Abstract
HTP (human thymidine phosphorylase), also known as PD-ECGF (platelet-derived endothelial cell growth factor) or gliostatin, has an important role in nucleoside metabolism. HTP is implicated in angiogenesis and apoptosis and therefore is a prime target for drug design, including antitumour therapies. An HTP structure in a closed conformation complexed with an inhibitor has previously been solved. Earlier kinetic studies revealed an ordered release of thymine followed by ribose phosphate and product inhibition by both ligands. We have determined the structure of HTP from crystals grown in the presence of thymidine, which, surprisingly, resulted in bound thymine with HTP in a closed dead-end com-plex. Thus thymine appears to be able to reassociate with HTP after its initial ordered release before ribose phosphate and induces the closed conformation, hence explaining the mechanism of non-competitive product inhibition. In the active site in one of the four HTP molecules within the crystal asymmetric unit, additional electron density is present. This density has not been previously seen in any pyrimidine nucleoside phosphorylase and it defines a subsite that may be exploitable in drug design. Finally, because our crystals did not require proteolysed HTP to grow, the structure reveals a loop (residues 406–415), disordered in the previous HTP structure. This loop extends across the active-site cleft and appears to stabilize the dimer interface and the closed conformation by hydrogen-bonding. The present study will assist in the design of HTP inhibitors that could lead to drugs for anti-angiogenesis as well as for the potentiation of other nucleoside drugs.
Keywords: angiogenesis, human thymidine phosphorylase, product inhibition, pyrimidine nucleoside phosphorylase, structure, X-ray crystallography
Abbreviations: 2-dR-1-P, 2-deoxy-D-ribose-1-phosphate; BVDU, (E)-5-(2-bromovinyl)-2′-deoxyuridine; EcTP, Escherichia coli thymidine phosphorylase; GST, glutathione S-transferase; HTP, human thymidine phosphorylase; LB, Luria–Bertani; PYNP, pyrimidine nucleoside phosphorylase; KIN59, 5′-O-tritylinosine; TPI, 5-chloro-6-[1-(2-iminopyrrolidinyl)methyl] uracil hydrochloride; PD-ECGF, platelet-derived endothelial cell growth factor; rmsd, root mean square deviation
INTRODUCTION
HTP (human thymidine phosphorylase; EC 2.4.2.4) belongs to the PYNP (pyrimidine nucleoside phosphorylase) family and catalyses the reversible phosphorolysis of 2′-deoxythymidine (dThd) to 2-dR-1-P (2-deoxy-D-ribose-1-phosphate) and thymine. Kinetic studies show an ordered binding mechanism for HTP with the phosphate being the first substrate to bind, while 2-dR-1-P is the last ligand to leave the active site following turnover [1,2]. HTP also has a deoxyribosyl transferase activity where the deoxyribosyl moiety is transferred from one pyrimidine base to another, increasing the importance of its key functional roles: the homoeostasis of the dNTP pool. Moreover, HTP is linked to both angiogenic and chemotactic properties and has been confirmed to be identical with PD-ECGF (platelet-derived endothelial cell growth factor) [3,4] and gliostatin [5]. It is still unclear how HTP is responsible for angiogenesis, but its enzymatic activity is essential for this [6–8], therefore it is most likely that the products from the catalysed reaction are responsible for the angiogenic property. Recently, it was inferred that 2-deoxy-D-ribose, a thymidine metabolite, induces angiogenesis by generating oxygen radical species that induce the secretion of molecules such as vascular endothelial cell growth factor [9]. It is thought that HTP-promoted vascularization via angiogenesis induces cancer growth, whereas its pharmacological inhibition might suppress tumour progression. Consistent with this relationship to tumour growth is the observation that HTP is overexpressed in a wide range of human cancers including breast, ovarian, colorectal and oesophageal tissues [10].
HTP has also been shown to inhibit tumour cell apoptosis, especially in cells damaged by hypoxia-induced stress for example [11,12]. Thus HTP is an important target for the development of drugs [10] that have potential use in the treatment of cancer and certain other diseases by two distinctive mechanisms. First, tumour cells depend on the salvage pathway for their proliferation and thus inhibiting this pathway could reduce tumour proliferation. Secondly, a number of nucleoside analogue drugs are metabolized by HTP [13,14]. Thus antitumour prodrugs such as doxifluoridine and capecitabine are activated [10], while the antiviral drug BVDU [(E)-5-(2-bromovinyl)-2′-deoxyuridine; brivudine] is inactivated by the action of HTP [15]. BVDU is converted by HTP into an inactive free base, (E)-5-(2-bromovinyl)uracil, which selectively inhibits a further enzyme, dihydropyrimidine dehydrogenase. Thus modulating HTP activity can be very useful in chemotherapeutic approaches to a number of disease states [16].
An HTP structure has been solved in complex with the substrate analogue, TPI {5-chloro-6-[1-(2-iminopyrrolidinyl)methyl] uracil hydrochloride} [27]. The structure showed a dimer with identical subunits of similar overall fold to EcTP (Escherichia coli thymidine phosphorylase), composed of an α domain connected by three loops to an α/β domain [17]. However, this previous HTP structure had several residues missing, as it was subjected to limited proteolysis with trypsin, which was reported to be the only means of obtaining well-ordered crystals [27]. In the present study, we report a non-trypsinized HTP structure (from residue 34 to 479) at 2.3 Å resolution (1 Å=0.1 nm) in a new crystal form grown under different conditions, which contained four molecules in the asymmetric unit. Although crystallized in the presence of one substrate (dThd), thymine (the main physiological product of the phosphorylation reaction) is present in the active site of three HTP subunits. Additionally, residues His150 and Ala239, which make a key hydrogen bond after binding of phosphate to HTP and were disordered in the previous HTP structure, show a different hydrogen-bond pattern from one monomer to the other.
The current structure thus adds significant further information that will be of value in drug design for HTP inhibitors based on product analogues for an enzyme that plays an important role in several biological processes, and in the metabolism of certain antiviral and antitumour nucleoside drugs.
EXPERIMENTAL
Protein purification
The pMOAL-10T expression plasmid (a gift from Dr R. Bicknell, Institute of Molecular Medicine, University of Oxford, Oxford, U.K.) [4] coding for an N-terminal GST (glutathione S-transferase)-tagged HTP (residues 12–482) was transformed into Rosetta(DE3)pLysS for protein expression. The cells were grown in a 60 ml LB (Luria–Bertani) broth starter culture supplemented with 50 μg/ml carbenicillin and 34 μg/ml chloramphenicol, overnight, with shaking at 37 °C. The culture was then diluted 1:100 into 6 litres of LB and grown at 37 °C until the D600 reached 0.7; isopropyl-β-D-thiogalactoside was added to 0.5 mM and the induction was carried out at 27 °C overnight. The cells were harvested by centrifugation and the pellets were resuspended in buffer A (50 mM Tris/HCl, pH 7.4, 200 mM NaCl, 1 mM dithiothreitol and 5 mM EDTA) containining 0.1 mM PMSF and 1% (v/v) Triton X-100. The cells were disrupted by sonication and the supernatant, following clarification by centrifugation, was applied to a 5 ml glutathione–Sepharose column (GST-trap column, Amersham Biosciences) pre-equilibrated with buffer A plus 0.1% (v/v) Triton X-100. GST–HTP was eluted with buffer A with 10 mM glutathione. The GST tag was cleaved overnight at 4 °C after additions of calcium chloride (2.5 mM) and thrombin (1 unit/mg of GST–HTP). The cleavage products were buffer exchanged to eliminate the glutathione and applied again to a glutathione–Sepharose column to trap cleaved GST or the uncleaved fusion protein. Fractions containing HTP were applied to a Superdex S200 gel-filtration column pre-equilibrated with buffer A without detergent and concentrated to 12 mg/ml.
Crystallization and data collection
Thymidine (2 mM) and inhibitor KIN59 (5′-O-tritylinosine; 1 mM) were added to HTP for initial screening in several Hampton, Wizard and Emerald kits in a total of 768 conditions. A Cartesian Technologies pipetting robot was used to set up 100+100 nl sitting drops in Greiner 96-well plates, which were placed in a TAP storage vault [18]. Crystallization plates were routinely imaged automatically by a Veeco visualization system. An initial crystal hit was obtained in poly(ethylene glycol) 1500 and glycerol after several days at 22 °C. After optimization, the best crystals with maximum dimensions of 70 μm×70 μm×70 μm were obtained from 20% (v/v) glycerol and 20% (v/v) poly(ethylene glycol) 3350. The crystallization experiments using the previously published conditions [27] were unsuccessful in our hands.
X-ray diffraction data were collected at 100 K in-house using a Rigaku MicroMax 007 generator radiation source and a MAR 345 imaging plate detector (MAR Research). Crystals showed a high variability in diffraction quality, mostly diffracting to a resolution limit of 3 Å. However a few crystals were better ordered with diffraction to 2.3 Å resolution. Images were indexed and integrated with DENZO and data were merged using SCALEPACK [19]. The space group is P21 and the unit cell parameters are a=103.1, b=76.1, c=99.6 (Å) and β=98.6°, with four molecules in the asymmetric unit. Detailed statistics for X-ray data collection are given in Table 1.
Table 1. Statistics for crystallographic structure determination.
Values in parentheses are outer shell data.
| Parameters | |
|---|---|
| Data collection details | |
| Data collection site | In house |
| Detector | MAR345 |
| Wavelength (Å) | 1.5418 |
| Resolution range (Å) | 30.0–2.3 (2.38–2.30) |
| Redundancy | 8.6 (8.0) |
| Completeness (%) | 100.0 (100.0) |
| Average I/σ(I) | 18.23 (2.9) |
| Rmerge* | 11.9 (69.5) |
| Refinement statistics | |
| Resolution range (Å) | 30–2.31 (2.37–2.31) |
| R-factor (Rwork/Rfree)† | 24.5/28.8 |
| rms bond length deviation (Å) | 0.009 |
| rms bond angle deviation (°) | 1.27 |
| Mean B-factor (Å2)‡ | 32.3/35.3/33.7 |
*Rmerge=Σ|I−〈I〉|/Σ〈I〉.
†R-factor=Σ|Fo−Fc|/ΣFo.
‡Main chain/side chain and water/all atoms.
The HTP crystal structure was solved by molecular replacement using MOLREP [20] with the co-ordinates of the previously solved HTP (PDB code 1UOU) as the search model. In spite of the high sequence identity, other molecular replacement programs did not yield any correct solution. Indeed, a pseudo-translation symmetry has been found in the HTP crystal which can make structure determination difficult [21]. Refinement was carried out using cycles of manual rebuilding with COOT [22], followed by NCS restrained refinement using REFMAC5 [23] with TLS [24]. Density for thymine was fitted as appropriate and refined. One of the four subunits in the asymmetric unit (molecule D) seemed partially disordered in one domain because presumably it was unliganded, thus proving difficult to refine; moreover, in the case of pseudosymmetry, high values of R-factors are often seen because of the preponderance of low intensity reflections [25]. The final refinement parameters of Rwork and Rfree were 24.5 and 28.8% respectively (Table 1).
Ramachandran plots generated from PROCHECK indicate that the model exhibits good stereochemistry, with 95% of the residues in the most favoured region and 4.7% in additionally allowed regions. Structural superpositions were performed with SHP [26], and Figures were drawn using PYMOL.
RESULTS AND DISCUSSION
Overall structure
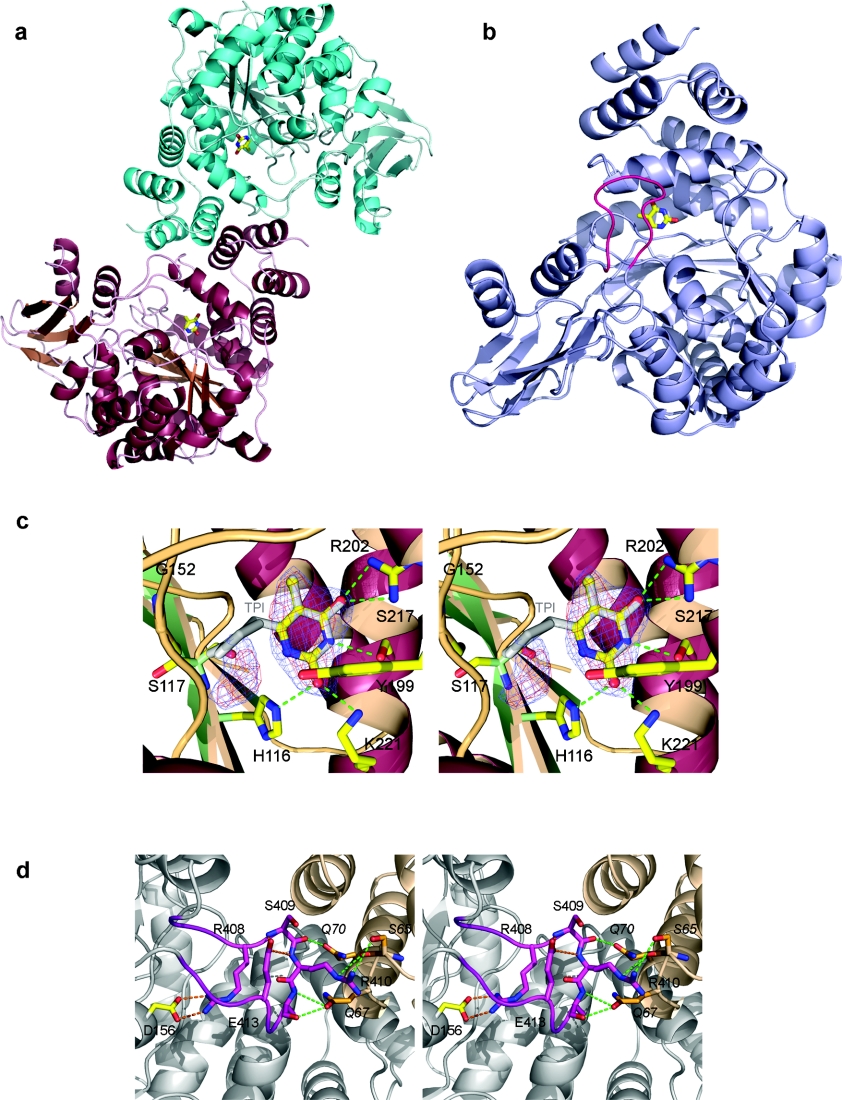
The crystal structure of HTP presented in the present study (Figures 1a and 1b) is of a different crystal form from the previous HTP–TPI complex, which belonged to the space group C2 and contained a single protomer in the asymmetric unit [27]. The crystals of the HTP–thymine complex, while also monoclinic, belong to the space group P21, with four molecules in the asymmetric unit, corresponding to two biological dimers (Figure 1a). These dimers have the same inter-subunit contacts as those observed for HTP–TPI, although in this latter case the dimer is positioned on a crystallographic 2-fold axis [27]. The dimer interface consists of a coiled coil formed by helices from each monomer (Figure 1a). The refinement was carried out with NCS applied to most of the residues of all four subunits, implying that they are almost identical in conformation. The rmsd (root mean square deviation) for Cαs from all overlapping residues from pairs of HTP molecules were: 0.29, 0.38, 0.43, 0.29, 0.35 and 0.20 (Å). Comparison of molecule A with the published HTP structure showed 99% of residues structurally equivalent to an rmsd for Cαs of 0.72 Å. Each HTP monomer consists of two domains, an α domain, composed of six α-helices that form the dimer interface and the substrate-binding site, while a large α/β domain has a central β-sheet surrounded by α-helices. Three loops between these domains allow the α and the α/β domain to counter-rotate to give a closed conformation. Indeed, HTP has been crystallized, as in the previous structure, in the closed conformation.
Figure 1. Crystal structure of HTP.
(a) Diagram showing the HTP dimer and the dimerization interface; the thymines present in the active sites are drawn as sticks. (b) Ribbon diagram showing the overall fold of the HTP monomer drawn in light blue. The loop region (405–415) is coloured in pink and the thymine is drawn as sticks. (c) Stereodiagram showing the hydrogen-bonding (green) of thymine to the HTP active site. The inhibitor TPI is superimposed on to the thymine. The final 2Fo−Fc electron density is contoured at 1σ and coloured in blue. The Fo−Fc omit map is contoured at 1.6σ and coloured in red. The additional electron density feature can be seen to the left of the thymine. Relative to (b), the active site is orientated looking towards the thymine through the 405–415 loop, with a 90° clockwise rotation. (d) Stereodiagram showing the interaction of the 405–415 loop coloured in pink, with the two subunits coloured in grey and wheat. Intra-chain hydrogen bonds are coloured in orange, whereas inter-chains hydrogen bonds are coloured in green. The molecule is rotated clockwise by 90° relative to the view in (b).
Our new crystal form is based on minimal crystallization conditions of poly(ethylene glycol) and glycerol and did not require any proteolytic cleavage to grow. Thus it was possible to build the missing loop (Figures 1b and 1d), together with some residues that were disordered in the earlier structure. While the proteolytically cleaved loop appears to be not essential for domain closure [27], it does potentially contribute to stabilization of the closed form. Indeed, the α and the α/β domain interact by hydrogen-bonding across the active-site cleft. Furthermore, the loop seems to stabilize the dimer by hydrogen bonds (Figure 1d). It is unclear if the loop region has a functional role in any of the other known important functions of HTP. Moreover, these residues are present in other PYNP structures, such as EcTP [17] or BsPYNP [Bacillus stearothermophilus (now Geobacillus stearothermophilus) PYNP] [28].
In the case of EcTP, it has been suggested that the domain closure movement brings substrates into close proximity, and isolates them from solvent so preventing hydrolysis, which would otherwise compete with phosphorolysis. Thus the loop could play a role in active-site integrity and stability of the closed form. Indeed, hydrogen bonds via residues Arg408, Ser409 and Arg410 are seen between the loop and the rest of the protein (Figure 1d).
Ligand binding
Refinement of the current HTP structure showed that the product thymine was present in three of the four subunits in the asymmetric unit (molecules A, B and C), rather than the substrate thymidine, which had been added at the start of the crystallizations. An additional electron density feature was also observed in molecule B close to the thymine molecule (Figure 1c). We were not able to fit any molecule used during HTP purification or crystallization into this density. Although an inorganic phosphate ion fitted well into the unbiased initial difference map and also refined satisfactorily, a later CNS annealed omit map was less convincing. Implications for the mechanism of product inhibition and use in drug design are discussed in later sections. The inhibitor KIN59, which has been shown to exhibit inhibitory activity against human and bacterial recombinant thymidine phosphorylases [29], was present under the crystallization conditions. While there were weak indications of electron density for KIN59 in some difference maps at the base of the α/β domain around residue 422, this was not further substantiated in subsequent refinements. Interestingly, in spite of an apparently only very low occupancy potential site for KIN59, its presence appeared essential in yielding good quality diffracting crystals of HTP. Indeed, well diffracting crystals of HTP have been reported to be difficult to grow. Thus Spraggon et al. [30] have reported HTP crystals for which diffraction was limited to 3.5 Å resolution. Norman et al. [27] were successful in solving the HTP structure to 2.1 Å resolution, although the enzyme required partial digestion, resulting in several disordered residues.
Conformational changes in HTP
EcTP is known to have to undergo conformational changes from an open form to a closed active form to facilitate the nucleophilic attack of the phosphate on to the sugar ring [31]. The HTP structure of Norman et al. [27] is in the closed conformation, suggesting that the substrate analogue TPI itself was able to cause closure of the protein [27]. In the structure reported in the present study, HTP is in closed conformation too, so we could argue that thymine by itself is sufficient to stabilize the closed form, and thus the 5-chlorouracil moiety of TPI is able also to act in this role. It has been suggested that the binding of phosphate triggers a partial closure consisting of an 8° rotation of the α/β domain with respect to the α domain, while subsequent binding of nucleoside substrate would be responsible for the fully closed conformation [28]. The two HTP structures would suggest that even if phosphate is able to trigger a partial domain closure, the thymine moiety is able to cause the full domain closure by itself. Kinetic studies have shown that phosphate binds before the pyrimidine [1,2], so if thymidine binds before the phosphate, phosphate would not be able to access its binding site because of the closed form. In this case, although the protein can adopt a closed conformation similar to the ‘active’ form as there is no phosphate present, no phosphorolysis would occur.
Structural and theoretical studies on EcTP suggested that few movements occur after phosphate binding, especially the formation of a key hydrogen bond between residues His119 and Gly208 (respectively His150 and Ala239 in HTP) [28]. These residues help to order the 115–120 loop that is disordered prior to phosphate binding. In our structure, a hydrogen bond is indeed present between His150 and Ala239 in molecules A and B, whereas in molecule C, the distance between the histidine nitrogen and the alanine carbonyl is doubled to 5.3 Å. Even though no density for phosphate is seen, the hydrogen bonds are present, suggesting that phosphate is not essential for this hydrogen bond to be formed. The unliganded subunit (molecule D) is less well ordered, compared with the other molecules in the asymmetric unit and has a closed conformation although no ligand is bound. The formation of this asymmetric dimer might indicate a low energy barrier between open/closed forms, or could simply be selected during crystallization.
Active site structure and mechanism of thymine product inhibition
Electron density for the thymine ring is very clear in three of the HTP monomers (Figure 1c). Its binding is similar to the 5-chlorouracil of TPI, in which the chlorine has replaced the methyl group of the thymine. The same hydrogen bonds are made between the thymine and residues Arg202, Ser217, Lys221 and His116 with small variation in bond length (0.1–0.2 Å) apart from His116 whose variation in length is around 0.5 Å. Moreover, Ser117 making hydrogen-bonding with the 2-iminopyrrolidinyl moiety is 0.6 Å closer to the N1 of the thymine. His116 and Ser117 both belong to the α/β domain, whereas the other residues involved in thymine/TPI binding belong to the α domain. This variation could be due either to the 2-iminopyrrolidinyl moiety of TPI (which could displace Ser117 and therefore His116), or due to the absence of the loop stabilizing the closed conformation.
Kinetic studies have shown an ordered binding mechanism with phosphate the first ligand to bind, while 2-dR-1-P is the last to dissociate. In our structure, thymine is present but 2-dR-1-P is not. If the normal reaction has occurred, we would not see thymine without 2-dR-1-P, as thymine must dissociate before the latter. So we can suggest that our structure contains thymine as a product inhibitor of the reaction. It has been shown that thymine can act as a non-competitive inhibitor [32], so it normally would be expected not to bind to the active site. However, our results indicate that the structural basis of thymine non-competitive inhibition is actually by re-associating with the unliganded enzyme and inducing a domain closure in HTP.
Thymidine was present under the crystallization conditions; thus clearly phosphorolysis has occurred to give the observed thymine, a product of the reaction, requiring the presence of inorganic phosphate. As no phosphate was added to the protein during purification or crystallization, this anion species was probably trapped during protein expression in E. coli and co-purified with the enzyme. A further, as yet unidentified, small molecule is found in the active site of molecule B, which probably also carried through the protein purification procedure. The centre of this electron density is situated around 3.5 Å from the thymine N1, but does not overlap with the TPI ring (Figure 1c). The density is not unequivocally related to any known substrate or a reaction product; nevertheless the hydrogen-bonding pattern and position may be incorporated into an inhibitor as part of a structure-based drug design effort. Indeed, residues Tyr199, Ser117 and Gly152 are proximal to this density (Figure 1c).
HTP is a key therapeutic target because of its involvement in angiogenesis and in many metastatic diseases. Some interesting chemotherapeutic approaches using doxifluridine, fluorouracil or capecitabine, for example, which are activated by HTP, are currently being developed. Cancer treatment targeting thymidine phosphorylase with these drugs strongly inhibited tumour growth in experiments of thymidine phosphorylase-transfected and thymidine phosphorylase-induced tumour xenografts in animals [10]. Knowledge of the three-dimensional structure of a protein together with a range of bound ligands can be extremely valuable in the design of new inhibitors and in the improvement of existing ones. The work reported in the present study is likely to be useful in this regard for several reasons: (i) we provide a more complete HTP structure than the existing one; (ii) thymine, a natural HTP inhibitor, is present in the active site, causing domain closure and hence explaining the non-competitive inhibition of thymine reported previously [32]. Finally, (iii) the presence of electron density close to residues Ser117, Gly152 and Tyr199 may be utilized in inhibitor design by forming hydrogen bonds to these side chains via addition of suitable substituents to an inhibitor based on the product thymine, for example. In order to further understand the HTP catalytic mechanism, determining other structures at different steps of the phosphorolysis or deoxyribosyl transferase reaction in complex with inhibitors, should be a priority.
Acknowledgments
We thank the EC for funding this work via project QLRT-2000-01004. Additional support was from the U.K. Medical Research Council and KU Leuven (Centers of Excellence; Krediet no. EF/05/15). We thank Robert Esnouf and Jun Dong for computer support.
References
- 1.Krenitsky T. A. Pentosyl transfer mechanisms of the mammalian nucleoside phosphorylases. J. Biol. Chem. 1968;243:2871–2875. [PubMed] [Google Scholar]
- 2.Schwartz M. Thymidine phosphorylase from Escherichia coli. Properties and kinetics. Eur. J. Biochem. 1971;21:191–198. doi: 10.1111/j.1432-1033.1971.tb01455.x. [DOI] [PubMed] [Google Scholar]
- 3.Usuki K., Saras J., Waltenberger J., Miyazono K., Pierce G., Thomason A., Heldin C. H. Platelet-derived endothelial cell growth factor has thymidine phosphorylase activity. Biochem. Biophys. Res. Commun. 1992;184:1311–1316. doi: 10.1016/s0006-291x(05)80025-7. [DOI] [PubMed] [Google Scholar]
- 4.Moghaddam A., Bicknell R. Expression of platelet-derived endothelial cell growth factor in Escherichia coli and confirmation of its thymidine phosphorylase activity. Biochemistry. 1992;31:12141–12146. doi: 10.1021/bi00163a024. [DOI] [PubMed] [Google Scholar]
- 5.Griffiths L., Stratford I. J. Platelet-derived endothelial cell growth factor thymidine phosphorylase in tumour growth and response to therapy. Br. J. Cancer. 1997;76:689–693. doi: 10.1038/bjc.1997.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moghaddam A., Zhang H. T., Fan T. P., Hu D. E., Lees V. C., Turley H., Fox S. B., Gatter K. C., Harris A. L., Bicknell R. Thymidine phosphorylase is angiogenic and promotes tumor growth. Proc. Natl. Acad. Sci. U.S.A. 1995;92:998–1002. doi: 10.1073/pnas.92.4.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyadera K., Sumizawa T., Haraguchi M., Yoshida H., Konstanty W., Yamada Y., Akiyama S. Role of thymidine phosphorylase activity in the angiogenic effect of platelet derived endothelial cell growth factor/thymidine phosphorylase. Cancer Res. 1995;55:1687–1690. [PubMed] [Google Scholar]
- 8.Sumizawa T., Furukawa T., Haraguchi M., Yoshimura A., Takeyasu A., Ishizawa M., Yamada Y., Akiyama S. Thymidine phosphorylase activity associated with platelet-derived endothelial cell growth factor. J. Biochem. (Tokyo) 1993;114:9–14. doi: 10.1093/oxfordjournals.jbchem.a124146. [DOI] [PubMed] [Google Scholar]
- 9.Brown N. S., Jones A., Fujiyama C., Harris A. L., Bicknell R. Thymidine phosphorylase induces carcinoma cell oxidative stress and promotes secretion of angiogenic factors. Cancer Res. 2000;60:6298–6302. [PubMed] [Google Scholar]
- 10.Toi M., Atiqur Rahman M., Bando H., Chow L. W. Thymidine phosphorylase (platelet-derived endothelial-cell growth factor) in cancer biology and treatment. Lancet Oncol. 2005;6:158–166. doi: 10.1016/S1470-2045(05)01766-3. [DOI] [PubMed] [Google Scholar]
- 11.Kitazono M., Takebayashi Y., Ishitsuka K., Takao S., Tani A., Furukawa T., Miyadera K., Yamada Y., Aikou T., Akiyama S. Prevention of hypoxia-induced apoptosis by the angiogenic factor thymidine phosphorylase. Biochem. Biophys. Res. Commun. 1998;253:797–803. doi: 10.1006/bbrc.1998.9852. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda R., Furukawa T., Kitazono M., Ishitsuka K., Okumura H., Tani A., Sumizawa T., Haraguchi M., Komatsu M., Uchimiya H., et al. Molecular basis for the inhibition of hypoxia-induced apoptosis by 2-deoxy-D-ribose. Biochem. Biophys. Res. Commun. 2002;291:806–812. doi: 10.1006/bbrc.2002.6432. [DOI] [PubMed] [Google Scholar]
- 13.Desgranges C., Razaka G., Rabaud M., Bricaud H., Balzarini J., De Clercq E. Phosphorolysis of (E)-5-(2-bromovinyl)-2′-deoxyuridine (BVDU) and other 5-substituted-2′-deoxyuridines by purified human thymidine phosphorylase and intact blood platelets. Biochem. Pharmacol. 1983;32:3583–3590. doi: 10.1016/0006-2952(83)90307-6. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Perez M. J., Priego E. M., Hernandez A. I., Camarasa M. J., Balzarini J., Liekens S. Thymidine phosphorylase inhibitors: recent developments and potential therapeutic applications. Mini Rev. Med. Chem. 2005;5:1113–1123. doi: 10.2174/138955705774933301. [DOI] [PubMed] [Google Scholar]
- 15.Balzarini J., Sienaert R., Liekens S., Van Kuilenburg A., Carangio A., Esnouf R., De Clercq E., McGuigan C. Lack of susceptibility of bicyclic nucleoside analogs, highly potent inhibitors of varicella-zoster virus, to the catabolic action of thymidine phosphorylase and dihydropyrimidine dehydrogenase. Mol. Pharmacol. 2002;61:1140–1145. doi: 10.1124/mol.61.5.1140. [DOI] [PubMed] [Google Scholar]
- 16.Matsushita S., Nitanda T., Furukawa T., Sumizawa T., Tani A., Nishimoto K., Akiba S., Miyadera K., Fukushima M., Yamada Y., et al. The effect of a thymidine phosphorylase inhibitor on angiogenesis and apoptosis in tumors. Cancer Res. 1999;59:1911–1916. [PubMed] [Google Scholar]
- 17.Walter M. R., Cook W. J., Cole L. B., Short S. A., Koszalka G. W., Krenitsky T. A., Ealick S. E. Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 Å resolution. J. Biol. Chem. 1990;265:14016–14022. doi: 10.2210/pdb1tpt/pdb. [DOI] [PubMed] [Google Scholar]
- 18.Walter T. S., Diprose J. M., Mayo C. J., Siebold C., Pickford M. G., Carter L., Sutton G. C., Berrow N. S., Brown J., Berry I. M., et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1996;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 21.Chook Y. M., Lipscomb W. N., Ke H. Detection and use of pseudo-translation in determination of protein structures. Acta Crystallogr. D Biol. Crystallogr. 1998;54:822–827. doi: 10.1107/s0907444997020064. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Winn M. D., Isupov M. N., Murshudov G. N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 25.Vajdos F. F., Yoo S., Houseweart M., Sundquist W. I., Hill C. P. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Protein Sci. 1997;6:2297–2307. doi: 10.1002/pro.5560061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuart D. I., Levine M., Muirhead H., Stammers D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 27.Norman R. A., Barry S. T., Bate M., Breed J., Colls J. G., Ernill R. J., Luke R. W., Minshull C. A., McAlister M. S., McCall E. J., et al. Crystal structure of human thymidine phosphorylase in complex with a small molecule inhibitor. Structure. 2004;12:75–84. doi: 10.1016/j.str.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Pugmire M. J., Ealick S. E. The crystal structure of pyrimidine nucleoside phosphorylase in a closed conformation. Structure. 1998;6:1467–1479. doi: 10.1016/s0969-2126(98)00145-2. [DOI] [PubMed] [Google Scholar]
- 29.Liekens S., Hernandez A. I., Ribatti D., De Clercq E., Camarasa M. J., Perez-Perez M. J., Balzarini J. The nucleoside derivative 5′-O-trityl-inosine (KIN59) suppresses thymidine phosphorylase-triggered angiogenesis via a noncompetitive mechanism of action. J. Biol. Chem. 2004;279:29598–29605. doi: 10.1074/jbc.M402602200. [DOI] [PubMed] [Google Scholar]
- 30.Spraggon G., Stuart D., Ponting C., Finnis C., Sleep D., Jones Y. Crystallization and X-ray diffraction study of recombinant platelet-derived endothelial cell growth factor. J. Mol. Biol. 1993;234:879–880. doi: 10.1006/jmbi.1993.1632. [DOI] [PubMed] [Google Scholar]
- 31.Pugmire M. J., Cook W. J., Jasanoff A., Walter M. R., Ealick S. E. Structural and theoretical studies suggest domain movement produces an active conformation of thymidine phosphorylase. J. Mol. Biol. 1998;281:285–299. doi: 10.1006/jmbi.1998.1941. [DOI] [PubMed] [Google Scholar]
- 32.Iltzsch M. H., el Kouni M. H., Cha S. Kinetic studies of thymidine phosphorylase from mouse liver. Biochemistry. 1985;24:6799–6807. doi: 10.1021/bi00345a011. [DOI] [PubMed] [Google Scholar]