Abstract
In this article, I summarize recent work from my group directed towards developing an improved model of intra and intermolecular interactions and applying this improved model to the prediction and design of macromolecular structures and interactions. Prediction and design applications can be of great biological interest in their own right, and also provide very stringent and objective tests which drive the improvement of the model and increases in fundamental understanding. I emphasize the results from the prediction and design tests that suggest progress is being made in high-resolution modelling, and that there is hope for reliably and accurately computing structural biology.
Keywords: computational biophysics, protein structure prediction, protein design
1. Introduction
The protein and design work in my group is carried out using a computer program called Rosetta. At the core of Rosetta are potential functions for computing the energies of interactions within and between macromolecules, and optimization methods for finding the lowest energy structure for an amino acid sequence (protein structure prediction) or a protein–protein complex, and for finding the lowest energy amino acid sequence for a protein or protein–protein complex (protein design). Both the potential functions and the search algorithms are continually being improved based on feedback from the prediction and design tests (see schematic in figure 1). There are considerable advantages in developing one computer program to treat these quite diverse problems: first, the different applications provide very complementary tests of the underlying physical model (the fundamental physics/physical chemistry is of course the same in all cases), and second, many problems of current interest, such as flexible backbone protein design and protein–protein docking with backbone flexibility involve a combination of the different optimization methods.
Figure 1.

Schematic diagram of Rosetta structure prediction and design efforts.
In the following sections, I summarize recent progress and highlights in each of the different areas and illustrate the development of the physical model. I will put particular emphasis on the results from each of the areas that suggest real progress is being made in high-resolution modelling.
(a) Design of protein structure
Over the past several years, we have used our computational protein design method to dramatically stabilize several small proteins by completely redesigning every residue of their sequences (Dantas et al. 2003), to redesign protein backbone conformation (Nauli et al. 2001), to convert a monomeric protein to a strand swapped dimer (Kuhlman et al. 2002), and to thermostabilize an enzyme (Korkegian et al. 2005). A highlight was the redesign of the folding pathway of protein G, a small protein containing two beta hairpins separated by an alpha helix. In the naturally occurring protein, the first hairpin is disrupted and the second hairpin is formed at the rate limiting step in folding, but in a redesigned variant in which the first hairpin was significantly stabilized and the second hairpin destabilized, the order of events is reversed: the first hairpin is formed and the second hairpin disrupted in the folding transition state (Nauli et al. 2002). The ability to rationally redesign protein folding pathways shows that our understanding of the determinants of protein folding has advanced considerably.
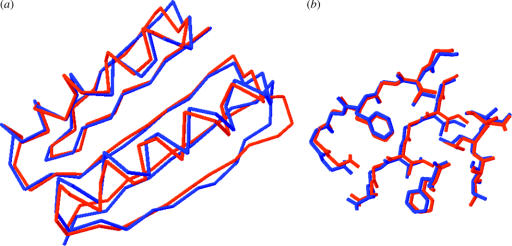
Particularly exciting more recently is the achievement of a grand challenge of computational protein design—the creation of novel proteins with arbitrarily chosen three dimensional structures. We developed a general computational strategy for creating such novel protein structures that incorporates full backbone flexibility into rotamer-based sequence optimization. This was accomplished by integrating ab initio protein structure prediction, atomic level energy refinement, and sequence design in Rosetta. The procedure was used to design a 93 residue protein called Top7 with a novel sequence and topology. Top7 was found experimentally to be monomeric and folded, and the X-ray crystal structure of Top7 is strikingly similar (r.m.s.d.=1.2 Å) to the design model (figure 2; Kuhlman et al. 2003). The successful design of a new globular protein fold and the very close correspondence of the crystal structure to the design model have broad implications for protein design and protein structure prediction, and open the door to the exploration of the large regions of the protein universe not yet observed in nature.
Figure 2.
Comparison of Top7 X-ray crystal structure (red) and design model (blue). (a) Calpha overlay; (b), detail of sidechain packing in the core.
(b) Design of protein–protein interactions
To explore the extension of these methods to protein–protein interactions, and in particular to the redesign of interaction specificity, we chose as a model system the high-affinity complex between Colicin E7 Dnase and its cognate inhibitory immunity protein. Novel Dnase–inhibitor protein pairs predicted to interact tightly with one another but not with the wild type proteins were generated using the physical model described above and a modification of our rotamer search-based computational design strategy incorporating elements of both positive and negative design. Experimental characterization demonstrated that the designed protein complexes have sub-nanomolar affinities, are functional and specific in vivo, and have more than an order of magnitude affinity difference between cognate and non-cognate pairs in vitro (Kortemme et al. 2004). The approach should be applicable to the design of interacting protein pairs with novel specificities for delineating and reengineering protein interaction networks in living cells.
In collaboration with Dr Barry Stoddard's and Dr Ray Monnat's research groups, we generated an artificial highly specific endonuclease by fusing domains of homing endonucleases I-DmoI and I-CreI through computational optimization of a new domain–domain interface between these normally non-interacting proteins. The resulting enzyme, E-DreI (Engineered I-DmoI/I-CreI), binds a long chimeric DNA target site with nanomolar affinity, cleaving it precisely at a rate equivalent to its natural parents (Chevalier et al. 2002). We are currently trying to develop a whole new generation of new endonucleases by redesigning the protein–DNA interface using an extension of our design methodology to protein–nucleic acid interfaces (Havranek et al. 2004).
In both of these systems, it has been possible to determine X-ray crystal structures of the designed complexes. As in the Top7 case, the actual structures are very close to the design models, which is an independent and important validation of the accuracy of our approach to high-resolution modelling.
(c) Prediction of protein structure
The picture of protein folding that motivates our approach to ab initio protein tertiary structure prediction is that sequence-dependent local interactions bias segments of the chain to sample distinct sets of local structures, and that non-local interactions select the lowest free-energy tertiary structures from the many conformations compatible with these local biases. In implementing the strategy suggested by this picture, we use different models to treat the local and non-local interactions. Rather than attempting a physical model for local sequence–structure relationships, we turn to the protein database and take the distribution of local structures adopted by short sequence segments (fewer than 10 residues in length) in known three-dimensional structures as an approximation to the distribution of structures sampled by isolated peptides with the corresponding sequences. The primary non-local interactions considered are hydrophobic burial, electrostatics, main-chain hydrogen bonding and excluded volume. Structures that are simultaneously consistent with both the local sequence structure biases and the non-local interactions are generated by minimizing the non-local interaction energy in the space defined by the local structure distributions using simulated annealing.
Rosetta has been tested in the biannual CASP protein structure prediction experiments in which predictors are challenged to make blind predictions of the structures of sequences whose structures have been determined but not yet published. Since CASP3 in 1998 Rosetta has consistently been the top performing method for ab initio prediction, as can be seen in the published reports of the independent assessors. For example, Rosetta was tested on 21 proteins whose structures had been determined but were not yet published in the CASP4 experiment. The predictions for these proteins, which lack detectable sequence similarity to any protein with a previously determined structure, were of unprecedented accuracy and consistency (Bonneau et al. 2002). Excellent predictions were also made in the CASP5 experiment (Bradley et al. 2003). Encouraged by these promising results, we generated models for all large protein families fewer than 150 amino acids in length (Bonneau et al. 2002). For CASP6 (December 2004), we developed improved methods for beta sheet protein prediction, and I was also delighted that many of the other top groups used the Rosetta software, which has been freely available (source code in addition to executable) for the past several years.
Since CASP4 I have been convinced that real progress in structure prediction (both de novo prediction and comparative modelling) would only come from progress in high-resolution refinement. While Rosetta predictions in CASP have been quite good on a relative scale, they have been poor on an absolute scale, with the topology roughly correct in favourable cases in at least one out of five submitted predictions but the high-resolution details for the most part completely wrong. Refinement of these rough models is critical for improving the accuracy of the models, and perhaps even more critically, for improving their reliability. The stability of proteins in large part derives from the close complementary packing of sidechains in the protein core, and hence evaluating the physical plausibility of a model requires modelling these interactions. Unfortunately, complementary sidechain packing is disrupted by changes in the backbone conformation of the magnitude of the errors in typical Rosetta low-resolution models. Hence, a major focus of our work in the past 5 years has been to develop high resolution all atom refinement methods which can drive the rough de novo models towards the native structure and thus transform our predictions from educated low-resolution guesses to confident high-resolution models. While we have been able make steady progress on both the sampling problem and the energy function, measurable progress on de novo prediction refinement has been small up until recently. However, the improved methods turned out to be very useful for both the design of Top7, described above, where they were critical in the backbone optimization step, and for the protein–protein docking method, described below, which utilizes the same energy function and much of the same optimization methodology.
A highlight of CASP6 for me was Target 281, the first de novo blind prediction which utilized our high-resolution refinement methodology to achieve close to high-resolution accuracy. As the sequence was relatively short (76 residues), during CASP we had time to apply our all atom refinement methodology not only to the native sequence but also to the sequence of many homologues. The centre of the lowest energy cluster of structures turned out to be remarkably close to the native structure (1.5 Å). The high-resolution refinement protocol decreased the r.m.s.d. from 2.2 to 1.5 Å and the sidechains pack in a somewhat native like manner in the protein core. Since last summer, we have used this protocol on a number of other very small proteins and results are very promising. There is still a huge amount to do on this very challenging problem, and improving refinement methods will continue to be a focus of our work for the next 5-year period. A very concrete problem of considerable practical importance is the closely related comparative modelling refinement problem: for proteins with sequence similarity to proteins of known structure, models can be built by essentially ‘copying’ the coordinates of the homologue, but most efforts to improve on this starting template structure have failed (we have had some success recently using evolutionary information to guide the sampling; Qian et al. 2004). Hence comparative models typically do not accurately represent the structural features that differ between the homologues, which is a serious shortcoming that impairs prediction of interaction specificity and other uses of the models. Thus, as we develop improved methods we will test them on both the de novo structure refinement problem and the comparative modelling problem. The goal is simple—to be able to produce sufficiently accurate models either with or without a starting template structure to allow structure-based biological insights without need for tedious and expensive experimental structure determination—or even more simply put, to solve the protein folding problem.
We have extended the Rosetta ab initio structure prediction strategy to the problem of generating models of proteins using limited experimental data. By incorporating chemical shift and Nuclear Overhouser effect (NOE) information (Bowers et al. 2000) and more recently dipolar coupling information (Rohl & Baker 2002) into the Rosetta structure generation procedure, it has been possible to generate much more accurate models than with ab initio structure prediction alone or using the same limited data sets with conventional NMR structure generation methodology. An exciting recent development is that the Rosetta procedure can also take advantage of unassigned NMR data and hence circumvent the difficult and tedious step of assigning NMR spectra (Meiler et al. 2003).
The Rosetta ab initio structure prediction method, the Rosetta-based NMR structure determination method, and a new method for comparative modelling (Rohl & Baker 2003) that uses the Rosetta de novo modelling approach to model the parts of a structure (primarily long loops) that cannot be accurately modelled based on a homologous structure template have all been implemented in a public server called Robetta which was one of the best all around fully automated structure prediction servers in the CASP5 and CASP6 tests (Chivian et al. 2005) and has a constant backlog of users worldwide.
(d) Prediction of protein–protein interactions
As described above, we have been working for a number of years on protein structure refinement, which is challenging because of the very large number of degrees of freedom. I became interested in the protein–protein docking problem because, with the approximation that the two partners do not undergo significant conformational changes during docking, the space to be searched is much smaller—only the 6 rigid body degrees of freedom in addition to the sidechain degrees of freedom, and thus it seemed like a good stepping stone to the harder structure refinement problem while being important in its own right.
We developed a new method to predict protein–protein complexes from the coordinates of the unbound monomer components (Gray et al. 2003) that employs a low-resolution, rigid-body, Monte Carlo search followed by simultaneous optimization of backbone displacement and sidechain conformations with the Monte Carlo minimization procedure and physical model used in our high-resolution structure prediction work. The simultaneous optimization of sidechain and rigid body degrees of freedom contrasts with most other current approaches which model protein–protein docking as a rigid body shape matching problem with the sidechains kept fixed. We have recently improved the method (RosettaDock) further (Wang et al. 2005) by developing an algorithm which allows efficient sampling of off rotamer sidechain conformations during docking.
The power of RosettaDock was highlighted in the very recent blind CAPRI protein–protein docking challenge which was held in December of 2004. In CAPRI, predictors are given the structures of two proteins known to form a complex, and challenged to predict the structure of the complex. RosettaDock predictions for targets without significant backbone conformational changes were quite striking, as shown in figure 3. Not only were the rigid body orientations of the two partners predicted nearly perfectly, but also almost all the interface sidechains were modelled very accurately. Importantly, these correct models clearly stood out as lower in energy than all other models we generated, which suggests the potential function is not too far off. These predictions were qualitatively better than predictions made using standard grid-based methods which keep protein sidechains fixed during docking.
Figure 3.
CAPRI protein–protein docking results. (a) (i): Energy spectrum of models generated in global docking calculations carried out before experimental structures were released; (ii) free energy landscape mapped out by starting trajectories at lowest energy points sampled in global docking runs. (b): comparison of predicted (blue) rigid body orientation with X-ray crystal structure (red and yellow). (c): close up of interface showing that in addition to the rigid body orientation also the detailed conformations of the sidechains were correctly predicted. The predicted models are those submitted to the CAPRI organizers and are the lowest energy models found in the global and local searches shown on the (a).
These very promising results suggest that the method may soon be useful for generating models of biologically important complexes from the structures of the isolated components, and more generally suggest that high-resolution modelling of structures and interactions is within reach. A clear goal for our monomeric structure prediction work is to approach the level of accuracy of these models.
2. Improvement of physical model
Our current approach to improving energy functions involves a combination of quantum chemistry calculations on simple model compounds, traditional molecular mechanics approaches, and protein structural analysis. We have used such an approach to develop an improved hydrogen bonding potential (Kortemme & Baker 2002; Morozov et al. 2004)—a particularly notable result is that the orientation dependence of the hydrogen bond in quantum chemistry calculations on formamide dimers is remarkably similar to that seen in sidechain–sidechain hydrogen bonds in protein structures, but quite different from that in current molecular mechanics force fields which neglect the covalent character of the hydrogen bond. Feedback from the prediction and design calculations has provided a continual impetus and guidance for improving the energy function, for example inadequacies in our treatment of protein–protien interactions have led to the recent development of a rotamer-based model for water-mediated hydrogen bonds (Jiang et al. 2005).
3. Plans for the next several years
It is exciting that our prediction and design methods have now reached the point where they can be applied to important biological problems. Particularly encouraging to me after quite a few years of work on high-resolution modelling are the close to atomic resolution predictions of the structures of complexes in CAPRI, the 1.5 Å de novo prediction in CASP6, and the close agreement of the Top7 and protein–protein interface design models with the experimentally determined X-ray crystal structures—taken together these results suggest that high-resolution modelling is really starting to work.
In the next several years we aim to improve and extend the methods still further and to apply them to problems of particular biological interest. Areas of particular focus are to improve the accuracy of high-resolution structure prediction (which will be required if the models are to be generally useful) by improving the underlying physical model and sampling methods, to predict and redesign protein–DNA interaction specificity, and to extend our protein design methodology to the design of enzymes which catalyse chemical reactions not catalysed by naturally occurring proteins. The long-range goal is to be able to compute structural biology.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Bioinformatics: from molecules to systems’.
References
- Bonneau R, Strauss C, Rohl C, Chivian D, Bradley P, Malmstrom L, Robertson T, Baker D. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 2002;322:65–71. doi: 10.1016/s0022-2836(02)00698-8. 10.1016/S0022-2836(02)00698-8 [DOI] [PubMed] [Google Scholar]
- Bowers P.M, Strauss C.E, Baker D. De novo protein structure determination using sparse NMR data. J. Biomol. NMR. 2000;18:311–318. doi: 10.1023/a:1026744431105. [DOI] [PubMed] [Google Scholar]
- Bradley P, et al. Rosetta predictions in CASP5: successes, failures, and prospects for complete automation. Proteins. 2003;53:457–468. doi: 10.1002/prot.10552. 10.1002/prot.10552 [DOI] [PubMed] [Google Scholar]
- Chevalier B.S, Kortemme T, Chadsey M.S, Baker D, Monnat R.J, Stoddard B.L. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. 10.1016/S1097-2765(02)00690-1 [DOI] [PubMed] [Google Scholar]
- Chivian D, Kim D.E, Malmstrom L, Schonbrun J, Rohl C.A, Baker D. Prediction of CASP6 structures using automated Robetta protocols. Proteins. 2005;61(Suppl. 7):157–166. doi: 10.1002/prot.20733. [DOI] [PubMed] [Google Scholar]
- Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A large scale test of computational protein design: folding and stability of nine completely redesigned globular proteins. J. Mol. Biol. 2003;332:449–460. doi: 10.1016/s0022-2836(03)00888-x. 10.1016/S0022-2836(03)00888-X [DOI] [PubMed] [Google Scholar]
- Gray J.J, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl C.A, Baker D. Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. 10.1016/S0022-2836(03)00670-3 [DOI] [PubMed] [Google Scholar]
- Havranek J.J, Duarte C.M, Baker D. A simple physical model for the prediction and design of protein-DNA interactions. J. Mol. Biol. 2004;344:59–70. doi: 10.1016/j.jmb.2004.09.029. 10.1016/j.jmb.2004.09.029 [DOI] [PubMed] [Google Scholar]
- Jiang L, Kuhlman B, Kortemme T, Baker D. A “solvated rotamer” approach to modeling water-mediated hydrogen bonds at protein–protein interfaces. Proteins. 2005;58:893–904. doi: 10.1002/prot.20347. [DOI] [PubMed] [Google Scholar]
- Korkegian A, Black M.E, Baker D, Stoddard B.L. Computational thermostabilization of an enzyme. Science. 2005;308:857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein–protein complexes. Proc. Natl Acad. Sci. USA. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T, Joachimiak L.A, Bullock A.N, Schuler A.D, Stoddard B.L, Baker D. Computational redesign of protein–protein interaction specificity. Nat. Struct. Mol. Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- Kuhlman B, O'Neill J.W, Kim D.E, Zhang K.Y, Baker D. Accurate computer-based design of a new backbone conformation in the second turn of protein L. J. Mol. Biol. 2002;315:471–477. doi: 10.1006/jmbi.2001.5229. 10.1006/jmbi.2001.5229 [DOI] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton G.C, Varani G, Stoddard B.L, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. 10.1126/science.1089427 [DOI] [PubMed] [Google Scholar]
- Meiler J, Baker D. Rapid protein fold determination using unassigned NMR data. Proc. Natl Acad. Sci. USA. 2003;100:15 404–15 409. doi: 10.1073/pnas.2434121100. 10.1073/pnas.2434121100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov A.V, Kortemme T, Tsemekhman K, Baker D. Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations. Proc. Natl Acad. Sci. USA. 2004;101:6946–6951. doi: 10.1073/pnas.0307578101. 10.1073/pnas.0307578101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauli S, Kuhlman B, Baker D. Computer-based redesign of a protein folding pathway. Nat. Struct. Biol. 2001;8:602–605. doi: 10.1038/89638. 10.1038/89638 [DOI] [PubMed] [Google Scholar]
- Qian B, Ortiz A.R, Baker D. Improvement of comparative model accuracy by free-energy optimization along principal components of natural structural variation. Proc. Natl Acad. Sci. USA. 2004;101:15 346–15 351. doi: 10.1073/pnas.0404703101. 10.1073/pnas.0404703101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohl C.A, Baker D. De novo determination of protein backbone structure from residual dipolar couplings using Rosetta. J. Am. Chem. Soc. 2002;124:2723–2729. doi: 10.1021/ja016880e. 10.1021/ja016880e [DOI] [PubMed] [Google Scholar]
- Rohl C.A, Baker D. Automated prediction of CASP-5 structures using the Robetta server. Proteins. 2003;53:524–533. doi: 10.1002/prot.10529. 10.1002/prot.10529 [DOI] [PubMed] [Google Scholar]
- Wang C, Schueler-Furman O, Baker D. Improved side-chain modeling for protein–protein docking. Protein Sci. 2005;14:1328–1339. doi: 10.1110/ps.041222905. [DOI] [PMC free article] [PubMed] [Google Scholar]