Abstract
Yeasts provide a powerful model system for comparative genomics research. The availability of multiple complete genome sequences from different fungal groups—currently 18 hemiascomycetes, 8 euascomycetes and 4 basidiomycetes—enables us to gain a broad perspective on genome evolution. The sequenced genomes span a continuum of divergence levels ranging from multiple individuals within a species to species pairs with low levels of protein sequence identity and no conservation of gene order. One of the most interesting emerging areas is the growing number of events such as gene losses, gene displacements and gene relocations that can be attributed to the action of natural selection.
Keywords: Saccharomyces cerevisiae, evolution, bioinformatics, genomics
1. Introduction
The rationale put forward in the late 1980s to justify sequencing the genome of the yeast Saccharomyces cerevisiae was twofold (Dujon 1996). First, yeast is an organism with high economic value in brewing, baking and biotechnology, and one of the primary model organisms for fundamental research into eukaryotic genetics and molecular biology. Second, its genome has many properties that made it an attractive target for early genome sequencing efforts. It has one of the smallest genome sizes among well-studied eukaryotes (only 14 million basepairs), a high gene density (72% of the genome codes for protein), few introns and little repetitive DNA—a combination of factors that made the sequencing relatively easy and cost-effective. The genome project was both doable and worth doing, with the result that S. cerevisiae became the first eukaryote to have a complete chromosome sequenced (Oliver et al. 1992), and later to have its whole genome sequenced (Goffeau et al. 1997).
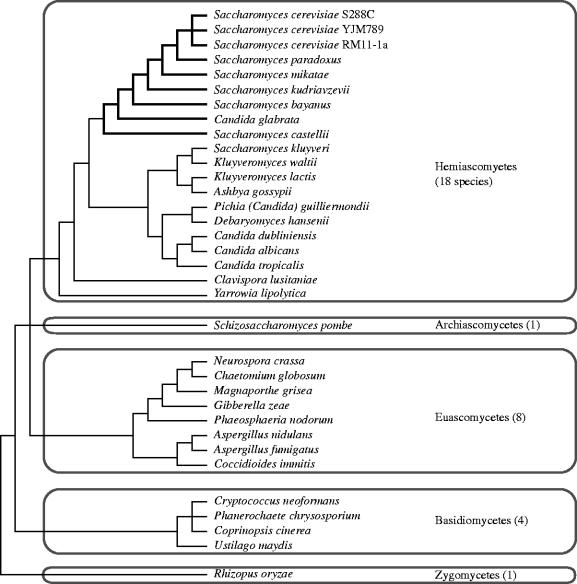
These same features have more recently led to yeast species emerging as a powerful eukaryotic model system for comparative genomics and studies of genome evolution. The group of fungi that includes S. cerevisiae—the hemiascomycetes—all have relatively small and non-repetitive genomes, and several are of biotechnological or medical interest. The result is that today we have genome sequences (either complete, or high-quality draft sequences) from 18 species of hemiascomycetes (figure 1). The genome of S. cerevisiae has now been sequenced three times: once from the laboratory strain S288C (Goffeau et al. 1997), once from the clinical isolate YJM789 (Gu et al. 2005), and once from the vineyard isolate RM11-1a (Broad Institute, unpublished; GenBank accession number AAEG01000000).
Figure 1.
The sequenced fungal genomes (June 2005). All these genomes have been sequenced to at least 3× coverage and the data are in the public domain (available through GenBank or from the Sanger Institute website). Thickened branches indicate the lineages that show whole-genome duplication. The phylogenetic tree is not drawn to scale and may contain errors. It is a composite drawn from several sources (Berbee & Taylor 1993; Cai et al. 1996; Fungal Genome Initiative Steering Committee 2003; Kurtzman & Robnett 2003; Rokas et al. 2003) and D. R. Scannell and K. H. W., unpublished results.
A sort of virtuous cycle has developed among the hemiascomycete genomes. The depth of knowledge that now exists about S. cerevisiae genes—such as thousands of microarray transcription experiments, and knockout phenotype information for every gene—makes it possible to make quite detailed inferences about the physiology of related yeasts based only on their genome sequences (Hittinger et al. 2004), which makes sequencing those genomes worthwhile. In return, information from the other species can be used to learn more about the S. cerevisiae genome—for example in detecting functional motifs in the regulatory regions of genes based on their evolutionary conservation (Cliften et al. 2001, 2003; Kellis et al. 2003)—which further elevates S. cerevisiae as a model organism. Thus, the combined value of the hemiascomycetes as a comparative genomics system is greater than the sum of its parts.
Complementing the recent progress that has been made with hemiascomycete genomes, significant strides have been made in sequencing genomes from the other major fungal clades. As shown in figure 1, we now have complete genome sequences from eight euascomycetes (filamentous ascomycetes such as Neurospora crassa), four basidiomycetes (including the mushroom Coprinopsis cinerea) and the archiascomycete Schizosaccharomyces pombe (Wood et al. 2002; Galagan et al. 2003; Dean et al. 2005; Loftus et al. 2005; and unpublished data available in GenBank). The euascomycete and basidiomycete genomes pose a somewhat greater technical challenge for sequencing than the hemiascomycetes owing to their larger sizes (typically 30–40 Mb) and greater number of introns, but we can see that they already present a rich resource for comparative genomics.
The hemiascomycete species whose genomes have been sequenced span a very broad evolutionary range (figure 1). It is difficult to put an absolute time-scale onto this phylogenetic tree because of the lack of fossil data for yeasts (Berbee & Taylor 1993). However, an intuitive feeling for the depth of the divergences among yeast species can be gained by comparing the average level of protein sequence difference between S. cerevisiae and other yeasts, to that between human and other animals (Dujon et al. 2004). By this measure, the divergence between S. cerevisiae and Candida glabrata is slightly greater than that between humans and fish, while the divergence between S. cerevisiae and Yarrowia lipolytica (the deepest-branching hemiascomycete whose genome has been sequenced) is about the same as that between human and the sea squirt Ciona (Dujon et al. 2004). Similarly, the divergences among the Saccharomyces sensu stricto species are comparable to those among different orders of mammal. These levels of divergence are surprisingly high and demonstrate the evolutionary depth that is encompassed by the available fungal genome sequences. They also suggest that we should be cautious about inferring that the function of a gene in one species will be exactly the same as that of its ortholog in S. cerevisiae (for an example of an exception, see Kadosh & Johnson 2001).
2. The yeast gene order browser
Nested within the phylogenetic tree of hemiascomycetes is a whole genome duplication (WGD) event (figure 1). This event was first detected by analysis of the S. cerevisiae genome sequence alone, which showed that in many places in the genome a series of genes on one chromosome had a series of paralogs on another chromosome, usually in the same order along the chromosome and with conserved transcriptional orientation (Wolfe & Shields 1997). We identified 55 such duplicated regions in the S. cerevisiae genome, and found that the centromere-to-telomere orientation was almost always conserved in each pair of regions. We interpreted this as evidence that entire duplicated chromosomes had existed at some stage during yeast's evolutionary past, and that the mosaic of duplicated regions currently identifiable were the result of subsequent interchromosomal rearrangements, principally reciprocal translocations (Wolfe & Shields 1997; Keogh et al. 1998; Seoighe & Wolfe 1998). Within any pair of duplicated regions, the paralogous genes were interspersed with many single-copy genes (figure 2), which indicated that large numbers of genes had been lost (deleted) from the genome after WGD. The limited amount of gene order information available at that time from other yeast species showed that some species, such as Kluyveromyces lactis and Saccharomyces kluyveri, had gene orders consistent with them having diverged from the S. cerevisiae lineage before WGD occurred in the latter, as shown in figure 1 (Keogh et al. 1998; Seoighe & Wolfe 1999). These findings were comprehensively confirmed in 2004 when the complete genome sequences of Ashbya gossypii (Dietrich et al. 2004), Kluyveromyces waltii (Kellis et al. 2004) and K. lactis (Dujon et al. 2004) were published. Each of these species shows a gene order similar to that inferred to have existed in an ancestor of S. cerevisiae immediately before the WGD happened.
Figure 2.

Illustration of our model of gene order evolution following whole-genome duplication (WGD). The box at the top shows a hypothetical region of chromosome containing ten genes numbered 1–10. After WGD, the whole region is briefly present in two copies. However, many genes subsequently return to single-copy state because there is no evolutionary advantage to maintaining both copies. In this example, only genes 1, 6 and 10 remain duplicated. However, the arrangement of these three homolog pairs in the post-WGD species (bottom) would be sufficient to allow the sister regions to be detected using that genome sequence alone. Also, the order of genes in sister regions in post-WGD species have well-defined relationships to the gene order that existed in the pre-WGD genome (top), which will also be similar to the gene order seen in any species that diverged from the WGD lineage before the WGD occurred. Modified from Montcalm & Wolfe (in press).
In order to visualize the synteny relationship among the sequenced yeast genomes, our laboratory recently developed a bioinformatics tool, the Yeast Gene Order Browser (YGOB; http://wolfe.gen.tcd.ie/ygob; Byrne & Wolfe 2005). YGOB was designed in particular to handle the 1:2 genomic relationship between ‘pre-WGD’ species and ‘post-WGD’ species (figure 3). In this shorthand notation, post-WGD means the group of species whose common ancestor underwent WGD, and pre-WGD means any species that diverged from the lineage leading to S. cerevisiae before the WGD happened. Under the WGD hypothesis, any genomic region in a pre-WGD species should have two counterparts in each post-WGD species; we refer to the paired regions in the post-WGD species as ‘sister regions’. At present YGOB includes genome sequence data from three post-WGD species (S. cerevisiae, S. castellii and C. glabrata), and four pre-WGD species (K. waltii, K. lactis, A. gossypii and S. kluyveri). The database underlying YGOB consists of sets of homologous genes from each species, corresponding to the columns of genes shown in figure 3. Each set (which we refer to as a ‘pillar’) can maximally consist of one gene from each pre-WGD species (i.e. orthologs) and two genes (a pair of paralogs) from each post-WGD species. The assignments of orthology in YGOB were based initially on BLAST results and on the annotations provided by the sequencers of each genome, but the pillars have been refined subsequently by extensive manual editing and curation. The strong conservation of gene order among this group of yeast species has the consequence that it is generally straightforward to identify the orthologs of any locus in each species, or to identify sites from which genes have been deleted (figure 3). The number of singleton genes (i.e. genes without a syntenic ortholog in at least one other species) in each species in YGOB is less than 11% (Byrne & Wolfe 2005), and many of these are in genomic regions close to telomeres (see below).
Figure 3.
Screenshot from the Yeast Gene Order Browser (YGOB; Byrne & Wolfe 2005). The image shows how gene order is related between two sister genomic regions in each of the post-WGD species S. cerevisiae (chromosome XI genes in dark blue and chromosome IX genes in light blue), S. castellii and C. glabrata, and the single homologous genomic region in the pre-WGD species A. gossypii, K. lactis and K. waltii. In this representation, each rectangle represents a gene and homologs are arranged as vertical columns. Arrows below the rectangles show transcriptional orientation. Gray bars connect genes that are adjacent but do not indicate the actual gene spacing on the chromosome. The large arrow indicates the rapidly evolving locus YKL023W discussed in the text.
For on-screen presentation of gene order information, the user selects one gene (from any species) to focus on. This gene is shown in the central pillar of the display, and other aspects of the display (such as gaps caused by gene deletions/insertions, and interchromosomal rearrangements in any species) are calculated relative to it. The user can walk along any chromosome in any species by sequentially focusing on neighbouring genes. Details of the data and visualization algorithms in YGOB are given in Byrne & Wolfe (2005).
One unanticipated outcome of developing YGOB was the discovery of several loci that are evolving very rapidly. One example is YKL023W, an S. cerevisiae gene whose function is unknown but which encodes a protein that co-purifies with the variant histone Htz1 (Krogan et al. 2003). None of the other published yeast genomes, except for the S. sensu stricto species, included a gene annotated as an ortholog of YKL023W. When we curated the original set of genomes in YGOB, we noticed that A. gossypii, K. lactis, K. waltii and C. glabrata were each annotated as having singleton genes in the interval between their orthologs of URA6 and CDC16, which corresponds to the position of YKL023W in S. cerevisiae (figure 3). None of these singletons hits any of the others in a BLASTP search (i.e. the BLASTP E-value between any of them is >10), but their coincident locations and identical transcriptional orientations suggested that they were in fact orthologs whose sequence had diverged too much to be detectable by BLAST. After noticing this we examined the S. castellii genome sequence and found that it too contains a large ORF (open reading frame) in the same region. The S. castellii gene does hit some of the others in BLASTP searches, but only very weakly. Multiple alignment of the proteins shows that only a short region at the N-terminus of this large protein is conserved among species, but this is sufficient to recognize that the genes are orthologs, which is why we show them in the same column in figure 3. Other examples of rapidly evolving loci are discussed in Wolfe (2004) and Byrne & Wolfe (2005).
3. Pairs of centromeres
We have also been able to use YGOB to investigate the evolutionary changes in chromosome number that occurred before and after the WGD (Montcalm & Wolfe in press). Genome duplication can occur either by autopolyploidization (doubling of the chromosome set of a species) or by allopolyploidization (hybridization between two species). In the former case the number of chromosomes should be doubled, and in the latter the new species should have the sum of the numbers of chromosomes in its parents. The process of gene loss sketched in figure 2 should not alter the chromosome number. The number of chromosomes in post-WGD species is approximately twice that in pre-WGD species (Keogh et al. 1998) but the arithmetic is not precise. In the pre-WGD species, there are 8 (K. waltii and S. kluyveri), 7 (A. gossypii) or 6 (K. lactis) chromosomes (Neuveglise et al. 2000; Dietrich et al. 2004; Dujon et al. 2004; Kellis et al. 2004). In the post-WGD species there are 16 (S. cerevisiae and the other sensu stricto species), or 13 (C. glabrata; Dujon et al. 2004). We suspect that the estimate of nine chromosomes in the post-WGD species S. castellii (Petersen et al. 1999) is an underestimate because it is based on pulsed-field electrophoresis (a technique that often tends to under-count); the genome sequence for this species (Cliften et al. 2003) is a 4× draft consisting of numerous contigs rather than complete chromosome sequences.
The number of chromosomes in a species is determined by the number of centromeres, so we used YGOB to examine the history of yeast centromeres and the protein-coding genes near them (figure 4; Montcalm & Wolfe in press). The 16 centromeres of S. cerevisiae can be arranged into eight pairs based on the sister relationships resulting from the WGD (Wong et al. 2002), and these are in conserved gene order relationships with the eight centromeres of K. waltii (Kellis et al. 2004). This observation shows that the WGD involved an 8-chromosome ancestral genome doubling (or two 8-chromosome species hybridizing) to form a 16-chromosome descendant. The other changes in chromosome number (reduction from 8 to 7 in A. gossypii; reduction from 8 to 6 in K. lactis; reduction from 16 to 13 in C. glabrata) can all be attributed to chromosome fusions. Each of these fusions seems to have been accompanied by the inactivation of one of the two centromeres in the newly formed dicentric chromosome (figure 4), without other changes in the local gene order (Montcalm & Wolfe in press). It is noteworthy that, despite the growing number of hemiascomycete genomes that have been sequenced, no examples of a gain of a centromere by a mechanism other than WGD have yet been observed.
Figure 4.
Relationships among centromeric regions of S. cerevisiae (Sc), C. glabrata (Cg), K. waltii (Kw), A. gossypii (Ag) and K. lactis (Kl) chromosomes. Each of the eight panels shows a group of genomic regions that are related by virtue of their gene contents close to a centromere (or a similar noncentromeric region). Black circles represent centromeres in pre-WGD species, and white circles represent centromeres of post-WGD species. X symbols indicate the absence of a centromere. Names indicate chromosome arms, or genes close to the centromere on each arm. Hooked lines indicate loss of relatedness. The assignment of the centromere of C. glabrata chromosome 9 to the group on the bottom-right is less certain than for the other centromeres, and is based on the linkage of CgCEN9 to the gene CAGL0I08107g, which is an ortholog of the genes KLLA0D01243g (K. lactis) and s56_22439 (K. waltii) which are close to the corresponding regions in those species. Gene names in K. lactis and C. glabrata have been shortened by writing A00627 instead of CAGL0A00627g, etc. Reproduced with permission from Montcalm & Wolfe (in press).
4. Evolution of gene content
‘Use it or lose it’ is one of the truisms of genome evolution. If a gene's function is not beneficial to the organism, there will be no natural selection against mutations that damage that gene. It will become a pseudogene and eventually disappear from the genome. For example, the plastid genomes of non-photosynthetic plants do not contain photosynthesis genes (Wolfe et al. 1992). Within fungi, the hemiascomycetes have lost several genes with functions related to splicing (Aravind et al. 2000) and to RNA interference and heterochromatin formation (Alexandersson & Sunnerhagen in press), as compared to euascomycetes and basidiomycetes. These losses are presumably attributable to the relative lack of introns and the simple centromeres in hemiascomycetes. Although the great majority of genes in any hemiascomycete species have homologs in each other species, a few examples of recent gene losses (i.e. losses within the hemiascomycetes) have come to light in the past year. Comparative genomics enables us to detect these losses, and they are particularly interesting because in some cases we can infer (or at least guess at) the physiological or ecological changes that rendered the genes unnecessary.
Most hemiascomycetes can grow on galactose as a sole carbon source, and in S. cerevisiae seven genes (the GAL genes) function exclusively in this pathway. Hittinger et al. (2004) showed that, although orthologs of the GAL genes are present in most of the sequenced yeast genomes, they have been lost independently in three or four lineages. In S. kudriavzevii all seven GAL loci are pseudogenes indicating very recent gene losses and hence a recent change in the metabolic capacity of this species, uniquely among the S. sensu stricto species. Likewise, the GAL genes are completely absent from three other yeasts (C. glabrata, A. gossypii and K. waltii). We do not really know what ecological changes permitted the loss of this pathway in each case, but it is perhaps most readily understood in the case of C. glabrata which (unlike most other hemiascomycetes) is a pathogen of mammals and is unlikely to encounter galactose in this environment.
C. glabrata has also lost the pathway of de novo synthesis of nicotinic acid (the kynurenine pathway; BNA genes), a loss that is directly linked to the pathogenesis of this species (Domergue et al. 2005). In both C. glabrata and S. cerevisiae, the histone deacetylase Sir2 is regulated by the availability of NAD+ because Sir2 consumes NAD+ when it modifies histones. Nicotinic acid is a precursor of NAD+, so S. cerevisiae can replenish its pool of NAD+ via de novo nicotinic acid synthesis, but C. glabrata cannot. Consequently C. glabrata Sir2 activity depends on the availability of external nicotinic acid. The pathogen appears to exploit this dependence as a way of activating a cell surface adhesion programme when it enters the urinary tract of a mammal. Urine is a poor source of nicotinic acid, so C. glabrata cells located in the urinary tract have low Sir2 activity. Consequently the adhesin (EPA) genes, whose transcription is normally repressed owing to Sir2-mediated histone modification, become activated and enable C. glabrata to adhere (Domergue et al. 2005). The loss of BNA genes in C. glabrata may be an example of a pathway loss that was advantageous rather than neutral.
Like redundant factory workers, genes can also be shed from a genome if their function is outsourced instead of being shut down completely. This has occurred in the uracil synthesis pathway in some hemiascomycetes (Gojkovic et al. 2004; Zameitat et al. 2004; Hall et al. 2005). In the deeper-branching hemiascomycetes (figure 5), and in all other eukaryotes, the enzyme dihydroorotate dehydrogenase (DHODase; product of the URA9 gene) is located in the mitochondrion and its activity is coupled to the mitochondrial respiratory chain because a quinone is the terminal electron acceptor. At one point during hemiascomycete evolution, however, an alternative DHODase enzyme was gained by means of horizontal gene transfer from a bacterium similar to Lactococcus lactis (Gojkovic et al. 2004; Hall et al. 2005). The new DHODase (product of the URA1 gene) is located in the cytosol. Because it uses fumarate as an electron acceptor, uracil biosynthesis can proceed even when the cell is not respiring. In some other yeasts such as S. kluyveri, the two enzymes coexist (figure 5). In S. cerevisiae, only URA1 is present and the ortholog of the ancestral eukaryotic gene URA9 has been completely lost.
Figure 5.
Gene displacement during evolution of the uracil biosynthesis pathway. The hemiascomycete species are indicated by the blue background. Red and green rectangles represent the URA9 and URA1 genes, respectively, and red and green X symbols on the phylogenetic tree represent losses of those genes. The inferred point of gain of URA1 is shown by the arrow. All three possible outcomes of this horizontal gene transfer (HGT) have occurred in different lineages descended from it: loss of URA9 (gene displacement, e.g. in S. cerevisiae); loss of URA1 (return to the original state, e.g. in C. glabrata); or retention of both genes (e.g. in S. kluyveri). Based on Gojkovic et al. (2004) and Hall et al. (2005).
Piskur & Langkjaer (2004) proposed that the resulting decoupling of uracil biosynthesis from respiration was an important step in allowing S. cerevisiae and its close relatives to develop the ability to grow almost completely without oxygen. This theme was expanded by Hall et al. (2005) who carried out a systematic search for genes horizontally transferred from bacteria into the S. cerevisiae lineage. They identified 10 S. cerevisiae genes that are candidates for having been gained by horizontal transmission, including URA1 as described above. Notably, for 7 of the 10 genes the most closely related prokaryotic sequences are in anaerobic bacteria. It is also interesting that 9 of the 10 putatively transferred genes are located near telomeres in S. cerevisiae.
A similar displacement, but over a longer evolutionary period, has occurred at the mating-type (MAT) loci of ascomycetes. Comparison of the genes flanking the loci that have been demonstrated genetically to be the MAT loci of Yarrowia lipolytica (a hemiascomycete), Neurospora crassa and Gibberella zeae (two euascomycetes) shows that these loci are positionally as well as functionally homologous. That is, the locus that specifies mating type has not moved in these genomes during all the time that has elapsed since hemiascomycetes and euascomycetes diverged from their common ancestor. The MAT genes in these three species are flanked by the gene APN2 on one side and SLA2 on the other (Butler et al. 2004). APN2 and SLA2 code for a DNA repair enzyme and a component of the cytoskeleton, respectively, neither of which appear to have any functional connection to mating type determination. Within the hemiascomycetes, several rearrangements of the genes flanking the MAT locus have occurred but a continuous line of descent can still be traced (Butler et al. 2004). The positional continuity of the MAT locus is remarkable because the actual contents of this locus have changed considerably. A gene for an alpha-domain protein (homologous to S. cerevisiae MATα1) is the only gene invariably present at the MAT locus in all ascomycetes. The euascomycetes have in addition genes for two HMG-domain proteins (mat-a1 and mat-A3 in Neurospora) and a coiled-coil protein (mat-A2), whereas S. cerevisiae encodes two homeodomain proteins (MATα2 and MATa1). How one type of DNA-binding protein displaced the other during the evolution of this locus remains a mystery.
5. Evolution of gene order
As implied in figure 2, and shown for one small region of the genome in figure 3, gene order is generally well conserved among the hemiascomycetes included in YGOB, once the effects of the WGD are taken into account (i.e. formation of sister regions and numerous gene deletions within each sister). In addition, synteny is interrupted by species-specific interchromosomal translocations. In designing YGOB we chose not to include species that are more distant from S. cerevisiae (for example, Candida albicans) because previous analyses have shown that gene order is noticeably more poorly conserved at this depth (Keogh et al. 1998; Llorente et al. 2000; Dujon et al. 2004).
In comparing gene order among species, we found a striking example of relocation of a set of genes during recent hemiascomycete evolution (figure 6; Wong & Wolfe 2005). The movement of these genes contrasts starkly with the syntenic stasis seen at most other loci. In S. cerevisiae, six of the eight genes involved in allantoin degradation form a physical gene cluster (called the DAL cluster). Allantoin is a degradation product of purines and can be used by S. cerevisiae as a non-preferred nitrogen source. The DAL genes are also clustered in the other S. sensu stricto species, and in S. castellii. Homologs of the six genes are present in the other hemiascomycetes but they are found at six separate chromosomal locations. The cluster originated at one particular point in the phylogenetic tree (on the branch leading to the common ancestor of S. cerevisiae and S. castellii) and is located in a subtelomeric region of the genome. Two of the clustered genes (DAL4 and DAL7) are duplicates of other genes located elsewhere in the S. cerevisiae genome. The other four DAL genes appear to have simply transposed to the cluster site, but we suspect that they originated by gene duplications (forming subtelomeric copies), followed by deletion of the original genes. The set of genomic rearrangements that produced the cluster seems so improbable (figure 6) that one can only conclude that random rearrangements that by chance moved the genes to a subtelomeric location were strongly favoured by natural selection, and that each incremental addition of one gene to the cluster was individually advantageous. We have found that the DAL cluster was formed approximately simultaneously with a reorganization of the first steps in the allantoin degradation pathway: species that have the DAL cluster obtain their allantoin by importing it from outside the cell (using the newly formed allantoin permease, Dal4) instead of by oxidation of urate (as is done in species without the cluster). This finding has suggested to us that, like the displacement of the DHODase gene (figure 5), the evolutionary genomic changes were the result of selection to minimize oxygen consumption during the evolution of the ‘petite-positive’ subset of hemiascomycetes (Piskur 2001; Wong & Wolfe 2005). This demonstration that natural selection can occasionally flex its muscles and reshape part of the genome suggests that the arrangement of genes elsewhere in fungal genomes might be less random than at first appears.
Figure 6.
Formation of the DAL gene cluster during hemiascomycete evolution. The six genes are unlinked in the species shown at the bottom of the phylogenetic tree (from S. kluyveri to Y. lipolytica). In S. castellii and the S. sensu stricto species, the six genes are adjacent and located close to a telomere. There is a rearrangement of gene order in the cluster in S. castellii relative to the sensu stricto species. Formation of the cluster involved apparent transposition of the genes DAL1, DAL2, DAL3 and DCG1, and duplication of FUR4 (to form DAL4) and MLS1 (to form DAL7). None of the DAL cluster genes are present in C. glabrata. Modified from Wong & Wolfe (2005).
6. Conclusions and prospects
Comparative genomics enables us to identify the parts of genomes that have changed during recent evolution, which gives us an indication of the evolutionary processes that are currently moulding the genome. Although the overall impression in most parts of the genome is one of strings of genes whose order is stable but that are occasionally disrupted by translocations or other types of rearrangement, the dynamic history glimpsed at some other loci shows that the genome is not a passive letter-rack of genes but an organelle whose form is shaped by its function. Increasing attention is being focused on subtelomeric regions because they seem to be particularly receptive to picking up genes that are newly transferred from bacteria or newly relocated from elsewhere in the genome. Whether this is owing to a mutational cause (e.g. a higher rate of illegitimate DNA recombination in subtelomeres) or a selective cause (e.g. preferential retention of newly integrated DNA if it is located at subtelomeres because it can be regulated effectively by means of histone modification) remains to be seen.
Acknowledgments
I am grateful to all members of my research group, particularly Kevin Byrne and Simon Wong, for their help. Research in my laboratory is supported by Science Foundation Ireland.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Bioinformatics: from molecules to systems’.
References
- Alexandersson, M. & Sunnerhagen, P. In press. Comparative genomics and gene finding in fungi. In Comparative genomics (ed. P. Sunnerhagen & J. Piskur). Berlin: Springer-Verlag.
- Aravind L, Watanabe H, Lipman D.J, Koonin E.V. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA. 2000;97:11 319–11 324. doi: 10.1073/pnas.200346997. 10.1073/pnas.200346997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee M.L, Taylor J.W. Dating the evolutionary radiations of the true fungi. Can. J. Bot. 1993;71:1114–1127. [Google Scholar]
- Butler G, Kenny C, Fagan A, Kurischko C, Gaillardin C, Wolfe K.H. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl Acad. Sci. USA. 2004;101:1632–1637. doi: 10.1073/pnas.0304170101. 10.1073/pnas.0304170101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K.P, Wolfe K.H. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Roberts I.N, Collins M.D. Phylogenetic relationships among members of the ascomycetous yeast genera Brettanomyces, Debaryomyces, Dekkera, and Kluyveromyces deduced by small-subunit rRNA gene sequences. Int. J. Syst. Bacteriol. 1996;46:542–549. doi: 10.1099/00207713-46-2-542. [DOI] [PubMed] [Google Scholar]
- Cliften P.F, Hillier L.W, Fulton L, Graves T, Miner T, Gish W.R, Waterston R.H, Johnston M. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 2001;11:1175–1186. doi: 10.1101/gr.182901. 10.1101/gr.182901 [DOI] [PubMed] [Google Scholar]
- Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen B.A, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. 10.1126/science.1084337 [DOI] [PubMed] [Google Scholar]
- Dean R.A, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. 10.1038/nature03449 [DOI] [PubMed] [Google Scholar]
- Dietrich F.S, et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. 10.1126/science.1095781 [DOI] [PubMed] [Google Scholar]
- Domergue R, Castano I, De Las Penas A, Zupancic M, Lockatell V, Hebel J.R, Johnson D, Cormack B.P. Nicotinic acid limitation regulates silencing of Candida adhesins during UTI. Science. 2005;308:866–870. doi: 10.1126/science.1108640. 10.1126/science.1108640 [DOI] [PubMed] [Google Scholar]
- Dujon B. The yeast genome project: what did we learn? Trends Genet. 1996;12:263–270. doi: 10.1016/0168-9525(96)10027-5. 10.1016/0168-9525(96)10027-5 [DOI] [PubMed] [Google Scholar]
- Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. 10.1038/nature02579 [DOI] [PubMed] [Google Scholar]
- Fungal Genome Initiative Steering Committee. Whitehead Institute/MIT Center for Genome Research; Cambridge, MA: 2003. Fungal Genome Initiative: a white paper for fungal comparative genomics (June 10, 2003) [Google Scholar]
- Galagan J.E, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. 10.1038/nature01554 [DOI] [PubMed] [Google Scholar]
- Goffeau A, et al. The yeast genome directory. Nature. 1997;387(Suppl.):5–105. [PubMed] [Google Scholar]
- Gojkovic Z, et al. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol. Genet. Genomics. 2004;271:387–393. doi: 10.1007/s00438-004-0995-7. 10.1007/s00438-004-0995-7 [DOI] [PubMed] [Google Scholar]
- Gu Z, David L, Petrov D, Jones T, Davis R.W, Steinmetz L.M. Elevated evolutionary rates in the laboratory strain of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2005;102:1092–1097. doi: 10.1073/pnas.0409159102. 10.1073/pnas.0409159102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Brachat S, Dietrich F.S. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. 10.1128/EC.4.6.1102-1115.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger C.T, Rokas A, Carroll S.B. Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc. Natl Acad. Sci. USA. 2004;101:14 144–14 149. doi: 10.1073/pnas.0404319101. 10.1073/pnas.0404319101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Johnson A.D. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. 10.1128/MCB.21.7.2496-2505.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. 10.1038/nature01644 [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren B.W, Lander E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. 10.1038/nature02424 [DOI] [PubMed] [Google Scholar]
- Keogh R.S, Seoighe C, Wolfe K.H. Evolution of gene order and chromosome number in Saccharomyces, Kluyveromyces and related fungi. Yeast. 1998;14:443–457. doi: 10.1002/(SICI)1097-0061(19980330)14:5<443::AID-YEA243>3.0.CO;2-L. 10.1002/(SICI)1097-0061(19980330)14:5%3C443::AID-YEA243%3E3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- Krogan N.J, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. 10.1016/S1097-2765(03)00497-0 [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P, Robnett C.J. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. 10.1016/S1567-1356(03)00012-6 [DOI] [PubMed] [Google Scholar]
- Llorente B, et al. Genomic exploration of the hemiascomycetous yeasts: 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 2000;487:101–112. doi: 10.1016/s0014-5793(00)02289-4. 10.1016/S0014-5793(00)02289-4 [DOI] [PubMed] [Google Scholar]
- Loftus B.J, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. 10.1126/science.1103773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcalm, L. J. & Wolfe, K. H. In press. Genome evolution in hemiascomycete yeasts. In The Mycota XIII (ed. A. J. P. Brown). Berlin: Springer-Verlag.
- Neuveglise C, Bon E, Lepingle A, Wincker P, Artiguenave F, Gaillardin C, Casaregola S. Genomic exploration of the hemiascomycetous yeasts: 9. Saccharomyces kluyveri. FEBS Lett. 2000;487:56–60. doi: 10.1016/s0014-5793(00)02280-8. 10.1016/S0014-5793(00)02280-8 [DOI] [PubMed] [Google Scholar]
- Oliver S.G, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. 10.1038/357038a0 [DOI] [PubMed] [Google Scholar]
- Petersen R.F, Nilsson-Tillgren T, Piskur J. Karyotypes of Saccharomyces sensu lato species. Int. J. Syst. Bacteriol. 1999;49:1925–1931. doi: 10.1099/00207713-49-4-1925. [DOI] [PubMed] [Google Scholar]
- Piskur J. Origin of the duplicated regions in the yeast genomes. Trends Genet. 2001;17:302–303. doi: 10.1016/s0168-9525(01)02308-3. 10.1016/S0168-9525(01)02308-3 [DOI] [PubMed] [Google Scholar]
- Piskur J, Langkjaer R.B. Yeast genome sequencing: the power of comparative genomics. Mol. Microbiol. 2004;53:381–389. doi: 10.1111/j.1365-2958.2004.04182.x. 10.1111/j.1365-2958.2004.04182.x [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams B.L, King N, Carroll S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. 10.1038/nature02053 [DOI] [PubMed] [Google Scholar]
- Seoighe C, Wolfe K.H. Extent of genomic rearrangement after genome duplication in yeast. Proc. Natl Acad. Sci. USA. 1998;95:4447–4452. doi: 10.1073/pnas.95.8.4447. 10.1073/pnas.95.8.4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe C, Wolfe K.H. Updated map of duplicated regions in the yeast genome. Gene. 1999;238:253–261. doi: 10.1016/s0378-1119(99)00319-4. 10.1016/S0378-1119(99)00319-4 [DOI] [PubMed] [Google Scholar]
- Wolfe K. Evolutionary genomics: yeasts accelerate beyond BLAST. Curr. Biol. 2004;14:R392–R394. doi: 10.1016/j.cub.2004.05.015. 10.1016/j.cub.2004.05.015 [DOI] [PubMed] [Google Scholar]
- Wolfe K.H, Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. 10.1038/42711 [DOI] [PubMed] [Google Scholar]
- Wolfe K.H, Morden C.W, Palmer J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl Acad. Sci. USA. 1992;89:10 648–10 652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Wolfe K.H. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 2005;37:777–782. doi: 10.1038/ng1584. 10.1038/ng1584 [DOI] [PubMed] [Google Scholar]
- Wong S, Butler G, Wolfe K.H. Gene order evolution and paleopolyploidy in hemiascomycete yeasts. Proc. Natl Acad. Sci. USA. 2002;99:9272–9277. doi: 10.1073/pnas.142101099. 10.1073/pnas.142101099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. 10.1038/nature724 [DOI] [PubMed] [Google Scholar]
- Zameitat E, Knecht W, Piskur J, Loffler M. Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri. FEBS Lett. 2004;568:129–134. doi: 10.1016/j.febslet.2004.05.017. 10.1016/j.febslet.2004.05.017 [DOI] [PubMed] [Google Scholar]