Abstract
Environmental sequencing, also dubbed metagenomics, is increasingly being used to obtain insights into organismal communities in diverse habitats, and has a variety of potential applications foreseeable in biotechnology and medicine. The first public large-scale data provide already a wealth of information hidden in vast amounts of fragmented pieces of DNA from unknown species residing in these environments. Comparative sequence analysis is essential for the interpretation of such data. However, different layers of complexity that are intrinsic to each sample require the establishment of some baselines for comparison: how to normalize for the differences in phylogenetic and functional diversity, how to avoid biases from incomplete data, and how to deal with differences in species dominance or genome sizes? Here we discuss a few of these items and delineate some simple discriminative sequence properties for four distinct habitats.
Keywords: comparison, diversity, environments, metagenomics
1. Introduction
After the delivery of the first completely sequenced bacterial genomes in 1995, environmental sequencing was already extensively discussed as a promising avenue (Stein et al. 1996), and the term ‘metagenome’ for the collective genomic information of a habitat appeared in the scientific literature as early as 1998 (Handelsman et al. 1998). Yet, until recently, it was mostly the sequencing of large amounts of 16/18S rRNA that gave the first insights into the species complexity within a number of different habitats (e.g. Rappe & Giovannoni 2003), whereby bacterial species seem by far the most abundant. All together, more than 120 000 sequences of 16S rRNAs from different prokaryotic species are currently captured in databases such as RDP (Cole et al. 2005). In contrast to the large numbers of species implied by their rRNA sequences, there are so far only a little more than 200 completely sequenced genomes published, and any in-depth analysis of building plans and functional repertoires is limited to those (mostly prokaryotic) species. Furthermore, the current genome sequences represent a biased view of living matter on earth, as they have been derived from a very few eukaryotic model organisms and from a variety of prokaryotes that can be cultivated and grown in a laboratory. However, cultivation is only possible (using standard conditions) for about 1% of all microbial species, and natural populations are greatly distorted under laboratory conditions (this is known as ‘great plate anomaly’ Staley & Konopka 1985). Only in 2004, the first large-scale metagenomics studies appeared (Tyson et al. 2004; Venter et al. 2004), which were cultivation-independent because they employed ‘shotgun’ approaches directly on environmental DNA preparations. To sub-clone the DNA, various strategies are being used, and especially the long-insert fosmid or BAC libraries are very promising for the future; they have already delivered the first results, either through random end-sequencing or through screening for and targeted sequencing of specific functional systems (Beja et al. 2002; Treusch et al. 2004).
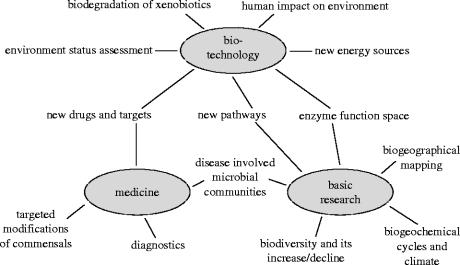
Whatever technology will be driving the data generation a few years from now, it is already clear that massive environmental sequencing is feasible and that it will generate a wealth of data for basic science, but also for more direct applications in many disciplines. The first areas that come to mind are biotechnology and medicine, with surveys for pathogens (Schmeisser et al. 2003) or the discovery of novel antibiotics and specific degradation pathways to be utilized, but applications are likely to be much more far-reaching (see figure 1 for a few of the potentials and hopes).
Figure 1.
Potential applications of environmental sequencing approaches.
Here, we will explore the first large-scale metagenomics datasets available, and discuss some of their properties and how they can be compared. In contrast to complete genomes, which are defined entities, all these data are incomplete so far to an almost unknown extent, perhaps analogous to the first EST data that were generated in the early 1990s, stimulating speculations on human gene numbers. More importantly, we will point to different layers of complexity that are imposed by differences in experimental and computational protocols and raise the question of how to compare the different datasets in a meaningful way. Despite these notes of caution, we claim that it is possible to extract specific information towards both the phylogenetic and functional characterization of microbial communities if one is aware of possible biases and formulates the questions accordingly.
2. Characterizing the first large-scale metagenomics datasets: apples and oranges
The first truly large-scale random shotgun sequencing data from an environment have been published only recently (Tyson et al. 2004), characterizing an underground biofilm under extremely acidic conditions (less than pH 1) in an iron mine drainage path. Just a month later, a much more complex environmental sample from surface water of the Sargasso sea has been reported (Venter et al. 2004), containing an order of magnitude more data (see table 1). This latter dataset alone comprises more predicted open reading frames (ORFs) than contained in all the completely sequenced genomes available at the time (although metagenomics ORFs are sometimes fragmented). Early in 2005, two more shotgun datasets have been released, from yet other, very different habitats, namely 116 Mbp from whalebone samples in more than 500 m water depth in two different oceans (hereafter whalefall), as well as 208 Mbp from surface soil on a Minnesota farm (Tringe et al. 2005; see table 1 for a summary). Several more datasets of up to 200 Mb are underway, as is a more data-rich and systematic sampling of ocean water.
Table 1.
Large-scale environmental sequencing projects: properties and scope.
| acid mine drainage | Sargasso seaa | farm soil | whale falls | |
|---|---|---|---|---|
| particle size filtering | none | >0.1 μm;<0.8 μm | none | none |
| number of subsamples | 1 | 4a | 1 | 3 |
| total amount sequenced–raw | 124 Mbp | 1687 Mbp | 208 Mbp | 116 Mbp |
| total amount sequenced–quality filtered | 76 Mbp | 1350 Mbp | 104 Mbpb | 78 Mbp |
| read average size–raw | 996 bp | 1015 bp | 1046 bp | 993 bp |
| read average size–quality filtered | 737 bp | 818 bp | 696 bp | 673 bp |
| fraction of reads failing any assembly | ∼20% | ∼40% | >99% | ∼55% |
| genomes reported as largely assembled | 5 | 3 | none | none |
| number of ORFs annotated | >12 000 | >1 000 000 | >180 000 | >120 000 |
| minimum number of species found | 5 | 1000 | 847c | 17c,d |
| estimated total number of species | n.r. | >1800 | >3000 | 25–150d |
| reference | (Tyson et al. 2004) | (Venter et al. 2004) | (Tringe et al. 2005) | (Tringe et al. 2005) |
not including data from the Sorcerer II expedition–these data (samples 5–7) were not considered in the original publication (Venter et al. 2004) for the pooled assembly; in addition, they were generated using a variety of different filtering protocols.
filtering here included removal of redundant reads generated by library amplification prior to cloning.
‘ribotypes’; species defined as having 97% identical rRNA sequences.
depending on sub-sample studied.
Although the resulting sequences are hard data, the experimental sampling protocols can be quite different, leading to considerable biases. For example, size filters have been used in the Sargasso sea that are likely to select against small viruses as well as against larger eukaryotic cells. This is simplifying the analysis of prokaryotic diversity, but has to be taken into account when re-analysing and comparing the data to other samples. Furthermore, as the data come from different laboratories, the protocols for read quality filtering, assembly and gene prediction can vary considerably, making it difficult to compare basic properties between different habitats such as the number of annotated ORFs or the degree of assembly. This will also have an impact on downstream analyses, such as determining the phylogenetic or functional composition.
Unfortunately (for details see table 1), not only the habitats, sampling procedures and the data treatments vary considerably but also the nature of the data itself. In some environments, certain species dominate, as exemplified in the acid mine drainage sample where five prokaryotes contribute greater than 80% of all the sequences obtained (notably, one of them, Leptospirillum, was the first sequenced member of an entire phylum, that of Nitrospira, illustrating the bias in classical genome sequencing).
On the contrary, the assembly rate of the much more complex soil data (less than 1%) indicates that a single species is unlikely to be abundant in this sample. It has been estimated that at least 1 Gbp (Tringe et al. 2005) would have to be sequenced before the most abundant species could be reasonably covered by assembling the reads. Thus, while the amount sequenced might have been sufficient to capture the major trends and functional repertoires in the acid mine drainage data, the coverage of the soil might still not be fully representative despite consisting of more than 200 Mbp of raw sequence.
Another factor to consider is the diversity of species within an environment, which is presumably much higher in 0.5 g of soil than even in hundreds of litres of ocean water (e.g. Torsvik et al. 2002). This is also reflected in higher estimates of species numbers: more than 3000 in the soil sample versus 1800 in the Sargasso sea samples (Venter et al. 2004; Tringe et al. 2005). In addition, the heterogeneity of a sample (0.5 g of soil harbours various differently populated subhabitats) and the number of individuals can only be estimated, yet will impact the data. The different constraints imposed by the environments are reflected in the genome sizes (estimates range from 2 to 6 Mbp in water and soil, respectively; Venter et al. 2004; Tringe et al. 2005). This all makes it difficult to extrapolate from individual ORFs to entire species in a sample and leaves a considerable uncertainty in ORF-based estimates. However, the elucidation of the phylogenetic composition of the communities in each sample remains one of the big scientific challenges in metagenomics. Is the current overrepresentation of proteobacteria in the set of completely sequenced genomes a result of their general abundance, or of a sampling bias? They certainly seem to dominate in the more complex samples of soil and surface water, but this might be a chicken-and-egg problem as we can possibly identify them better than other phyla, knowing more about them already.
3. Phylogenetic versus functional differences between metagenomes
While several metagenome properties are obvious, or easy to obtain (e.g. table 1), other features such as the phylogenetic spectrum or the functional repertoire of a sample are more difficult to compute due to the different nature of the samples. A simplifying, best-hit similarity analysis of the ORFs should nevertheless give some rough trends (table 2), although even there major biases could have been introduced. For example, virus genes tend to evolve quickly and their homologues will be easily overlooked, and the size filter used for the Sargasso sea data introduces an extra bias against viruses in this particular sample. Furthermore, many of the predicted ORFs do not have any obvious homologue in the public databases so far. For the most complex soil data, as many as 47% of the reads do not show any obvious hit and even in the sample for which most ORFs have an homology assignment, that of the Sargasso sea, more than a quarter of all ORFs seem entirely novel. This fraction could easily be enriched in viruses, or hitherto undescribed archaea, making the estimates in table 2 even less reliable. What the data do confirm is that the bacterial domain contributes by far the most ORFs in complex environments, and also that extreme habitats can indeed differ. Given the diverse phylogenetic backgrounds, another hope is that the metagenomics data can reveal the adaptation of the communities to their environments; some of this has already been characterized by looking at individual samples (Tyson et al. 2004; Venter et al. 2004) and indeed the first comparative study revealed different features of the environments that impose constraints on the genomes, e.g. the dominant energy sources available in different environments, or different concentrations of ions (Tringe et al. in press).
Table 2.
Summary of BLAST similarity searches, showing the distribution of best hits across the three domains of life (and viruses/phages). (Only open reading frames of at least 300 bp were considered. Database searched: UniREF (08/2004). ORFs generating no hits or hits below 80 bits were counted under ‘no homology’. Assembly depth correction: ORFs from highly covered parts of the assembly were given proportionally more weight, because they represent more abundant species in the environment. The analysis was repeated with other parameters, and for longer, more reliable ORFs (greater than or equal to 450 nt), similar results were obtained. When lowering the threshold for accepting homologies from 80 to 60 bits in the BLAST scoring scheme, ca 20% more assignments were possible, but they are likely to include a considerable number of false positives.)
| best hit prokaryotic (%) | best hit archaeal (%) | best hit eukaryotic (%) | best hit phage/virus (%) | no homology (%) | |
|---|---|---|---|---|---|
| farm soil | 48.7 | 2.3 | 1.1 | 0.2 | 47.7 |
| Sargasso sea | 69.5 | 2.0 | 2.4 | 0.3 | 25.8 |
| whale falls | 61.4 | 1.3 | 1.2 | 0.2 | 35.9 |
| acid mine drainage | 26.6 | 42.5 | 0.5 | 0.1 | 30.3 |
4. Base composition as a property that discriminates metagenomes from different habitats
As we still know very little about metagenomes, there might be many other basic community properties that can differ substantially, imposing further challenges for comparative analyses. For example, in the absence of any phylogenetic information, the base composition of DNA fragments should be an indicator of unexpected distortions or differences. It has long been known that organisms and phyla differ in their overall base composition (for review see Karlin et al. 1998; Bentley & Parkhill 2004). This has been studied at several levels of detail—ranging from simple compositional measures such as GC content or dinucleotide frequencies, to codon usage, and higher order measures such as hexanucleotide frequencies (White et al. 1993; Elhai 2001).
The distributions of GC content values for all four environmental genomics datasets were expected to cover a wide range of values because they each consist of a complex mixture of many species. However, both the soil DNA and the surface water seem to have relatively narrow ranges of GC content values (Foerstner et al. 2005). While it certainly cannot be excluded that this narrow distribution of GC content values is due to sampling or cloning biases, the datasets do contain sequences from hundreds of species from a wide variety of bacterial phyla, and so no major biases are immediately obvious. It is not yet fully understood what drives the evolution of GC content, although a number of correlations with environmental parameters have been reported (and sometimes disputed). These include temperature, oxygen availability and other rather indirect factors such as the average genome size (which correlates weakly with GC content and is itself probably related to environmental factors; see the following references for discussions on these and other factors: McEwan et al. 1998; Hurst & Merchant 2001; Moran 2002; Naya et al. 2002; Rocha & Danchin 2002; Bentley & Parkhill 2004; Musto et al. 2004). The validity and relative contribution of the above factors remain largely unclear and leave room for other, yet unknown, selective pressures that may force the GC content within a community to be more similar than expected. The GC content does have an impact on codon usage and thus on the proteins encoded in the metagenomes, as exemplified by the differences in amino acid compositions of the predicted proteins. The interplay of these compositional differences and environment-specific functional constraints remains to be elucidated.
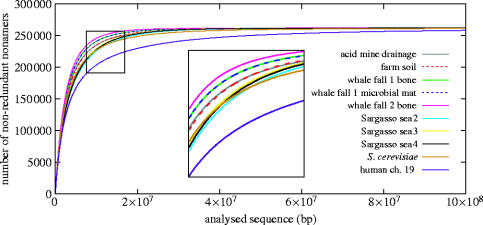
While the above theories provide ways to discuss and interpret the observed distinct GC patterns in the samples, for other compositional features we have fewer explanations. For example, a complexity analysis using nucleotide nonamer frequencies (the largest oligomers for which the majority of permutations are still present in large genomes and samples) revealed some unexpected similarities between samples. We simulated the accumulation of distinct nonamers for each of eight environmental (sub)-samples by selecting the sequencing reads in random order, and repeated the procedure with bakers' yeast and human chromosome 19 as controls (figure 2). Sequences with low complexity (i.e. high repeat density) should show a flatter accumulation curve, as is observed for the human chromosome. The data implicitly indicate a slightly higher gene density in environmental samples than in Saccharomyces cerevisiae (where it is 72%), confirming the high prokaryotic gene content of the samples. The detailed behaviour of the samples in this simulation cannot be easily explained. Although the subsamples tend to cluster together, whalefall DNA seems to be more complex than soil, although the latter has the highest species diversity. It is tempting to link the nonamer occurrence simply to GC content and claim that the numbers of non-redundant nonamers is limited by unbalanced AT–GC distributions. Yet many other factors might contribute as we are only now starting to understand the metagenomes and the biases of the approach for deciphering them.
Figure 2.
DNA complexity analysis. The curves show the simulated accumulation of nonamer occurrences (each distinct nonamer is counted only once), generated by random sampling of nonamers from the environmental sequences. As controls, the genome of Saccharomyces cerevisiae, and the human chromosome 19 were similarly sampled. The maximum number of 262 144 (49) distinct nonamers was reached in each environmental sample after analysing a total sequence length in the order of 108 bp.
5. Conclusions
It is clear that environmental genomics approaches represent an entirely new quality of sequencing projects in terms of scope and complexity. This comes along with unique features and pitfalls, and poses various new challenges for the analysis and interpretation of the data. Simple technical differences in the sample preparation and subsequent analysis might have a much larger impact on the resulting data than is the case for current genome projects, where the assembly of only a single entity (a genome) and external information such as physical maps can give some feedback on the original quality. In metagenome assemblies, shared phages or recently horizontally transferred fragments of DNA might cause species to merge artificially. Thus, as with the deposition of raw sequencing traces in genome projects, resources that allow for the deposition of intermediate steps of the data treatment (such as details on quality filtering and assembly, e.g. Salzberg et al. 2004) become important. This enables other scientists to follow the treatment of the raw data, as various different questions in the promising avenue of metagenomics probably each require different approaches to the data. The maintenance and extension of such data resources should not be underestimated when applying for funds, as only a comparative analysis of many different habitats under many conditions will provide the context sufficient for understanding each individual sample. All these technical hurdles and problems are clearly outweighed by the enormous potentials of the metagenomics approach. Despite the early struggle to understand and dissect the different layers of complexity, comparative metagenome analysis is well suited to tackle many new, exciting questions, from finding a surprising new gene variant to estimating the total number of genes and species on earth. The practical impacts are equally promising and the application areas summarized in table 1 can be easily extended.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Bioinformatics: from molecules to systems’.
These authors contributed equally to the study.
References
- Beja O, Suzuki M.T, Heidelberg J.F, Nelson W.C, Preston C.M, Hamada T, Eisen J.A, Fraser C.M, DeLong E.F. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature. 2002;415:630–633. doi: 10.1038/415630a. 10.1038/415630a [DOI] [PubMed] [Google Scholar]
- Bentley S.D, Parkhill J. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. 10.1146/annurev.genet.38.072902.094318 [DOI] [PubMed] [Google Scholar]
- Cole J.R, Chai B, Farris R.J, Wang Q, Kulam S.A, McGarrell D.M, Garrity G.M, Tiedje J.M. The ribosomal database project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. 10.1093/nar/gki038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai J. Determination of bias in the relative abundance of oligonucleotides in DNA sequences. J. Comput. Biol. 2001;8:151–175. doi: 10.1089/106652701300312922. 10.1089/106652701300312922 [DOI] [PubMed] [Google Scholar]
- Foerstner K.U, Von Mering C, Hooper S.D, Bork P. Environments shape the nucleotide composition of genomes. EMBO Rep. 2005;6:1208–1213. doi: 10.1038/sj.embor.7400538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J, Rondon M.R, Brady S.F, Clardy J, Goodman R.M. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. 10.1016/S1074-5521(98)90108-9 [DOI] [PubMed] [Google Scholar]
- Hurst L.D, Merchant A.R. High guanine–cytosine content is not an adaptation to high temperature: a comparative analysis amongst prokaryotes. Proc. R. Soc. B. 2001;268:493–497. doi: 10.1098/rspb.2000.1397. 10.1098/rspb.2000.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S, Campbell A.M, Mrazek J. Comparative DNA analysis across diverse genomes. Annu. Rev. Genet. 1998;32:185–225. doi: 10.1146/annurev.genet.32.1.185. 10.1146/annurev.genet.32.1.185 [DOI] [PubMed] [Google Scholar]
- McEwan C.E, Gatherer D, McEwan N.R. Nitrogen-fixing aerobic bacteria have higher genomic GC content than non-fixing species within the same genus. Hereditas. 1998;128:173–178. doi: 10.1111/j.1601-5223.1998.00173.x. 10.1111/j.1601-5223.1998.00173.x [DOI] [PubMed] [Google Scholar]
- Moran N.A. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. 10.1016/S0092-8674(02)00665-7 [DOI] [PubMed] [Google Scholar]
- Musto H, Naya H, Zavala A, Romero H, Alvarez-Valin F, Bernardi G. Correlations between genomic GC levels and optimal growth temperatures in prokaryotes. FEBS Lett. 2004;573:73–77. doi: 10.1016/j.febslet.2004.07.056. 10.1016/j.febslet.2004.07.056 [DOI] [PubMed] [Google Scholar]
- Naya H, Romero H, Zavala A, Alvarez B, Musto H. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J. Mol. Evol. 2002;55:260–264. doi: 10.1007/s00239-002-2323-3. 10.1007/s00239-002-2323-3 [DOI] [PubMed] [Google Scholar]
- Rappe M.S, Giovannoni S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. 10.1146/annurev.micro.57.030502.090759 [DOI] [PubMed] [Google Scholar]
- Rocha E.P, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. 10.1016/S0168-9525(02)02690-2 [DOI] [PubMed] [Google Scholar]
- Salzberg S.L, Church D, DiCuccio M, Yaschenko E, Ostell J. The genome assembly archive: a new public resource. PLoS Biol. 2004;2:E285. doi: 10.1371/journal.pbio.0020285. 10.1371/journal.pbio.0020285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser C, et al. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 2003;69:7298–7309. doi: 10.1128/AEM.69.12.7298-7309.2003. 10.1128/AEM.69.12.7298-7309.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J.T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. 10.1146/annurev.mi.39.100185.001541 [DOI] [PubMed] [Google Scholar]
- Stein J.L, Marsh T.L, Wu K.Y, Shizuya H, DeLong E.F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V, Ovreas L, Thingstad T.F. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. 10.1126/science.1071698 [DOI] [PubMed] [Google Scholar]
- Treusch A.H, Kletzin A, Raddatz G, Ochsenreiter T, Quaiser A, Meurer G, Schuster S.C, Schleper C. Characterization of large-insert DNA libraries from soil for environmental genomic studies of Archaea. Environ. Microbiol. 2004;6:970–980. doi: 10.1111/j.1462-2920.2004.00663.x. 10.1111/j.1462-2920.2004.00663.x [DOI] [PubMed] [Google Scholar]
- Tringe S.G, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Tyson G.W, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. 10.1038/nature02340 [DOI] [PubMed] [Google Scholar]
- Venter J.C, et al. Environmental genome shotgun sequencing of the Sargasso sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. 10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- White O, Dunning T, Sutton G, Adams M, Venter J.C, Fields C. A quality control algorithm for DNA sequencing projects. Nucleic Acids Res. 1993;21:3829–3838. doi: 10.1093/nar/21.16.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]