3 ligand poly(I:C) RNA. Poly(I:C) RNA stimulation of MCs increased mRNA and protein synthesis of IL-6, IL-1β, M-CSF, IL-8/CXCL8, RANTES/CCL5, MCP-1/CCL2, and ICAM-I; it also increased anti-proliferative and proapoptotic effects, the latter of which was decreased by inhibiting caspase-8. In microdissected glomeruli of normal and non-HCV membranoproliferative glomerulonephritis biopsies, TLR3 mRNA expression was low. In contrast TLR3 mRNA expression was significantly increased in hepatitis C-positive glomerulonephritis and was associated with enhanced mRNA for RANTES/CCL5 and MCP-1/CCL2. We hypothesize that immune complexes containing viral RNA activate mesangial TLR3 during HCV infection, thereby contributing to chemokine/cytokine release and effecting proliferation and apoptosis. Thus, TLR3 expression on renal cells, and especially MCs, may establish a link between viral infections and glomerular diseases.
Hepatitis C virus infection (HCV) is a major problem worldwide, frequently complicated by a virus-associated glomerulonephritis. During the course of infection, immune complexes and viral RNA reach the mesangium.1,2 The recent identification of Toll-like receptors (TLRs) has shown that the innate immune system can recognize conserved pathogen-associated molecular patterns through TLRs expressed on immune cells but also on a number of nonimmune cells.3–5 TLRs recognize molecular patterns associated with microbial pathogens and induce an immune response.3,4,6 Eleven members of the TLR family (TLR1 to TLR11) have so far been identified in mice and 10 in humans, each recognizing a distinct component of infectious agents.7–9 TLR3 recognizes dsRNA of viral origin as exemplified by polyriboinosinic:polyribocytidylic acid [poly(I:C) RNA], a synthetic analog of viral dsRNA.10,11 Human TLR3 is expressed in dendritic cells (DCs), fibroblasts, and intestinal epithelial cells, playing physiological roles in anti-viral innate immunity.10,12–14 In contrast to most TLRs, signaling of TLR3 is independent of the common TLR adaptor protein MyD88 and involves the adaptor protein Trif (TICAM-1). The identification of the TLR3-TICAM-1 pathway in mammalian cells provides a link between dsRNA and inflammatory cytokine production, including interferons, interleukin (IL)-6, and IL-12, some of which participate in anti-viral response.9,15 Furthermore IL-6 and IL-12 influence numerous aspects of the immune response. In contrast to TLR2 and TLR4, TLR3 is a predominantly intracellular endosomal receptor in DCs16 but is also expressed on the surface of fibroblasts and intestinal epithelial cells.13,16 In addition to the anti-viral immune responses,17 TLR3 activation can induce apoptosis in 293 cells by recruiting RIP kinases and caspases.18,19
Early organ screens by Zarember and Godowski20 showed that TLR3 expression is not restricted to leukocytes but also occurs in nonimmune organs, including the kidney. We have found a robust expression of TLR3 in mesangial cells (MCs) in vivo and in culture and have demonstrated TLR3 to activate MCs in response to poly(I:C) RNA resulting in the production of chemokines and cytokines but also in considerable MC apoptosis—depending on cytokine prestimulation. We postulate that TLR3 may be important for the clearance of viral RNA reaching the glomerular mesangium, possibly serving in a housekeeping manner under normal conditions. During pathological conditions such as viral infections, viral RNA alone or as part of immune complexes could reach the mesangium and trigger glomerular inflammation, resulting, eg, in HCV-associated glomerulonephritis.2,21 In support of this hypothesis, we found increased mRNA levels for TLR3 and for proinflammatory cytokines and chemokines in microdissected glomeruli from biopsies of hepatitis C-associated but not idiopathic membranoproliferative glomerulonephritis and propose that TLR3 expression in MCs may play a role in some forms of glomerulonephritis.
Materials and Methods
Preparation of Human Tissue
Human tissue was used following the guidelines of the Ethics Committee of the Medical Faculty of the University of Heidelberg (Heidelberg, Germany). The selection criteria for biopsies with membranoproliferative glomerulonephritis were as follows. All archival biopsies with the histopathological diagnosis of membranoproliferative glomerulonephritis, known HCV status, and sufficient available material for mRNA expression analysis were obtained from a European multicenter study for gene expression analysis (the European Renal cDNA Bank) and from the Department of Cellular and Molecular Pathology at the German Cancer Research Center collected in the years 1999 to 2004. The histological and mRNA expression analyses were performed in a blinded manner without knowledge of a concomitant HCV infection. Subsequently, the results were grouped according to the HCV status, and statistical evaluation was performed.
Immunohistochemistry
Immunohistochemistry for TLR3 was conducted on 5-μm frozen tissue sections from tumor nephrectomies after fixation in acetone at −10°C for 10 minutes as described previously.22 Staining for TLR3 was performed using a mIgGIk antibody (Ab) specific for human TLR3, obtained from eBioscience (San Diego, CA). Tissue sections were incubated with the mIgGIk Ab for 18 hours at 4°C. A rabbit anti-mouse IgG Ab (Z259, diluted 1:40; DAKO, Glostrup, Denmark) was applied at 22°C for 1 hour, and alkaline phosphatase-specific mouse mAb (diluted 1:40) was then applied at 22°C for 1 hour. All dilutions were performed in phosphate-buffered saline (PBS) (pH 7.6). For staining, sections were exposed for 15 minutes to a solution of sodium nitrite (28 mmol/L), new fuchsin (basic fuchsin, 21 mmol/L), naphthol-AS-B1-phosphate (0.5 mol/L), dimethylformamide (64 mmol/L), and levanisol (5 mmol/L) in 50 mmol/L Tris-HCl buffer (pH 8.4) containing 164 mmol/L NaCl. Negative controls generated with nonimmune control Ab and without primary Ab did not show any staining (data not shown).
Eight biopsies from HCV-positive membranoproliferative glomerulonephritis and nine biopsies from HCV-negative membranoproliferative glomerulonephritis were available for CD68 immunohistochemistry. Immunohistochemical staining for CD68-positive cells was performed on 3-μm sections of formaldehyde-fixed and paraffin-embedded tissue. The tissues were deparaffinized with xylene and rehydrated through graded concentrations of ethanol. After rehydration, pretreatment with 0.05% protease XXIV (Sigma-Aldrich, Taufkirchen, Germany) at 37°C for 20 minutes was performed. The primary Ab obtained from DAKO was a mouse anti-human mAb directed against the CD68-positive subpopulation of macrophages. An alkaline phosphatase anti-alkaline phosphatase detection system was applied for visualization. Controls, omitting the first Ab or replacing the first Ab by a nonimmune IgG for each paraffin block tested, were negative.
Glomerular Sclerosis Index
As previously described23 a semiquantitative score was used to evaluate the degree of glomerular sclerosis on periodic acid-Schiff-stained sections. Glomerulosclerosis was defined as follows: 0, no sclerosis; 0.5, sclerosis of less than 25% of capillary loops; 1, sclerosis of 26 to 50% of the capillary loops; 2, sclerosis of 51 to 75% of the capillary loops; 3, sclerosis of more than 75% of the capillary loops. Glomerulosclerosis score was calculated as the sum of all specific injury indices, whereby the index of glomeruli with degree 0.5 was multiplied by 0.5, that of degree 1 × 1, that of degree 2 × 2, and that of degree 3 × 3.
Cell Culture of Human MCs
Immortalized human MCs were grown as described previously.24 For enzyme-linked immunosorbent assay (ELISA) and RNA extraction, MCs were incubated with or without tumor necrosis factor (TNF)-α (25 ng/ml), IL-1β (10 ng/ml), and interferon (IFN)-γ (20 ng/ml) alone or in combination for 24 hours, washed with PBS, incubated in culture medium (containing 10% fetal calf serum) for 6 hours and washed again with PBS. Subsequently, MCs were incubated with culture medium alone (control) or culture medium containing poly(I:C) RNA, poly(I:C) DNA, or CpG oligonucleotide as indicated. Aliquots of the supernate culture medium were removed for ELISA analysis at the times indicated. For analysis of mRNA levels, extraction of total RNA was performed using an RNeasy Mini Kit (Qiagen, Hilden, Germany) with additional DNase digestion.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR) Analysis
Semiquantitative and quantitative RT-PCR analysis was done as described.25 Manual microdissection of fresh human biopsies or laser microdissection of glomerular tissue from formalin-fixed archival biopsies was performed as described before.26 For quantitative RT-PCR, 2 μg of isolated total RNA underwent random primed reverse transcription using a modified Moloney murine leukemia virus reverse transcriptase (Superscript; Life Technologies, Karlsruhe, Germany). In parallel, 2-μg aliquots were processed without reverse transcription to control for contaminating genomic DNA. Real-time RT-PCR was performed on a TaqMan ABI 7700 sequence detection system (PE Applied Biosystems, Weiterstadt, Germany). GAPDH was used as reference gene. All water controls were negative for target and housekeeper. Sequences, with the following gene bank accession numbers, served for the design of the predeveloped TaqMan assay reagents (PDAR) or primers and probe, purchased from Applied Biosystems: NM_003265/U88879 (human TLR3), NM_000600 (human IL-6), XM_010760 (human IL-1β), Z11686 (human IL-8/CXCL8), AF043341 (human RANTES/CCL5), X14768 (human MCP-1/CCL2), and M33197 (human GAPDH).
RNase Protection Assay
Protection assays were performed as previously described.24 As specific probe for human TLR3, a 350-bp fragment of human TLR3 cDNA (NM_003265) was amplified using the primer 5′-CGG GAT CCC TAAAGG GTC TGT CTC AC-3′ (sense) and 5′-GGA ATT CTG GCT TGA CAG CTC AGGG-3′ (anti-sense) and cloned into the pCR2.1-TOPO vector (Invitrogen, Karlsruhe, Germany). Linearization of the plasmid with EcoRV allowed labeling of a 426-bp-long TLR3 probe. Total RNA (25 μg) from human MCs were used to analyze the expression of TLR3 mRNA by MCs. After polyacrylamide gel electrophoresis, protected fragments were analyzed using a Storm 840 PhosphorImaging system (Amersham Biosciences Corp., Arlington Heights, IL) and ImageQuant software (version 5.2; Molecular Dynamics, Sunnyvale, CA).
Fluorescence-Activated Cell Sorting (FACS) Analysis
MCs were cultured under standard conditions or stimulated with a combination of the proinflammatory cytokines TNF-α (25 ng/ml), IL-1β (10 ng/ml), and IFN-γ (20 ng/ml) for 24 hours. For FACS analysis human MCs were detached with PBS and 10 mmol/L ethylenediaminetetraacetic acid (pH 8) and stained for TLR3 using a specific mouse mAb (eBioscience) and a phycoerythrin-conjugated rabbit polyclonal secondary Ab against mouse F(ab)2 fragment (R0439; DAKO, Carpinteria, CA) as described.27 For intracellular staining, MCs were treated with Cytofix/Cytoperm reagent (Pharmingen, La Jolla, CA) followed by mAb against TLR3 and secondary Ab incubations in saponin buffer. The TLR3 signal was analyzed using a FACSCalibur with CellQuest analysis software (Becton-Dickinson, Heidelberg, Germany). Appropriate IgG isotype preparations (mouse IgG1 clone MOPC 21; Sigma-Aldrich, Germany) were used to control for nonspecific staining.
Inflammation Microarray Analysis
Total RNA was isolated from cultured human MCs using a commercially available silica-gel based isolation protocol (RNeasy Mini kit, Qiagen). RNA quantity was controlled by OD measurement, and RNA quality was visualized on a 1% MOPS agarose gel. Total RNA (5 μg) was reverse-transcribed in a 20-μl reaction with 1 μl of SuperScript II (200 U/μl, Invitrogen) and 10 pmol of T7-(dT)24 primer (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGA-(dT)24–3′) in 5× first-strand buffer (Invitrogen), 2 μl of 100 mmol/L dithiothreitol (Invitrogen), 1 μl of 10 mmol/L dNTP (Invitrogen), 1 μl of RNase inhibitor (RNasin, 40 U/μl; Promega, Mannheim, Germany) at 42°C for 1 hour. The second-strand synthesis was performed at 16°C for 2 hours in the presence of Escherichia coli enzymes, DNA polymerase I (10 U, Invitrogen), DNA ligase (10 U, Invitrogen), RNase H (2 U/μl; Roche, Basel, Switzerland), and 1× second-strand buffer (Invitrogen). The double-stranded cDNA was blunt-ended using 10 U of T4 DNA polymerase (Invitrogen), purified by phenol/chloroform extraction and transcribed in the presence of unlabeled-ribonucleotides using the Ambion MEGAscript T7 kit (Ambion, Huntington, UK). The cRNA was purified using RNeasy minicolumn (RNeasy kit, Qiagen) followed by an OD quantity measurement. Quality of the amplified products was examined by 1% agarose gel. Amplified RNA (20 μg) was used for chemical labeling of random guanosine nucleotides using the Bio-ULS (Universal Linkage System) Blitz reagents (MetriGenix, Gaithersburg, MD). Here, biotin is chemically attached to the RNA in a 30-minute labeling procedure at 80°C. Afterward unincorporated biotin label is removed using proprietary biotin affinity columns (KREApure columns, MetriGenix). Biotin-labeled RNA (10 μg) was applied for hybridization in a volume of 70 μl. The MGX-4D inflammation array was loaded and hybridized for 4 hours at 42°C. Incubation, hybridization, and washing procedures were performed on the MGX2000 hybridization station, and chemiluminescent images were captured on the MGX1200CL charge-coupled device-camera detection station (MetriGenix). The array contains 80 oligonucleotide probes associated to inflammatory processes and 40 to 50 bases in length, eight housekeeping genes, and eight hybridization and staining controls.
Normalization of the spot signal values was performed automatically after background correction by comparison with the values obtained from a spike-in control (Atcab1 = Arabidopsis thaliana Cab mRNA for photosystem I chlorophyll A/B-binding protein) as described previously by Kessler and co-workers.28 Before hybridization, the product from the amplified sample was mixed with spike-in controls (for sample normalization) in sample dilution buffer and denatured for 5 minutes at 90°C. The normalized values were calculated as ratios of the total signal for each individual gene and the mean value of the total signal intensity for the gene selected as normalization gene.
ELISA
ELISA for IL-6, IL-1β, IL-8/CXCL8, RANTES/CCL5, MCP-1/CCL2, and IFN-β were performed on cell culture supernatants using commercial assay kits (Quantikine; R&D Systems, Minneapolis, MN) and following the manufacturer’s instructions.
Proliferation Assays
To assess the proliferative activity of human MCs, MTT assays were performed as described.29 Aliquots of 20 × 103 cells in 100 μl of culture medium were cultured in 96-well microtiter plates for 24 hours under standard conditions to yield firmly attached and stably growing cells. After discarding the supernatants, 100 μl of culture medium with or without a combination of TNF-α (25 ng/ml), IL-1β (10 ng/ml), and IFN-γ (20 ng/ml) were added, and the cells were incubated for 24 hours, washed with PBS, incubated in culture medium for 6 hours, and washed again with PBS. Subsequently, MCs were stimulated with or without poly(I:C) RNA in different concentrations for 24 hours. The supernatants were removed, and 50 μl of a 1 mg/ml solution of MTT (Sigma-Aldrich) were added. After a 3-hour incubation at 37°C, formazan crystals were dissolved by the addition of 50 μl of isopropanol. Absorbance was measured at 550 to 630 nm as a reference using a Dynatech (Denkendorf, Germany) MR7000 ELISA reader. For each experiment at least five wells were analyzed per experimental condition.
Cell Death Assays
For analysis of effects of poly(I:C) RNA on apoptosis of MCs, cells were pretreated as above for 24 hours, washed with PBS, incubated in culture medium for 6 hours with or without caspase-8 inhibitor Ac-IETD-CHO (5 μg/ml; Biomol, Hamburg, Germany), and washed again with PBS. Subsequently, MCs were stimulated with or without poly(I:C) RNA (5 μg/ml) with or without caspase-8 inhibitor (5 μg/ml) or with poly(I:C) DNA (5 μg/ml) for 24 hours. To determine the percentage of apoptotic cells, flow cytometric cell cycle analysis using propidium iodide staining was performed as described.30 Caspase-8 assays were performed using commercial assay kits (Promega, Madison, WI) and following the manufacturer’s instructions.
Statistical Analysis
Values are provided as mean ± SEM. Statistical analysis was performed by unpaired t-test and Mann-Whitney U-test for biopsy data. Significant differences are indicated for P values <0.05 (*) or 0.01 (**), respectively.
Results
TLR3 Is Expressed in Normal Human Kidney
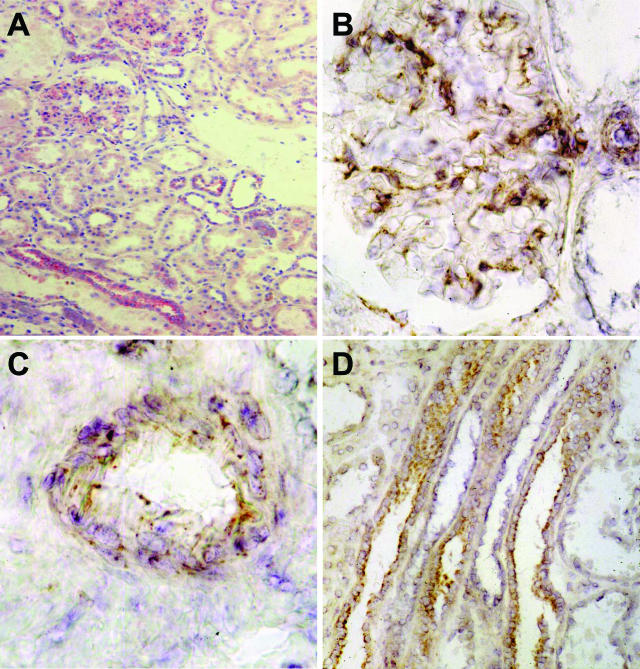
Frozen sections obtained from unaffected areas of kidneys removed for renal tumors stained positive for TLR3 in glomeruli, in distal-collecting tubules, and in vascular smooth muscle cells (Figure 1A). TLR3 staining followed a predominant intracellular localization (Figure 1B). In addition smooth muscle cells of arterioles and mid-sized arteries (Figure 1C) and epithelium of the collecting tubules (Figure 1D) showed a positive signal for TLR3. The signal in the collecting tubules was most consistent with luminal membrane localization.
Figure 1.
Immunohistochemistry for TLR3 in human kidney. A: Immunohistochemistry for TLR3 in frozen sections of human kidney show a positive signal in glomeruli, vascular smooth muscle cells, and distal-collecting tubules. B: Glomeruli show a positive mesangial staining for TLR3 with a predominant intracellular pattern. C and D: Smooth muscle cells of mid-sized arteries (C) and luminal epithelial cells of collecting tubules (D) stain positive for TLR3, in the latter with a luminal membrane localization. Original magnifications: ×100 (A); ×400 (B).
Cultured Human MCs Express TLR3
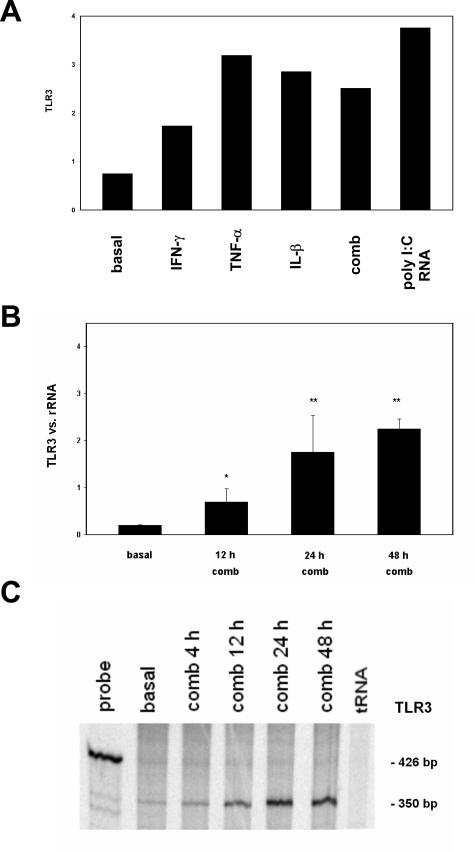
Because MCs play a role in glomerular disease processes and staining for TLR3 resulted in a mesangial pattern, we examined cultured human MCs for expression of TLR3. RNA was prepared from cells growing under standard conditions as well as from cells that had been stimulated with the cytokines TNF-α, IL-1β, and IFN-γ for different time intervals (12, 24, 48 hours) to simulate a proinflammatory milieu as would occur during immune-mediated glomerular disease. By quantitative RT-PCR, specific products for TLR3 mRNA were amplified from both unstimulated and stimulated cells. The low basal expression for TLR3 could be increased with each of the cytokines TNF-α, IL-1β, and IFN-γ alone or in combination (Figure 2A). The combination of the three cytokines was essentially equipotent to individual components. Furthermore, the TLR3 ligand poly(I:C) RNA (5 μg/ml) also up-regulated the mRNA of TLR3 (Figure 1A). The expression of mRNA for TLR3 on cultured MCs increased with time of stimulation up to 48 hours (Figure 2B). To confirm the quantitative RT-PCR data for TLR3, RNase protection assays were performed, and expression of TLR3 was found to be low under basal conditions and increased after stimulation for up to 48 hours with proinflammatory cytokines (Figure 2C). Semiquantitative PCR with primer sequences for TLR3 to detect splicing isoforms31 showed one single band under basal conditions and after cytokine treatment for 24 hours (results not shown). To test whether the human MC line might express other TLRs, we screened for mRNA of TLR1 through TLR10 by real-time RT-PCR under basal and cytokine-treated conditions. In addition to the expression of TLR3, human MCs expressed TLR1 and TLR4, mostly after stimulation with proinflammatory cytokines (results not shown). The expression for TLR2, TLR5, TLR6, TLR7, and TLR9, at basal conditions and after stimulation, was too low to allow evaluation, and no expression was found for TLR8 and TLR10 (data not shown).
Figure 2.
Expression of TLR3 by human MCs. Human MCs were cultured under standard conditions (basal) or after stimulation with proinflammatory cytokines TNF-α, IL-1β, and IFN-γ for the time indicated before RNA extraction. Real-time RT-PCR was performed using primers specific for human TLR3 as indicated in Materials and Methods. A: Basal expression of TLR3 was increased after stimulation with IFN-γ, TNF-α, IL-1β, the combination of these cytokines (comb), and poly(I:C) RNA (5 μg/ml) for 24 hours. B: TLR3 expression of human MCs. Results are mean ± SEM of two independently performed experiments and rRNA served as the reference gene. C: TLR3 expression of human MCs determined by RNase protection assay with 20 μg of total RNA used for each reaction as described in Materials and Methods. Under basal conditions (basal) a weak band for TLR3 was detectable, which increased markedly after stimulation for up to 48 hours with the cytokine combination (comb). The probe used exceeds the actual band for TLR3 because of the flanking sequences of 426 bp. The size of the bands is indicated on the right. Semiquantitative PCR showed only one isoform for TLR3 in MCs (data not shown).
Expression of TLR3 Protein on MCs
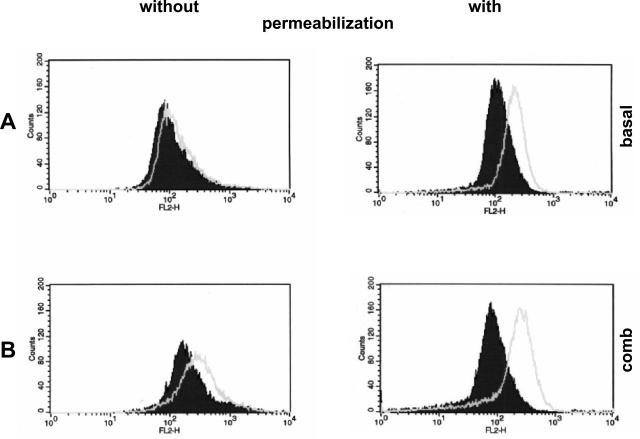
To confirm expression of TLR3 protein on human MCs, FACS analysis was conducted with a TLR3-specific mAb.10 A barely detectable surface staining for TLR3 was found on MCs under basal conditions (Figure 3A) and TLR3 staining increased after a 24-hour cytokine stimulation (Figure 3B). Because TLR3 has been described as an intracellular receptor,16 MCs were permeabilized before TLR3 staining and FACS analysis. This resulted in a robust TLR3 staining of MCs under basal conditions (Figure 3A) and a further enhancement after prestimulation with cytokines (Figure 3B). Comparable results were obtained with poly(I:C) RNA stimulation for intracellular TLR3 by FACS analysis (results not shown).
Figure 3.
Expression of TLR3 protein on MCs. The localization of TLR3 protein on cultured MCs was analyzed by flow cytometry without and with prior permeabilization. A weak surface TLR3 signal was found on MCs under basal conditions (A) that increased after preincubation with a combination of the cytokines TNF-α, IL-1β, and IFN-γ for 24 hours (B). A stronger signal for TLR3 was detected after the cells were permeabilized under basal conditions with a further increase after the combined TNF-α, IL-1β, and IFN-γ treatment (B). Open histograms represent the fluorescence activity after incubation with monoclonal anti-TLR3 Ab. Filled histograms demonstrate the signal of the appropriate isotype control. Results shown are from one of three independent experiments that showed reproducible staining patterns.
Effect of TLR3 Ligand Poly(I:C) RNA on Chemokine and Cytokine mRNA Levels
To test the effect of TLR3 activation by poly(I:C) RNA mimicking viral RNA, we performed an mRNA expression screen using a microchip analysis containing 80 probes for inflammation-associated genes. MCs were pretreated with the cytokine combination of TNF-α, IL-1β, and IFN-γ for 24 hours to up-regulate TLR3 expression. After washing-off the cytokine-containing culture medium, MCs were left in culture medium for 6 hours, washed again, and then incubated with or without addition of poly(I:C) RNA (5 μg/ml) to the culture medium before extraction of RNA and expression analysis of the 96 gene probes present on the microchip (inflammatory plus housekeeping genes). Thirty-one genes showed a signal above background. The results are shown in Table 1. The genes for IL-6, IL-1β, IL-8/CXCL8, M-CSF, and ICAM-1 were up-regulated whereas the signal for the genes transforming growth factor-β1, transforming growth factor-βR2, STAT1, MHC class I, CD27BP, interferon α-inducible protein 27, β-actin, dynamitin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hypoxanthine phosphoribosyltransferase 1 (HPRT-1), peptidylprolyl isomerase A, ribosomal s5, and K-ALPHA-1 decreased after poly(I:C) RNA treatment. The up-regulated genes comprise mostly proinflammatory cytokines and adhesion molecules, whereas those down-regulated include cell cycle-associated molecules as well as so called housekeeping molecules.
Table 1.
Expression Screen for mRNA in Cultured MCs Using a Microchip Analysis Containing 80 Probes for Inflammation Associated Genes
| Genes
| |
|---|---|
| Up-regulated | Down-regulated |
| IL-6 (+26 %) | TGF-β1 (−73 %) |
| IL-8 (+629 %) | TGF-βR2 (−64 %) |
| IL-1β (+260 %) | STAT1 (−70 %) |
| CSF-1 (+101 %) | MHC, class I, A (−45 %) |
| ICAM-1(+41 %) | CD27BP (−66 %) |
| Interferon, α-inducible protein 27 (−60 %) | |
| Actin β (−64 %) | |
| Dynamitin mRNA (−68 %) | |
| GAPDH (−49 %) | |
| HPRT-1 (−59 %) | |
| Cyclophilin A (−68 %) | |
| Ribosomal s5 (−49 %) | |
| Tubulin, α, ubiquitous (−76 %) | |
MCs were pretreated with the cytokine combination of TNF-α, IL-1β, and IFN-γ for 24 hours to up-regulate TLR3 expression. After washing-off the cytokine-containing culture medium, MCs were left in culture medium for 6 hours, washed again, and then incubated for 24 hours with or without addition of poly(I:C) RNA (5 μg/ml) to the culture medium before extraction of RNA and expression analysis of the gene probes present on the microchip. The genes indicated on the left side are up-regulated; the genes on the right side are down-regulated. The genes indicated in italics are housekeeping genes. The changes in mRNA level after poly(I:C) RNA treatment as compared to the control are given as percentage.
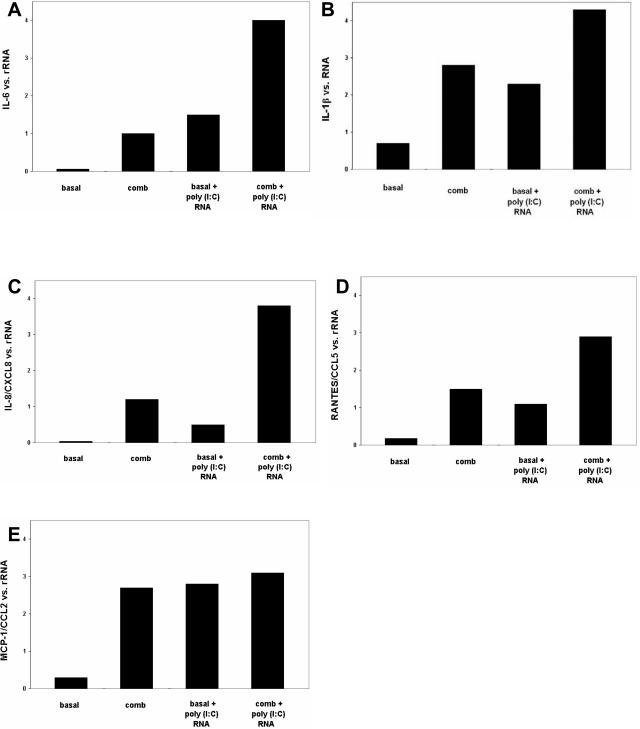
We confirmed the effects of poly(I:C) RNA on selected chemokine and cytokine mRNA levels by real-time RT-PCR. MCs showed barely detectable basal expression for IL-6 mRNA. Expression was induced by stimulation with proinflammatory cytokines. Incubation with poly(I:C) RNA increased IL-6 expression under basal conditions, an effect that was markedly potentiated when MCs were pretreated with proinflammatory cytokines (Figure 4A) to enhance their TLR3 expression. Similar results were obtained for IL-1β (Figure 4B). As to be expected basal expression for mRNA of IL-8/CXCL8 (Figure 4C), RANTES/CCL5 (Figure 4D), and MCP-1/CCL2 (Figure 4E) was low and was markedly increased after stimulation with proinflammatory cytokines. Exposure of MCs maintained under basal conditions to poly(I:C) RNA also increased mRNA levels for IL-8/CXCL8, RANTES/CCL5, and MCP-1/CCL2. Although RANTES/CCL5 mRNA levels were further enhanced by pretreatment followed by poly(I:C) RNA levels of mRNA for IL-8/CXCL8 and MCP-1/CCL2 showed little additional increase with poly(I:C) RNA when cells had been pretreated with the combination of cytokines. Our data suggest a positive effect on cytokine and chemokine synthesis after poly(I:C) RNA stimulation. However, we cannot differentiate if this represents a synergistic or only additive effect.
Figure 4.
Effect of incubation with poly(I:C) RNA on chemokine and cytokine mRNA levels. MCs were preincubated with or without a combination of proinflammatory cytokines (IFN-γ, TNF-α, IL-1β) for 24 hours, washed with PBS, incubated in culture medium for 6 hours, washed again with PBS, and incubated with or without poly(I:C) RNA (5 μg/ml) for 24 hours before RNA extraction. Levels of mRNA expression for IL-6 (A), IL-1β (B), IL-8/CXCL8 (C), RANTES/CCL5 (D), and MCP-1/CCL2 (E) were analyzed by real-time RT-PCR. Poly(I:C) DNA (5 μg/ml) had no effect on cytokine and chemokine mRNA levels (data not shown).
Effect of Poly(I:C) RNA on Cytokine and Chemokine Release by MCs
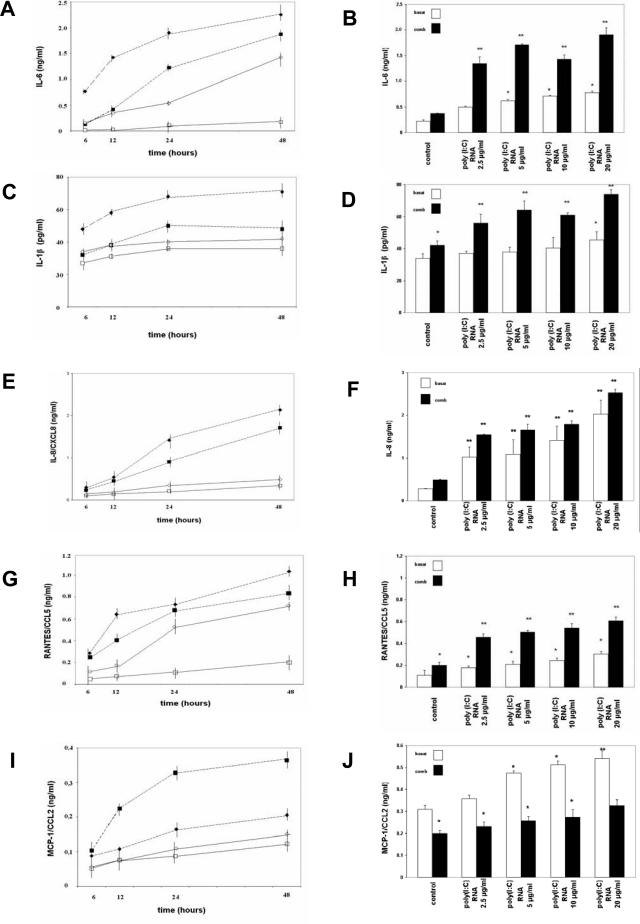
When MCs were incubated with poly(I:C) RNA (5 μg/ml) for up to 48 hours, release of IL-6 (Figure 5A), IL-1β (Figure 5C), IL-8/CXCL8 (Figure 5E), RANTES/CCL5 (Figure 5G), and MCP-1/CCL2 (Figure 5I) increased both under basal conditions and further after prestimulation with proinflammatory cytokines. Stimulation of MCs with poly(I:C) RNA (5 μg/ml) led to an increase in synthesis of IL-6, IL-1β, and RANTES/CCL5 under basal conditions with further enhancement when MCs were pretreated with TNF-α, IL-1β, and IFN-γ. MCP-1/CCL2 synthesis was lower when MCs were pretreated with cytokines. Stimulation with the TLR3 ligand poly(I:C) RNA for 24 hours increased IL-6 (Figure 5B), IL-1β (Figure 5D), IL-8/CXCL8 (Figure 5F), RANTES/CCL5 (Figure 5H), and MCP-1/CCL2 (Figure 5J) release of human MCs in a concentration-dependent manner in a range from 2.5 to 20 μg/ml poly(I:C) RNA, an effect enhanced by pretreatment with the cytokine combination. MCP-1/CCL2 synthesis in response to increasing concentrations of poly(I:C) RNA was lower when MCs were pretreated with cytokines. This is consistent with the observation that MCP-1 stimulation (in contrast to for example RANTES stimulation) in MCs is a rapid (30 minutes) and short-lived (24 hours) effect. In contrast, the regulation of RANTES in MCs occurs slower and peaks much later (24 to 48 hours).24
Figure 5.
Time- and concentration-dependent influence of poly(I:C) RNA on release of IL-6, IL-1β, IL-8/CXCL8, RANTES/CCL5, and MCP-1/CCL2 by MCs. MCs were pretreated under basal (squares) or cytokine combination conditions (triangles) as described in Materials and Methods and after washing they were incubated with (filled symbols, discontinued line) or without (open symbols, continuous line) poly(I:C) RNA (5 μg/ml) for different time intervals (6, 12, 24, or 48 hours). The concentrations of IL-6, IL-1β, IL-8/CXCL8, RANTES/CCL5, and MCP-1/CCL2 were determined by ELISA. Incubation of MCs with poly(I:C) RNA results in a concentration-dependent increase of IL-6 (A, B), IL-1β (C, D), IL-8/CXCL8 (E, F), RANTES/CCL5 (G, H), and MCP-1/CCL2 (I, J) release under both basal (open bars) and combination prestimulation (filled bars). No IFN-β release was observed (data not shown). Poly(I:C) DNA (5 μg/ml) had no effect on cytokine and chemokine release (data not shown). Values are means ± SEM of two independently performed series of experiments.
No IFN-β release was observed, both under basal and prestimulation conditions and after stimulation with poly(I:C) RNA (data not shown). Incubation with poly(I:C) DNA (5 μg/ml), both without and with cytokine pretreatment or CpG DNA (2 μmol/L), had no effect on cytokine and chemokine release (data not shown). The effect of a blocking anti-TLR3 Ab10 was tested for IL-6, RANTES/CCL5, and MCP-1/CCL2. Poly(I:C) RNA-induced cytokine and chemokine release after prestimulation with the cytokine combination was reduced when MCs were incubated with an anti-TLR3 Ab (10 μg/ml) 6 hours before and during poly(I:C) RNA stimulation (IL-6, ∼90% inhibition; RANTES/CCL5, ∼75%; MCP-1/CCL2, ∼65%; data not shown).
Effect of Poly(I:C) RNA on MC Proliferation and MC Apoptosis
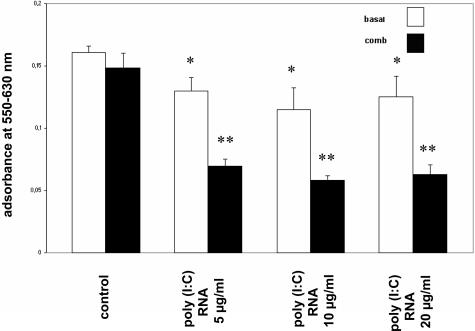
The microchip analysis indicated that genes associated with cell proliferation were down-regulated after exposure of MCs to poly(I:C) RNA. We therefore examined MC proliferation under the different experimental conditions. As shown in Figure 6, exposure of MCs maintained under basal conditions to increasing concentrations of poly(I:C) RNA only resulted in a modest (20%) decrease in cell proliferation as determined by the MTT assay. In contrast pretreatment of MCs with the cytokine combination followed by exposure to poly(I:C) RNA markedly reduced cell proliferation (∼60%).
Figure 6.
Effect of poly(I:C) RNA on MC proliferation. Exposure of MCs to poly(I:C) RNA in increasing concentrations (5, 10, 20 μg/ml) resulted in a decrease in MC proliferation. Pretreatment with the cytokine combination followed by exposure to poly(I:C) RNA markedly reduced cell proliferation (filled bars). MCs were analyzed for cell proliferation with the MTT assay as described in Materials and Methods. Each bar represents the mean ± SEM of five parallel incubations for each condition. Comparable results were obtained in three series of independent experiments.
Because of the decreased cell proliferation, the potential role of TLR3 for MC survival was studied in cell death assays using flow cytometric cell-cycle analysis with propidium iodide staining (Table 2; results are means ± SEM of three individual experimental series). Cell-cycle analysis revealed a background of 12.8 ± 4.6% apoptotic cells under basal conditions. When MCs were incubated in culture medium and subsequently stimulated with poly(I:C) RNA (5 μg/ml) for 24 hours, the percentage of apoptotic cells was increased to 28.4 ± 6.5%. After preincubation with the cytokine combination for 24 hours, 15.5 ± 6.1% of the cells belonged to a population with a sub-G1 DNA content that was not different from the 12.8 ± 4.6% under basal conditions. Preincubation with cytokines for 24 hours and subsequent stimulation with poly(I:C) RNA (5 μg/ml) for 24 hours increased the amount of MCs displaying a sub-G1 DNA content to 34.5 ± 9.6%, consistent with a marked increase in apoptosis. When MCs were incubated with caspase-8 inhibitor (5 μg/ml) 6 hours before and during poly(I:C) RNA stimulation, the proapoptotic effect of poly(I:C) RNA was mitigated [basal + poly(I:C) RNA + caspase-8 inhibitor, 18.1 ± 3.9%; comb + poly(I:C) RNA + caspase-8 inhibitor, 21.3 ± 5.1%]rsqb] (Table 2). Stimulation with poly(I:C) DNA (5 μg/ml) revealed 15.4 ± 3.6% apoptotic cells under basal conditions and 16.4 ± 6.2% after cytokine pretreatment and showed no difference to untreated cells. Caspase-8 activity was indeed increased by the cytokine combination and increased further by poly(I:C) RNA, an effect that dose dependently was prevented by the caspase-8 inhibitor at 0.5 μg/ml, 2.5 μg/ml, 5 μg/ml, and 10 μg/ml (data not shown).
Table 2.
Effect of Poly(I:C) RNA and Poly(I:C) DNA on MC Apoptosis
| Percentage of apoptotic cells
| ||
|---|---|---|
| Without | With | |
| Poly (I:C) RNA | Poly (I:C) RNA | Poly (I:C) DNA |
| Caspase-8 inhibitor | ||
| − 12.8 ± 4.6 | Basal 28.4 ± 6.5* | 15.4 ± 3.6 |
| + 12.7 ± 3.9 | 18.1 ± 3.9 | |
| − 15.5 ± 6.1 | Comb 34.5 ± 9.6 | 16.4 ± 6.2 |
| + 16.3 ± 4.3 | 21.3 ± 5.1 | |
DNA profiles of human MCs were analyzed by flow cytometry after staining with propidium iodide. Table 2 shows the percentage of apoptotic cells containing subG1 DNA without or with stimulation with poly(I:C) RNA without or with caspase-8 inhibitor or poly(I:C) DNA and without or with prior treatment with the combination of cytokines. The results shown are mean ± SEM of three independently performed series of experiments.
Expression of TLR3 and Chemokine mRNA in Microdissected Glomeruli from Human Kidney Biopsies
Because MCs express TLR3 and activation of TLR3 by poly(I:C) RNA up-regulates proinflammatory chemokines and cytokines, we used quantitative RT-PCR to assess mRNA expression of TLR3 and of selected chemokines, ie, RANTES/CCL5 and MCP-1/CCL2, in manually or laser-microdissected glomeruli from biopsies of patients with hepatitis C-positive and hepatitis C-negative membranoproliferative glomerulonephritis. Hepatitis C infection was confirmed or excluded by serological testing for HCV Ab and antigen in all analyzed patients with the histopathological diagnosis of membranoproliferative glomerulonephritis. In the cases with positive HCV serology, the presence of the virus was confirmed by PCR. Cryoglobulinemia was detectable in 5 of 10 HCV-positive patients (patients 1, 3, 5, 7, and 9; Table 3) and in 1 of 9 HCV-negative patients (patient 8) with membranoproliferative glomerulonephritis.
Table 3.
Clinical Data at Biopsy and Glomerular Histopathological Findings of Microdissected Kidney Biopsies
| Biopsy group | Patient number | Age (years) | Gender (f/m) | Serum creatinine (mg/dl) at biopsy | Proteinuria (g/d) at biopsy | Glomerular histopathology | CD68+ cells/ glomerulus | Glomerulo- sclerosis index |
|---|---|---|---|---|---|---|---|---|
| Pretransplant donor biopsies | ||||||||
| 1 | 26 | m | 0.9 | <0.2 | normal | n.a. | n.a. | |
| 2 | 49 | m | <1.1 | <0.2 | normal | n.a. | n.a. | |
| 3 | 53 | f | <1.1 | <0.2 | normal | n.a. | n.a. | |
| 4 | 66 | m | <1.1 | <0.2 | normal | n.a. | n.a. | |
| 5 | 50 | m | 0.9 | <0.2 | normal | n.a. | n.a. | |
| 6 | 51 | f | 0.73 | <0.2 | normal | n.a. | n.a. | |
| Means ± SEM | 49 ± 12 | 2:4 | 0.9 ± 0.1 | <0.2 | ||||
| HCV-positive MPGN | ||||||||
| 1 | 28 | m | 1.5 | 3.4 | MPGN type I | 35 | 0 | |
| 2 | 56 | m | 1.6 | 23 | MPGN type I | n.a. | n.a. | |
| 3 | 19 | f | 0.6 | n.d. | MPGN type I | n.a. | n.a. | |
| 4 | 62 | f | 1.6 | 1.2 | MPGN type I | 19.8 | 0 | |
| 5 | 61 | f | 1.2 | 4 | MPGN type I, segmental necrosis and GS | 16.5 | 49.98 | |
| 6 | 48 | m | 1.3 | 5.5 | MPGN type I | 39.2 | 0 | |
| 7 | 50 | f | 2.4 | 1.5 | MPGN type I, reactive FSGS | 12.7 | 15.3 | |
| 8 | 63 | f | 1.2 | 1 | MPGN type I | 12.3 | 0 | |
| 9 | 39 | f | 1.3 | 0.3 | MPGN type I, reactive FSGS | 25.1 | 58.32 | |
| 10 | 41 | m | 2.7 | 2.5 | MPGN type I, reactive FSGS | 42.3 | 33.33 | |
| Means ± SEM | 46 ± 14 | 6:4 | 1.5 ± 0.6 | 4.7 ± 6.6 | 25.36 ± 4.2 | 22.42 ± 9.4 | ||
| HCV-negative MPGN | ||||||||
| 1 | 34 | f | n.a. | 9.9 | MPGN type I, reactive FSGS | 36.1 | 78.54 | |
| 2 | 82 | f | 2.88 | 0.3 | MPGN type I, reactive FSGS | 9.5 | 129.97 | |
| 3 | 36 | m | 2.55 | 0.3 | MPGN type I, reactive FSGS | 9.5 | 18.18 | |
| 4 | 68 | f | 3.5 | n.d. | MPGN type I | 12.5 | 37.5 | |
| 5 | 52 | m | 2.25 | 2.5 | MPGN type I, reactive FSGS | 5.8 | 126.64 | |
| 6 | 63 | m | 1.6 | 18 | MPGN type I, reactive FSGS | 9.1 | 30.76 | |
| 7 | 52 | f | 1.9 | 11 | MPGN type I | 27.2 | 0 | |
| 8 | 53 | f | 1.4 | 0.4 | MPGN type I | 30.2 | 0 | |
| 9 | 55 | m | 2.2 | 2.9 | MPGN type I, focal global GS | 2.9 | 153.32 | |
| Means ± SEM | 55 ± 13 | 5:4 | 2.2 ± 0.6 | 5.0 ± 5.9 | 15.87 ± 3.9 | 71.86 ± 20.69 |
Patient’s age, sex, serum creatinine (mg/dl), and proteinuria (g/day) at biopsy as well as histopathological findings of glomerular lesions are summarized. Microdissected glomeruli from pretransplant living donor biopsies were compared with glomeruli from hepatitis C-positive or hepatitis C-negative membranoproliferative glomerulonephritis. Mean values ± SEM for each group are provided. MPGN, membranoproliferative glomerulonephritis; FSGS, focal and segmental glomerulosclerosis; n.a., no tissue available; n.d., not determined.
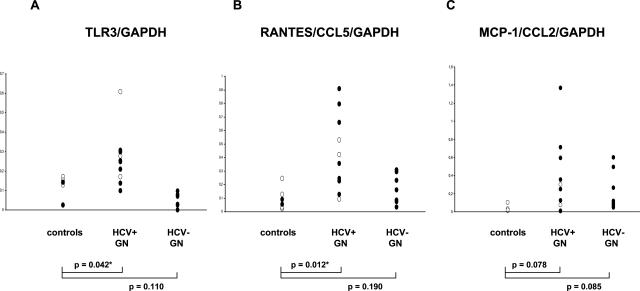
Information on patient characteristics and biopsy pathology are provided in Table 3. Consistent with the results obtained with cultured MCs and with immunohistochemistry of normal kidney, mRNA for TLR3 was present in glomeruli from normal human kidneys, ie, pretransplant donor biopsies (n = 6). Glomerular expression of TLR3 was significantly increased in biopsies from patients with hepatitis C-positive membranoproliferative glomerulonephritis (n = 10) as compared to controls or hepatitis C-negative membranoproliferative glomerulonephritis (n = 9) (Figure 7A). Low levels of mRNA for RANTES/CCL5 (Figure 7B) and MCP-1/CCL2 (Figure 7C) were detectable in microdissected glomeruli of healthy donor kidneys and hepatitis C-negative membranoproliferative glomerulonephritis and were up-regulated in patients with hepatitis C-positive membranoproliferative glomerulonephritis. Thus glomeruli from hepatitis C-positive membranoproliferative glomerulonephritis showed signs of activation of TLR3 and of chemokines and such activation was not present in HCV-negative forms of membranoproliferative glomerulonephritis despite comparable degrees of glomerular pathology (Table 3).
Figure 7.
Levels of mRNA for TLR3 and for chemokines in microdissected glomeruli from human kidney biopsies. Expression of mRNA for TLR3 (A) and selected chemokines, ie, RANTES/CCL5 (B) and MCP-1/CCL2 (C), was analyzed in microdissected glomeruli from human biopsies obtained for routine diagnosis from patients with hepatitis C-positive membranoproliferative glomerulonephritis. Biopsies from living kidney transplant donors served as controls and showed a basal level of mRNA for TLR3. Glomerular expression of TLR3 was significantly increased in most biopsies from patients with hepatitis C-positive membranoproliferative glomerulonephritis. In contrast TLR3 mRNA levels were not increased in glomeruli from hepatitis C-negative idiopathic membranoproliferative biopsies. Low levels of mRNA for RANTES/CCL5 and MCP-1/CCL2 were detectable in microdissected glomeruli of healthy donor kidneys and hepatitis C-negative membranoproliferative glomerulonephritis and were up-regulated in patients with hepatitis C-positive membranoproliferative glomerulonephritis (open symbols, manual microdissection; filled symbols, laser microdissection; controls, living kidney transplant donors (n = 6); HCV+GN, hepatitis C-positive membranoproliferative glomerulonephritis (n = 10), HCV−GN, hepatitis C-negative membranoproliferative glomerulonephritis (n = 9). P values refer to group comparison by unpaired t-test.
To further assess if the renal pathology was comparable in both groups, we performed an immunohistochemical staining for CD68-positive cells in the glomeruli and examined the glomerulosclerosis scores in all available biopsies, as previously described in detail.23 A total of 17 paraffin-embedded sections (eight HCV-positive; nine HCV-negative) was available for this analysis. The detailed histopathological analysis (included in Table 3) revealed no significant differences between both groups: immunohistochemical staining for CD68-positive cells showed 25.36 ± 4.25 CD68-positive cells/glomerulus in the HCV-positive and 15.87 ± 3.99 CD68-positive cells/glomerulus in the HCV-negative biopsies (P = 0.074); the glomerulosclerosis index was 22.42 ± 9.4 in HCV-positive biopsies and 71.86 ± 20.7 in HCV-negative biopsies (P = 0.09).
The small and not statistically significant differences in glomerular CD68-positive cell infiltrate cannot explain the differences in mRNA levels for TLR3 because there was no correlation between these parameters (coefficient of determination, R2 = 0.1077; data not shown). The differences could perhaps be due to performance of renal biopsies at an earlier stage in patients with known HCV infection compared to patients with idiopathic membranoproliferative glomerulonephritis. However, we have no information on the duration of renal disease before biopsy in the patient groups analyzed.
Discussion
Viral dsRNA has been recognized as a major ligand for TLR3 on DCs and is considered to serve as an anti-viral system by generating α- and β-interferons. In addition TLR3 has also been linked to induction of autoimmune disease via activation of DCs and macrophages. TLR3 is, however, also expressed in nonimmune cells such as intestinal epithelial cells and fibroblasts.13,16 Furthermore, an initial organ expression screen showed TLR3 mRNA in kidney,20 and Tsuboi and colleagues32 showed TLR3 expression in cultured murine tubular epithelial cells. In this context our finding of TLR3 immunostaining in normal human kidney is of special interest. Immunohistochemistry showed TLR3 expression in glomeruli with a mesangial pattern and in vascular smooth muscle cells of preglomerular vessels. This is of interest because MCs are considered to be modified vascular smooth muscle cells.33 In MCs and vascular smooth muscle cells, the pattern of cellular staining is consistent with a predominant intracellular localization of TLR3. Furthermore, specific staining for TLR3 was also noted in collecting duct epithelial cells. Interestingly in this polarized epithelium, the staining pattern for TLR3 was most consistent with tubular luminal membrane localization where it might serve as a defense mechanism against ascending viral infections. This would be a function similar to that of TLR4 and TLR11 in bacterial urinary infections, a hypothesis deserving further investigations.
Because MCs play a role in a variety of glomerular diseases and especially in immune-mediated forms, we concentrated on TLR3 in MCs. The mesangium is only separated from the glomerular capillary blood flow by a fenestrated endothelium and thus is percolated by plasma, including larger blood components such as lipids and immune globulins, immune complexes as well as DNA and RNA fragments. Thus, during viral and bacterial diseases, viral or bacterial components including RNA or DNA could reach MCs either alone or in the form of immune complexes. For example, such immune complexes containing viral RNA play an important role in triggering the glomerulonephritis and vasculitis of hepatitis C.2,21
First, we examined if TLR3 mRNA and protein could be induced on MCs by a combination of proinflammatory cytokines. TNF-α, IL-1β, and IFN-γ were chosen because these cytokines are up-regulated during glomerular diseases.34 Indeed, these cytokines alone and their combination resulted in a time-dependent up-regulation of TLR3 mRNA and protein expression. This is consistent with the IFN-mediated up-regulation of TLR3 in macrophages, DCs, and epithelial cells.35–37 Furthermore, the TLR3 ligand poly(I:C) RNA increased mRNA and protein expression of TLR3 on the MCs. These findings are in agreement with similar results reported by Alexopoulou and colleagues11 and indicate that TLR3 ligands can up-regulate their own receptors. By immunohistochemistry and by FACS analysis, TLR3 was localized mostly intracellularly but also to a small extent on the cell surface of MCs. The surface TLR3 staining could be clearly distinguished from the appropriate isotype Ab control. In DCs TLR3 has been localized almost exclusively to intracellular compartments, especially endosomes.12,16 Considerable surface expression has, however, also been described in fibroblasts and intestinal epithelial cells.13,16
To test the functionality of TLR3 on MCs we exposed them to poly(I:C) RNA as a mimetic for viral dsRNA. For an initial screen we used an inflammation microchip containing 80 genes associated with an inflammatory response. Interestingly most of the genes up-regulated on the chip belong to proinflammatory cytokines and chemokines that have been previously shown to be regulated in MCs and to play a role in response to various glomerular injuries.38–40 Also ICAM-1 was up-regulated in MCs by poly(I:C) RNA, a finding of interest because this adhesion molecule is up-regulated and plays a significant role in glomerulonephritis.41 Furthermore, M-CSF mRNA levels were increased by poly(I:C) RNA exposure of MCs. M-CSF generation by MCs under basal and immune complex-stimulated conditions42,43 and glomerular CSF expression glomerulonephritis44 have been reported in and correlate with renal macrophage accumulation as well as glomerular injury and proteinuria.45 Furthermore recent studies support the importance of M-CSF in the generation of immunecomplex-mediated glomerulonephritis.44–46 In this context the increase in mRNA for M-CSF in MCs exposed to poly(I:C) RNA may be of interest for viral disease-induced glomerulonephritis.
We confirmed the increase of selected cytokines (IL-6 and IL-1β) and chemokines (IL-8/CXCL8, MCP-1/CCL2, and RANTES/CCL5) by quantitative RT-PCR and by ELISA, with the exception of MCP-1. The difference for MCP-1 is most likely due to the rapid down-regulation of this early response gene on stimulation.24 These genes are up-regulated by poly(I:C) RNA under basal conditions and even further when MCs were prestimulated with the cytokine combination of TNF-α, IL-1β, and IFN-γ. As to be expected the cytokine combination alone already increased mRNA levels for IL-6, IL-1β, IL-8/CXCL8, RANTES/CCL5, and MCP-1/CCL2 with a further increase by poly(I:C) RNA exposure, data also supported by ELISA determinations. Poly(I:C) DNA or CpG-DNA did not stimulate any of the cytokines or chemokines arguing in favor of a TLR3-mediated effect by poly(I:C) RNA. The absence of a response to CpG-DNA is consistent with the failure to demonstrate TLR9 in human MCs (present result) and in murine MCs.47 Finally mediation of the poly(I:C) RNA effects by TLR3 is supported by our observation that a blocking Ab for TLR3 markedly reduced the effects of poly(I:C) RNA on cytokine generation. All of the cytokines and chemokines up-regulated by poly(I:C) RNA in MCs have been implicated in viral disease associated forms of glomerulonephritis.38,48–51
The effects of poly(I:C) RNA were not restricted to the stimulation of proinflammatory genes but also included an anti-proliferative, proapoptotic response. Such a proapoptotic effect of TLR3 activation by poly(I:C) RNA has also been reported by Zhang and colleagues19 in 293 cells. In this context it may be of interest that signaling of TLR3 differs from that of other TLRs in that it is independent of the adaptor protein MyD88 but dependent on the adaptor protein Trif.18 Three signaling endpoints for TLR3-Trif signaling are recognized at present. These include: 1) activation of AP-1 and nuclear factor-κB and expression of a wide variety of proinflammatory genes18,52; 2) activation of the transcription factor IRF-3 resulting in expression of type I interferons52; and 3) a pathway leading to apoptosis,52 involving caspase-8 activation.53
Our data provide evidence that at least two of these signaling pathways are activated by poly(I:C) RNA in MCs. The proinflammatory genes that we found elevated are all regulated in a major way by the transcription factors AP-1 and nuclear factor-κB. The second pathway involving IRF-3 and generation of type I interferons could not be evaluated because MCs do not generate type I interferons. The third pathway leading to apoptosis of TLR3 signaling was activated by poly(I:C) RNA in MCs as evidenced by the down-regulation of various proliferation genes in the microchip analysis and the direct demonstration of apoptosis, which could be mitigated by caspase-8 inhibitor. Both, the anti-proliferative and proapoptotic effects of poly(I:C) RNA depended on preincubation of the MCs with the cytokines. These results provide further evidence that the effects of poly(I:C) RNA on proliferation and apoptosis may be mediated by TLR3. In the context of the MCs in vivo this could indicate that dsRNA by itself—without a proinflammatory cytokine milieu—would not influence MC survival but that MC apoptosis might occur after exposure of MCs to proinflammatory cytokines, as would be the case during viral diseases.
How could all these in vitro findings potentially relate to the in vivo situation under either normal physiological conditions or during diseases with circulating viral RNA or immune complex-containing viral RNA, such as hepatitis C-triggered glomerulonephritis? The physiological role of TLR3 on MCs could be that of a housekeeper removing viral RNA that reaches the mesangium. The expression of TLR3 would be low, as would be the amount of viral RNA reaching the mesangium and only negligible amounts of potentially proinflammatory cytokines and chemokines would be generated. Under conditions of viral infection with immune stimulation, enhanced levels of IFN-γ, TNF-α, and IL-1β would up-regulate TLR3 on MCs, and the increased amounts of viral RNA reaching MCs would result in the generation of chemokines such as IL-8/CXCL8, RANTES/CCL5, and MCP-1/CCL2. The chemokines would initially attract neutrophils (IL-8/CXCL8),54 followed by macrophages (RANTES/CCL5, MCP-1/CCL2).50 M-CSF would sustain and activate the monocytes46 to macrophages and ICAM-1 favor their glomerular infiltration.41 The potential role of IL-6 in this hypothetical scenario is less clear but could influence the overall immune response and especially the Th2 response and determine potential chronicity.55–57
Although the effects of poly(I:C) RNA on chemokine and cytokine generation by cytokine-conditioned MCs would fit into the general scheme of glomerular immune injury, how about the anti-proliferative, proapoptotic effects of poly(I:C) RNA observed in our study? Whether or not apoptosis prevails in vivo is dependent on the local balance of pro- and anti-apoptotic factors present in the mesangium at any given time during the glomerular disease process and cannot be conclusively evaluated in cell culture. In any case MC apoptosis plays an important role in glomerulonephritis in vivo.58
Intrigued by our results on TLR3 expression in MCs and the effects of poly(I:C) RNA on MCs in culture, we wondered whether TLR3 was also regulated in glomeruli isolated from human renal biopsies. Unfortunately, the available TLR3 Ab only worked on frozen kidney tissue, which was not available from routine biopsies. We therefore used real-time RT-PCR for TLR3 on either microdissected glomeruli or on laser-microdissected glomerular tissue from formalin-fixed biopsies.26 Furthermore, we checked for IL-8/CXCL8, RANTES/CCL5, and MCP-1/CCL2 because these had been up-regulated by poly(I:C) RNA in cultured MCs. The validity of these methods has been carefully established by our group.26 We have chosen biopsies from living kidney transplant donors as controls, from hepatitis C-negative idiopathic membrano-proliferative glomerulonephritis and from patients with hepatitis C-associated glomerulonephritis. Consistent with the results obtained with cultured MCs and by immunohistochemistry of normal kidney, mRNA for TLR3 was present in glomeruli from pretransplant donor biopsies and was increased in most biopsies from patients with hepatitis C-positive membranoproliferative glomerulonephritis but not from hepatitis C-negative membranoproliferative glomerulonephritis. This could not be related to differences in the degree of histopathological changes between the hepatitis C-positive and -negative membranoproliferative glomerulonephritis (Table 3).
Because we observed poly(I:C) RNA stimulation of cytokines and chemokines in MC culture, we evaluated mRNA levels for RANTES/CCL5 and MCP-1/CCL2 in parallel in the microdissected glomeruli. Intriguingly these chemokines were also up-regulated in microdissected glomeruli from hepatitis C-positive membranoproliferative glomerulonephritis as compared to either transplant donor biopsies and or biopsies from patients with hepatitis C-negative membranoproliferative glomerulonephritis. These results would be consistent with the hypothesis that glomerular/mesangial TLR3 could be activated by viral RNA in the course of viral infection and could thereby contribute to the disease process by chemokine/cytokine release, enhanced ICAM expression and effects on proliferation and apoptosis. Although our results only show an association, we consider this a novel and attractive hypothesis for viral disease-associated glomerulonephritis or even for viral disease-triggered exacerbation of other forms of glomerular injury, eg, systemic lupus erythematosus. The latter hypothesis is supported by data from our laboratory showing a marked worsening of the chronic lupus-like nephritis of MRL-Fas(lpr) mice injected with poly(I:C) RNA.59 TLR3 could also play a role in other forms of viral disease-associated glomerulonephritis and vasculitis, eg, IgA nephritis, a hypothesis that deserves further evaluation. The presence of TLR3 by immunohistology on vascular smooth muscle cells of arterial vessels raises the interesting question of their role and potential contribution to vasculitis and perhaps even atherosclerosis. In any case the novel finding of functional expression of TLR3 on intrinsic renal cells, and especially on MCs, may indicate a different role of TLR3 in these cells and establish a novel link between viral infections and glomerular diseases.
Acknowledgments
We thank Claudia Schmidt, Ingrid Bayer, and Dr. Eva Kiss for expert technical assistance.
Footnotes
Address reprint requests to Prof. Dr. Detlef Schlöndorff, Medizinische Poliklinik, Klinikum der LMU, Pettenkoferstrasse 8a, 80336 München, Germany. E-mail: detlef.schloendorff@med.uni-muenchen.de.
Supported by the Ludwig-Maximilians-University Munich (FöFoLe 231 to M.W.), the Deutsche Forschungsgemeinschaft (BA 2137/1-1 to B.B. and D.S.), the Deutsche Forschungsgemeinschaft (SFB405/Projekt B10 to H.-J.G.), and biopsies were available from the ERCB (European Renal cDNA Bank) generated with support by EU-FP V (chronic kidney disease QLRT-2001-01215).
M.W., H.S., and B.B. contributed equally to this study.
References
- Agnello V, Chung RT, Kaplan LM. A role for hepatitis C virus infection in type II cryoglobulinemia. N Engl J Med. 1992;327:1490–1495. doi: 10.1056/NEJM199211193272104. [DOI] [PubMed] [Google Scholar]
- Meyers CM, Seef LB, Stehman-Breen CO, Hoofnagle JH. Hepatitis C and renal disease: an update. Am J Kidney Dis. 2003;42:631–657. doi: 10.1016/s0272-6386(03)00828-x. [DOI] [PubMed] [Google Scholar]
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of non-clonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhang G, Hayden MS, Greenblatt MB, Bussey C, Flavell RA, Ghosh S. A toll-like receptor that prevents infection by uropathogenic bacteria. Science. 2004;303:1522–1526. doi: 10.1126/science.1094351. [DOI] [PubMed] [Google Scholar]
- Anders HJ, Banas B, Schlöndorff D. Signaling danger: toll-like receptors and their potential roles in kidney disease. J Am Soc Nephrol. 2004;15:854–867. doi: 10.1097/01.asn.0000121781.89599.16. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Kikkawa S, Kohase M, Miyake K, Seya T. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem Biophys Res Commun. 2002;293:1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Muzio M, Bosisio D, Polentarutti N, D‘amico G, Stoppacciaro A, Mancinelli R, van‘t Veer C, Penton-Rol G, Ruco LP, Allavena P, Mantovani A. Differential expression and regulation of toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J Immunol. 2000;164:5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. 2000;68:7010–7017. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin A, Mazzoni A, Spitzer JH, Wyllie DH, Dower SK, Segal DM. Regulation of Toll-like receptors in human monocytes and dendritic cells. J Immunol. 2001;166:249–255. doi: 10.4049/jimmunol.166.1.249. [DOI] [PubMed] [Google Scholar]
- Akira S. Toll-like receptors and innate immunity. Adv Immunity. 2001;78:1–56. doi: 10.1016/s0065-2776(01)78001-7. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Funami K, Tanabe M, Oshiumi H, Shingai M, Seto Y, Yamamoto A, Seya T. Subcellular localization of Toll-like receptor 3 in human dendritic cells. J Immunol. 2003;171:3154–3162. doi: 10.4049/jimmunol.171.6.3154. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Funami K, Oshiumi H, Seya T. Toll-like receptor 3: a link between toll-like receptor, interferon and viruses. Microbiol Immunol. 2004;48:147–154. doi: 10.1111/j.1348-0421.2004.tb03500.x. [DOI] [PubMed] [Google Scholar]
- Barton GM, Medzhitov R. Toll signaling: RIPping off the TNF pathway. Nat Immunol. 2004;5:472–474. doi: 10.1038/ni0504-472. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu LG, Han KJ, Wie X, Shu HB. PIASy represses TRIF-induced ISRE and NF-kappaB activation but not apoptosis. FEBS Lett. 2004;16:97–101. doi: 10.1016/j.febslet.2004.05.081. [DOI] [PubMed] [Google Scholar]
- Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J Immunol. 2002;168:554–561. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- Uchiyama-Tanaka Y, Mori Y, Kishimoto N, Nose A, Kijima Y, Nagata T, Umeda Y, Masaki H, Matsubara H, Iwasaka T. Membranous glomerulonephritis associated with hepatitis C virus infection: case report and literature review. Clin Nephrol. 2004;61:144–150. doi: 10.5414/cnp61144. [DOI] [PubMed] [Google Scholar]
- Gröne HJ, Simon M, Gröne EF. Expression of vascular endothelial growth factor in renal vascular disease and renal allograft. J Pathol. 1995;177:259–267. doi: 10.1002/path.1711770308. [DOI] [PubMed] [Google Scholar]
- Adams J, Kiss E, Arroyo ABV, Bonrouhi M, Sun Q, Li Z, Gretz N, Schnitger A, Zouboulis CC, Wiesel M, Wagner J, Nelson PJ, Gröne HJ. 13-cis retinoic acid inhibits development and progression of chronic allograft nephropathy. Am J Pathol. 2005;167:285–298. doi: 10.1016/S0002-9440(10)62973-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas B, Luckow B, Moller M, Klier C, Nelson PJ, Schadde E, Brigl M, Halevy D, Holthofer H, Reinhart B, Schlondorff D. Chemokine and chemokine receptor expression in a novel human mesangial cell line. J Am Soc Nephrol. 1999;10:2314–2322. doi: 10.1681/ASN.V10112314. [DOI] [PubMed] [Google Scholar]
- Porubsky S, Schmid H, Bonrouhi M, Kretzler M, Malle E, Nelson PJ, Gröne HJ. Influence of native and hypochlorite-modified low-density lipoprotein on gene expression in human proximal tubular epithelium. Am J Pathol. 2004;164:2175–2187. doi: 10.1016/S0002-9440(10)63775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CD, Gröne HJ, Gröne EF, Nelson PJ, Schlöndorff D, Kretzler M. Laser microdissection and gene expression analysis on formaldehyde-fixed archival tissue. Kidney Int. 2002;61:125–132. doi: 10.1046/j.1523-1755.2002.00112.x. [DOI] [PubMed] [Google Scholar]
- Mack M, Kleinschmidt A, Brühl H, Klier C, Nelson PJ, Cihak J, Plachy J, Stangassinger M, Erfle V, Schlöndorff D. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med. 2000;6:769–775. doi: 10.1038/77498. [DOI] [PubMed] [Google Scholar]
- Kessler N, Ferraris O, Palmer K, Marsh W, Steel A. Use of the DNA flow-thru chip, a three-dimensional biochip, for typing and subtyping of influenza viruses. J Clin Microbiol. 2004;42:2173–2185. doi: 10.1128/JCM.42.5.2173-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeg K, Reimann J, Kabelitz D, Hardt C, Wagner H. A rapid colorimetric assay for the determination of IL-2-producing helper T cell frequencies. J Immunol Methods. 1985;77:237–246. doi: 10.1016/0022-1759(85)90036-5. [DOI] [PubMed] [Google Scholar]
- Berger T, Brigl M, Herrmann JM, Vielhauer V, Luckow B, Schlöndorff D, Kretzler M. The apoptosis mediator mDAP-3 is a novel member of a conserved family of mitochondrial proteins. J Cell Sci. 2000;113:3603–3612. doi: 10.1242/jcs.113.20.3603. [DOI] [PubMed] [Google Scholar]
- Yang E, Shin JS, Kim H, Park HW, Kim MH, Kim SJ, Choi IH. Cloning of TLR3 isoform. Yonsei Med J. 2004;45:359–361. doi: 10.3349/ymj.2004.45.2.359. [DOI] [PubMed] [Google Scholar]
- Tsuboi N, Yoshikai Y, Matsuo S, Kikuchi T, Iwami K, Nagai Y, Takeuchi O, Akira S, Matsuguchi T. Roles of toll-like receptors in C-C chemokine production by renal tubular epithelial cells. J Immunol. 2002;169:2026–2033. doi: 10.4049/jimmunol.169.4.2026. [DOI] [PubMed] [Google Scholar]
- Ardaillou R. Biology of glomerular cells in culture. Cell Biol Toxicol. 1996;12:257–261. doi: 10.1007/BF00438155. [DOI] [PubMed] [Google Scholar]
- Kim YS, Zheng S, Yang SH, Kim HL, Lim CS, Chae DW, Chun R, Lee JS, Kim S. Differential expression of various cytokine and chemokine genes between proliferative and non-proliferative glomerulonephritides. Clin Nephrol. 2001;56:199–206. [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O‘Connel R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Heinz S, Haehnel V, Karaghiosoff M, Schwarzfischer L, Muller M, Krause SW, Rehli M. Species-specific regulation of Toll-like receptor 3 genes in men and mice. J Biol Chem. 2003;278:21502–21509. doi: 10.1074/jbc.M301476200. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Sareneva T, Julkunen I, Matikainen S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun. 2001;2:349–355. doi: 10.1038/sj.gene.6363791. [DOI] [PubMed] [Google Scholar]
- Papayianni A. Cytokines, growth factors, and other inflammatory mediators in glomerulonephritis. Renal Failure. 1996;18:725–740. doi: 10.3109/08860229609047702. [DOI] [PubMed] [Google Scholar]
- Timoshanko JR, Kitching AR, Iwakura Y, Holdsworth SR, Tipping PG. Contributions of IL-1beta and IL-1alpha to crescentic glomerulonephritis in mice. J Am Soc Nephrol. 2004;15:910–918. doi: 10.1097/01.asn.0000115704.86897.f4. [DOI] [PubMed] [Google Scholar]
- Niemir ZI, Stein H, Ciechanowicz A, Olejniczak P, Dworacki G, Ritz E, Waldherr R, Czekalski S. The in situ expression of interleukin-8 in the normal human kidney and in different morphological forms of glomerulonephritis. Am J Kidney Dis. 2004;43:983–998. doi: 10.1053/j.ajkd.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Moon KC, Park SY, Kim HW, Hong HK, Lee HS. Expression of intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 in human crescentic glomerulonephritis. Histopathology. 2002;41:158–165. doi: 10.1046/j.1365-2559.2002.01446.x. [DOI] [PubMed] [Google Scholar]
- Hora K, Satriano JA, Santiago A, Mori T, Stanley ER, Shan Z, Schlondorff D. Receptors for IgG complexes activate synthesis of monocyte chemoattractant peptide 1 and colony-stimulating factor 1. Proc Natl Acad Sci USA. 1992;89:1745–1749. doi: 10.1073/pnas.89.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satriano JA, Hora K, Shan Z, Stanley ER, Mori T, Schlondorff D. Regulation of monocyte chemoattractant protein-1 and macrophage colony-stimulating factor-1 by IFN-gamma, tumor necrosis factor-alpha, IgG aggregates, and cAMP in mouse mesangial cells. J Immunol. 1993;150:1971–1978. [PubMed] [Google Scholar]
- Matsuda M, Shikata K, Makino H, Sugimoto H, Ota Z. Glomerular expression of macrophage colony-stimulating factor and granulocyte-macrophage colony-stimulating factor in patients with various forms of glomerulonephritis. Lab Invest. 1996;75:403–412. [PubMed] [Google Scholar]
- Wada T, Naito T, Griffiths RC, Coffman TM, Kelley VR. Systemic autoimmune nephritogenic components induce CSF-1 and TNF-alpha in MRL kidneys. Kidney Int. 1997;52:934–941. doi: 10.1038/ki.1997.415. [DOI] [PubMed] [Google Scholar]
- Lenda DM, Stanley ER, Kelley VR. Negative role of colony-stimulating factor-1 in macrophage, T cell and B cell mediated autoimmune disease in MRL-Fas(lpr) mice. J Immunol. 2004;173:4744–4754. doi: 10.4049/jimmunol.173.7.4744. [DOI] [PubMed] [Google Scholar]
- Anders HJ, Vielhauer V, Eis V, Linde Y, Kretzler M, Perez de Lema G, Strutz F, Bauer S, Rutz M, Wagner H, Gröne HJ, Schlöndorff D. Activation of toll-like receptor-9 induces progression of renal disease in MRL-Fas(lpr) mice. FASEB J. 2004;18:534–536. doi: 10.1096/fj.03-0646fje. [DOI] [PubMed] [Google Scholar]
- Fukatsu A, Matsuo S, Tamai H, Sakamoto N, Matsuda T, Hirano T. Distribution of interleukin-6 in normal and diseased human kidney. Lab Invest. 1999;65:61–66. [PubMed] [Google Scholar]
- Mezzano S, Burgos ME, Olavarria F, Caorsi I. Immunohistochemical localization of IL-8 and TGF-beta in streptococcal glomerulonephritis. J Am Soc Nephrol. 1997;8:234–241. doi: 10.1681/ASN.V82234. [DOI] [PubMed] [Google Scholar]
- Haberstroh U, Pocock J, Gomez-Guerrero C, Helmchen U, Hamann A, Gutierrez-Ramos JC, Stahl RA, Thaiss F. Expression of the chemokines MCP-1/CCL2 and RANTES/CCL5 is differentially regulated by infiltrating inflammatory cells. Kidney Int. 2002;62:1264–1276. doi: 10.1111/j.1523-1755.2002.kid572.x. [DOI] [PubMed] [Google Scholar]
- Natori Y, Sekiguchi M, Ou Z, Natori Y. Gene expression of CC chemokines in experimental crescentic glomerulonephritis (CGN). Clin Exp Immunol. 1997;109:143–148. doi: 10.1046/j.1365-2249.1997.4271321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171:4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Brown Z, Strieter RM, Chensue SW, Ceska M, Lindley I, Neild GH, Kunkel SL, Westwick J. Cytokine-activated human mesangial cells generate the neutrophil chemoattractant, interleukin-8. Kidney Int. 1991;40:86–90. doi: 10.1038/ki.1991.184. [DOI] [PubMed] [Google Scholar]
- Kluth DC, Rees AJ. New approaches to modify glomerular inflammation. J Nephrol. 1999;12:66–67. [PubMed] [Google Scholar]
- Diehl S, Chow CW, Weiss L, Palmetshofer A, Twardzik T, Rounds L, Serfling E, Davis RJ, Anguita J, Rincon M. Induction of NFATc2 expression by interleukin 6 promotes T helper type 2 differentiation. J Exp Med. 2002;196:39–49. doi: 10.1084/jem.20020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst SM, Wilkinson TS, McLoughlin RM, Jones S, Horiuchi S, Yamamoto N, Rose-John S, Fuller GM, Topley N, Jones SA. IL-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity. 2001;14:705–714. doi: 10.1016/s1074-7613(01)00151-0. [DOI] [PubMed] [Google Scholar]
- Hughes J, Cailhier JF, Watson S, Savill JS. Apoptosis in glomerulonephritis. Rheum Dis Clin North Am. 2004;30:655–676. doi: 10.1016/j.rdc.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Patole P, Gröne HJ, Segerer S, Ciubar R, Belemezova E, Henger A, Kretzler M, Schlöndorff, Anders HJ. Viral double-stranded RNA aggravates lupus nephritis trough Toll-like receptor 3 on glomerular mesangial cells and antigen-presenting cells. J Am Soc Nephrol. 2005;16:1326–1338. doi: 10.1681/ASN.2004100820. [DOI] [PubMed] [Google Scholar]