Abstract
In recent decades, the prevalence of Neisseria meningitidis isolates with reduced susceptibility to penicillins has increased. The intermediate resistance to penicillin (Peni) for most strains is due mainly to mosaic structures in the penA gene, encoding penicillin-binding protein 2. In this study, susceptibility to β-lactam antibiotics was determined for 60 Swedish clinical N. meningitidis isolates and 19 reference strains. The penA gene was sequenced and compared to 237 penA sequences from GenBank in order to explore the total identified variation of penA. The divergent mosaic alleles differed by 3% to 24% compared to those of the designated wild-type penA gene. By studying the final 1,143 to 1,149 bp of penA in a sequence alignment, 130 sequence variants were identified. In a 402-bp alignment of the most variable regions, 84 variants were recognized. Good correlation between elevated MICs and the presence of penA mosaic structures was found especially for penicillin G and ampicillin. The Peni isolates comprised an MIC of >0.094 μg/ml for penicillin G and an MIC of >0.064 μg/ml for ampicillin. Ampicillin was the best antibiotic for precise categorization as Pens or Peni. In comparison with the wild-type penA sequence, two specific Peni sites were altered in all except two mosaic penA sequences, which were published in GenBank and no MICs of the corresponding isolates were described. In conclusion, monitoring the relationship between penA sequences and MICs to penicillins is crucial for developing fast and objective methods for susceptibility determination. By studying the penA gene, genotypical determination of susceptibility in culture-negative cases can also be accomplished.
Neisseria meningitidis (meningococci) is a widespread human pathogen causing meningitis and septicemia (16). During the last decades, there have been several reports from different countries of N. meningitidis with reduced susceptibility to penicillin G (6, 14). This is of general concern since penicillin is the first-line antibiotic for treatment of meningococcal disease (17). The intermediate resistant isolates, Peni, have previously been defined by MICs of >0.064 μg/ml to ≤1.0 μg/ml by using Etest (8) and MICs of >0.064 μg/ml to 0.5 μg/ml by using the agar dilution method (7, 9). This intermediate resistance has been reported to be due mainly to alterations in the structure of penicillin-binding protein 2 (PBP2), encoded by the penA gene (3, 20). The penA genes of susceptible isolates, the so-called wild-type penA gene (1,745 bp total in MC58 [23]), seem to be highly conserved in their DNA sequence. However, the genes of Peni isolates are fairly variable and highly divergent from the wild-type penA gene (3, 9, 21). These variations have been suggested to be due to genetic exchange through transformation between N. meningitidis and nonpathogenic commensal neisserial species, for example, Neisseria flavescens (19). Due to this transformation, the penA gene of Peni isolates has a mosaic structure, consisting of regions that are essentially identical to those in susceptible isolates and regions that are 14% to 23% divergent in sequence (20). The polymorphisms are located mainly in the last two-thirds of the gene that encodes about 400 amino acids at the C-terminal part of the protein (5).
Sweden is a country with low incidence of meningococcal disease at present, i.e., an incidence of 0.5 to 0.9 cases per 100,000 inhabitants in 1997 to 2005 (The Swedish Institute for Infectious Disease Control, http://gis.smittskyddsinstitutet.se/mapapp/build/11-109000/table/Meningococci_eng_year_all.html, accessed 23 March 2006). There has been an increase in the number of invasive N. meningitidis isolates with reduced susceptibility to penicillin G during the last decade. Consequently, in 2005, 23% of the Swedish invasive isolates comprised the Peni phenotype (MIC > 0.064 μg/ml) compared to 5% in 1996 (12).
The reports of increasing numbers of circulating N. meningitidis strains with the Peni phenotype emphasize the need for fast and objective methods for the determination of susceptibility to penicillins. Hence, the aims of the present study were to explore the total reported and presently identified variation in the penA gene and to describe the detailed association between N. meningitidis penA sequences and the MICs of mainly different penicillins. This would also provide a way to determine penicillin susceptibility of culture-negative cases of meningococcal meningitis/septicemia as well as to approach the development of an objective control system for both phenotypic and genetic penicillin susceptibility testing.
Consequently, in the present study, the total variation of the penA gene in N. meningitidis was examined. In addition, the MICs of penicillin G, penicillin V, ampicillin, cefuroxime, and cefotaxime were determined to explore the correlation with altered penA genes. Penicillin G was chosen, as being considered a first-line antibiotic, along with penicillin V as well as ampicillin to evaluate whether the Peni phenotype would be easier to distinguish when their MICs were compared to penA sequences. Another reason to include penicillin V, which normally is not used for treatment of meningococcal disease, was that all commensal or carrier strains of N. meningitidis are exposed to a high pressure of antibiotics in the society, including penicillin V. Cefotaxime was included as a representative of effective cephalosporins and cefuroxime as a less satisfactory cephalosporin.
MATERIALS AND METHODS
Bacterial isolates and clinical samples.
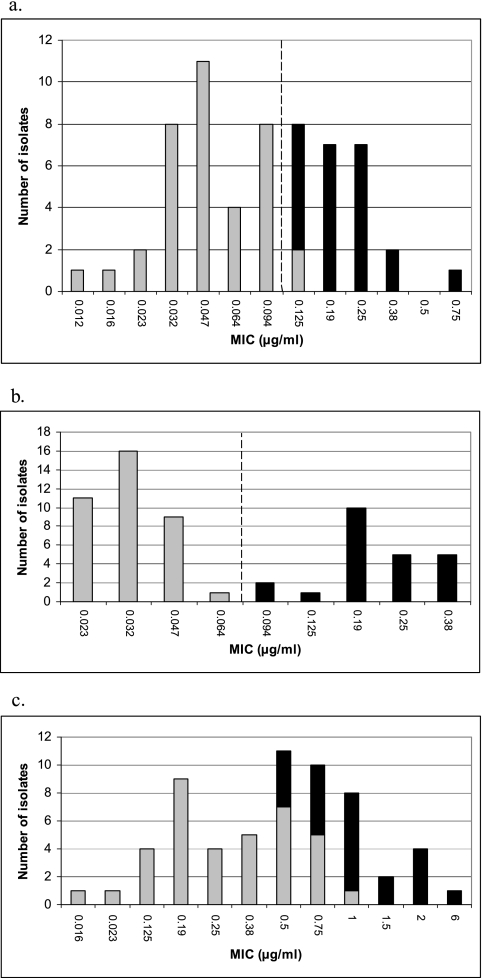
Sixty clinical isolates, invasive (n = 55) and carrier isolates (n = 5), of N. meningitidis collected in Sweden between 1996 and 2004 and 17 N. meningitidis strains, previously used in an antibiotic susceptibility study performed by the European Monitoring Group on Meningococci (EMGM) (26), were examined. The clinical isolates were selected to represent all of the different MICs of penicillin G found in Sweden (Fig. 1a). Additional isolates comprising MICs in close association to the phenotypical breakpoint for Peni were also included. Of the 60 clinical isolates, 27 were phenotypically determined to be susceptible to penicillin G (MIC ≤ 0.064 μg/ml) and 33 comprised a reduced susceptibility (Fig. 1a). For comparison, two N. meningitidis reference strains, i.e., MC58 (23) and OR173/87 (10), and one Neisseria gonorrhoeae reference strain (CCUG 15821) were included in the study.
FIG. 1.
MICs of penicillin G (a), ampicillin (b), and penicillin V (c) for 60 selected clinical N. meningitidis isolates collected in Sweden between 1996 and 2004. Gray bars indicate wild-type penA genes, and black bars indicate mosaic structures in the penA gene. The broken line in panels a and b indicates the suggested breakpoint for Peni isolates (MIC > 0.094 μg/ml for penicillin G and MIC > 0.064 μg/ml for ampicillin).
Five cerebrospinal fluid (CSF) samples derived from patients suffering from meningococcal meningitis were also examined in order to analyze whether the protocol was suitable for direct sequencing from CSF.
Phenotypical antibiotic susceptibility testing.
The MICs of penicillin G, penicillin V, ampicillin, cefuroxime, and cefotaxime were determined using the Etest method (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar (Becton, Dickinson and Company, Franklin Lakes, NJ) supplemented with 5% sheep blood at 37°C in 5% CO2 for 16 to 18 h. The breakpoints used for penicillin G were described by Hughes et al. (susceptible [S], MIC ≤ 0.064 μg/ml; resistant [R], MIC > 1 μg/ml) (8). For ampicillin, the breakpoint was described by Jorgensen et al., i.e., an MIC of <0.25 μg/ml (S) (9). For the remaining antibiotics, the breakpoints were as follows: penicillin V, MIC ≤ 1 μg/ml (S) and MIC > 1 μg/ml (R); cefuroxime, MIC ≤ 0.25 μg/ml (S) and MIC > 1 μg/ml (R); and cefotaxime, MIC ≤ 0.064 μg/ml (S) and MIC > 1 μg/ml (R) (in accordance with the Swedish Reference Group for Antibiotics, http://www.srga.org, accessed 23 March 2006).
Isolation of genomic DNA.
Isolation of bacterial DNA from N. meningitidis isolates was performed using a MagNA Pure LC system with DNA isolation kit III (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. For isolation of DNA from the five CSF samples, the samples (100 to 500 μl) were initially centrifuged (8000 × g for 10 min) and then processed in the same way as the bacterial isolates. The DNA preparations were stored at 4°C prior to PCR.
penA PCR.
The PCR was performed as previously described by Arreaza and Vázquez (5), with minor modifications. The 50-μl PCR mixture contained 1.25 U AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA) and 1 μl of the genomic DNA template. One of the primers, Mod-Gcdown3, was slightly modified compared to the primer used by Arreaza and Vázquez (Table 1) (5). The PCR conditions were as previously described except that an initial enzyme activation step at 94°C for 10 min was included and, for the CSF samples, there were 40 cycles of amplification instead of 30. MC58 was used as a positive control and distilled water as a negative control. The PCR products were analyzed by electrophoresis through a 2% agarose gel and by ethidium bromide staining. DNA molecular weight marker VI (Roche Diagnostics, Mannheim, Germany) was included on each gel.
TABLE 1.
Primers used in PCR and sequencing of the penA gene
| Primera | Sequence (5′→3′) | Use(s) | Nucleotide locationb | Source and/or reference |
|---|---|---|---|---|
| Gcup2 | TTTGCACACGTCATCGGATTTAC | PCR and sequencing | 523-542 | 5 |
| Mod-Gcdown3 | CGGGGATATAACTGCGGCCGTC | PCR and sequencing | 187-166 bp downstream of the stop codon of the penA gene | Shortened Gcdown3; 5 |
| Fo | TATACCGCACTGACGCACGAC | Sequencing | 1301-1323 | 5 |
| Ro | GCCGTCGTGCGTCAGTGC | Sequencing | 1326-1309 | 5 |
| AA-1c | ATCGAACAGGCGACGATGTC | Sequencing | 1237-1256 | 3 |
| PenA-R2c | GCCTGTTTTTCAAAGCTGACC | Sequencing | 1358-1338 | Present study |
Synthesized by Scandinavian Gene Synthesis AB, Köping, Sweden.
According to the nucleotide sequence of the penA gene in MC58 (23).
For some of the isolates, the primers Fo and Ro did not work due to polymorphism in the annealing site and AA-1 and PenA-R2 were used instead.
DNA sequencing.
The PCR products were purified using the High Pure PCR product purification kit (Roche Diagnostics, Mannheim, Germany) and then cycle sequenced. The primers used for the cycle sequencing PCR are shown in Table 1. Each sequencing reaction (10 μl) contained 4 μl of a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom), 1.6 pmol primer, and 1 μl purified PCR product. The cycle sequencing PCR consisted of 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Subsequently, the products were purified using ethanol-sodium acetate precipitation and resuspended in 10 μl formamide (Applied Biosystems, Warrington, United Kingdom) according to the manufacturer's instructions. The nucleotide sequences were determined using an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA). The sequence of each strand of each compiled sequence was determined.
Sequence analysis.
Multiple-sequence alignments (1,149 unambiguously aligned nucleotides) of the final part of the penA gene and the corresponding deduced amino acid sequences were performed using the software BioEdit (version 5.0.9) and by manual adjustment. For comparison, penA gene sequences from 26 different Peni meningococcal isolates with MICs of penicillin G ranging from 0.094 μg/ml to 1.28 μg/ml and 211 penA gene sequences from meningococcal isolates without depicted MICs, deposited in GenBank, were also included. In fact, all penA sequences with a minimum length of the final 1,143 bp deposited in GenBank (1 March 2006) were included in the alignment. A shorter multiple-sequence alignment of 402 bp in the end of the penA gene sequences, which has been suggested to be sufficient for the identification of mosaic-structured penA genes (EMGM Working Group on Antibiotics and M.-K. Taha, personal communication), was also performed.
For the identification of different sequence variants, phylogenetic trees were constructed with TREECON (version 1.3b) software as previously described (24).
RESULTS
Total variability in penA gene.
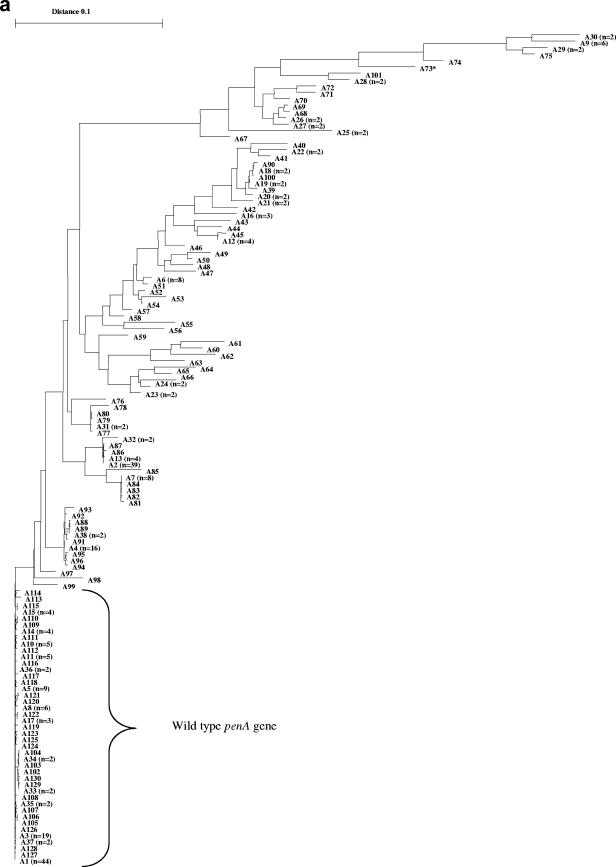
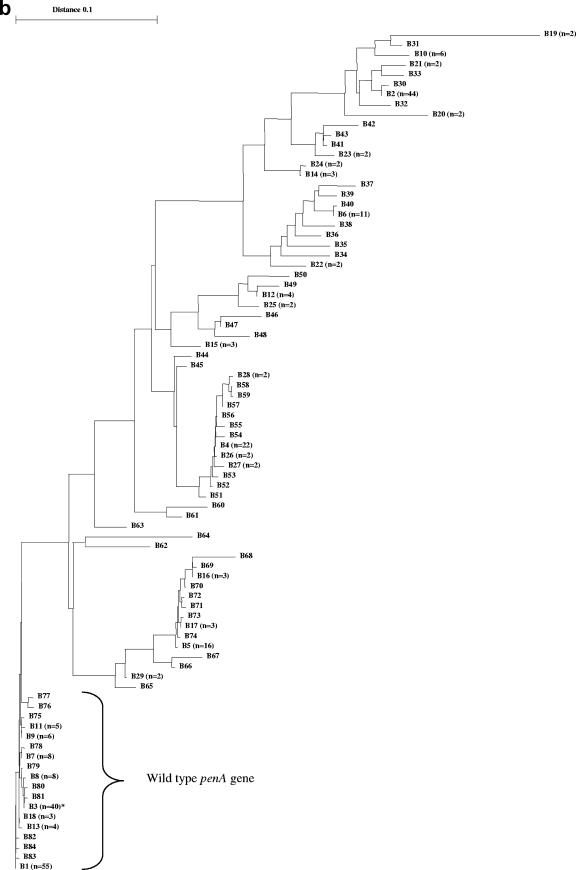
In total, 321 meningococcal penA gene sequences were examined. The phylogenetic tree of the different sequence variants (based on the 1,149-bp alignment) identified one highly homogeneous group, with only a few nucleotide polymorphisms (Fig. 2a). Within this homogeneous group, 138 sequences were found. Between these 138 sequences, there were a maximum of 1% nucleotide difference and no recognizable mosaic structures, and all of the Pens isolates were found in this group. Based on these observations, the penA gene sequences included in this group were designated wild-type penA gene sequences. Among the remaining penA gene sequences (n = 183), 22% of the nucleotides differed between the two most divergent sequences (A30 and A97) (Fig. 2a). All of these sequences (n = 183) comprised obvious mosaic structures; however, the extent of mosaic structures varied substantially between the sequences, spanning from over about 100 bp to the whole sequence examined.
FIG. 2.
(A) Phylogenetic tree based on a 1,149-bp alignment of penA gene sequences of N. meningitidis (n = 321). (B) Phylogenetic tree based on a 402-bp alignment of penA gene sequences of N. meningitidis (n = 321). In both trees, one homogeneous group, comprising wild-type penA genes, was identified. The remaining sequences in both trees were highly divergent and displayed obvious mosaic patterns. One single sequence (*, accession no. AY127670) was identified as a mosaic allele when the larger segment was examined but as the wild type when only 402 bp was examined.
When the 1,149-bp multiple-sequence alignment of the mosaic-structured penA genes (n = 183) was studied, 87 different nucleotide sequence variants, which coded for 74 PBP2 amino acid sequence variants, were identified (Fig. 2a). In the alignment, a total of 409 polymorphic nucleotide sites were identified. When the deduced amino acid sequences (383 unambiguously aligned amino acids) were studied, a total of 103 polymorphic amino acid sites were identified. In the 402-bp multiple-sequence alignment of the mosaic sequences (n = 182; one sequence was excluded due to lack of mosaic structure in this shorter segment), 66 different nucleotide sequence variants, which coded for 36 PBP2 amino acid sequence variants, were identified (Fig. 2b). One hundred forty-two polymorphic nucleotide sites and 36 polymorphic amino acid sites were identified (134 unambiguously aligned amino acids in total). For the wild-type penA sequences (n = 138), 43 different nucleotide sequence variants and 17 PBP2 amino acid sequence variants were found when the 1,149-bp alignment was studied and 18 nucleotide and 7 PBP2 variants were found when the 402-bp alignment was studied. Thirty-eight polymorphic nucleotide sites and 16 polymorphic amino acid sites were identified in the 1,149-bp alignment, and 15 polymorphic nucleotide sites and 6 polymorphic amino acid sites were identified in the 402-bp alignment.
The sequences of the divergent mosaic alleles differed by 3% to 24% compared to the wild-type sequence, represented by MC58, when 1,149 bp of the penA gene was studied. When we studied 402 bp, the difference was 7% to 23% compared to the wild-type sequence.
penA gene versus MIC.
Of the clinical isolates with the Peni phenotype (n = 33), all isolates with an MIC of >0.125 μg/ml (n = 17) and 75% of the isolates with an MIC of 0.125 μg/ml of penicillin G had mosaic structures in the penA gene (Fig. 1a). For ampicillin, all clinical isolates with an MIC of >0.064 μg/ml (n = 23) had mosaic-structured penA genes (Fig. 1b). For penicillin V, a somewhat lower correlation between MICs and mosaic-structured penA genes was found (Fig. 1c). For cefuroxime, no absolute correlation was found either; all isolates with an MIC of >0.094 μg/ml (n = 21) showed mosaic structures in the penA gene, but mosaic structures were also found in one of three isolates with an MIC of 0.094 μg/ml and one of eight isolates with an MIC of 0.064 μg/ml. All of the isolates were fully susceptible to cefotaxime, and hence, no correlation with penA gene sequences was possible to determine.
In addition, all 11 “EMGM strains” with reduced susceptibility to penicillin G also had mosaic structures in the penA gene. For ampicillin and penicillin V, all isolates with an MIC of >0.047 μg/ml (n = 11) and an MIC of >0.38 μg/ml (n = 11), respectively, had mosaic structures in the penA gene. For cefuroxime, all isolates with an MIC of >0.125 μg/ml (n = 11) showed mosaic structures, and as for the clinical isolates, no correlation was possible to determine between the low MICs of cefotaxime and penA gene sequences.
Covariation between elevated MICs of penicillin G, penicillin V, ampicillin, and, to some extent, cefuroxime could be observed. The low MICs of cefotaxime of the isolates were unaffected by the elevated MICs for the other antibiotics.
Based on the reported correlation between MIC and penA gene sequence in the present study, Peni isolates could be defined as the ones comprising an MIC of >0.094 μg/ml for penicillin G and an MIC of >0.064 μg/ml for ampicillin by using the Etest method.
All of the 26 penA sequences, collected from GenBank, from Peni strains proved to have mosaic-structured penA genes. Consequently, a total of 70 isolates with both the Peni phenotype and mosaic structures in the penA gene were examined. Among these isolates, 34 different penA nucleotide sequence variants and 31 different PBP2 amino acid sequence variants were identified when the 1,149-bp alignment was examined. In the 402-bp alignment, 27 penA nucleotide sequence variants and 15 PBP2 variants were identified. No obvious correlation between individual MICs and any specific penA sequence variants or cluster of variants was found. Consequently, no penA mosaic allele was predominant to give a particular MIC.
Peni-specific sites.
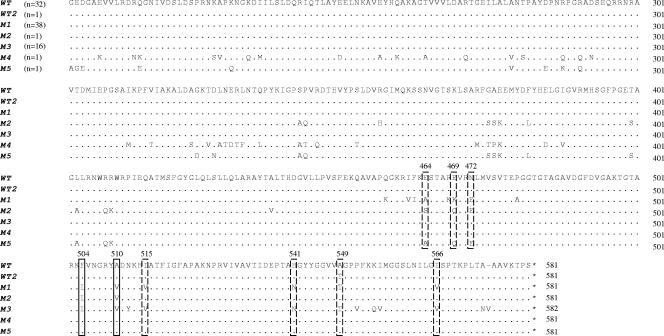
According to previous studies (2, 21, 22), in comparison with that of the wild-type penA gene, between five and nine specific nucleotides are altered in the penA mosaic allele of all Peni isolates. All of these polymorphic nucleotides result in alterations in the corresponding encoded amino acids, which are located in the C-terminal part of PBP2. In the present study, none of these five to nine specific Peni sites was altered in any of the identified divergent penA mosaic alleles. Two of the specific Peni sites were altered in all except two (accession no. AF515100 and AY127670) of the penA mosaic-structured sequences. These sequences lacked mosaic patterns in the end of the penA gene, i.e., in the last 250 bp and 550 bp of the gene, respectively, and hence, none of the specific Peni sites was altered (Fig. 3).
FIG. 3.
Multiple-sequence alignment of seven different partial PBP2 amino acid sequences, which include amino acids 202 to 581 of the native protein in MC58 (23), comprising different numbers of the amino acid alterations (in boxes) found in Peni isolates (2, 21, 22). The two consistently lined boxes indicate the alterations found in all of the phenotypically confirmed Peni isolates. The wild-type sequences (WT) are represented by MC58. WT2 is a wild-type sequence containing one of the alterations (accession no. AY292992). Mosaic sequence 1 (M1) comprises all of the alterations, M2 lacks alterations 515, 541, 549, and 566, and M3 lacks alterations 464, 469, and 472. M4 (accession no. AY127670) comprises wild-type structure in the final 550 bp of the penA gene, and M5 (accession no. AF515100) contains wild-type structure in the final 250 bp. For confirmation of the sequences, one representative of each sequence variant was PCR amplified and sequenced twice. No confirmation was possible to perform for M4 and M5, which were downloaded from GenBank.
Cerebrospinal fluid samples.
Of the five CSF samples, four were culture negative and hence could not be phenotypically analyzed regarding antibiotic susceptibility. The single culture-positive isolate was susceptible to penicillin G. The penA gene of N. meningitidis in the five CSF samples was successfully amplified and sequenced. One of the culture-negative samples had an altered penA gene with mosaic structure.
DISCUSSION
The penA genes of Peni isolates are fairly variable, in most cases, all over the last two-thirds of the gene. The large amount of more or less evenly spread alterations indicates that the mosaic-structured penA genes are most likely due to many different genetic events and/or genetic exchange with many different donors.
In the present study, two different sequence alignments were made, one of the final 1,149 bp of the penA gene and one of 402 bp in the end of the gene. One single sequence was not properly categorized as a penA mosaic allele in the 402-bp alignment because it lacked mosaic patterns in the final 550 bp of the gene. However, when the longer alignment was examined, this sequence (accession no. AY127670) was correctly categorized as a mosaic penA allele (Fig. 2a and b). This clearly illustrates the risks associated with studying only a shorter segment of a gene.
The results of phenotypic antibiotic susceptibility testing of N. meningitidis are hard to value in, for example, interlaboratory comparisons. This is due mainly to significant differences in the media and critical MICs used in different laboratories (26). In the present study, we found a good correlation between elevated MICs and the presence of mosaic-structured penA genes, especially for penicillin G and ampicillin but to a somewhat smaller extent for penicillin V and cefuroxime. Based on our findings, using Etest and penA gene sequencing, Peni isolates could be defined as the ones comprising an MIC of >0.094 μg/ml for penicillin G and an MIC of >0.064 μg/ml for ampicillin. However, these are not clinical breakpoints but epidemiologic definitions. In addition, it is important to note that the number of tested clinical isolates is rather limited (n = 60) and that the difference between MICs for penicillin G and ampicillin is very small in the present study. In an earlier study (4), Arreaza and coworkers proposed an MIC of >0.064 μg/ml to be a suitable breakpoint for penicillin G. Their breakpoint is slightly lower than ours, but again, the results of the phenotypic antibiotic susceptibility testing are hard to compare between laboratories.
Both in the present study and in earlier studies (4, 9), the correlation between elevated MICs and the presence of mosaic-structured penA genes was even higher for ampicillin than for penicillin G (Fig. 1). Determining the MIC of ampicillin might therefore be a sharper way to categorize N. meningitidis isolates into the Pens or Peni group.
PCR-based methods are increasingly being used for diagnosing meningococcal infection and for characterizing bacteria (10, 11, 13, 25). The protocol for penA sequencing, used in the present study, also proved to be effective for culture-negative CSF samples. Hence, in addition to the already existing genetic methods, the present penA sequencing can be used to accomplish further characterization of culture-negative samples from patients suffering from meningococcal disease. Another advantage with the penA sequencing protocol is that it can also be utilized for N. gonorrhoeae. In the N. gonorrhoeae population, only a small part is still fully susceptible to penicillins and altered penA genes have been described previously (1, 18).
Previous studies have proposed that in comparison with those of the wild-type penA gene, between five and nine specific nucleotide positions, and the corresponding amino acids, are altered in all Peni isolates (2, 21, 22). In our penA sequence collection, none of those specific Peni sites was altered in any of the identified divergent penA mosaic alleles. Two of the alterations (Phe504→Leu504 and Ala510→Val510) were found in all mosaic-structured penA genes, except for one sequence having no mosaic patterns in the final 550 bp of the gene and one having no mosaic patterns in the final 250 bp (Fig. 3). However, we have no evidence that these two isolates really belong to the Peni group, since we have only the penA sequences and no MICs. In addition, one of the clinical Peni isolates had no mosaic patterns in the final 220 bp of the gene and hence lacked three of the alterations found in all other mosaic-structured sequences (Ile515→Val515, His541→Asn541, and Ile566→Val566), except for the two discrepant sequences mentioned above. Besides the lack of absolute correlation between alterations in Peni-specific sites and mosaic structure, it is worth mentioning that one of the sites was also altered in a wild-type sequence (accession no. AY292992) (Fig. 3).
When individual MICs and penA sequence variants were compared within the group of Peni isolates, no obvious correlation was found for any of the antibiotics. This lack of correlation could be due to the fact that we studied larger segments of the gene instead of trying to identify “hot spots” particularly important for penicillin susceptibility, e.g., active sites that directly affect the affinity to penicillin. In addition, a limited effect on the MIC in individual isolates due to other factors, such as mtr, penB, penC, and ponA (15, 18), which influence the penicillin susceptibility in N. gonorrhoeae, cannot be excluded. The somewhat lower correlation between the elevated MIC of penicillin V and the mosaic penA gene sequence could also be due to these or similar factors. Since penicillin V is a less potent antibiotic, compared to penicillin G and ampicillin, for N. meningitidis, other pharmacokinetic mechanisms may also be involved. In addition to the genetic factors mentioned above, the drawbacks of phenotypic determination of susceptibility must be taken into account. The results of both the agar dilution method and Etest are highly dependent on the media used (26). A previous study has shown that the sources and batches of the ingredients, e.g., blood, in the medium can also affect the MICs (22). It is therefore very important to standardize the media used, especially to enable interlaboratory comparisons.
In conclusion, an up-to-date description of penA gene variability and the relation between penA gene sequence and the MIC of penicillins made it possible to identify mosaic structures clearly associated with reduced susceptibility. The level of correlation slightly varied between different antibiotics, and in the present study, ampicillin proved to be the best antibiotic for precise categorization of N. meningitidis isolates as Pens or Peni. By studying the penA gene, genotypical determination of susceptibility in culture-negative cases and hence further characterization of these samples can be accomplished.
Acknowledgments
This study was supported by grants from the Örebro County Council Research Committee and the Foundation for Medical Research at Örebro University Hospital, Örebro, Sweden.
We also want to thank Helena Eriksson for assistance with MIC determination.
REFERENCES
- 1.Ameyama, S., S. Onodera, M. Takahata, S. Minami, N. Maki, K. Endo, H. Goto, H. Suzuki, and Y. Oishi. 2002. Mosaic-like structure of penicillin-binding protein 2 gene (penA) in clinical isolates of Neisseria gonorrhoeae with reduced susceptibility to cefixime. Antimicrob. Agents Chemother. 46:3744-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antignac, A., I. G. Boneca, J.-C. Rousselle, A. Namane, J.-P. Carlier, J. A. Vázquez, A. Fox, J.-M. Alonso, and M.-K. Taha. 2003. Correlation between alterations of the penicillin-binding protein 2 and modifications of the peptidoglycan structure in Neisseria meningitidis with reduced susceptibility to penicillin G. J. Biol. Chem. 278:31529-31535. [DOI] [PubMed] [Google Scholar]
- 3.Antignac, A., P. Kriz, G. Tzanakaki, J.-M. Alonso, and M.-K. Taha. 2001. Polymorphism of Neisseria meningitidis penA gene associated with reduced susceptibility to penicillin. J. Antimicrob. Chemother. 47:285-296. [DOI] [PubMed] [Google Scholar]
- 4.Arreaza, L., C. Salcedo, B. Alcalá, M. J. Uría, R. Abad, R. Enríquez, and J. A. Vázquez. 2004. Sequencing of Neisseria meningitidis penA gene: the key to success in defining penicillin G breakpoints. Antimicrob. Agents Chemother. 48:358-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arreaza, L., and J. A. Vázquez. 2001. Molecular approach for the study of penicillin resistance in Neisseria meningitidis, p. 107-119. In A. J. Pollard and M. C. J. Maiden (ed.), Meningococcal disease: methods and protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 6.Bowler, L. D., Q.-Y. Zhang, J.-Y. Riou, and B. G. Spratt. 1994. Interspecies recombination between the penA genes of Neisseria meningitidis and commensal Neisseria species during the emergence of penicillin resistance in N. meningitidis: natural events and laboratory simulation. J. Bacteriol. 176:333-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI/NCCLS M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 8.Hughes, J. H., D. J. Biedenbach, M. E. Erwin, and R. N. Jones. 1993. E test as susceptibility test and epidemiologic tool for evaluation of Neisseria meningitidis isolates. J. Clin. Microbiol. 31:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen, J. H., S. A. Crawford, and K. R. Fiebelkorn. 2005. Susceptibility of Neisseria meningitidis to 16 antimicrobial agents and characterization of resistance mechanisms affecting some agents. J. Clin. Microbiol. 43:3162-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mölling, P., S. Jacobsson, A. Bäckman, and P. Olcén. 2002. Direct and rapid identification and genogrouping of meningococci and porA amplification by LightCycler PCR. J. Clin. Microbiol. 40:4531-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newcombe, J., K. Cartwright, W. H. Palmer, and J. McFadden. 1996. PCR of peripheral blood for diagnosis of meningococcal disease. J. Clin. Microbiol. 34:1637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olcén, P., H. Fredlund, P. Mölling, and M. Unemo. 2006. Annual report concerning serogroup, serotype, genosubtype and antibiotic sensitivity for Swedish Neisseria meningitidis isolates and results with direct PCR for diagnosis of acute bacterial meningitis, 2005. National Reference Laboratory for Pathogenic Neisseria, Örebro, Sweden.
- 13.Olcén, P., P.-G. Lantz, A. Bäckman, and P. Rådström. 1995. Rapid diagnosis of bacterial meningitis by a seminested PCR strategy. Scand. J. Infect. Dis. 27:537-539. [DOI] [PubMed] [Google Scholar]
- 14.Oppenheim, B. A. 1997. Antibiotic resistance in Neisseria meningitidis. Clin. Infect. Dis. 24(Suppl. 1):S98-S101. [DOI] [PubMed] [Google Scholar]
- 15.Orús, P., and M. Viñas. 2001. Mechanisms other than penicillin-binding protein-2 alterations may contribute to moderate penicillin resistance in Neisseria meningitidis. Int. J. Antimicrob. Agents 18:113-119. [DOI] [PubMed] [Google Scholar]
- 16.Peltola, H. 1983. Meningococcal disease: still with us. Rev. Infect. Dis. 5:71-91. [DOI] [PubMed] [Google Scholar]
- 17.Quagliarello, V. J., and W. M. Scheld. 1997. Treatment of bacterial meningitis. N. Engl. J. Med. 336:708-716. [DOI] [PubMed] [Google Scholar]
- 18.Ropp, P. A., M. Hu, M. Olesky, and R. A. Nicholas. 2002. Mutations in ponA, the gene encoding penicillin-binding protein 1, and a novel locus, penC, are required for high-level chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46:769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spratt, B. G., Q.-Y. Zhang, D. M. Jones, A. Hutchison, J. A. Brannigan, and C. G. Dowson. 1989. Recruitment of a penicillin-binding protein gene from Neisseria flavescens during the emergence of penicillin resistance in Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 86:8988-8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt, B. G. 1994. Resistance to antibiotics mediated by target alterations. Science 264:388-393. [DOI] [PubMed] [Google Scholar]
- 21.Stefanelli, P., A. Carattoli, A. Neri, C. Fazio, and P. Mastrantonio. 2003. Prediction of decreased susceptibility to penicillin of Neisseria meningitidis strains by real-time PCR. J. Clin. Microbiol. 41:4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taha, M.-K., M. L. Zarantonelli, A. Neri, R. Enriquez, J. A. Vázquez, and P. Stefanelli. 2006. Interlaboratory comparison of PCR-based methods for detection of penicillin G susceptibility in Neisseria meningitidis. Antimicrob. Agents Chemother. 50:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 24.Unemo, M., P. Olcén, J. Albert, and H. Fredlund. 2003. Comparison of serologic and genetic porB-based typing of Neisseria gonorrhoeae: consequences for future characterization. J. Clin. Microbiol. 41:4141-4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urwin, R., E. B. Kaczmarski, M. Guiver, A. J. Fox, and M. C. J. Maiden. 1998. Amplification of the meningococcal porB gene for non-culture serotype characterization. Epidemiol. Infect. 120:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vázquez, J. A., L. Arreaza, C. Block, I. Ehrhard, S. J. Gray, S. Heuberger, S. Hoffmann, P. Kriz, P. Nicolas, P. Olcén, A. Skoczynska, L. Spanjaard, P. Stefanelli, M.-K. Taha, and G. Tzanakaki. 2003. Interlaboratory comparison of agar dilution and Etest methods for determining the MICs of antibiotics used in management of Neisseria meningitidis infections. Antimicrob. Agents Chemother. 47:3430-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]