Abstract
The endoplasmic reticulum (ER) quality control processes recognize and remove aberrant proteins from the secretory pathway. Several variants of the plasma protein fibrinogen are recognized as aberrant and degraded by ER-associated protein degradation (ERAD), thus leading to hypofibrinogenemia. A subset of patients with hypofibrinogenemia exhibit hepatic ER accumulation of the variant fibrinogens and develop liver cirrhosis. One such variant named Aguadilla has a substitution of Arg375 to Trp in the γ-chain. To understand the cellular mechanisms behind clearance of the aberrant Aguadilla γ-chain, we expressed the mutant γD domain in yeast and found that it was cleared from the ER via ERAD. In addition, we discovered that when ERAD was saturated, aggregated Aguadilla γD accumulated within the ER while a soluble form of the polypeptide transited the secretory pathway to the trans-Golgi network where it was targeted to the vacuole for degradation. Examination of Aguadilla γD in an autophagy-deficient yeast strain showed stabilization of the aggregated ER form, indicating that these aggregates are normally cleared from the ER via the autophagic pathway. These findings have clinical relevance in the understanding of and treatment for ER storage diseases.
Fibrinogen, a large plasma protein synthesized in hepatocytes, plays a critical role in blood coagulation. Mature circulating fibrinogen is a symmetric dimeric molecule in which each half consists of three polypeptide chains: Aα, Bβ, and γ. The two halves of the molecule associate into a trinodal D-E-D structure with the N termini of all six chains in the central E domain, the C termini of Bβ and γ forming the two globular D domains, and the C termini of the Aα-chains forming a less compact structure associated with the E domain.1,2 Assembly of all six chains into the fibrinogen molecule occurs within the hepatocyte endoplasmic reticulum.3–5 Individual unassembled fibrinogen chains are retained within the endoplasmic reticulum (ER) and ultimately degraded in a proteasome-dependent manner, classifying them as substrates of ER-associated protein degradation (ERAD).5,6
The identification of the genetic defects associated with low levels of circulating fibrinogen (hypofibrinogenemia) has shown that mutations within any of the three chains can lead to misfolding of that chain, recognition of the aberrant polypeptide by ER quality control, and subsequent degradation.7 However, studies of individuals carrying various mutant forms of fibrinogen chains have shown that a subset of those individuals with hypofibrinogenemia also exhibit hepatic ER accumulation of fibrinogen that appears to lead to liver disease.8–10 For example, we previously described one family harboring a novel γ-chain mutation, Arg375Trp, termed fibrinogen Aguadilla, in which all carriers present with hypofibrinogenemia, but only a subset have liver inclusions.10 Biopsies from diseased livers showed aggregates of fibrinogen within the ER lumen of hepatocytes.10 This disease pathology, ie, both the loss-of-function hypofibrinogenemia and the gain-of-function liver disease, is similar to α-1-antitrypsin deficiency, another ER storage disease.
Individuals who are homozygous for the Z variant of the serum protein α-1-antitrypsin (α-1-protease inhibitor, A1Pi) exhibit low levels of A1Pi and the resulting loss-of-function disease pathology of early onset emphysema.11 In addition, 12 to 15% of A1PiZ homozygous juveniles also display the gain-of-function disease pathology of liver cirrhosis,12–15 and studies of adults indicate a risk of cirrhosis in 30 to 50% of homozygous individuals.16 It has been previously demonstrated, in both mammalian and yeast cell systems, that the intracellular fate of A1PiZ is ERAD.17,18 However, when this pathway is saturated such that it is insufficient to rid the ER of misfolded A1PiZ, aggregates accumulate within the ER.19 Interestingly, the clearance of these A1PiZ aggregates from the ER is dependent on the autophagic pathway to the vacuole.19 These findings correlate with the observation that there is an increased number of autophagosomes in hepatocytes of A1PiZ homozygous individuals with liver disease.20 Together, these data suggest that liver disease associated with α-1-antitrypsin deficiency may be due to different inherited efficiencies in ER quality control or possibly to a second mutation within the ERAD or autophagy pathways.
Because of the similarity between the pathology of α-1-antitrypsin deficiency and hypofibrinogenemia-associated liver disease, we reasoned that the fibrinogen Aguadilla Argγ375Trp variant would have an intracellular fate similar to that of A1PiZ. Thus, to investigate the mechanism involved in the fibrinogen Aguadilla-related ER storage disease, we decided to express the fibrinogen γD domain with the Arg375Trp mutation in our yeast model system. The rationale for the use of the γD domain comes from previous studies showing that when expressed in Pichia pastoris, the normal γD domain is correctly folded into a stable configuration with both its calcium binding site and polymerization pocket intact.21 Should the Aguadilla mutation affect folding of the fibrinogen γD domain, we anticipate that the quality control mechanisms that contribute to clearance of the aberrant polypeptide may be identified in our yeast system. Knowledge of these quality control mechanisms could begin to answer questions regarding the molecular mechanisms driving the pathology of hypofibrinogenemia and liver disease.
Materials and Methods
Strains, Plasmids, Media, and Antisera
Electrocompetent Escherichia coli strain HB101 (F−, mcrB, mrr, hsdS20(rB−, mB−), recA13, supE44, ara14, galK2, lacY1, proA2, rpsL20(Smr), xyl5, λ−, leu, and mtl) was purchased from Invitrogen Corp. (Carlsbad, CA). The P. pastoris strain used in this study was GS115/his4 (Invitrogen). The Saccharomyces cerevisiae strains used were wild type22 : BY4742/MATα, his3Δ1, leu2Δ0, lysΔ0, and ura3Δ0; BY4742: pep4Δ22/MATα, his3Δ1, leu2Δ0, lysΔ0, ura3Δ0, and YPL154c::KANMX (Invitrogen); BY4742: atg14Δ19/MATα, his3Δ1, leu2Δ0, lysΔ0, ura3Δ0, and YBR128c::KANMX4; the proteasome mutant23 : pre1-1, pre2-2/MATα, leu2-3, 112, his3-11, 15, ura3(Δ5), GAL+, can(s), pre1-1, and pre2-2; and the wild-type isogenic parent23 : WCG4/MATα, leu2-3, 112, his3-11, 15, ura3(Δ5), and GAL+ (kindly provided by Dieter Wolf, University of Stuttgart, Germany).
P. pastoris yeast cells were cultured in synthetic media containing 1.34% yeast nitrogen base without amino acids and ammonium sulfate, 2% peptone, 1% yeast extract, 100 mmol/L potassium phosphate, pH 6.0, 400 ng/ml biotin, and 1% glycerol as a carbon source or with 1% methanol to induce expression of γD fibrinogen. S. cerevisiae yeast cells were cultured in synthetic media containing 0.67% yeast nitrogen base with amino acids, 0.5% casamino acids, and 2% dextrose as a carbon source or with 2% galactose to induce expression of γD fibrinogen. Antibodies used included rabbit anti-human fibrinogen (DakoCytomation California Inc., Carpinteria, CA) and horseradish peroxidase-conjugated goat anti-rabbit (U.S. Biochemical Co., Cleveland, OH), monoclonal mouse anti-yeast phosphoglycerate kinase (Invitrogen), rabbit anti-Kar2p24 (generously provided by Jeffrey Brodsky, University of Pittsburgh, Pittsburgh, PA), and sheep anti-mouse IgG horseradish peroxidase-conjugated antibody (GE Health Care Bio-Sciences, Little Chalfont, UK).
Construction of Expression Vectors
The integrative expression vector, pPIC9 (Invitrogen), carrying the coding regions spanning amino acid residues 143 to 411 of the human fibrinogen γ-gene25 (hereafter referred to as the fibrinogen γD domain) was kindly supplied by Dr. Tatiana Ugarova (Cleveland Clinic Foundation, Cleveland, OH). The fragment encoding the fibrinogen γD domain was cloned in frame with the yeast α-factor secretion signal and propeptide and driven by an alcohol oxidase 1 methanol-inducible promoter. The fibrinogen Aguadilla mutation, Argγ375Trp,10 was inserted using mutagenic primers (available on request) with a QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA), and the mutation was verified using automated sequencing. Using polymerase chain reaction and primers that introduced unique restriction sites, the α-factor pre-pro and γ-sequences were subcloned into an S. cerevisiae expression vector with a regulatable promoter26 : either the inducible GAL1 promoter p426Gal1 (2μ, Ampr, and URA3; American Type Culture Collection, Manassas, VA) or the repressible MET25 promoter p426MET25 (2μ, Ampr, and URA3; American Type Culture Collection). Correct cloning was verified by restriction digest and automated sequencing.
Transformations and Plasmid Preparation
The Cell-Porator E. coli Pulser (Invitrogen) was used to electroporate competent E. coli. Plasmids were isolated from bacterial transformants using the Quantum Prep Plasmid Miniprep kit (Bio-Rad, Hercules, CA). P. pastoris transformations were performed using the spheroplasting protocol within the Pichia Expression kit M instruction manual (Invitrogen), and transformants were selected in medium lacking histidine. Southern blot was used to choose transformants with only a single copy27 of the integrative pPIC9 expression vector. S. cerevisiae transformations were performed using a standard lithium acetate procedure,28 and transformants were isolated after growth in selective medium containing 2% dextrose.
ERAD Assays
The colony-blot immunoassays for both S. cerevisiae and P. pastoris were a modification of a previously described procedure.19 In brief, exponentially growing yeast were resuspended to a final optical density (OD; at 600 nm) of 0.001OD/μl to control for cell number. A 3-μl aliquot of these yeast were spotted onto a nitrocellulose disk overlaid on medium containing 1% methanol to induce expression in P. pastoris of the γD off the AOX1 promoter, medium containing 2% galactose to induce expression in S. cerevisiae of the γD off the GAL1 promoter, or synthetic medium lacking methionine to induce expression in S. cerevisiae of the γD off the MET25 promoter. After incubation at 30°C for 18 hours, cells were lysed with solution 1 (0.2 mol/L NaOH, 0.1% sodium dodecyl sulfate (SDS), and 0.05% 2-mercaptoethanol) for 1 hour at room temperature, and blots were then assayed as previously described.29 Immunoreactive proteins were detected by developing the blots with SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL) and Super RX X-ray film (Fuji, Tokyo, Japan). The density of immunoreactive protein at each colony spot was quantified using Molecular Analyst (Bio-Rad).
For pulse-chase protein radiolabeling to assay for γD degradation in P. pastoris, cultures were grown at 30°C in medium containing 1% methanol to induce expression for 24 hours before analysis. Using a protocol based on those previously described,30,31 cells were resuspended at 4 OD/ml in medium lacking histidine, cysteine, and methionine. They were incubated at 30°C for 45 minutes, then pulsed with 25 μCi/OD EasyTag Express Protein Labeling Mix (PerkinElmer Inc., Boston, MA) for 10 minutes, and chased with 1 mg/ml cold cysteine and methionine. Aliquots were removed at the desired time points. Cell lysates were prepared and immunoprecipitated with rabbit polyclonal antibody to human fibrinogen. The purified proteins were separated by 10% reducing SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography, and signal intensities were quantified using Molecular Analyst (Bio-Rad).
The cycloheximide chase analyses of S. cerevisiae were a modification of a previously described procedure.19 In brief, overnight cultures were incubated in medium containing 2% galactose for 4 to 8 hours at 30°C. To control for cell number, cultures were then resuspended to a final OD600 of 2 OD/ml in medium containing 2% dextrose plus 200 μg/ml cycloheximide. They were incubated at 30°C with shaking, 500-μl aliquots were collected, and the cells were harvested by centrifugation at the indicated time points. At each time point, the medium was reserved and represented the secreted material. The cell pellets were resuspended in 50 μl of lysis buffer (160 mmol/L Tris, pH 6.8, 4% SDS. 0.2% bromophenol blue, 200 mmol/L dithiothreitol [DTT], and 20% glycerol) and incubated at 95°C for 2 minutes, and 0.5-mm acid-washed glass beads were added. The cells were disrupted via four sequential 60-second bursts on a Vortex mixer at the highest setting followed by cooling on ice for 60 seconds. Finally, an additional 50-μl aliquot of lysis buffer and the reserved media were added, resulting in 0.0017 OD/μl final concentration. Then, 20-μl aliquots of lysate/media were combined with 10 μl of sample buffer (0.125 mol/L Tris, pH 6.8, 4% SDS, 0.004% bromophenol blue, 10% 2-mercaptoethanol, and 20% glycerol), heated to 95°C for 10 minutes, and resolved by 10% reducing SDS-PAGE. The γD domain was detected by immunoblot analysis, and results were quantified as described above.
Protein Characterization Studies and Sequencing
P. pastoris cultures were grown in medium containing 1% methanol for 72 hours at 30°C. Cell pellets were lysed by vortexing with acid-washed glass beads as described above but in 50 mmol/L Tris-HCl, pH 7.4, and 1 mmol/L EDTA, washed twice with 1% Triton X-100 and then three times with water. The remaining pellet was solubilized in 8 mol/L urea, 100 mmol/L Tris-HCl, pH 8.0, and 15 mmol/L DTT and reduced for 16 hours at room temperature.
The high and low molecular weight γD species were separated by reverse phase high performance liquid chromatography (HPLC) on a 5-μm Jupiter C4 column (Phenomenex, Torrence, CA) with a 0.05% trifluoroacetic acid/acetonitrile gradient similar to that used for reduced human fibrinogen chains.32 Peaks were collected and assayed at the Protein Microchemistry Facility at the University of Otago, New Zealand for N-terminal protein sequencing using established methods.33 A 20-μl sample of each peak crest was also analyzed by electrospray ionization mass spectrometry (ESI MS) on a VG Platform II quadrupole analyzer (Micromass, Manchester, UK) as previously described.32
For peptide mapping, HPLC peaks were dried, redissolved in 50 μl of 50 mmol/L ammonium carbonate and digested with 2 μg of trypsin for 16 hours at 37°C. After being dried under vacuum with P2O5 and redissolved in 50 μl of 50% acetonitrile and 0.1% formic acid, 20 μl of each digest was analyzed by ESI MS.
Cell Fractionation
S. cerevisiae cultures were grown in medium containing 2% galactose for 40 hours at 30°C to induce expression of γD. Yeast were pelleted, and the medium was reserved and concentrated fourfold for use as the extracellular, secreted fraction. Spheroplasts were formed by incubating cells with 10 mg/ml lyticase in 1.2 mol/L sorbitol and 0.1 mol/L K2HPO4, pH 7.2, and spheroplasts and the periplasm fraction were separated by centrifugation through a cushion of 0.8 mol/L sucrose, 1.5% Ficoll, and 20 mmol/L HEPES, pH 7.4, at 4000 × g for 10 minutes at 4°C. After the spheroplasts were washed with 1.2 mol/L sorbitol and 0.1 mol/L K2HPO4, pH 7.2, they were resuspended in ice-cold Spheroplast lysis buffer (0.1 mol/L sorbitol, 50 mmol/L KOAc, 2 mmol/L EDTA, 20 mmol/L HEPES, pH 7.4, 1 mmol/L DTT, and 1 mmol/L phenylmethylsulfonyl fluoride) and lysed by 10 strokes with a dounce homogenizer on ice. Resulting cell lysates were layered over a cushion of 1 mol/L sucrose, 50 mmol/L potassium acetate, 20 mmol/L HEPES, pH 7.4, and 1 mmol/L DTT and subjected to 6500 × g centrifugation for 10 minutes at 4°C. The top fraction, above the interface, was collected and subjected to further centrifugation at 22,000 × g for 10 minutes at 4°C. The resulting pellet was the microsome fraction, and the supernatant/cytosol was further cleared of small vesicles by centrifugation at 100,000 × g for 1 hour at 4°C. Protease protection assays were performed on the microsome fraction by exposing washed microsomes either to 0.4 mg/ml trypsin in buffer 88 (20 mmol/L HEPES, pH 6.8, 150 mmol/L KOAc, 250 mmol/L sorbitol, and 5 mmol/L MgOAc) or to 0.4 mg/ml trypsin and 2% Triton X-100 in buffer 88 for 30 minutes on ice. Specific proteins were then visualized by immunoblot analysis.
Sucrose Density Gradient Analysis
Microsomes were isolated as described above from S. cerevisiae producing γD during 40 hours of induction. The microsomes were lysed in buffer 88 that had been supplemented with 0.5% Triton X-100 and complete protease inhibitor cocktail (Sigma, St. Louis, MO) by incubation on ice for 2 hours, with brief vortex mixing every 30 minutes. The total protein content of the lysates was determined using Coomassie Plus Protein Assay (Pierce). An aliquot of 1 mg of total protein from each lysate was loaded onto a 5 to 60% sucrose gradient34,35 and centrifuged in a Beckman (Beckman Coulter, Inc., Fullerton, CA) SW50 at 145,000 × g for 20 hours at 4°C. Fractions of 250 μl were collected, and proteins were concentrated by precipitation in 10% trichloroacetic acid. Specific proteins were visualized by immunoblot analysis.
Endoglycosidase H Analysis
Microsomes were prepared as described above and lysed on ice for 30 minutes in denaturing buffer (50 mmol/L sodium acetate, pH 5.2, 0.5% 2-mercaptoethanol, and 0.5% SDS). Samples were heated to 95°C for 3 minutes and then centrifuged 14,000 × g for 5 minutes at 4°C to remove membranes. An equal volume of reaction buffer (50 mmol/L sodium acetate, pH 5.2, 0.5 mmol/L phenylmethylsulfonyl fluoride, and 0.5% NP-40) was added to the supernatant, and the sample was split in two. One sample received 0.05 units of endoglycosidase H (Roche Diagnostics, Indianapolis, IN), both samples were incubated at 37°C for 1 hour, and proteins were visualized by immunoblot analysis.
Results
Aguadilla Variant of γ-Fibrinogen Selectively Degraded in Yeast
The ∼30-kd C-terminal domain of the γ-chain of human fibrinogen (Val143 through Val411) cloned in-frame behind the yeast α-factor pre-pro sequence was used for these studies because this γD domain had previously been shown to correctly fold and function.21,25 Furthermore, the site of the Aguadilla mutation is within the γD domain, and the Arg375Trp mutation is believed to alter folding such that it would be recognized by ER quality control.10 Site-directed mutagenesis was used to generate the fibrinogen Aguadilla mutation. The wild-type and Aguadilla γD were subcloned into the P. pastoris shuttle vector as well as the appropriate shuttle vectors for expression in S. cerevisiae.
To examine the fates of wild-type and Aguadilla γD, both were expressed in P. pastoris and S. cerevisiae, and expression was examined using a colony-blot immunoassay that allows for quantification of the steady-state level of γD using an antibody to fibrinogen. In both P. pastoris and S. cerevisiae, the steady-state level of Aguadilla γD was less than that seen for the wild-type γD (Figure 1A), suggesting degradation of the mutant that mimics the phenotype of individuals who carry the Aguadilla mutation, ie, low levels of fibrinogen.10
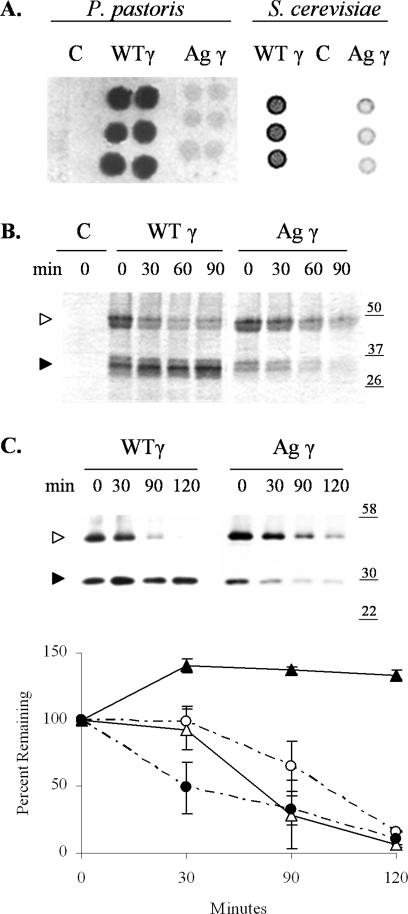
Figure 1.
Aguadilla γD degraded in P. pastoris and S. cerevisiae. A: Representative colony-blot immunoassays of single-copy transformants of P. pastoris and high-expression transformants of S. cerevisiae, each expressing wild-type γD (WTγ) or Aguadilla γD (Agγ) or containing a vector control (C). B: A representative pulse-chase immunoprecipitation of single-copy transformants of P. pastoris expressing wild-type γD (WTγ) or Aguadilla γD (Agγ) or containing the vector control (C). Protein standards are noted to the right in kilodaltons. A high molecular weight (◃) and low molecular weight (▸) form of γD are indicated. C: A representative cycloheximide chase immunoblot of a S. cerevisiae wild-type strain expressing either wild-type γD (WTγ) or Aguadilla γD (Agγ) at high levels off the p426Gal1 vector. A graph of the means ± SEM, from at least three experiments is shown with wild-type γD LMW (solid triangles and solid line), wild-type γD HMW (open triangles and solid line), Aguadilla γD LMW (solid circles and dashed line), and Aguadilla γD HMW (open circles and dashed line).
To determine whether the low level of Aguadilla γD was due to degradation, kinetic studies were undertaken in both yeast systems: pulse-chase protein radiolabeling studies were performed in P. pastoris, and cycloheximide chase analyses were performed in S. cerevisiae (Figure 1, B and C). In both yeast systems, using either chase protocol, two molecular weight species were observed. Furthermore, the low molecular weight (LMW) form of the wild-type γD was relatively stable over the chase period, whereas the LMW form of the mutant Aguadilla γD species was clearly degraded by both P. pastoris and S. cerevisiae.
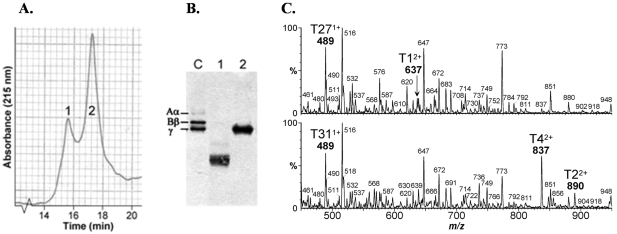
The γD fragment encodes a ∼30-kD polypeptide21 that correlates well with the LMW species, but the presence of a high molecular weight (HMW) species running at approximately 48 kd on reducing SDS-PAGE was puzzling. To characterize this material, lysates from P. pastoris were fractionated by reverse phase HPLC (Figure 2A). Analysis of individual peaks by Western blot showed that the LMW species eluted just ahead of the more hydrophobic HMW form (Figure 2B). Direct ESI MS analysis of peak 1 established a mass of 30,470 D, which was in excellent agreement with the theoretical mass of 30,471 D expected for mature γD. Repeated analysis of the HMW material was unfortunately less informative. However, an N-terminal protein sequence of APVNTTTEDET- was established, indicating that the α-factor pre sequence had been cleaved, but the pro sequence was still intact. The anticipated molecular mass of the γD domain with an N-terminal pro sequence is 38 kd, and the three N-linked oligosaccharides of the propeptide36 increase the predicted molecular mass to approximately 47 kd, close to the observed value of ∼48 kd. Furthermore, tryptic peptide mass mapping confirmed the primary structures of the two components (Figure 2C). Maps of the LMW material showed the expected ions arising solely from the γD domain, including the C-terminal peptide. These ions were also present in the HMW digests, together with unique ions arising from the propeptide sequence.
Figure 2.
Structural analysis of intracellular forms of γD. A: Reverse phase HPLC profile of P. pastoris lysate solubilized in 8 mol/L urea, 100 mmol/L Tris-HCl, pH 8.0, and 15 mmol/L DTT. B: Western blot of peaks 1 and 2 together with a fibrinogen control (lane C); the mass of the γ-chain of fibrinogen is 48 kd. C: ESI MS tryptic peptide maps of the LMW γD species (top spectrum) and HMW γD species (bottom spectrum). Both maps show the same expected C-terminal QAGDV ion at 489 m/z, indicating normal transcription termination. However, the N-terminal ion of γD (YVVQIHDITGK; 637 m/z) is missing in the HMW map and replaced by an 837 m/z ion composed of the same sequence together with the C-terminal portion of the propeptide (EAEA YVVQIHDITGK). The 890 m/z ion (EEGVSLEK) is unique to the HMW material and is also predicted to originate from the pro peptide.
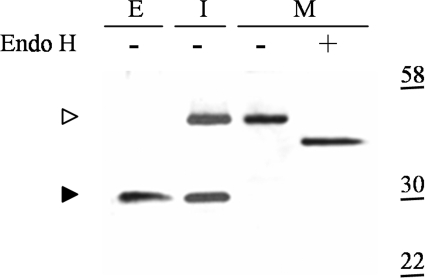
The absence of a pre sequence indicated that all of the γD material had entered the rough ER, the site of the signal peptidase, whereas the presence of the pro sequence indicated material that had not progressed to the trans-Golgi network (TGN), the site of the KEX2 protease.37,38 We addressed whether the pro sequence was glycosylated by isolating a membrane fraction enriched for ER-derived microsomes and treating the γD species found within the microsome fraction with endoglycosidase H, an enzyme that specifically cleaves N-linked carbohydrates that were added in the ER. The results seen in Figure 3 show that the microsome-enriched membrane fraction contains only the HMW species and furthermore that the HMW γD is sensitive to endoglycosidase H, indicating the γD had entered the ER and that the pro sequence had been glycosylated.
Figure 3.
HMW γD harbors a glycosylated pro sequence. A representative immunoblot of fractions prepared from the S. cerevisiae wild-type strain expressing high levels of wild-type γD as described in Materials and Methods, with visualization of the extracellular (E) secreted material found in the media, the intracellular (I) whole-cell lysates that includes the secreted γD in the periplasm, and a membrane fraction enriched for ER-derived microsomes (M). The microsome fraction was tested for the presence of N-linked core carbohydrates by processing the sample with endoglycosidase H (EndoH) (+) as described in Materials and Methods. Protein standards are noted to the right in kilodaltons. The HMW (◃) and LMW (▸) forms of γD are indicated.
These findings allowed a more complete interpretation of the data shown in Figure 1, B and C. The HMW ER species of wild-type γD with the pro sequence intact appeared to be either slowly degraded or, more likely, in transit to the TGN where pro sequence cleavage occurs. The stable species of wild-type γD was the LMW form with the pro sequence cleaved, suggesting it was within or beyond the TGN. Studies in both yeast systems demonstrated that the LMW wild-type γD was efficiently secreted, as shown for S. cerevisiae in Figure 3. Conversely, both species (LMW and HMW) of the mutant Aguadilla γD were reduced over time, with no evidence of secreted material (our unpublished data), indicating selective degradation.
Aguadilla γD Degradation Is Proteasome-Dependent
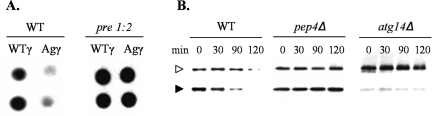
ERAD has been demonstrated to play a role in the degradation of fibrinogen chains that are aberrant or do not properly assemble within the ER.3,6,39 Thus, we hypothesized that ERAD would be the mechanism for degradation of the Aguadilla γD domain. The role of ERAD was examined in the pre1-1, pre2-2 strain of S. cerevisiae, a proteasome-deficient strain in which ERAD substrates are stabilized.18,23 The wild-type and Aguadilla γD cassettes, cloned into a Met25 low-level expression vector, were transformed into the isogenic wild-type and proteasome mutant strains, and the steady-state level was monitored by colony blot immunoassay (Figure 4A). Stabilization of the Aguadilla γD was clearly seen in the pre1-1, pre2-2 proteasome mutant strain, demonstrating proteasome-dependent degradation and thus implicating ERAD.
Figure 4.
Aguadilla γD degradation is proteasome, vacuole, and autophagy dependent. A: Representative colony-blot immunoassay for both the wild-type isogenic parent (WT) and the proteasome mutant (pre1:2) strains of S. cerevisiae expressing either wild-type γD (WTγ) or Aguadilla γD (Agγ) at low levels off the p426Met25 vector. B: Representative immunoblots from a cycloheximide chase analysis of the wild-type isogenic parent (WT), pep4Δ vacuolar protease-deficient strain, and atg14Δ autophagy-deficient strain of S. cerevisiae expressing Aguadilla γD (Agγ) at high levels off the p426Gal1 vector. The HMW (◃) and LMW (▸) forms of γD are indicated.
Degradation of the LMW Aguadilla γD Is Vacuole-Dependent
Our studies in the proteasome mutant illustrate that Aguadilla γD, when expressed at low levels, is cleared from the cell by ERAD. But at high levels of expression, do overflow pathways handle the excess aberrant Aguadilla polypeptide? As shown in Figure 1C, both the HMW and LMW species of the Aguadilla γD were degraded over time, with no build-up of the LMW γD species. Degradation of the LMW species of Aguadilla γD suggested that the mutant Aguadilla polypeptide was selected for degradation after arriving in the TGN and after having the pro sequence removed. The selection of aberrant polypeptides within the TGN for trafficking to the vacuole and subsequent degradation has been seen when ERAD has been overwhelmed40 or when mutant proteins either escape or are not recognized by ER quality control.41–45 In a previous study, we found that when overexpressed, soluble A1PiZ was transported through the biosynthetic pathway from the ER to the Golgi and at the TGN was selectively trafficked via the endosome to the vacuole for degradation.19 Thus, we next asked whether LMW Aguadilla γD that had arrived in the TGN was recognized as aberrant and selectively trafficked to the vacuole for degradation.
The role of the vacuole in degrading Aguadilla γD was examined by cycloheximide chase analysis in a S. cerevisiae pep4Δ mutant,46 a knock-out of vacuolar proteinase A, an enzyme required for posttranslational precursor maturation of a number of vacuolar proteases. Consequently, the pep4Δ mutant lacks most vacuolar protease activity, resulting in significant stabilization of most substances delivered to the vacuole. We found that for Aguadilla γD, the LMW pro sequence cleaved form was stabilized in the pep4Δ strain (Figure 4B), demonstrating that the Aguadilla γD was recognized as aberrant by quality control mechanisms within the TGN and selectively trafficked to the vacuole for degradation. The HMW form of Aguadilla γD was also stabilized in the pep4Δ strain (Figure 4B), suggesting vacuole-dependent degradation of this species as well, and possibly delivery to the vacuole via the autophagic pathway.
Degradation of the HMW Aguadilla γD Is Autophagy-Dependent
We recently demonstrated that the aggregation-prone, misfolded protein A1PiZ47,48 forms aggregates within the ER of our yeast model system and that the aggregates are delivered from the ER to the vacuole in an autophagy-dependent manner.19 Because of their metastable state and the mechanism by which the D domains of fibrinogen γ-chains polymerize in mature fibrinogen,7 we suggested that γD may be aggregation prone and form aggregates within the ER and that clearance of these aggregates may be autophagy-dependent.
To test our hypothesis, the S. cerevisiae atg14Δ strain was used. Atg14p is a unique protein in a phosphatidylinositol 3-kinase complex that is required for autophagy49,50 and is known to be required for the clearance of A1PiZ aggregates from the ER.19 Using cycloheximide chase analyses, we found that HMW Aguadilla γD was stabilized in the atg14Δ strain (Figure 4B). Interestingly, there was less of the LMW Aguadilla γD at the 0 time point in the atg14Δ strain, possibly because there was less Aguadilla γD that exited the ER via the overflow pathway and transited to the TGN where the pro sequence is cleaved. This may be because the deletion of ATG14 constitutively induces the unfolded protein response,19 allowing for more efficient ERAD and removal of the mutant Aguadilla γD. However, despite the constitutive induction of the unfolded protein response, an accumulation of HMW Aguadilla γD was seen. Given that defective autophagy leads to the accumulation of A1PiZ aggregates,19 the stabilization of the HMW Aguadilla γD in the atg14Δ strain suggested autophagy-dependent accumulation of HMW Aguadilla γD aggregates.
Aggregated HMW Aguadilla γD Degradation Is Autophagy-Dependent
Two possible intracellular locations for the accumulation of Aguadilla γD aggregates are suggested from studies of other aggregation-prone ERAD substrates. First, the aggregates may form in the ER and are then delivered to the vacuole via ER-derived autophagic vesicles, as seen for A1PiZ.19 Alternatively, Aguadilla γD aggregates could form in the cytoplasm with subsequent autophagic delivery to the vacuole. For example, cystic fibrosis transmembrane regulator, a known ERAD substrate, is retro-translocated to the cytosol but when inefficiently degraded by the proteasome, forms aggresomes that are cleared by autophagy.51–53 Therefore, we asked whether Aguadilla γD was present in the cytosol or sequestered exclusively within a membrane fraction. High levels of Aguadilla γD were expressed in the atg14Δ strain for 40 hours, after which the cells were collected, and the microsome and cytosol fractions were isolated. Each fraction was examined by immunoblot analysis for the presence of the cytosolic marker protein phosphoglycerate kinase (PGK), an ER luminal marker protein BiP, and fibrinogen. PGK was seen only in the cytosol fraction, as expected. Conversely, HMW Aguadilla γD along with the ER luminal chaperone BiP resided exclusively within the microsome fraction (Figure 5A). To determine whether the γD was protected in the lumen of the microsomes or exposed on the exterior of the microsomes, the membrane fractions were treated with trypsin. Trypsin activity was verified by treating microsomes with trypsin in the presence of Triton X-100 to allow trypsin access to both interior and exterior pools of γD (Figure 5A). These data demonstrate that HMW Aguadilla γD had not retro-translocated from the ER but rather accumulated within it.
Figure 5.
Autophagy plays a role in clearing aggregated γD from the ER. A: Cytosol (C) and microsome (M) fractions were prepared from the atg14Δ strain expressing high levels of Aguadilla γD as described in Materials and Methods. The microsome fractions were untreated (−) or treated with trypsin (+) or with trypsin and Triton X-100 (++). Fractions were examined by immunoblot analysis using antisera to the cytosolic marker protein, PGK, fibrinogen (Fib), or the ER luminal chaperone, BiP. B: Representative immunoblots of fractions from a 5% (top, fraction 1) to 60% (bottom, fraction 20) sucrose density gradient separation of microsome lysates prepared from wild-type (WT) and atg14Δ strains expressing high levels of Aguadilla γD. A short exposure (se) of lane 20 is shown to the right of the blots, and densitometry indicates that there is approximately five times more γD aggregate in the atg14Δ strain compared with that found in the wild-type strain.
Finally, to address whether the HMW Aguadilla γD forms aggregates within the ER, lysates of microsomes isolated from both wild-type and atg14Δ cells that had been induced to express high levels of Aguadilla γD were separated by sucrose gradient (see Materials and Methods). Analysis of the wild-type microsomes revealed that some Aguadilla γD was present in fractions 5 and 6, a density expected for soluble γD, whereas the majority of the polypeptide was found at the bottom of the gradient in fraction 20, a density expected for aggregated γD (Figure 5B, WT). The microsomes from the autophagy-defective strain atg14Δ contained no detectable soluble Aguadilla γD and a fivefold greater amount of aggregated γD in fraction 20 (Figure 5B, atg14Δ). Together, these findings demonstrate that Aguadilla γD aggregates within the ER and that removal of these aggregates is autophagydependent.
Discussion
Understanding the molecular mechanisms of cellular protein quality control is important in addressing ER storage diseases, such as the liver disease associated with antitrypsin deficiency or hypofibrinogenemia. The striking similarities in pathologies of individuals carrying either A1PiZ or the fibrinogen Aguadilla mutation led us to hypothesize that the molecular mechanisms behind the clearance of these aberrant polypeptides would be comparable. To investigate this question, we used our yeast model system and studied the fate of the mutant Aguadilla γD. A1PiZ had been previously shown to be an ERAD substrate with proteasome-dependent degradation,18 and here we demonstrate that Aguadilla γD is also a proteasome-dependent ERAD substrate (Figure 4A). However, when either of these ERAD substrates is expressed at high levels, such that the ERAD pathway is challenged, overflow pathways act to clear the ER of the excess aberrant polypeptides. One such pathway is used by excess amounts of soluble Aguadilla γD and soluble A1PiZ, because both transit the secretory pathway and are selectively trafficked from the TGN to the vacuole for degradation (Figure 4B; A1PiZ19 ). Furthermore, we have shown that autophagy is required as an additional overflow pathway when excess aberrant polypeptides form aggregates within the ER (Aguadilla γD in Figures 4B and 5B; A1PiZ19 ). It is likely that the decreased amount of LMW Aguadilla γD seen at the 0 time point in the atg14Δ strain is due to the constitutive activity of the unfolded protein response generating increased levels of ER chaperones and elevated ERAD activity.19 Another possible explanation is that the accumulated Aguadilla γD aggregates in the ER of the atg14Δ strain offer a point for nucleated growth of aggregates, recruiting the majority of the soluble Aguadilla γD present, thus reducing the pool of HMW Aguadilla γD reaching the TGN for processing into LMW γD. Either way, ER quality control must recognize and dispose of two pools of the aberrant Aguadilla γD in the ER: a soluble pool and an aggregated pool.
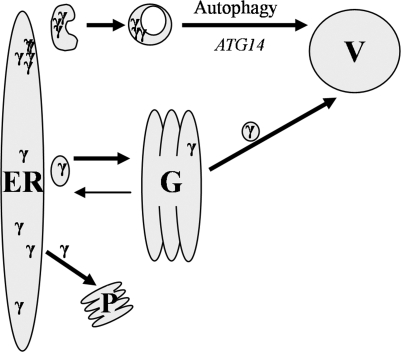
These studies provide evidence for the model presented in Figure 6, in which ERAD is the initial step in ER quality control, removing the aberrant Aguadilla γD for proteasome-dependent degradation. When ERAD is overwhelmed, overflow pathways are used, with the soluble Aguadilla γD transiting the secretory path and on arrival in the TGN selectively trafficking to the vacuole. In addition, and of importance to the study of ER storage diseases, Aguadilla γD that aggregates within the ER is cleared from the ER in an autophagy-dependent manner with subsequent vacuolar degradation.
Figure 6.
Proposed model for Aguadilla γD quality control. Aguadilla γD is targeted to ERAD and thus exits the ER by retrotranslocation with subsequent degradation by the proteasome (P). When overexpressed, excess soluble Aguadilla γD (γ) exits the ER by vesicle transport, transits the Golgi (G), and is sorted to the vacuole (V) for degradation; excess Aguadilla γD that aggregates (γγγγ) within the ER is sent to the vacuole via autophagy.
This model, supported by this study and our previous studies with A1PiZ, is directly applicable to the elucidation of the molecular mechanisms of the gain-of-function liver disease associated with hypofibrinogenemia, anti-trypsin deficiency, and perhaps other ER storage diseases. For example, within a population of individuals either homozygous for A1PiZ or heterozygous for fibrinogen Aguadilla, only a subpopulation develops liver disease due to the accumulation of aggregates of aberrant protein in the ER of their hepatocytes.10,15,54 Based on our model, it is tempting to speculate that the subsets of individuals who develop liver disease are predisposed to less efficient ER quality control mechanisms, perhaps through a second mutation in a gene important for the prevention or clearance of protein aggregates within the ER. Although we cannot exclude the possibility that other quality control pathways exist, our data suggest that accumulation or autophagy-dependent removal of the aggregates from the ER may be the limiting step that leads to liver disease.
Our model is supported further by the findings from studies of the cellular mechanisms at play in other ER storage diseases. For example, in work to understand the pathology of chronic degeneration and inclusion bodies seen in motoneurons of individuals with amyotrophic lateral sclerosis, Tarabal et al55 note a parallel between α-1-antitrypsin deficiency and the results of their studies. These researchers demonstrated that motoneurons under chronic excitotoxicity display protein aggregation in the ER, autophagic response, and degenerative changes in the motoneurons.55 Furthermore, mutant myocilins, associated with autosomal dominant juvenile- and adult-onset primary open angle glaucoma, were shown by Liu and Vollrath56 to be highly aggregation prone and to form aggregates in the ER of human trabecular meshwork cells leading to eventual cell death. Finally, autophagy has been shown to play a critical role in the clearance of aggregated mutant vasopressin from the ER, the presence of which leads to the stress-induced neuronal death seen in conjunction with the loss-of-function autosomal dominant familial neurohypophyseal diabetes insipidus.57
The number and severity of ER storage diseases points to the importance of understanding the molecular mechanisms underlying the formation and clearance of aggregated aberrant polypeptides within the ER, and yeast model systems provide a means to address these questions and test possible therapeutic agents. Yeast are being used to examine the molecular mechanisms driving the pathology of several other conformational diseases, including cystic fibrosis, prion-related disease, Huntington’s disease, and Alzheimer’s disease.58 Finally, the results presented here and in previous studies with aggregated A1PiZ demonstrate a role for autophagy in clearing aggregated aberrant polypeptides from the ER, findings with important medical applications.
Acknowledgments
We thank Drs. Tatiana Ugarova, Dieter Wolf, and Jeffrey Brodsky for supplying reagents. We also thank Dr. Jeffrey Brodsky for critical reading of the manuscript.
Footnotes
Address reprint requests to Ardythe A. McCracken, Biology Department, University of Nevada, Reno, NV 89557. E-mail: mccracke@unr.edu.
Supported by the National Science Foundation (grant MCB-011079 to A.A.M.).
References
- Mosesson MW. Fibrinogen and fibrin structure and function. J Thromb Haemost. 2005;3:1894–1904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- Weisel JW. Fibrinogen and fibrin. Adv Protein Chem. 2005;70:247–299. doi: 10.1016/S0065-3233(05)70008-5. [DOI] [PubMed] [Google Scholar]
- Roy S, Yu S, Banerjee D, Overton O, Mukhopadhyay G, Oddoux C, Grieninger G, Redman CM. Assembly and secretion of fibrinogen: degradation of individual chains. J Biol Chem. 1992;267:23151–23158. [PubMed] [Google Scholar]
- Huang S, Cao Z, Chung DW, Davie EW. The role of betagamma and alphagamma complexes in the assembly of human fibrinogen. J Biol Chem. 1996;271:27942–27947. doi: 10.1074/jbc.271.44.27942. [DOI] [PubMed] [Google Scholar]
- Redman CM, Xia H. Fibrinogen biosynthesis: assembly, intracellular degradation, and association with lipid synthesis and secretion. Ann NY Acad Sci. 2001;936:480–495. [PubMed] [Google Scholar]
- Xia H, Redman CM. The degradation of nascent fibrinogen chains is mediated by the ubiquitin proteasome pathway. Biochem Biophys Res Commun. 1999;261:590–597. doi: 10.1006/bbrc.1999.1081. [DOI] [PubMed] [Google Scholar]
- Brennan SO, Fellowes AP, George PM. Molecular mechanisms of hypo- and afibrinogenemia. Ann NY Acad Sci. 2001;936:91–100. doi: 10.1111/j.1749-6632.2001.tb03496.x. [DOI] [PubMed] [Google Scholar]
- Callea F, Brisigotti M, Fabbretti G, Bonino F, Desmet VJ. Hepatic endoplasmic reticulum storage diseases. Liver. 1992;12:357–362. doi: 10.1111/j.1600-0676.1992.tb00589.x. [DOI] [PubMed] [Google Scholar]
- Brennan SO, Wyatt J, Medicina D, Callea F, George PM. Fibrinogen brescia: hepatic endoplasmic reticulum storage and hypofibrinogenemia because of a gamma284 Gly→Arg mutation. Am J Pathol. 2000;157:189–196. doi: 10.1016/s0002-9440(10)64530-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan SO, Maghzal G, Shneider BL, Gordon R, Magid MS, George PM. Novel fibrinogen gamma375 Arg→Trp mutation (fibrinogen aguadilla) causes hepatic endoplasmic reticulum storage and hypofibrinogenemia. Hepatology. 2002;36:652–658. doi: 10.1053/jhep.2002.35063. [DOI] [PubMed] [Google Scholar]
- Crystal RG. Alpha 1-antitrypsin deficiency, emphysema, and liver disease: genetic basis and strategies for therapy. J Clin Invest. 1990;85:1343–1352. doi: 10.1172/JCI114578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveger T. The natural history of liver disease in alpha 1-antitrypsin deficient children. Acta Paediatr Scand. 1988;77:847–851. doi: 10.1111/j.1651-2227.1988.tb10767.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Whitman I, Molmenti E, Moore K, Hippenmeyer P, Perlmutter DH. A lag in intracellular degradation of mutant alpha-1-antitrypsin correlates with the liver disease phentotype in homozygous PiZZ individuals. Proc Natl Acad Sci USA. 1994;91:9014–9018. doi: 10.1073/pnas.91.19.9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas DA, Mahadeva R. α1-Antitrypsin polymerization and the serpinopathies: pathobiology and the prospects for therapy. J Clin Investig. 2002;110:1585–1590. doi: 10.1172/JCI16782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter DH. Alpha1-antitrypsin deficiency: liver disease associated with retention of a mutant secretory glycoprotein in the endoplasmic reticulum. Bross P, Gregersen N, editors. Totowa: NJ, Humana Press; Protein Misfolding and Disease, Principles and Protocols. 2003:pp 39–56. doi: 10.1385/1-59259-394-1:39. [DOI] [PubMed] [Google Scholar]
- Eriksson S. A 30-year perspective on alpha 1-antitrypsin deficiency. Chest. 1996;110:237S–242S. doi: 10.1378/chest.110.6_supplement.237s. [DOI] [PubMed] [Google Scholar]
- Teckman JH, Burrows J, Hidvegi T, Schmidt B, Hale PD, Perlmutter DH. The proteasome participates in degradation of mutant alpha 1-antitrypsin Z in the endoplasmic reticulum of hepatoma-derived hepatocytes. J Biol Chem. 2001;276:44865–44872. doi: 10.1074/jbc.M103703200. [DOI] [PubMed] [Google Scholar]
- Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci USA. 1999;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse KB, Brodsky JL, McCracken AA. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble A1PiZ and another for aggregates of A1PiZ. Mol Biol Cell. 2005;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teckman JH, Perlmutter DH. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol. 2000;279:G961–G974. doi: 10.1152/ajpgi.2000.279.5.G961. [DOI] [PubMed] [Google Scholar]
- Yee VC, Pratt KP, Cote HC, Trong IL, Chung DW, Davie EW, Stenkamp RE, Teller DC. Crystal structure of a 30 kDa C-terminal fragment from the gamma chain of human fibrinogen. Structure. 1997;15:125–138. doi: 10.1016/s0969-2126(97)00171-8. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko VK, Yakubenko VP, Hertzberg KM, Grieninger G, Ugarova TP. The alternatively spliced alpha(E)C domain of human fibrinogen-420 is a novel ligand for leukocyte integrins alpha(M)beta(2) and alpha(X)beta(2). Blood. 2001;98:2448–2455. doi: 10.1182/blood.v98.8.2448. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare JJ, Rayment FB, Ballantine SP, Sreekrishna K, Romanos MA. High-level expression of tetanus toxin fragment-C in Pichia-pastoris strains containing multiple tandem integrations of the gene. Bio-Technology. 1991;9:455–460. doi: 10.1038/nbt0591-455. [DOI] [PubMed] [Google Scholar]
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Techniques and protocols: high-efficiency transformation of yeast. Dickerson MM, editor. New York: Cold Spring Harbor Press; Methods in Yeast Genetics. 1997:pp 99–102. [Google Scholar]
- McCracken AA, Karpichev IV, Ernaga JE, Werner ED, Dillin AG, Courchesne WE. Yeast mutants deficient in ER-associated degradation of the Z variant of alpha-1-protease inhibitor. Genetics. 1996;144:1355–1362. doi: 10.1093/genetics/144.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Kruse KB. Selective protein degradation in the yeast exocytic pathway. Mol Biol Cell. 1993;4:729–736. doi: 10.1091/mbc.4.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Lawrence JG, Caplan AJ. Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- Brennan SO. Electrospray ionisation analysis of human fibrinogen. Thromb Haemost. 1997;78:1055–1058. [PubMed] [Google Scholar]
- Hubbard MJ, Mchugh NJ, Carne DL. Isolation of ERp29, a novel endoplasmic reticulum protein, from rat enamel cells: evidence for a unique role in secretory-protein synthesis. Eur J Biochem. 2000;267:1945–1956. doi: 10.1046/j.1432-1327.2000.01193.x. [DOI] [PubMed] [Google Scholar]
- Kabani M, Kelley SS, Morrow MW, Montgomery DL, Sivendran R, Rose MD, Gierasch LM, Brodsky JL. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol Biol Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt BZ, Perlmutter DH. Grp78, Grp94 and Grp170 interact with alpha-1-antitrypsin mutants that are retained in the endoplasmic reticulum. Am J Physiol Gastrointest Liver Physiol. 2005;289:G444–G455. doi: 10.1152/ajpgi.00237.2004. [DOI] [PubMed] [Google Scholar]
- Caplan S, Green R, Rocco J, Kurjan J. Glycosylation and structure of the yeast MF alpha 1 alpha-factor precursor is important for efficient transport through the secretory pathway. J Bacteriol. 1991;173:627–635. doi: 10.1128/jb.173.2.627-635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell DH, Huang S, Davie EW. Processing of the carboxyl 15-amino acid extension in the alpha-chain of fibrinogen. J Biol Chem. 1993;268:10351–10355. [PubMed] [Google Scholar]
- Brennan SO, Hammonds B, George PM. Aberrant hepatic processing causes removal of activation peptide and primary polymerisation site from fibrinogen Canterbury (A alpha 20 Val → Asp). J Clin Invest. 1995;96:2854–2858. doi: 10.1172/JCI118356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Redman CM. Differential degradation of the three fibrinogen chains by proteasomes: involvement of Sec61p and cytosolic Hsp70. Arch Biochem Biophys. 2001;390:137–145. doi: 10.1006/abbi.2001.2374. [DOI] [PubMed] [Google Scholar]
- Spear ED, Ng DT. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol Biol Cell. 2003;14:2756–2767. doi: 10.1091/mbc.E02-11-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E, Davidson AR, Kaiser CA. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkeri H, Makarow M. Different degradation pathways for heterologous glycoproteins in yeast. FEBS Lett. 1998;429:162–166. doi: 10.1016/s0014-5793(98)00586-9. [DOI] [PubMed] [Google Scholar]
- Jorgensen MU, Emr SD, Winther JR. Ligand recognition and domain structure of Vps10p, a vacuolar protein sorting receptor in Saccharomyces cerevisiae. Eur J Biochem. 1999;260:461–469. doi: 10.1046/j.1432-1327.1999.00176.x. [DOI] [PubMed] [Google Scholar]
- Coughlan CM, Walker JL, Cochran JC, Wittrup KD, Brodsky JL. Degradation of mutated bovine pancreatic trypsin inhibitor (BPTI) in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J Biol Chem. 2004;279:15289–15297. doi: 10.1074/jbc.M309673200. [DOI] [PubMed] [Google Scholar]
- Arvan P, Zhao X, Ramos-Castaneda J, Chang A. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 2002;3:771–780. doi: 10.1034/j.1600-0854.2002.31102.x. [DOI] [PubMed] [Google Scholar]
- Jones EW, Zubenko GS, Parker RR. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics. 1982;102:665–677. doi: 10.1093/genetics/102.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z alpha 1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- Dafforn TR, Mahadeva R, Elliott PR, Sivasothy P, Lomas DA. A kinetic mechanism for the polymerization of alpha1-antitrypsin. J Biol Chem. 1999;274:9548–9555. doi: 10.1074/jbc.274.14.9548. [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J Cell Biol. 2002;158:761–772. doi: 10.1083/jcb.200112050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3:388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- Teckman JH, Qu D, Perlmutter DH. Molecular pathogenesis of liver disease in alpha1-antitrypsin deficiency. Hepatology. 1996;24:1504–1516. doi: 10.1002/hep.510240635. [DOI] [PubMed] [Google Scholar]
- Tarabal O, Caldero J, Casas C, Oppenheim RW, Esquerda JE. Protein retention in the endoplasmic reticulum, blockade of programmed cell death and autophagy selectively occur in spinal cord motoneurons after glutamate receptor-mediated injury. Mol Cell Neurosci. 2005;29:283–298. doi: 10.1016/j.mcn.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Liu Y, Vollrath D. Reversal of mutant myocilin non-secretion and cell killing: implications for glaucoma. Hum Mol Genet. 2004;13:1193–1204. doi: 10.1093/hmg/ddh128. [DOI] [PubMed] [Google Scholar]
- Castino R, Davies J, Beaucourt S, Isidoro C, Murphy D. Autophagy is a prosurvival mechanism in cells expressing an autosomal dominant familial neurohypophyseal diabetes insipidus mutant vasopressin transgene. FASEB J. 2005;19:1021–1023. doi: 10.1096/fj.04-3162fje. [DOI] [PubMed] [Google Scholar]
- Coughlan CM, Brodsky JL. Use of yeast as a model system to investigate protein conformational diseases. Mol Biotechnol. 2005;30:171–180. doi: 10.1385/MB:30:2:171. [DOI] [PubMed] [Google Scholar]