Abstract
A protein in RAW 264.7 macrophages, which became phosphorylated in response to LPS (lipopolysaccharide), was identified as the RNA-binding protein called DAZAP1 [DAZ (deleted in azoospermia)-associated protein 1]. The phosphorylation of this protein was prevented by specific inhibition of MKK1 [MAPK (mitogen-activated protein kinase) kinase 1], indicating that it was phosphorylated via the classical MAPK cascade. Further experiments showed that DAZAP1 was phosphorylated stoichiometrically in vitro by ERK2 (extracellular-signal-regulated protein kinase 2) at two Thr-Pro sequences (Thr269 and Thr315), and that both sites became phosphorylated in HEK-293 (human embryonic kidney 293) cells in response to PMA or EGF (epidermal growth factor), or RAW 264.7 macrophages in response to LPS. Phosphorylation induced by each stimulus was prevented by two structurally distinct inhibitors of MKK1 (PD184352 and U0126), demonstrating that DAZAP1 is a physiological substrate for ERK1/ERK2. The mutation of Thr269 and Thr315 to aspartate or the phosphorylation of these residues caused DAZAP1 to dissociate from its binding partner DAZ. DAZ interacts with PABP [poly(A)-binding protein] and thereby stimulates the translation of mRNAs containing short poly(A) tails [Collier, Gorgoni, Loveridge, Cooke and Gray (2005) EMBO J. 24, 2656–2666]. In the present study we have shown that DAZ cannot bind simultaneously to DAZAP1 and PABP, and suggest that the phosphorylation-induced dissociation of DAZ and DAZAP1 may allow the former to stimulate translation by interacting with PABP.
Keywords: AU-rich element (ARE), deleted in azoospermia (DAZ), deleted-in-azoospermia-associated protein 1 (DAZAP1), extracellular-signal-regulated protein kinase (ERK), mRNA stability, mRNA translation
Abbreviations: ARE, AU-rich element; AREBP, ARE-binding protein; DAZ, deleted in azoospermia; DAZAP1, DAZ-associated protein 1; DAZL, DAZ-like protein; EGF, epidermal growth factor; ERK, extracellular-signal-regulated protein kinase; GST, glutathione S-transferase; HA, haemagglutinin; HEK-293, human embryonic kidney 293; hnRNP, heterogeneous nuclear ribonucleoprotein; IL, interleukin; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MKK, MAPK kinase; PABP, poly(A)-binding protein; RRM, RNA-recognition motif; TNFα, tumour necrosis factor α; Tpl2, tumour progression locus 2
INTRODUCTION
Inflammation is an important process in the defence of invasion by pathogens, and is mediated in part by the production of pro-inflammatory cytokines, such as TNFα (tumour necrosis factor-α), IL-6 (interleukin-6) and IL-8. Normal expression of cytokines is vital for the inflammatory response, but their overexpression can lead to a number of inflammatory diseases, such as rheumatoid arthritis [1], inflammatory bowel disease [2] and certain neurodegenerative disorders [3]. It is therefore crucial that the production of cytokines is tightly regulated.
The expression of cytokines is controlled at multiple levels, including transcriptional and post-transcriptional mechanisms, protein translation and secretion. Several intracellular signalling pathways are switched on in response to pathogen-derived molecules, such as bacterial LPS (lipopolysaccharide), through the activation of Toll-like receptors [4]. Engagement of Toll-like receptor 4 by LPS triggers the activation of numerous protein kinases, including transforming-growth-factor-β-activated kinase 1 [5,6] and COT/Tpl2 (tumour progression locus 2) [7,8]. In turn, transforming-growth-factor-β-activated kinase 1 induces the activation of NF-κB (nuclear factor κB), as well as c-Jun N-terminal kinase and p38 MAPK (mitogen-activated protein kinase) [9], whereas COT/Tpl2 activates the classical MAPK cascade [7,8]. These pathways all contribute to pro-inflammatory cytokine production at one or more of the different levels mentioned above. For example, it has been suggested that the classical MAPK cascade is involved in regulating TNFα production at the post-transcriptional level [7,8].
Two post-transcriptional mechanisms involve regulation of the rate of turnover of mRNA and regulation of the translation of these messages via cis elements found along the mRNA, both of which are affected by trans-acting factors. One such regulatory element is the ARE (AU-rich element) found in the 3′-untranslated regions of a number of pro-inflammatory cytokines, such as TNFα [10], IL-6 [11] and IL-8 [12], and immediate early genes, which has been shown to confer mRNA instability [13]. The destabilizing effects of the AREs are suppressed when macrophages are stimulated by LPS. To investigate how LPS exerts this effect, much effort has been directed towards identifying relevant AREBPs (ARE-binding proteins) [14–17]. In some instances, AREBPs, such as KSRP (K-homology splicing regulatory protein), have been reported to recruit the exosome, a complex of 3′–5′ exonucleases involved in the degradation of ARE-containing mRNAs [18]. However, it is also believed that other AREBPs might stabilize mRNA by competing with other ARE-binding factors that destabilize mRNAs [19–21]. Taken together, these findings suggest that AREs can act as RNA instability determinants and/or translational modulators.
The control of protein translation is also crucial to post-transcriptional regulation. For example, by binding to the poly(A) tails, PABP [poly(A)-binding protein] not only stabilizes mRNAs by protecting them from deadenylation, but also initiates protein translation via interaction with eIF4G (eukaryotic protein synthesis initiation factor 4G), a protein which interacts with the eIF4E that binds to the 5′ cap structure of mRNA. Through these interactions, a model has been proposed in which PABP protects against degradation and also contributes to the circularization of mRNAs, which is thought to enhance their stability and promote their translation [22,23].
The ability of LPS to stabilize mRNAs and stimulate their translation is likely to be mediated by the phosphorylation of proteins that regulate these processes. This is supported by the observation that relatively specific inhibitors of p38 MAPK decrease the production of TNFα and other cytokines [24], and that mice which are deficient in COT/Tpl2, and therefore cannot activate the MAPKs ERK1 (extracellular-signal-regulated protein kinase 1) and ERK2, do not produce TNFα in response to LPS [7]. Although several AREBPs that are substrates for the p38 MAPK–MAPKAP-K2 (MAPK-activated protein kinase 2) pathway have been identified, such as TTP (tri-tetraproline) [25] and hnRNP A0 (heterogeneous nuclear ribonucleoprotein A0) [17], which may regulate mRNA stability/translation, the identity of the key substrates for ERK1/ERK2 that contribute to such regulation is still lacking. In the present study, we identify an AREBP, DAZAP1 [DAZ (deleted in azoospermia)-associated protein 1], as a novel physiological substrate for ERK1/ERK2 and demonstrate that its phosphorylation induces dissociation from DAZ, a protein which has been shown to initiate protein translation by interacting with PABP [26].
EXPERIMENTAL
Materials
PD184352 [26a], a specific inhibitor of MKK1 (MAPK kinase 1) and MKK2, was synthesized by an improved method [27]. U0126 was purchased from Calbiochem (Nottingham, U.K.), Escherichia coli LPS, PMA and DMSO were from Sigma (Poole, Dorset, U.K.), Complete™ protease inhibitor tablets were from Roche (Lewes, Sussex, U.K.), EGF (epidermal growth factor) and cell culture media were from Gibco (Paisley, Renfrewshire, Scotland, U.K.), precast Bis-Tris gradient SDS polyacrylamide gels, running buffer and transfer buffer were from Invitrogen (Paisley, Renfrewshire, Scotland, U.K.), and Protein G–Sepharose, glutathione–Sepharose and ECL (enhanced chemiluminescence) reagent were from Amersham (Little Chalfont, Bucks., U.K.). All the peptides utilized in this study were synthesized by Dr Graham Bloomberg (Department of Biochemistry, University of Bristol, Bristol, U.K.). Activated ERK2 was produced and assayed as described previously [28].
Plasmids
DAZAP1b was amplified from IMAGE clone 4549444 with the GC-rich PCR system (Roche) using the oligonucleotides MP1233 and MP1235 shown below. DAZAP1b differs slightly from DAZAP1; they share the same amino acid sequence from residues 1 to 349, but DAZAP1b has a distinct and longer C-terminal region (terminating at residue 408) compared with DAZAP1a (terminating at residue 379); these two species presumably arising from alternative splicing. The resulting fragment was cloned into pCR2.1 (Invitrogen), sequenced and then ligated into pCMV-HA-1 to form pCMV-HA-DAZAP1b or into pGEX6P-1 to make pGEX6P-1-DAZAP1b. pCMV-HA-DAZAP1b(155–407) was made in a similar way using oligonucleotides MP1820 and MP1235. The T269D mutation was introduced using oligonucleotides NM120 and NM121 with the Quikchange® site-directed mutagenesis kit (Stratagene), whereas the T315D mutation was introduced using oligonucleotides NM122 and NM123. The T269A and T315A mutations were made using oligonucleotides MP1436/MP1437 and MP1438/MP1439 respectively. PABP-CI (PABP cytoplasmic I) was amplified in a similar manner from IMAGE clone 5597273, then cloned with the oligonucleotides NM163 and NM164. It was subcloned into pEBG6P to form pEBG6P-1-PABP-CI. DAZ was amplified from IMAGE clone 5297459 with oligonucleotides MP1845 and MP1846, as described above, and subcloned into pCMV-FLAG-1 to form pCMV-FLAG-DAZ.
The oligonucleotides used were: MP1233, GCG GAT CCA ACA ACT CGG GCG CCG ACG AG; MP1235, GCG GAT CCC TAG CGT CGG TAG GGG TGG AAC; MP1436, TCC TAC ATC GTG TCC GCC CCT CCT GGA GGC TTT; MP1437, AAA GCC TCCA GGA GGG GCG GAC ACG ATG TAG GA; MP1438, CCT CCT CCA CCA GCC GCT CCC GGG GCA GCA CCT; MP1439, AGG TGC TGC CCC GGG AGC GGC TGG TGG AGG AGG; MP1820, GCG GAT CCG GTT TTG GAT TTA TTA CTT TCG AGG ACG AAC AAT; MP1845, GCG GAT CCA TGT CTG CTG CAA ATC CTG AGA CTC C; MP1846, GCG CGG CCG CTC AGT CTC TTC TCT GGA TTA AAC AGA CAA GAT AC; NM120, TCC TAC ATC GTG TCC GAC CCT CCT GGA GGC TT; NM121, AAG CCT CCA GGA GGG TCG GAC ACG ATG TAG GA; NM122, CCT CCT CCA CCA GCC GAT CCC GGG GCA GCA CC, NM123, GGT GCT GCC CCG GGA TCG GCT GGT GGA GGA GG; NM163, GCG GAT CCA ACC CCA GTG CCC CCA GCT ACC CCA T; NM164, GCG CGG CCG CTT AAA CAG TTG GAA CAC CGG TGG CAC TG.
Expression of DAZAP1 in E. coli
The plasmid pGEX6P-1-DAZAP1, encoding a GST (glutathione S-transferase) fusion protein of full-length human DAZAP1, was expressed in BL21 cells and induced with 50 μM IPTG (isopropyl β-D-thiogalactoside) and grown overnight at 26 °C. The bacterial extracts were purified at 4 °C by affinity chromatography on glutathione–Sepharose 4B (Amersham Pharmacia Biotech). The bound fusion protein was eluted with 20 mM glutathione in 150 mM Tris/HCl (pH 7.5), dialysed overnight in 150 mM Tris/HCl (pH 7.5), 0.1% (v/v) 2-mercaptoethanol and 50% sucrose, and stored at −80 °C.
Antibodies
The peptides SYIVST*PPGG and VPPPPAT*PGAA (where T* is phosphothreonine), corresponding to residues 264–273 and 309–319 of human DAZAP1, were coupled to both BSA and keyhole-limpet haemocyanin, then injected into sheep at Diagnostics Scotland (Edinburgh, Scotland, U.K.). The antisera were affinity purified on CH–Sepharose to which the relevant phosphorylated peptide had been coupled covalently. Antibodies capable of recognizing the unphosphorylated and phosphorylated forms of DAZAP1 equally well were raised against GST–DAZAP1, and the antisera affinity purified on CH–Sepharose, to which GST–DAZAP1 had been coupled covalently, followed by passage through GST–Sepharose to remove anti-GST antibodies. The antibodies that recognize HA (haemagglutinin) and GST were also raised in sheep and used for immunoblotting. Anti-HA antibodies coupled covalently to agarose and anti-FLAG antibodies were purchased from Sigma, and antibodies that recognized the active phosphorylated forms of ERK1 and ERK2 were from Cell Signalling Technologies (Hitchin, Herts., U.K). Rabbit-, mouse- and sheep-specific secondary antibodies conjugated to horseradish peroxidase were purchased from Pierce (Rockford, IL, U.S.A.).
Cell culture, transfection and cell lysis
HEK-293 (human embryonic kidney 293) cells and murine RAW 264.7 macrophages (purchased from the European Tissue Culture Collection) were maintained in a 95% air/5% CO2 atmosphere at 37 °C. HEK-293 cells and RAW 264.7 macrophages were cultured in DMEM (Dulbecco's modified Eagle's medium) containing 10% foetal calf serum (HEK-293 cells) or heat-inactivated foetal calf serum (RAW 264.7 cells) plus 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. The HEK-293 cells were transfected with the indicated amounts of the different DNA constructs using poly(ethyleneimine) [29]. The cells were lysed in buffer composed of 50 mM Tris/HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1 mM sodium orthovanadate, 10 mM sodium β-glycerophosphate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose, 1% (v/v) Triton X-100, 0.1% (v/v) 2-mercaptoethanol and Complete™ protease inhibitor cocktail (1 tablet/50 ml of cell lysis buffer). The lysates were centrifuged at 13000 g for 15 min at 4 °C and the supernatants were collected. Protein concentrations were determined using the Bradford method. Lysates (1 mg of extract) were incubated for 10 min at 37 °C with 10 units of benzonuclease to digest RNA and DNA (Novagen).
Immunoprecipitation and immunoblotting
Cell extract (1 mg) was incubated for 2 h at 4 °C with 10 μl of anti-HA antibody–agarose or 10 μl of glutathione–Sepharose 4B. The beads were washed twice with 1 ml of 50 mM Tris/HCl (pH 7.5)/150 mM NaCl and twice with 1 ml of 50 mM Tris/HCl (pH 7.5)/0.5 M NaCl. The beads were heated in SDS for 3 min at 90 °C and, after pelleting the beads, the supernatant was subjected to SDS/PAGE on 4–12% gradient gels. After transfer on to nitrocellulose, the membrane was immunoblotted with anti-HA, anti-FLAG or anti-GST antibodies, each at 1 μg/ml, using the ECL® system.
Phosphorylation of DAZAP1 by ERK2
Bacterially expressed GST–DAZAP1 was incubated for 30 min at 30 °C alone or with 2 units/ml ERK2 in presence of 0.1 mM ATP and 10 mM MgCl2. The reaction was stopped by the addition of SDS to denature the samples, and the samples were subjected to SDS/PAGE as described in the Results section.
RESULTS
Identification of two proteins that become phosphorylated via the classical MAPK cascade in LPS-stimulated macrophages
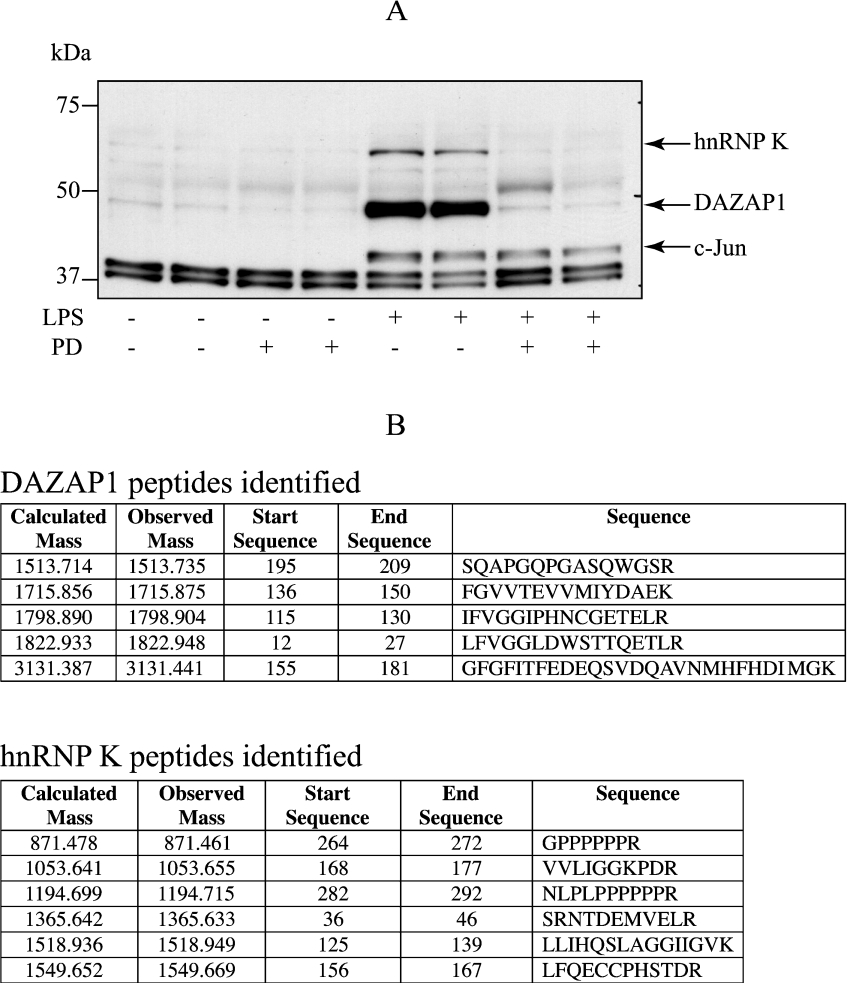
While re-investigating the multisite phosphorylation of the transcription factor c-Jun [30], we identified two proteins, distinct from c-Jun, that were present in c-Jun immunoprecipitates and were recognized ‘non-specifically’ by another antibody that detects c-Jun only when it is phosphorylated at Thr91 (Figure 1A). However, in contrast with the LPS-induced phosphorylation of c-Jun at Thr91, which is catalysed by the MAPK family member c-Jun N-terminal kinase and therefore unaffected by PD184352, a specific inhibitor of MKK1, the LPS-induced phosphorylation of the other two proteins was prevented by this inhibitor (Figure 1A). Control experiments in which the antibody used to immunoprecipitate c-Jun was omitted then revealed that, unlike c-Jun, the other two proteins still adhered to Protein G–Sepharose, demonstrating that they were not c-Jun-interacting proteins (results not shown).
Figure 1. Identification of two novel proteins phosphorylated in response to LPS in RAW 264.7 macrophages.
(A) Macrophages were incubated for 1 h with or without 2 μM PD184352 and then stimulated for 30 min with LPS (100 ng/ml) and lysed. c-Jun, immunoprecipitated from the lysates, was denatured in SDS, subjected to SDS/PAGE and transferred on to nitrocellulose. The membranes were immunoblotted with an antibody that recognizes c-Jun phosphorylated at Thr91. This detected not only c-Jun, but also two other proteins indicated by arrows that were identified in (B) as DAZAP1 and hnRNP K. (B) RAW 264.7 cell extract (2 mg of protein) was incubated for 1 h at 4 °C with Protein G–Sepharose (60 μl) and the immunoprecipitation carried out as described in the Experimental section. The stained protein bands corresponding to the proteins recognized by the anti-phospho-c-Jun(Thr91) antibody in (A), were excised, alkylated with iodoacetamide and digested with trypsin. The resultant peptides were analysed by MALDI-TOF/TOF (matrix-assisted laser-desorption ionization–tandem time-of-flight) MS or LCMS (liquid chromatography MS) as described previously [44].
The two proteins were purified from the RAW 264.7 cell extracts by affinity chromatography with Protein G–Sepharose followed by SDS/PAGE, and were identified as DAZAP1 and hnRNP K by tryptic mass fingerprinting (Figure 1B). It has previously been established that hnRNP K is a physiological substrate for ERK1/ERK2 [31], the two MAPKs that are activated by MKK1, the target of PD184352. In contrast, DAZAP1 was a novel downstream target for this pathway and therefore subjected to a more detailed analysis.
DAZAP1 is phosphorylated at Thr269 and Thr315 by ERK2 in vitro and binds to the ARE of the mRNA encoding TNFα
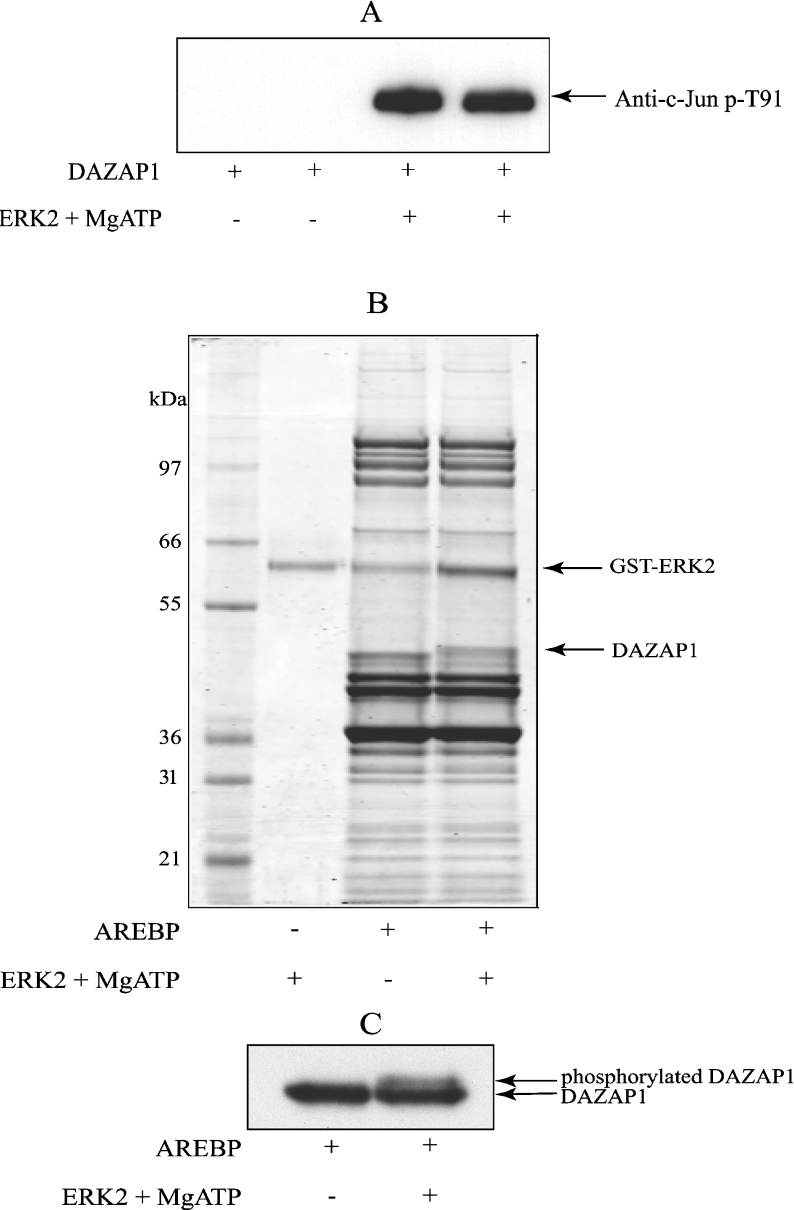
MAPKs nearly always phosphorylate serine or threonine residues that are followed by a proline, suggesting that the c-Jun Thr91 phospho-specific antibody might be recognizing a phospho-Thr-Pro sequence present in DAZAP1 and that the phosphorylation of this site might be catalysed by ERK1/ERK2. We therefore expressed and purified DAZAP1 and examined whether it could be phosphorylated by ERK2 in vitro. As shown in Figure 2(A), this proved to be the case, as incubation with ERK2 and MgATP led to the recognition of this protein by the anti-Thr91 antibody (Figure 2A).
Figure 2. Phosphorylation of DAZAP1 in vitro by the MAPK ERK2.
(A) Bacterially expressed GST–DAZAP1 was phosphorylated with ERK2, subjected to SDS/PAGE and immunoblotted with the anti-phospho-Thr91 antibody as in Figure 1(A). (B, C) AREBPs were purified from 5 mg of RAW 264.7 macrophage extract by affinity chromatography [17], then phosphorylated as described in (A), subjected to SDS/PAGE, and the proteins were revealed by staining with colloidal Coomassie Blue (B) or immunoblotted with an anti-DAZAP-1 antibody that recognizes DAZAP-1 phosphorylated and unphosphorylated forms equally well (C).
We have previously identified DAZAP1 as a protein that binds specifically to the ARE of the mRNA encoding TNFα [17]. In order to examine whether DAZAP1 could still be phosphorylated by ERK2 when bound to this ARE, we incubated the immobilized AREBPs from RAW 264.7 macrophages with MgATP and ERK2. As shown in Figure 2(B), this led to a decrease in the mobility of DAZAP1 during SDS/PAGE, indicative of stoichiometric phosphorylation at one or more sites. MS analysis established that the band whose mobility was decreased by phosphorylation with ERK2 was indeed DAZAP1 (Figure 2B, and results not shown). Immunoblotting also showed that DAZAP1 underwent a decrease in electrophoretic mobility after phosphorylation by ERK2 (Figure 2C). Strikingly, DAZAP1 was the only major AREBP that underwent this band shift. After phosphorylation by ERK2, DAZAP1 was still retained by the ARE-conjugated beads, indicating that phosphorylation did not disrupt its ability to interact with this element (results not shown).
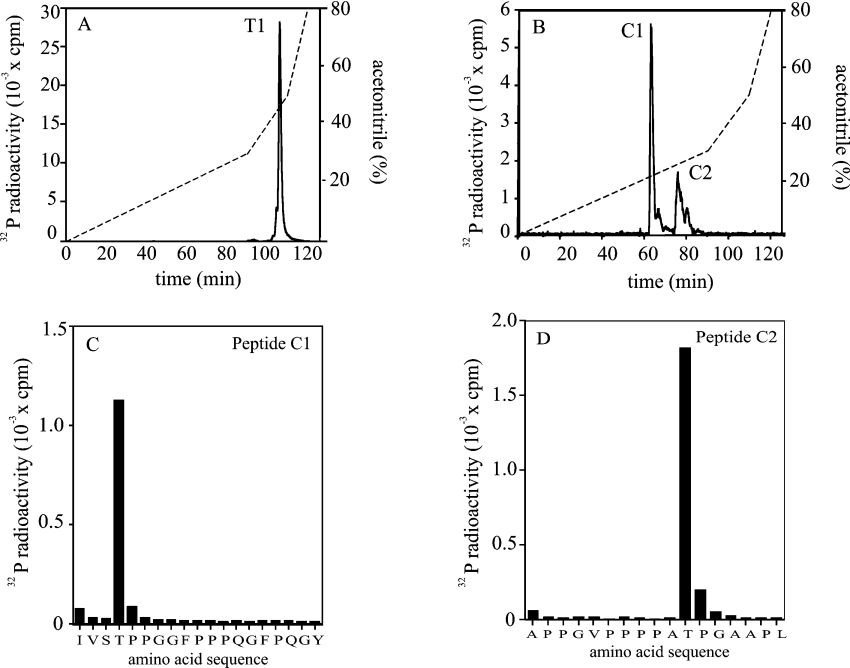
DAZAP1 that had been phosphorylated maximally by ERK2 was digested with trypsin and the resulting peptides were separated on a C18 column. One 32P-labelled peptide was detected, which was eluted at nearly 50% acetonitrile (Figure 3A), indicating that it was very large. As the size precluded identification of the site(s) of phosphorylation by MS, it was re-digested with chymotrypsin. Chromatography was carried out again on the C18 column and now resolved two 32P-labelled peptides, denoted C1 and C2 (Figure 3B). Analysis by MS showed that C1 and C2 were the peptides comprising residues 266–284 and 305–321 of DAZAP1, each containing one covalently bound phosphate (results not shown). Solid-phase sequencing revealed that the sites of phosphorylation in peptides C1 and C2 were located at Thr269 and Thr315 respectively (Figures 3C and 3D).
Figure 3. Identification of the residues on DAZAP1 phosphorylated by ERK2.
GST–DAZAP1 was maximally phosphorylated with GST–ERK2, as in Figure 2, and subjected to SDS/PAGE. The band corresponding to 32P-labelled DAZAP1 was revealed by staining with Coomassie Blue, excised and digested with trypsin. (A) The tryptic phosphopeptides were separated by HPLC on a Vydac C18 column equilibrated in 0.1% (v/v) trifluoroacetic acid. The column was developed with an acetonitrile gradient in 0.1% (v/v) trifluoroacetic acid (broken line). Radioactivity is indicated by the continuous line. (B) The major 32P-labelled peptide T1 from (A) was re-digested with chymotrypsin and re-chromatographed on the C18 column as in (A). Peptide C1 (C) and peptide C2 (D) were subjected to solid-phase sequencing to determine the sites of phosphorylation [45]. Amino acid sequences shown are those inferred from MS analysis.
DAZAP1 is phosphorylated in cells by ERK1/ERK2 at Thr269 and Thr315
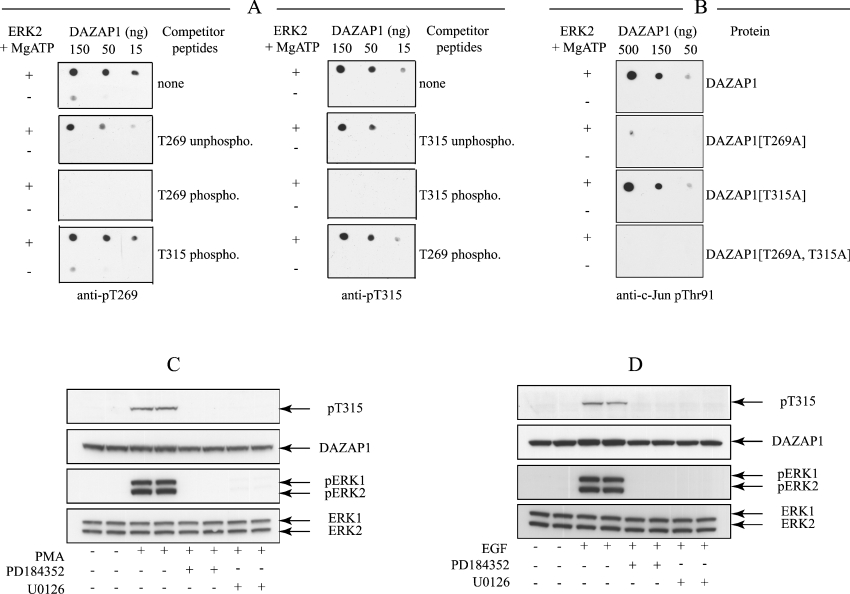
In order to investigate whether DAZAP1 was phosphorylated at Thr269 and Thr315 in cells, we generated phospho-specific antibodies that recognize these phosphorylated epitopes. The anti-phospho-Thr269 antibody recognized DAZAP1 only after phosphorylation by ERK2 and was neutralized by the phosphopeptide immunogen, but not by the unphosphorylated form of the peptide or a distinct peptide containing the Thr315 phosphorylated site (Figure 4A). Similarly, the anti-phospho-Thr315 antibody recognized DAZAP1 only after phosphorylation by ERK2 and was neutralized by the phosphopeptide immunogen, but not by the unphosphorylated form of the peptide or a distinct peptide containing the Thr269 phosphorylated site (Figure 4A). The antiphospho-Thr269 antibody was used in the presence of the unphosphorylated form of the peptide immunogen, because, in its absence, it showed slight cross-reactivity with unphosphorylated DAZAP1 (Figure 4A, top left-hand panel). Interestingly, the antibody that recognized c-Jun phosphorylated at Thr91 only recognized DAZAP1 phosphorylated at Thr269, as shown by experiments using variants of DAZAP1 in which either Thr269 or Thr315 had been replaced by an alanine residue. (Figure 4B). The recognition of DAZAP1 by this antibody in LPS-stimulated macrophages (Figure 1A) therefore establishes that DAZAP1 is phosphorylated by ERK1/ERK2 at Thr269 in these cells. Similarly, the anti-phospho-Thr315 antibody also recognized DAZAP1 in PMA- (Figure 4C) or EGF-stimulated (Figure 4D) HEK-293 cells. In each case phosphorylation was prevented by PD184352 or U0126, the latter being another MKK1 inhibitor that is structurally unrelated to PD184352. Thus DAZAP1 becomes phosphorylated at both Thr269 and Thr315 by ERK1/ERK2 in response to agonists that activate the classical MAPK cascade.
Figure 4. DAZAP1 is phosphorylated in cells at Thr269 and Thr315 in response to PMA or EGF.
(A) GST–DAZAP1 (0.3 mg/ml) was maximally phosphorylated with ERK2 in the presence (+) or absence (−) of MgATP. The indicated amounts of each preparation were spotted on to a nitrocellulose membrane and immunoblotted with the antibodies that recognized DAZAP1 phosphorylated at either Thr269 (anti-pT269) or Thr315 (anti-pT315) in the absence (none) or presence of 10 μg/ml of the unphosphorylated (unphospho) or phosphorylated (phospho) peptide antigen or the phosphorylated peptide antigen raised against the other phosphorylation site on DAZAP1. (B) The wild-type or mutant forms of DAZAP1 shown were phosphorylated as in (A), spotted on to nitrocellulose membranes and immunoblotted with the phospho-specific antibody that recognises c-Jun phosphorylated at Thr91 (anti-c-Jun pThr91) in the presence of 10 μg/ml of the unphosphorylated form of the phosphopeptide immunogen [30]. (C, D) HEK-293 cells were pre-incubated for 1 h without or with 2 μM PD184352 or 10 μM U0126, then stimulated for 30 min with 100 ng/ml PMA (C) or for 10 min with 100 ng/ml EGF (D) and lysed. Cell lysates were denatured in SDS, subjected to SDS/PAGE, transferred on to nitrocellulose membranes and immunoblotted with an antibody that recognized DAZAP1 phosphorylated at Thr315 (pT315) and with an antibody that recognises the phosphorylated and unphosphorylated forms of DAZAP1 equally well (DAZAP1). Further samples of the cell lysates were immunoblotted using antibodies that recognized phosphorylated ERK1 and ERK2 (pERK1, pERK2) or the phosphorylated and unphosphorylated forms of ERK1/ERK2 equally well (ERK1, ERK2). Similar results were obtained three different experiments.
Phosphorylation of DAZAP1 induces its dissociation from DAZ
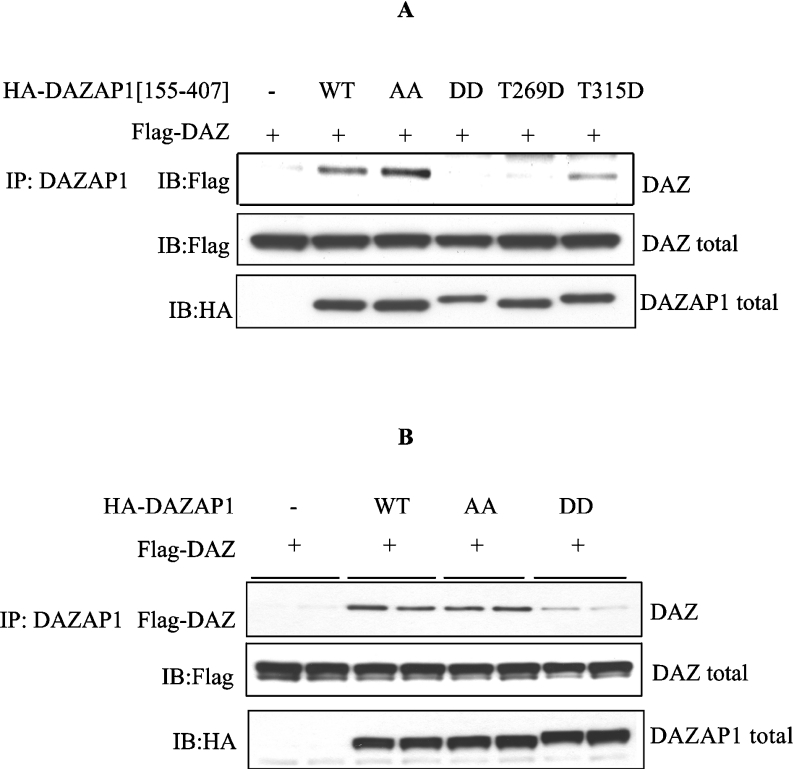
DAZAP1 was originally identified as a binding partner for DAZ, a protein whose gene is frequently deleted in cases of male sterility [32]. Since the phosphorylation of DAZAP1 did not disrupt its ability to interact with the ARE, as described above, we next examined whether phosphorylation affected its ability to bind to DAZ. Similar to DAZAP1, DAZ is an RNA-binding protein containing RRMs (RNA-recognition motifs), and such RNA-binding motifs can sometimes act as protein–protein interaction motifs [33,34]. Therefore initial experiments were carried out using an N-terminally truncated form of DAZAP1 [DAZAP1-(155–407)], which lacks its RNA-binding motifs, but retains the region thought to interact with DAZ [32,35]. In these experiments, we co-transfected FLAG–DAZ along with forms of HA–DAZAP1 in which Thr269 and/or Thr315 had been mutated to Ala to prevent phosphorylation or to Asp to mimic phosphorylation by introducing a negative charge at the same position. After immunoprecipitation of unmutated DAZAP1(155–407) from the cell extracts, DAZ was present in the anti-HA immunoprecipitates, confirming that DAZ interacts with DAZAP1 outwith the RNA-binding regions [32]. DAZ was also present in the anti-HA immunoprecipitates of DAZAP1(155–407) in which the two phosphorylation sites of DAZAP1 were mutated to Ala, but was absent when these residues had been mutated to Asp (Figure 5A). Further experiments using mutants of DAZAP1 in which either Thr269 or Thr315 was changed to Asp showed that each single mutation greatly reduced the interaction with DAZ, although the Thr269→Asp mutation had the strongest effect (Figure 5A). The combined mutation of Thr269 and Thr315 to Asp also decreased significantly the interaction between full-length DAZAP1 and DAZ (Figure 5B). Taken together, these experiments suggested that the phosphorylation of DAZAP1 might induce its dissociation from DAZ.
Figure 5. A phospho-mimetic form of DAZAP1 does not interact with DAZ.
(A) HEK-293 cells were transfected without (−) or with 2.5 μg of FLAG-DAZ plasmid DNA (+), 2.5 μg of DNA expressing unmodified HA–DAZAP1(155–407) (WT), HA–DAZAP1(155–407) in which Thr269 and Thr315 had both been mutated to Ala (AA), HA–DAZAP1(155–407) in which Thr269 and Thr315 had both been mutated to Asp (DD) or singly mutated forms of DAZAP1 in which either Thr269 or Thr315 had been mutated to Asp (T269D and T315D). After 36 h, DAZAP1 was immunoprecipitated (IP) from 1 mg of cell extract protein with an anti-HA antibody and immunoblotted (IB) with anti-HA and anti-FLAG antibodies. (B) As described for (A), except that the cells were transfected with full-length forms of DAZAP1.
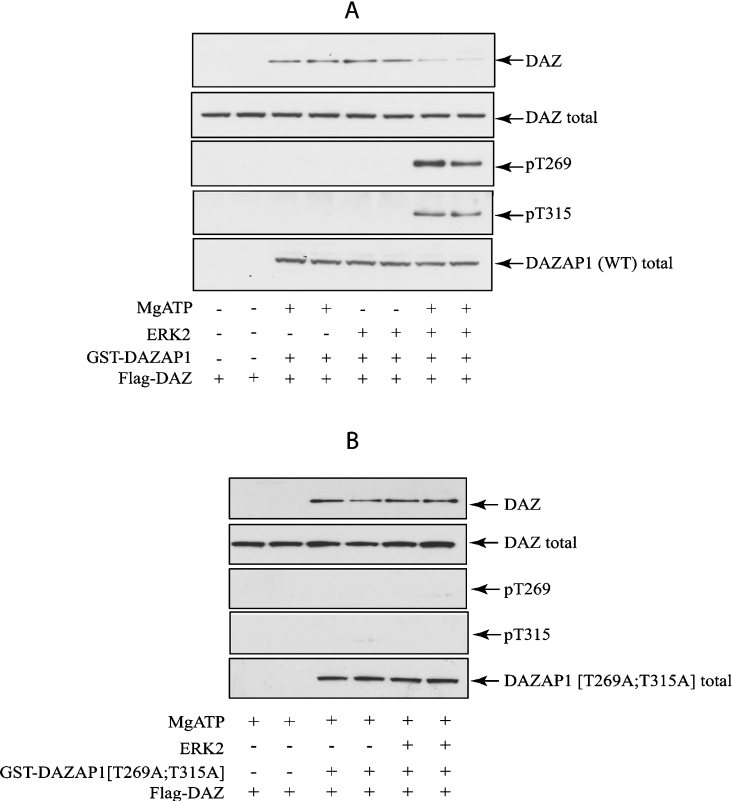
In order to investigate whether phosphorylation did indeed trigger the dissociation of these two proteins, vectors expressing GST–DAZAP1 or GST–DAZAP1(T269A/T315A) and FLAG–DAZ were co-transfected into HEK-293 cells. After purification on glutathione–Sepharose, the immobilized DAZAP1 and its associated DAZ were incubated with MgATP and ERK2. This caused a substantial release of DAZ from DAZAP1 in comparison with control incubations performed in the absence of MgATP and/or ERK2 (Figure 6A). However, this dissociation did not occur when the non-phosphorylatable form of DAZAP1(T269A/T315A) was co-transfected with DAZ (Figure 6B).
Figure 6. Phosphorylation of full-length DAZAP1 by ERK2 impairs its binding to DAZ.
HEK-293 cells were transfected with 1 μg of DNA encoding FLAG–DAZ and with 0.5 μg of DNA encoding GST–DAZAP1(WT) (A) or a non-phosphorylatable form GST–DAZAP1(T269A/T315A) (B). After 36 h, the cells were lysed and the extract (1 mg of protein) was incubated for 2 h at 4 °C with glutathione–Sepharose (20 μl). The beads were washed, and phosphorylated with ERK2 in the presence (+) or absence (−) of MgATP, before being washed again and denatured in SDS. Protein released from the beads was subjected to SDS/PAGE, transferred on to nitrocellulose membranes and immunoblotted. The GST–DAZAP1 in the extract (DAZAP1 total) was revealed with an anti-GST-antibody (DAZAP1) and FLAG–DAZ with an anti-FLAG antibody (DAZ). The membranes were also immunoblotted with the phospho-specific antibody that recognises DAZAP1 phosphorylated at Thr315 (pT315) and with the phospho-specific antibody that not only recognizes c-Jun phosphorylated at Thr91, but also DAZAP1 phosphorylated at Thr269 (pT269).
Evidence that DAZAP1 cannot bind simultaneously to both DAZ and PABP1
It has recently been reported that DAZ also interacts with PABP1, thereby stimulating the translation of mRNAs containing short poly(A) tails in Xenopus oocytes [26]. It was therefore of interest to investigate whether DAZ could interact with DAZAP1 and PABP1 simultaneously. DNA expressing epitope-tagged versions of each protein were therefore co-transfected into HEK-293 cells, and PABP1 or DAZAP1 was immunoprecipitated from the extracts. Interestingly, DAZAP1 was absent from PABP1 immunoprecipitates (Figure 7A) and PABP1 was absent from DAZAP1 immunoprecipitates (Figure 7B), although DAZ was present in both. These results, which imply that the binding of DAZAP1 and PABP to DAZ is mutually exclusive, are considered further in the Discussion.
Figure 7. DAZAP1 and PABP exist as distinct complexes with DAZ.
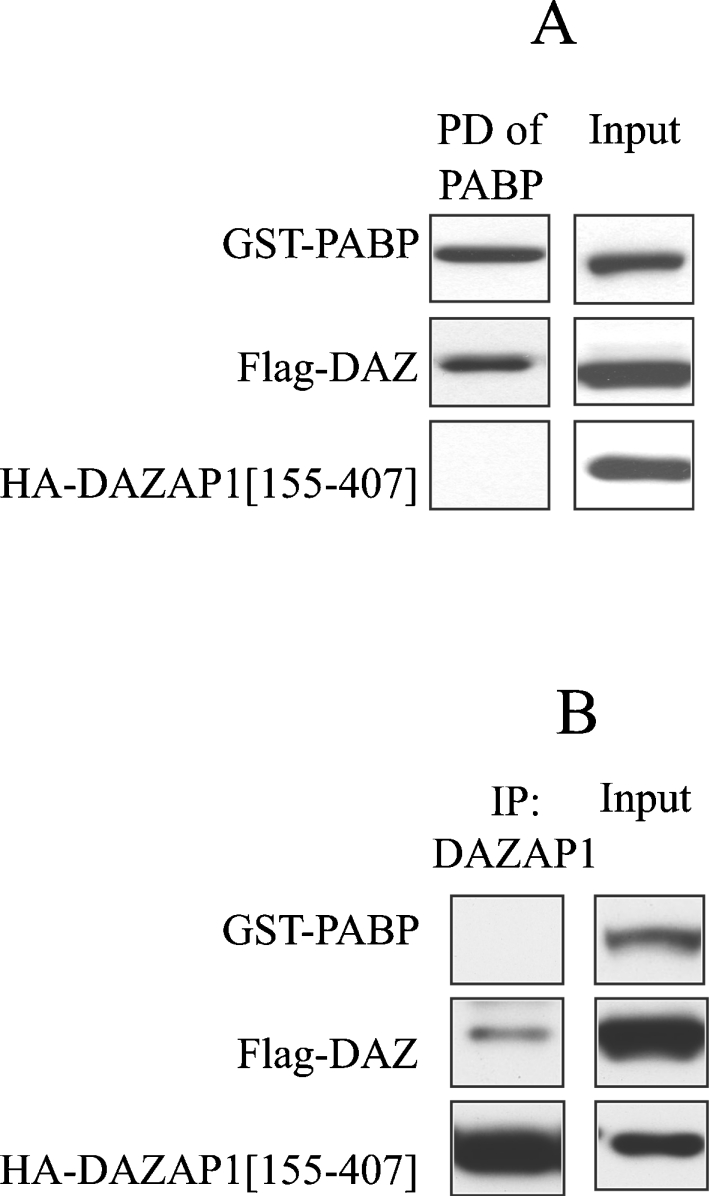
(A) HEK-293 cells were transfected with 0.2 μg of DNA encoding FLAG–DAZ, 1 μg of DNA encoding GST–PABP or 1 μg of DNA encoding HA–DAZAP1(155–407). After 48 h, the cells were lysed, and 1 mg of extract protein was incubated for 1 h with 10 μl of glutathione–Sepharose beads to pull down (PD) GST–PABP. After extensive washing (see the Experimental section), the proteins were released from the beads by denaturation in SDS and immunoblotted with anti-HA antibody to detect DAZAP1, anti-GST antibody to detect PABP or anti-FLAG antibody to detect DAZ. (B) As described for (A), except that DAZAP1 was immunoprecipitated (IP) with an anti-HA antibody.
DISCUSSION
In the present study, we identified DAZAP1 as a new physiological substrate for ERK1/ERK2 because of its cross-reactivity with an antibody that recognizes the Thr91 phosphorylation site on c-Jun and by its ‘non-specific’ binding to Protein G–Sepharose. We have found that approx. 2% of the proteins in cell extracts are pulled down by Protein G–Sepharose, many of which we have identified by MS as RNA-binding proteins (results not shown). Indeed the ‘sticky’ nature of these proteins has been well documented, and procedures for minimizing their interaction with chromatographic matrices have been published [36]. Interestingly, we have previously identified DAZAP1 as one of the more abundant proteins that interacts specifically with the ARE of the TNFα mRNA [17] and the c-Fos mRNA (S. Rousseau, unpublished work), and other investigators have shown that its RRMs bind preferentially to poly(U) RNA homopolymers compared with poly(G) or poly(A) [32]. These findings raise the possibility that DAZAP1 might play a role in the regulation of mRNA stability/translation by the classical MAPK cascade.
DAZAP1 was first identified as a protein that interacts with DAZ and the structurally related DAZL (DAZ-like protein) [32]. DAZ and DAZL, as well as another family member called BOULE, are all highly expressed in germ cells. Indeed, the DAZ gene is located on the Y chromosomes of humans, great apes and Old World monkeys [37–39], and is frequently deleted in male sterility [40]. There is evidence that members of this family play a role in stimulating protein translation. For example, in Drosophila, BOULE is essential for the translation of the mRNA encoding Twine, a Cdc25-like phosphatase involved in the regulation of meiosis [41]. Moreover, DAZ and DAZL have been found associated with polyribosomes that are actively engaged in protein synthesis [42]. Furthermore, DAZ, DAZL and BOULE have all been shown to interact with PABP in Xenopus oocytes and thereby to stimulate the translation of mRNAs with short poly(A) tails [26].
The key finding of our present study is that the phosphorylation of DAZAP1 at Thr269 and Thr315 does not disrupt its interaction with the ARE, but induces its dissociation from DAZ. This is consistent with the location of the phosphorylation sites, which are in a proline-rich region located C-terminal to the RRMs. Deletion of the RRMs disrupts RNA binding [32], but not the interaction with DAZ (Figure 5), indicating that the sites of phosphorylation may be in the vicinity of the DAZ interaction site [32,35].
The observation that phosphorylation of DAZAP1 induces its dissociation from DAZ suggested that this might regulate the function of either or both proteins. For example, it could be envisaged that the DAZ–DAZAP1 complex confers mRNA instability by binding to the ARE and/or prevents DAZ from interacting with other binding partners. Following the phosphorylation of DAZAP1, the ensuing release of DAZ may reduce mRNA instability and now allow DAZ to interact with other proteins involved in promoting translation, such as PABP. Additionally, the interaction of DAZ with PABP may further confer mRNA stability by promoting mRNA circularization (see the Introduction). Consistent with this hypothesis, we found that DAZ could not bind simultaneously to both DAZAP1 and PABP (Figure 7), and others have reported that, in contrast with PABP, DAZ and DAZL, DAZAP1 is absent from polyribosomes [35].
DAZ and DAZL have so far only been found in germ cells, whereas BOULE has also been found in the nervous system of Drosophila [43]. In contrast, DAZAP1 is expressed not only in the tissues where members of the DAZ family are present, but also in many other tissues [35], as demonstrated by our original identification of DAZAP1 in RAW 264.7 macrophages [17] (Figure 1A). As no other protein with DAZ motifs has been found by searching databases (results not shown), this raises the interesting question as to the identity of the binding partner(s) for DAZAP1 in macrophages and other cells which do not express DAZL. To understand if and how the ERK1/ERK2-catalysed phosphorylation of DAZAP1 is involved in regulating the stability and/or translation of ARE-containing mRNAs encoding pro-inflammatory cytokines, which is the major interest of our laboratory (see the Introduction), it is clearly now critical to find its binding partners in macrophages.
Acknowledgments
We thank the Protein Production and Antibody Purification teams, Division of Signal Transduction Therapy, University of Dundee (co-ordinated by Hilary McLauchlan and James Hastie), for expression and purification of proteins and affinity purification of antibodies, Dr Mark Peggie for making several DNA constructs, and the Sequencing Service, School of Life Sciences, University of Dundee (www.dnaseq.co.uk), for DNA sequencing. We are grateful to the U.K. Medical Research Council, The Royal Society, AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co, Merck KGaA and Pfizer for financial support.
References
- 1.Feldmann M., Maini R. N. TNF defined as a therapeutic target for rheumatoid arthritis and other autoimmune diseases. Nat. Med. 2003;9:1245–1250. doi: 10.1038/nm939. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart D. C., Wiedenmann B., Dignass A. U. Biologic therapy of inflammatory bowel disease. Z. Gastroenterol. 2003;41:1017–1032. doi: 10.1055/s-2003-42924. [DOI] [PubMed] [Google Scholar]
- 3.Allan L. M., Ballard C. G., Burn D. J., Kenny R. A. Prevalence and severity of gait disorders in Alzheimer's and non-Alzheimer's dementias. J. Am. Geriatr. Soc. 2005;53:1681–1687. doi: 10.1111/j.1532-5415.2005.53552.x. [DOI] [PubMed] [Google Scholar]
- 4.Janssens S., Beyaert R. Role of Toll-like receptors in pathogen recognition. Clin. Microbiol. Rev. 2003;16:637–646. doi: 10.1128/CMR.16.4.637-646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature (London) 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 6.Lee J., Mira-Arbibe L., Ulevitch R. J. TAK1 regulates multiple protein kinase cascades activated by bacterial lipopolysaccharide. J. Leukocyte Biol. 2000;68:909–915. [PubMed] [Google Scholar]
- 7.Dumitru C. D., Ceci J. D., Tsatsanis C., Kontoyiannis D., Stamatakis K., Lin J. H., Patriotis C., Jenkins N. A., Copeland N. G., Kollias G., Tsichlis P. N. TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell (Cambridge, Mass.) 2000;103:1071–1083. doi: 10.1016/s0092-8674(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 8.Eliopoulos A. G., Wang C. C., Dumitru C. D., Tsichlis P. N. Tpl2 transduces CD40 and TNF signals that activate ERK and regulates IgE induction by CD40. EMBO J. 2003;22:3855–3864. doi: 10.1093/emboj/cdg386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung P. C., Campbell D. G., Nebreda A. R., Cohen P. Feedback control of the protein kinase TAK1 by SAPK2a/p38α. EMBO J. 2003;22:5793–5805. doi: 10.1093/emboj/cdg552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kontoyiannis D., Pasparakis M., Pizarro T. T., Cominelli F., Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 11.Neininger A., Kontoyiannis D., Kotlyarov A., Winzen R., Eckert R., Volk H. D., Holtmann H., Kollias G., Gaestel M. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 2002;277:3065–3068. doi: 10.1074/jbc.C100685200. [DOI] [PubMed] [Google Scholar]
- 12.Winzen R., Gowrishankar G., Bollig F., Redich N., Resch K., Holtmann H. Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol. Cell. Biol. 2004;24:4835–4847. doi: 10.1128/MCB.24.11.4835-4847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang T., Kruys V., Huez G., Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 14.Dean J. L., Wait R., Mahtani K. R., Sully G., Clark A. R., Saklatvala J. The 3′ untranslated region of tumor necrosis factor α mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 16.Lai W. S., Carballo E., Strum J. R., Kennington E. A., Phillips R. S., Blackshear P. J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousseau S., Morrice N., Peggie M., Campbell D. G., Gaestel M., Cohen P. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 2002;21:6505–6514. doi: 10.1093/emboj/cdf639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C. Y., Gherzi R., Ong S. E., Chan E. L., Raijmakers R., Pruijn G. J., Stoecklin G., Moroni C., Mann M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell (Cambridge, Mass.) 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 19.Fan X. C., Steitz J. A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myer V. E., Fan X. C., Steitz J. A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng S. S., Chen C. Y., Xu N., Shyu A. B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachs A. B., Sarnow P., Hentze M. W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell (Cambridge, Mass.) 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 23.Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 24.Lee J. C., Laydon J. T., McDonnell P. C., Gallagher T. F., Kumar S., Green D., McNulty D., Blumenthal M. J., Heys J. R., Landvatter S. W., et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature (London) 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 25.Chrestensen C. A., Schroeder M. J., Shabanowitz J., Hunt D. F., Pelo J. W., Worthington M. T., Sturgill T. W. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 26.Collier B., Gorgoni B., Loveridge C., Cooke H. J., Gray N. K. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Sebolt-Leopold J. S., Dudley D. T., Herrera R., Van Becelaere K., Wiland A., Gowan R. C., Tecle H., Barrett S. D., Bridges A., Przybranowski S., et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nat. Med. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- 27.Shpiro N., Marquez R. An improved synthesis of the potent MEK inhibitor PD 184352. Synth. Commun. 2005;35:2265. [Google Scholar]
- 28.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton S., Davis R. J., McLaren A., Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2003;22:3876–3886. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A. L., Shokat K. M., Hentze M. W., Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 32.Tsui S., Dai T., Roettger S., Schempp W., Salido E. C., Yen P. H. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 33.Ding J., Hayashi M. K., Zhang Y., Manche L., Krainer A. R., Xu R. M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J. H., Hahm B., Kim Y. K., Choi M., Jang S. K. Protein–protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 2000;298:395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- 35.Dai T., Vera Y., Salido E. C., Yen P. H. Characterization of the mouse Dazap1 gene encoding an RNA-binding protein that interacts with infertility factors DAZ and DAZL. BMC Genomics. 2001;2:6. doi: 10.1186/1471-2164-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryder U., Sproat B. S., Lamond A. I. Sequence-specific affinity selection of mammalian splicing complexes. Nucleic Acids Res. 1990;18:7373–7379. doi: 10.1093/nar/18.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxena R., Brown L. G., Hawkins T., Alagappan R. K., Skaletsky H., Reeve M. P., Reijo R., Rozen S., Dinulos M. B., Disteche C. M., Page D. C. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat. Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 38.Shinka T., Nakahori Y. The azoospermic factor on the Y chromosome. Acta Paediatr. Jpn. 1996;38:399–404. doi: 10.1111/j.1442-200x.1996.tb03514.x. [DOI] [PubMed] [Google Scholar]
- 39.Yen P. H. Putative biological functions of the DAZ family. Int. J. Androl. 2004;27:125–129. doi: 10.1111/j.1365-2605.2004.00469.x. [DOI] [PubMed] [Google Scholar]
- 40.Reijo R., Lee T. Y., Salo P., Alagappan R., Brown L. G., Rosenberg M., Rozen S., Jaffe T., Straus D., Hovatta O., et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat. Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 41.Maines J. Z., Wasserman S. A. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat. Cell Biol. 1999;1:171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- 42.Tsui S., Dai T., Warren S. T., Salido E. C., Yen P. H. Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol. Reprod. 2000;62:1655–1660. doi: 10.1095/biolreprod62.6.1655. [DOI] [PubMed] [Google Scholar]
- 43.Joiner M. L., Wu C. F. Nervous system function for the testis RNA-binding protein boule in Drosophila. J. Neurogenet. 2004;18:341–363. doi: 10.1080/01677060490477435. [DOI] [PubMed] [Google Scholar]
- 44.Pozuelo Rubio M., Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell D. G., Morrice N. M. Identification of protein phosphorylation sites by a combination of mass spectrometry and solid phase Edman sequencing. J. Biomol. Tech. 2002;13:119–130. [PMC free article] [PubMed] [Google Scholar]