Abstract
The lack of a robust small-animal model for hepatitis C virus (HCV) has hindered the discovery and development of novel drug treatments for HCV infections. We developed a reproducible and easily accessible xenograft mouse efficacy model in which HCV RNA replication is accurately monitored in vivo by real-time, noninvasive whole-body imaging of gamma-irradiated SCID mice implanted with a mouse-adapted luciferase replicon-containing Huh-7 cell line (T7-11). The model was validated by demonstrating that both a small-molecule NS3/4A protease inhibitor (BILN 2061) and human alpha interferon (IFN-α) decreased HCV RNA replication and that treatment withdrawal resulted in a rebound in replication, which paralleled clinical outcomes in humans. We further showed that protease inhibitor and IFN-α combination therapy was more effective in reducing HCV RNA replication than treatment with each compound alone and supports testing in humans. This robust mouse efficacy model provides a powerful tool for rapid evaluation of potential anti-HCV compounds in vivo as part of aggressive drug discovery efforts.
Human liver disease caused by hepatitis C virus (HCV) has emerged as a major challenge to public health, affecting an estimated 175 million people worldwide (2). Greater than 50% of infections lead to chronic liver disease with a risk of developing liver cirrhosis and hepatocellular carcinoma (1). Infection with HCV has also been identified as the most common indication for liver transplantation in the United States and Europe (9). Treatment options for chronic HCV infection are limited to a combination of pegylated alpha interferon (IFN-α) and ribavirin (RB) (11), which is only partially effective and is often associated with troublesome side effects. Patients with genotype 1 HCV, the predominant genotype worldwide, are the most resistant to IFN-α and RB treatment (14). Thus, development of novel therapies for HCV is greatly needed, yet has progressed slowly.
HCV is an enveloped, positive-strand RNA virus and a member of the family Flaviviridae (10). Efficient replication of an HCV subgenomic replicon in in vitro cell culture has provided a valuable tool for molecular characterization of HCV, investigating virus host interactions, screening antiviral compounds, and developing new drug targets (4, 26, 29). Recently, an in vitro cell culture system capable of producing an infectious genotype 2a HCV has also been described (25, 39, 41). While the cell-based assays have provided a useful tool for screening compounds, they have not proved sufficient to predict the activity of compounds in vivo (21, 35, 38). The limited pipeline of new HCV antivirals may, in part, be attributed to the absence of robust small-animal models, which are typically used for simultaneously assessing drug action, efficacy, and toxicity. Difficulty in developing animal models is largely a result of the narrow host range of HCV, which infects only humans and chimpanzees. Over the past few years, several small-animal models such as the HCV-Trimera and chimeric scid-Alb/uPA Hepatech mouse models have been developed and demonstrated to be useful for studying HCV infection and drug evaluation (8, 17, 19, 21, 30, 40). However, low throughput, technical difficulty, and high cost significantly limit the utility of these models for drug discovery. Recently, a mouse model that permits the evaluation of the HCV NS3/4A serine protease target only has been described (33). The disadvantages of this model include the following (i) only antiviral compounds targeting HCV protease can be evaluated;(ii) the target is not in the context of the native replisome and thus may not accurately reflect the conformation of the target in infected cells; and (iii) the viral vectors used to deliver the protease construct may affect the cell biology of the host, which may, in turn, influence the dynamics of the assay.
In this report we describe the development, validation, and application of a simple, reproducible noninfectious HCV mouse efficacy model for evaluating antiviral compounds against multiple viral targets. The model utilizes a mouse-adapted replicon-containing Huh-7 human hepatoma cell line expressing a luciferase reporter linked to the HCV subgenome. These cells can be implanted subcutaneously (SC) or directly into the liver of gamma (γ)-irradiated SCID mice. The replicon used in this model expresses the HCV nonstructural proteins that comprised the replisome and is transfected into human hepatoma Huh-7 cells (32). The replication level of HCV RNA replicon in individual mice was monitored by measuring luciferase activity using a noninvasive whole-body, real-time Xenogen IVIS imaging system (Fig. 1A). Both the SC and liver models were validated by demonstrating a statistically significant reduction in the viral RNA replication levels after treatment with IFN-α 2b (7) or a small-molecule HCV NS3/4A protease inhibitor (BILN 2061) (23). These results indicate that the model recapitulates the clinical activity of these compounds and has potential for contributing to the rapid evaluation of novel treatments and combination therapies for HCV.
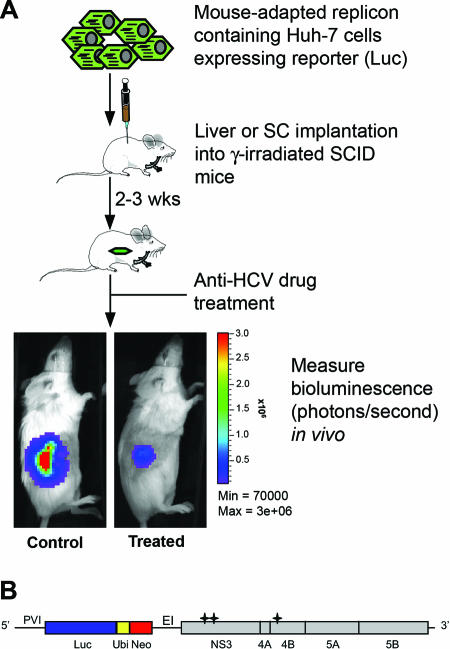
FIG. 1.
(A) Schematic of the HCV replication animal model. Mouse-adapted replicon-containing T7-11 cells expressing a high level of luciferase (Luc) were implanted subcutaneously or directly into the liver parenchyma of γ-irradiated SCID mice. Drug evaluation was performed 2 to 3 weeks after cell implantation. The HCV replication level was monitored quantitatively by measuring the bioluminescence signal using the IVIS imaging system. (B) Structure of the subgenomic replicon rep114/ET derived from pFK I389 LucNS3-3′/5.1 (22) and pFK I341PI Luc NS3-3′/ET (27). 5′, HCV 5 ′ nontranslated region; 3′, 3′ HCV nontranslated region; PVI, poliovirus IRES; Luc, firefly luciferase; Ubi, ubiquitin cleavage site; Neo, neomycin phosphotransferase; EI, encephalomyocarditis virus IRES; *, adaptive mutations E1202G, T1280I, and K1846T.
MATERIALS AND METHODS
Animals.
Female C.B17 SCID (SCID), SCID/bg, Nu/Nu, and BALB/c mice (5 to 13 weeks old) were obtained from Charles River Laboratories (Wilmington, MA). All studies were conducted in an American Association for Laboratory Animal Science-certified facility under the direction of the Institutional Animal Care and Use Committee. The mice were irradiated at 3 Gy (∼3.2 min).
Cell cultures.
Human hepatoma cell line Huh-7 (32) was maintained in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) containing high glucose, 10% fetal bovine serum, l-glutamine, and nonessential amino acids at 37°C in 5% CO2. G418 (geneticin; Invitrogen, Carlsbad, CA) was added at a final concentration of 0.5 mg/ml for replicon-containing cells.
Plasmid construction.
The replicon rep114/ET was constructed by replacing the 1.6-kb XbaI-NotI fragment from pFKI341PILucNS3-3′/ET (27) with the 2.6-kb XbaI-NotI fragment from pFKI389LucUbiNeoNS3-3′/5.1 (22).
RNA preparation and electroporation.
Plasmids were linearized with ScaI, and RNA was synthesized with a MEGAscript kit (Ambion, Austin, TX). RNA was extracted and purified using TRIzol reagent following the manufacturer's instructions (Invitrogen, Carlsbad, CA). RNA transfection was performed as described previously (15).
Subcloning the adapted S6.1, S6.1-6 and T7-11 cell lines. (i) S6.1 cells.
Five million Huh 5-2 cells (22) were implanted subcutaneously into the right flank of 10 female SCID/bg mice. The three largest tumors were harvested 37 days postimplantation of cells and mashed with the frosted ends of two sterilized glass slides. A total of 50 to 100 μl of mashed cells was inserted subcutaneously with a trocar into each of 10 anesthetized SCID/bg mice. This in vivo passaging process was repeated twice from tumors harvested on days 22 and 28. Twenty-four days after the final implantation, three of the larger tumors were harvested, and tumor cells were expanded in cell culture to create the pooled mouse-adapted S6.1 tumor cells.
(ii) S6.1-6 cells.
A total of 5 μg of in vitro transcribed rep114/ET RNA was electroporated into S6.1 cells. After electroporation, stable cell clones were selected in medium containing 0.5 mg/ml G418. Luciferase expression of G418-resistant colonies was evaluated (27), and the colony with highest expression was expanded to create the cell line S6.1-6.
(iii) T7-11 cells.
Five million S6.1-6 cells in 0.2 ml of Hank's balanced salt solution (HBSS) were implanted subcutaneously in female C.B17 SCID mice, which had been irradiated (3 Gy) 4 hours before cell implantation. A tumor emitting strong bioluminescence was excised 27 days later, and the bioluminescent portion of the tumor was cultured in vitro in the presence of 0.5 mg/ml G418. Luciferase expression of G418-resistant colonies was evaluated using an IVIS imaging system (Xenogen Corporation, Alameda, CA), and one colony with high expression was expanded to create the cell line T7-11. The T7-11 cell line was stably maintained in vitro in Dulbecco's modified Eagle medium containing 0.5 mg/ml G418 prior to implantation. Consistent in vivo engraftment was seen using the in vitro cultured T7-11 cells up to passage 20.
Analysis of HCV RNA by Northern hybridization.
Total RNA was extracted from tumor tissue samples with TRIzol reagent (Invitrogen, Carlsbad, CA). Ten micrograms of total RNA was fractionated on a 1% agarose gel containing 2.2 M formaldehyde and transferred onto a nylon membrane (Ambion, Austin, TX), which was hybridized with a 32P-labeled negative-sense Riboprobe (Promega, Madison, WI) complementary to the 3′ end of NS5A and the 5′ end of NS5B.
Quantitative real-time RT-PCR.
Real time reverse transcription-PCR (RT-PCR) assays were performed as described previously (34). Briefly, the primers specific to the HCV 5′ untranslated region and to the human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Applied Biosystems, Foster City, CA) as a normalization control were used. The HCV 5′ untranslated region sense primer sequence is 5′-TCTTCACGCAGAAAGCGTCTA-3′, and the antisense primer is 5′-CGGTTCCGCAGACCACTATG-3′; the probe labeled at the 5′ end with a FAM (6-carboxyfluorescein) fluorophore reporter molecule and at the 3′ end with a TAMRA (6-carboxytetramethylrhodamine) quencher molecule is 5′-TGAGTGTCGTGCAGCCTCCAGGA-3′. Amplification and fluorescence detection were performed on an Applied Biosystems 7500 Real-Time PCR System. Forty-five cycles were used according to the manufacturer's instructions; the thermal cycle conditions were 48°C for 30 min, 95°C for 10 min, and 45 repetitions of 95°C for 15 s, followed by 60°C for 1 min. HCV cDNA standards were used starting at 106 copies of virus and decreasing in 10-fold serial dilutions.
Immunohistochemical analysis.
Sections (4 μm thick)were prepared from either formalin-fixed or 4% paraformaldehyde-fixed, paraffin-embedded tumors. For detection of cleaved poly(ADP-ribose) polymerase (PARP) and activated caspase 3, a polyclonal rabbit anticleaved PARP antibody (BioSource, Camarillo, CA) and a polyclonal rabbit anticleaved caspase 3 antibody (CalBiochem, San Diego, CA) were employed along with biotinylated anti-rabbit secondary antibody (Jackson Labs, West Grove, PA). For detection of viral protein NS5B expression, a polyclonal rabbit anti-NS5B antibody (Chiron Corporation, Emeryville, CA) (12) and a fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin antibody (Invitrogen, Carlsbad, CA) were used. For detection of human mitochondria expression, a monoclonal mouse anti-mitochondria antibody (Chemicon, Temecula, CA) was employed along with biotinylated anti-mouse secondary antibody (Jackson Labs, West Grove, PA). Slides were stained using a Discovery XT slide staining instrument and compatible ancillary reagents (Ventana Medical Systems, Tucson, AZ) for diaminobenzidine chromogen detection.
In vivo studies. (i) Cell implantation.
Female SCID mice were shaved at the site of cell implantation and gamma-irradiated with 3 Gy either 1 day prior to or on the same day as cell implantation. For subcutaneous implantation, 5 million cells in 0.2 ml of HBSS were implanted at the right flank. Mice were euthanized once mean tumor volume reached a size of >2,000 mm3 for the group or >3,000 mm3 for individual mice. For direct intrahepatic implant, mice were anesthetized, and one liver lobe was exteriorized through a small incision. One to two million cells in 20 to 50 μl of HBSS were injected directly into the liver parenchyma.
(ii) Whole-body in vivo imaging.
Anesthetized mice were injected intraperitoneally with 150 mg/kg luciferin (Xenogen Corporation, Alameda, CA). After 10 to 20 min, mice were imaged using a charge-coupled-device camera in the Xenogen IVIS imaging system. The bioluminescence signal intensity was quantified using Living Image Software (Xenogen Corporation, Alameda, CA).
(iii) Drug treatment.
IFN-α 2b (Intron A; Schering-Plough, New Jersey), BILN 2061, and recombinant interleukin-2 (Aldesleukin/Proleukin; Chiron Corporation, Emeryville, CA) were injected subcutaneously in the suprascapular area.
(iv) Statistical analysis.
Differences in bioluminescence signals between control and treatment groups of mice were compared using a two-tailed Mann-Whitney test. Differences in signals among the control, treatment, and rebound groups of mice and among combination groups were compared using a nonparametric Kruskal-Wallis test. A P value of <0.05 was considered significant.
RESULTS
Mouse-adapted human hepatoma Huh-7 cell lines harboring a replicon with a strong luciferase signal are required for in vivo detection of HCV RNA replication.
Successful transplantation of heterologous cells that replicate and express a viral genome or subgenome into a recipient animal requires tolerance for both the human cells and replicon RNA. Our initial effort to implant G418-selected Huh 5-2 cells (5-2) containing a genotype 1b subgenomic replicon in vitro (22) was suboptimal in that the cells grew very slowly in immunodeficient SCID/bg mice, and the bioluminescence from the luciferase reporter was quickly lost. To generate a mouse-adapted replicon-containing Huh-7 cell line with improved growth characteristics in vivo, 5-2 cells from subcutaneous tumors were serially passaged in SCID/bg mice. Following three passages, a subclone (S6.1) that produced subcutaneous tumors in approximately 2 weeks was isolated. In comparison, the parental 5-2 cell line produced tumors in over 4 weeks. Nevertheless, a replicon RNA needed to be reintroduced into S6.1 cells for in vivo study since the original replicon RNA had been rapidly eliminated from S6.1 cells during serial passaging.
In order to achieve a minimum 1- to 2-log dynamic range of signal from the luciferase reporter in this in vivo model, the replicon contained in the 5-2 cells (22) was reengineered to improve luciferase expression and RNA replication. This new construct, rep114/ET (Fig. 1B), contained a poliovirus internal ribosome entry site (IRES) upstream of the luciferase gene, a neor selective marker (G418r), and a combination of highly adaptive mutations (E1202G, T1280I, and K1846T) in the HCV nonstructural (NS) region that increased RNA replication (27). To generate a stable cell line, rep114/ET RNA was transfected into mouse-adapted S6.1 cells. The G418-resistant cell clone expressing the highest level of luciferase signal, S6.1-6, was identified and evaluated in SCID/bg mice. Following subcutaneous implantation of S6.1-6 cells, the HCV RNA level was higher than that in 5-2 cells but still decreased rapidly, as evidenced by loss of bioluminescence. This rapid decrease in RNA level was most likely not due to the absence of the selectable marker in vivo because the replicon RNA level is stable in in vitro cell culture in the absence of G418 for greater than 1 month.
Based on our hypothesis that the partially suppressed mouse immune system played a role in the rejection of human cells and replicon RNA, mice were further immunosuppressed by γ irradiation. Different strains of mice (athymic Nu/Nu, SCID, and SCID/bg) were either exposed or not exposed to γ irradiation prior to subcutaneous implantation of S6.1-6 cells and evaluated for bioluminescence and growth properties. The result showed that γ-irradiated SCID and SCID/bg mice were the best hosts, maintaining a signal approximately 1 log over background after 7 days, while the signal in nonirradiated mice rapidly declined to baseline (data not shown). Therefore, γ-irradiated SCID mice were used as hosts for all subsequent studies.
We reasoned that the host would select cells that could maintain the replicon in vivo for an extended period of time since the signal in S6.1-6 cells was not sustained sufficiently for long-term studies. Therefore, tumors exhibiting the highest luciferase signal a month after implantation of S6.1-6 cells were harvested and mashed up to isolate cells that were cultured in the presence of G418 to identify the G418-resistant, luciferase-positive colonies. These cells were then implanted and allowed to form tumors. One clone, T7-11, which exhibited increased bioluminescence more than 2 logs over background 3 weeks after SC implantation into γ-irradiated SCID mice and maintained this high level for at least another 2 weeks, was selected for further study (Fig. 2A). The initial decline of bioluminescence around 10 days after implantation was most likely due to cell rejection by the host, which was also seen in other replicon-containing Huh-7 cell lines tested. The tumor growth rate of T7-11 cells was measured and found to be similar to its parental S6.1-6 cells (Fig. 2B). To evaluate the stability of the HCV replicon RNA in T7-11 cells in liver, which is the primary target organ for HCV infection, cells were directly injected into the parenchyma of liver. The tumor grew within the liver lobe, and bioluminescence more than 1 log above background was observed 3 weeks after implantation. Lower bioluminescence in liver compared to SC was mainly due to lower number of cells implanted and possibly due to attenuation of the signal by implantation of the cells deeper in mouse tissue. The result demonstrated the utility of T7-11 cells for both subcutaneous and intrahepatic HCV RNA replication models.
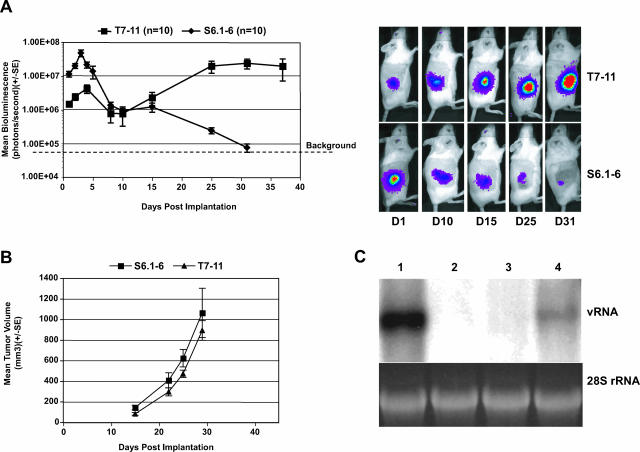
FIG. 2.
Correlation between mean bioluminescence in vivo and replication of subgenomic RNA in T7-11 cell subcutaneous model. (A) Mean bioluminescence of mouse-adapted replicon-containing T7-11 cells and parental S6.1-6 cells in gamma-irradiated SCID mice. Mean bioluminescence was determined for each group (n = 10 per group) at different times after subcutaneous implantation of T7-11 and S6.1-6 cells. The images of representative mice from both groups at days 1, 10, 15, 25, and 31 are also shown. (B) Tumor growth rate of T7-11 and S6.1-6 cells. Mean tumor volumes (0.5 × width × width × length = mm3) from both groups measured at days 15, 22, 25, and 29 are plotted. The graph is representative of four independent experiments. Error bars indicate standard error. (C) Detection of HCV viral RNA in tumor tissue. Total RNA was isolated from subcutaneous tumors which were excised 26 days after the implantation of T7-11 cells and analyzed by Northern blot analysis (10 μg per lane). Blots were hybridized with 32P-labeled RNA probes corresponding to the HCV NS5 region. A total of 1 × 108 genomes of in vitro transcribed HCV RNA (vRNA) served as a marker and control for the hybridization reaction (lane 1), and 28S rRNA served as a control for the amount of RNA present in each sample analyzed. Lanes 2 and 3 were total RNA extracted from naïve Huh-7 and S6.1 cells as negative controls. Lane 4 is RNA from a T7-11 tumor. SE, standard error.
Previous reports showed that the luciferase activity in this genotype 1b HCV replicon accurately reflected the RNA replication level in vitro (27). To rule out the possibility that the luciferase signal in T7-11 cells in vivo was not associated with HCV RNA replication, the viral RNA and protein levels in tumors were examined by Northern blot analysis and immunofluorescence assay (IFA), respectively. The results showed that positive-strand HCV RNA was detected in T7-11 tumor tissue harvested 26 days after subcutaneous implantation (Fig. 2C, lane 4). Consistent with this result, viral protein NS5B was detected (data not shown). The association between bioluminescence and RNA replication level in vivo was also determined by calculating the amount of HCV RNA by quantitative real-time RT-PCR relative to the photons emitted per tumor. Approximately 500 copies of viral RNA correlated to one photon/second in bioluminescence per tumor. These data indicated that T7-11 cells supported constitutive HCV RNA replication and protein expression in vivo and bioluminescence reflects RNA replication accurately.
Validation of subcutaneous and intrahepatic models for drug evaluation.
Confidence in an animal model can be acquired by demonstrating that antiviral compounds active in humans are also efficacious in the model. We therefore validated the models using IFN-α 2b (IFN-α) (7) and a small-molecule HCV NS3/4A protease inhibitor, BILN 2061 (23), both of which have shown anti-HCV activity in patients although acting by different mechanisms.
Efficacy of IFN-α and BILN 2061 in γ-irradiated SCID mice subcutaneously implanted with T7-11 cells.
IFN-α efficacy studies with T7-11-implanted mice were initiated when the bioluminescence was approximately 2 logs above background. Mice were treated with 7,500 or 15,000 IU/mouse/day IFN-α starting 19 days after implantation. Control animals received human albumin. As shown in Fig. 3A, the mean bioluminescence signal in the low-dose group (7,500 IU/mouse/day) was reduced 5- and 10-fold after 3 and 7 days of treatment, respectively, compared to the control group (P < 0.01). At the same time points, a 10- and 25-fold reduction of signal was observed for the high-dose group relative to the control group (P < 0.001). The data clearly demonstrated that IFN-α treatment had a dose-dependent effect on HCV RNA level. Since the tumor growth kinetics did not change in the treated group compared to the untreated group (Fig. 3B), this result suggested that the reduction of bioluminescence was most likely due to a direct antiviral effect of IFN-α on RNA replication.
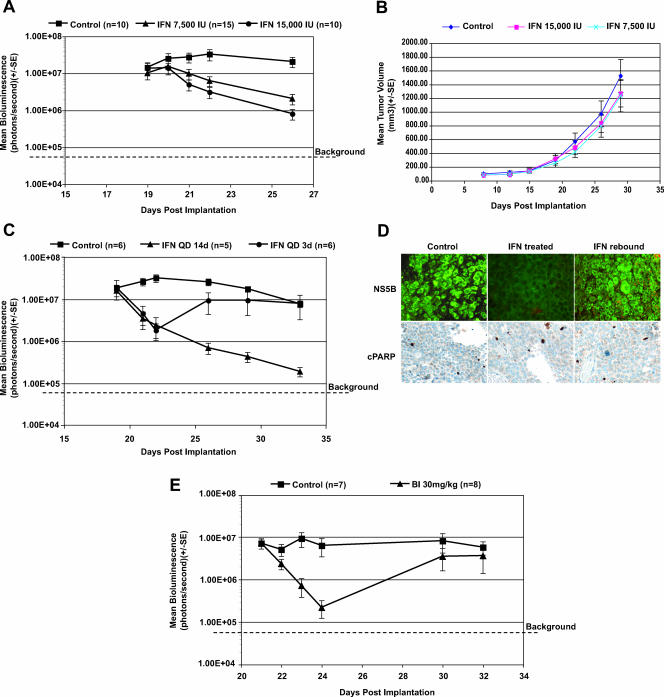
FIG. 3.
Antiviral activity of IFN-α 2b and NS3/4A protease inhibitor BILN 2061 (BI) in the subcutaneous T7-11 model. (A) Dose-dependent IFN anti-HCV effect. Subcutaneous IFN treatment was initiated on day 19 after cell implantation and was administered once daily for 7 days: control, 0.1 ml of 100 μg/ml human albumin in HBSS; 7,500 IU group, 7,500 IU of IFN per day per 18 g of mouse; 15,000 IU group, 15,000 IU of IFN per day per 18 g of mouse. n = 10 to 15 mice per group. (B) Tumor growth rate in subcutaneous T7-11 model. Mean tumor volumes (0.5 × width × width × length = mm3) are plotted from control, 7,5000 IU of IFN, and 15,000 IU of IFN groups measured at days 8, 12,15, 19, 22, 26, and 29. (C) IFN treatment was given as described in panel A with 15,000 IU/mouse/day in two groups: IFN QD 14 days, once daily administration for 2 weeks; IFN QD 3 days, once daily administration for 3 days. Individual mice were imaged on day 19 before treatment, followed by days 21, 22, 26, 29, and 33. (D) Immunohistochemical analysis of tumor tissues from untreated, IFN-treated (IFN QD 14 days), and IFN rebound (IFN QD 3 days) groups 33 days postimplantation as described in panel C. The expression of viral protein NS5B was detected with a polyclonal antibody against NS5B. Cell death was detected with cPARP antibody as described in Materials and Methods. (E) Antiviral effect of protease inhibitor BILN 2061. BILN 2061 treatment was given subcutaneously at 30 mg/kg once daily for 3 days starting on day 21 postimplantation. BILN 2061 treatment was then withdrawn on day 24. Individual mice were imaged prior to each day's treatment on days 21, 22, 23, 24, 30, and 32. Error bars indicate standard error (SE). QD, once daily. BI, BILN 2061.
To demonstrate that the decrease in bioluminescence in the IFN-treated mice was not due to IFN-induced killing of the replicon-containing cells, administration of IFN-α was discontinued after 3 days of treatment. As shown in Fig. 3C, rebound in bioluminescence was clearly observed in the treated group 4 days after IFN-α (15,000 IU/mouse) was discontinued, and the mean bioluminescence returned to the level of the untreated group (P > 0.05). In contrast, the mean bioluminescence continued to decrease in the group that continued to receive IFN-α treatment for 2 weeks (P < 0.01) (Fig. 3C). To further examine the effect of treatment on tumor cells, tumor tissue was excised for immunohistochemical and tumor morphology analysis at the end of the study. As expected, immunohistochemical analysis of activated caspase 3 and cleaved PARP (cPARP), two markers of cell death, showed that both activated caspase 3 (data not shown) and cPARP staining were present in only a small percentage of tumor cells in all three groups (Fig. 3D, lower panel) compared to the rabbit IgG-negative control group for cPARP (data not shown). Staining with an antibody against Ki67, a marker of cell proliferation, also indicated that there was no difference between the IFN-α-treated group and the control group (data not shown). In contrast, significant expression of viral NS5B protein was detected by IFA in control and rebound groups, whereas NS5B protein was mostly undetectable in the IFN-α-treated group (Fig. 3D upper panel) compared to the negative control naïve Huh-7 cells (data not shown). Consistent with both IFA and in vivo imaging data, the level of HCV RNA as measured by quantitative RT-PCR (34) was approximately 10-fold higher in untreated than treated mice. Moreover, human recombinant interleukin-2, a known immunomodulator that lacks anti-HCV activity (6, 37), had no effect on bioluminescence in T7-11-implanted mice treated for 5 days at 1 mg/kg, a dose that has shown antitumor activity in renal cell cancer models (13) (data not shown). Taken together, these results demonstrated that the reduction of bioluminescence observed in this model after IFN-α treatment resulted from direct inhibition of viral replication and not a cytotoxic or cytostatic effect on the T7-11 tumor cells.
The feasibility of using this model to evaluate the efficacy of a target-specific inhibitor of HCV was also examined. BILN 2061, an NS3/4A protease inhibitor, was delivered subcutaneously to maintain a sustained plasma exposure for more than 24 h. BILN 2061 was administered at 30 mg/kg once daily starting on day 21 after tumor cell implantation while signal was 2 logs above background. The results shown in Fig. 3E indicated that the bioluminescence signal decreased more than 1 log within 3 days compared to the control group (P = 0.0006). As expected, 6 days after treatment stopped, the bioluminescence signal returned to the levels similar to those in the untreated control group (P = 0.5338) (Fig. 3E). These data demonstrated that the compound effectively reduced HCV RNA in vivo.
Efficacy of IFN-α and BILN 2061 in an intrahepatic model of HCV replication.
To evaluate the efficacy of IFN-α in liver, T7-11 cells were directly injected into the parenchyma of one lobe of the liver in γ-irradiated SCID mice. To demonstrate engraftment of T7-11 tumor cells into the mouse liver, hematoxylin and eosin staining as well as human mitochondrial staining of liver tissue sections was performed (Fig. 4A). IFN-α treatment at 15,000 IU/mouse/day was initiated 27 days postimplantation when the luciferase signal was more than 1 log above background. Bioluminescence decreased more than 10-fold in the IFN-α-treated group compared to control after 2 days of dosing (Fig. 4B) (P < 0.0001). A slight rebound was observed 5 days after removal of treatment (Fig. 4B). The antiviral effect of BILN 2061 on HCV RNA replication was also examined. BILN 2061 was administered subcutaneously at 30 mg/kg once daily starting on day 25 after tumor cell implantation. A 10-fold decline in bioluminescence within 3 days was observed in the treated group compared to the untreated control group (P < 0.0001) (Fig. 4C). Taken together, the effect of IFN-α and BILN 2061 in the liver model was very similar to that observed in the subcutaneous model (Fig. 3C and E), demonstrating the utility of both models for evaluation of anti-HCV compounds.
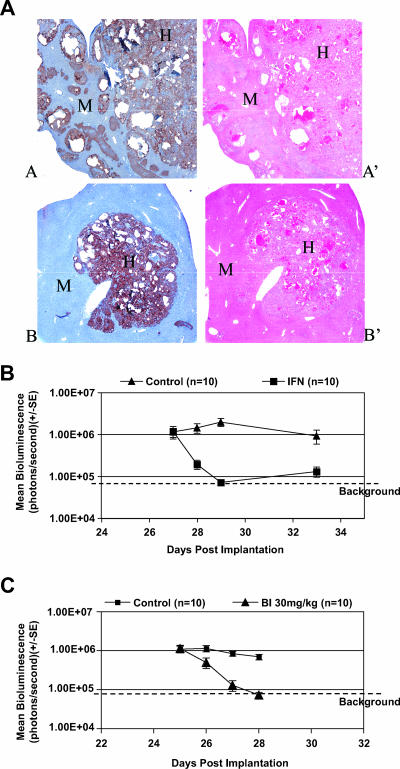
FIG. 4.
Anti-HCV activity of IFN-α 2b and BILN 2061 in the intrahepatic mouse efficacy model. (A) Histology of a chimeric liver. Hematoxylin and eosin staining (A′ and B′) allows identification of the human hepatoma T7-11 cells (H) within mouse (M) parenchyma. Clusters of human T7-11 cells can also be visualized with antibody specific for human mitochondria (A and B) shown in brown. (B) Antiviral effect of IFN-α 2b (IFN). T7-11 cells were implanted directly into the parenchyma of liver in γ-irradiated SCID mice. IFN treatment was given SC once daily for 2 days starting on day 27 postimplantation. A dose of 15,000 IU/mouse/day of IFN was given to the treated group, and 10 μg/mouse/day of human albumin was given to the control group. (C) Antiviral effect of BILN 2061 (BI). A treatment of 30 mg/kg of BILN 2061 was initiated once daily subcutaneously for 3 days starting on day 25 postintrahepatic implantation. Individual mice were imaged daily prior to each day's treatment on days 25 through 28. Error bars indicate standard error (SE).
Efficacy of combination therapy with BILN 2061 and IFN-α.
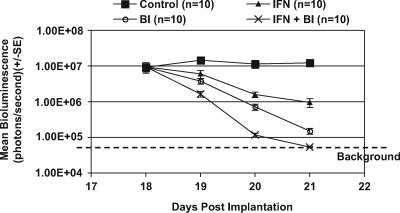
Given the results from the efficacy studies of IFN-α and BILN 2061, the subcutaneous model was utilized to examine whether the individual effect of BILN 2061 or IFN-α on HCV RNA replication could be improved by a combination of both agents. Treatment regimens were initiated on day 18 after subcutaneous implantation of T7-11 cells. Three groups of mice were administered 30 mg/kg BILN 2061 or 15,000 IU of IFN-α or a combination of both agents daily. Two days after treatment began, the mean bioluminescence signal decreased 7- or 16-fold in mice treated with either IFN-α or BILN 2061, respectively, compared to the control group (P < 0.01) (Fig. 5). In contrast, an approximately 100-fold reduction in bioluminescence was observed in mice 2 days after treatment with the BILN 2061/IFN-α combination therapy (P < 0.001) (Fig. 5), indicating that the combination therapy inhibited HCV RNA replication more efficiently than treatment with either active single agent alone.
FIG. 5.
Antiviral effect of protease inhibitor BILN 2061 (BI) in combination with IFN-α 2b (IFN) in the T7-11 subcutaneous efficacy model. T7-11 cells were implanted subcutaneously into γ-irradiated SCID mice. BILN 2061 at 30 mg/kg and/or IFN at 15,000 IU per mouse was administrated once daily for 3 days starting on day 18 after cell implantation. Individual mice were imaged daily prior to each day's treatment on days 18 through 21.
DISCUSSION
Anti-HCV compounds are often advanced into clinical trials based on demonstration of in vitro activity in cell-based assays and an acceptable pharmacokinetic (PK) and safety profile in the absence of in vivo efficacy. One drawback of this approach is that it is difficult to accurately predict the exposure necessary to achieve in vivo activity based on in vitro systems. In addition, determination that a compound does have sufficient theoretical exposure in the target organ of a normal animal based on PKs does not guarantee the efficacy of that compound in an animal containing the drug target. Major limitations of current small-animal efficacy models include low throughput, the requirement for highly specialized surgical skills, high cost, and access to human liver and HCV-infected sera. Therefore, only a very limited number of compounds can be evaluated. Additionally, because these systems use live infectious virus, specialized containment is required, and researchers are unnecessarily exposed to HCV, for which there is only a partially effective therapy available at the present time. The mouse HCV RNA replication model described here provides a noninfectious, reproducible, time- and cost-effective tool for evaluating the efficacy, PK, and pharmacodynamic (PD) relationship of potential HCV therapies simultaneously in vivo. Since our initial efforts failed to produce a replicon cell line that could persist in vivo sufficiently to generate a usable efficacy model, we reconsidered previous studies in which it was shown that RNA adaptation and/or cell adaptation were advantageous in the selection of conditions most supportive for viral replication in cell culture systems (3, 5, 15, 20, 22, 27, 28, 31, 36). By taking advantage of the host's natural ability to apply selective pressure on the replicon-containing cells passaged in vivo, we were able to select T7-11 cells, which supported high levels of HCV RNA replication in vivo for several weeks.
We found that the immune status of the host mouse strain significantly influenced the magnitude and duration of the HCV RNA replication in vivo. Whole-body γ irradiation causes marked depletion of hematopoietic cells, which in turn impairs functional immune response. A partial reconstitution of the host immune system typically occurs after 2 weeks (18). γ-Irradiated SCID and SCID/bg mice best supported HCV replicon-containing cells but for only approximately 1 week postimplantation. Since a rapid decrease in bioluminescence signal was immediately observed in nonirradiated SCID and SCID/bg mice that had T and B lymphocyte deficiencies (24), the results suggested that viral replication was either directly or indirectly inhibited by the residual host immunity associated with the innate immune responses. Whole-body γ irradiation of genetically immunodeficient mice resulted in the optimal conditions for maintaining viral and tumor cell replication.
Interestingly, the bioluminescence signal in mice bearing T7-11 cells was able to recover after approximately 2 weeks, while the signal in mice bearing S6.1-6 cells continued to decline, coincident with the expected reconstitution of innate immune response. Since the tumor growth rates of S6.1-6 and T7-11 cells in γ-irradiated mice were similar, the loss of bioluminescence signal in S6.1-6 cells can best be explained by the reduction in HCV RNA replication. Preliminary data showed that RNA replication in T7-11 cells was five- to eightfold more resistant to human IFN-α in vitro than that in parental S6.1-6 cells, suggesting that the antiviral responses in the T7-11 cells appeared to be attenuated, a condition that would account for the observed increased stability of replicon RNA in vivo. This decreased sensitivity to IFN-α was associated with adaptive mutations acquired at the cellular level, not at the viral RNA level. This was demonstrated by transient transfection of an identical replicon RNA (rep114/ET) into either S6.1 or cured T7-11 cells cleared of replicon RNA by IFN-α treatment in vitro. Only cured T7-11 cells maintained the IFN-α insensitivity phenotype as determined by the 50% effective concentration of IFN-α in vitro (unpublished data). However, both T7-11 and S6.1 cells possessed functional Jak/STAT antiviral signaling pathways, as demonstrated by immunoblot analysis of expression of phosphorylated Stat1 proteins. This result suggested that downstream effectors of Jak/STAT might potentially be impaired in T7-11 cells (unpublished data).
Validation of the animal efficacy model was performed using human IFN-α and protease inhibitor BILN 2061, two agents that have demonstrated anti-HCV activity in patients. Dose-dependent reduction of bioluminescence and rebound of signal after withdrawal of treatments clearly demonstrated the expected antiviral activity of these two agents. The careful examination of tumors from these studies indicated that the decline of bioluminescence resulted from a specific antiviral effect rather than a cytotoxic effect of the treatment. A good correlation was observed between the mouse model and what is seen in the clinic with respect to the in vivo therapeutic effects of these two agents (7, 16, 23). Importantly, we showed that the combination IFN-α/BILN 2061 treatment significantly improved the antiviral effect on HCV RNA replication compared with either single agent, strongly suggesting that the combination therapy of small-molecule anti-HCV inhibitor and IFN-α could be a promising therapeutic approach to the treatment of HCV. The results suggest that combination therapies (i.e., protease and polymerase inhibitors), which have not yet been tested in humans, can be assessed in this model.
Major advantages of the models described in this report include the following: (i) the simultaneous evaluation of multiple parameters such as efficacy and PKs/PDs can be performed since HCV replication in individual animals can be frequently monitored in vivo using the IVIS imaging system; (ii) the models can achieve statistical significance since large numbers of animals in multiarm studies can be implanted within a short period of time; (iii) using a noninfectious system supports a high-throughput mode for compound evaluation; and (iv) combination therapies can be rapidly assessed for one or more antiviral targets. Importantly, our findings showed that IFN-α and BILN 2061 efficacy studies were comparable in both subcutaneous and liver models. Thus, the subcutaneous model provides a powerful tool for initial rapid in vivo compound screening and evaluation of tolerability and efficacy of compounds, while the intrahepatic model, which is a closer approximation of the human disease, can be used to more effectively evaluate the PKs/PDs and safety of drugs prior to evaluation in clinical trials.
Our data are the first to show that Huh-7 cells containing an HCV replicon can be stably adapted to grow in immunodeficient mice for more than 1 month. The workable time for drug evaluation is approximately 2 to 3 weeks. Preliminary data suggest that the stability of the viral RNA in the T7-11 cells in vivo is dependent upon the cell adaptations rather than the genetic constitution of the replicon contained within the mouse-adapted cell line (unpublished data). Therefore, while the data presented in this report are based on a mouse-adapted Huh-7 cell line harboring a subgenomic genotype 1b replicon, it is worth noting that this approach may be applied to other HCV genotypes using the appropriate replicons (infectious or noninfectious) and may potentially be applied to other viruses.
Acknowledgments
We thank S. Chey, J. Kline, E. Chan, K. Aardalen, J. Barajas, and Q. Tung for technical assistance; J. Burrows, Z. J. Ni, and D. Menezes for helpful discussion; L. Long for statistical data analysis; and B. Jallal, Q. L. Choo, and T. Abrams for critically reading the manuscript and helpful comments. We appreciate the support of K. Bair, M. Houghton, and R. Rippouli.
REFERENCES
- 1.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1999. Global surveillance and control of hepatitis C. Report of a WHO consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J. Viral Hepat. 6:35-47. [PubMed] [Google Scholar]
- 3.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., A. Kaul, and S. Sparacio. 2003. Replication of the hepatitis C virus in cell culture. Antiviral Res. 60:91-102. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulestin, A., K. Sandres-Saune, L. Alric, B. Pipy, M. Dubois, J. P. Vinel, and J. Izopet. 2005. Evolution of hepatitis C virus quasispecies during therapy with IL2 combinated to alpha interferon and ribavirin. Antivir. Ther. 10:499-504. [PubMed] [Google Scholar]
- 7.Brass, C. A. 1998. Efficacy of interferon monotherapy in the treatment of relapsers and nonresponders with chronic hepatitis C infection. Clin. Ther. 20:388-397. [DOI] [PubMed] [Google Scholar]
- 8.Bright, H., A. R. Carroll, P. A. Watts, and R. J. Fenton. 2004. Development of a GB virus B marmoset model and its validation with a novel series of hepatitis C virus NS3 protease inhibitors. J. Virol. 78:2062-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, R. S. 2005. Hepatitis C and liver transplantation. Nature 436:973-978. [DOI] [PubMed] [Google Scholar]
- 10.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 11.Di Bisceglie, A. M., J. McHutchison, and C. M. Rice. 2002. New therapeutic strategies for hepatitis C. Hepatology 35:224-231. [DOI] [PubMed] [Google Scholar]
- 12.Eckart, M. R., M. Selby, F. Masiarz, C. Lee, K. Berger, K. Crawford, C. Kuo, G. Kuo, M. Houghton, and Q. L. Choo. 1993. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem. Biophys. Res. Commun. 192:399-406. [DOI] [PubMed] [Google Scholar]
- 13.Fan, K., M. Zhou, M. K. Pathak, D. J. Lindner, C. Z. Altuntas, V. K. Tuohy, E. C. Borden, and T. Yi. 2005. Sodium stibogluconate interacts with IL-2 in anti-Renca tumor action via a T cell-dependent mechanism in connection with induction of tumor-infiltrating macrophages. J. Immunol. 175:7003-7008. [DOI] [PubMed] [Google Scholar]
- 14.Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967-972. [DOI] [PubMed] [Google Scholar]
- 15.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinrichsen, H., Y. Benhamou, H. Wedemeyer, M. Reiser, R. E. Sentjens, J. L. Calleja, X. Forns, A. Erhardt, J. Cronlein, R. L. Chaves, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2004. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127:1347-1355. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, E. C., B. Hsi, M. Hirota-Tsuchihara, J. Ruland, C. Iorio, F. Sarangi, J. Diao, G. Migliaccio, D. L. Tyrrell, N. Kneteman, and C. D. Richardson. 2003. Modified apoptotic molecule (BID) reduces hepatitis C virus infection in mice with chimeric human livers. Nat. Biotechnol. 21:519-525. [DOI] [PubMed] [Google Scholar]
- 18.Huang, P., A. Taghian, A. Allam, J. Freeman, M. Duffy, and H. Suit. 1996. The effect of whole-body irradiation of nude mice on the tumor transplantability and control probability of a human soft tissue sarcoma xenograft. Radiat. Res. 145:337-342. [PubMed] [Google Scholar]
- 19.Ilan, E., J. Arazi, O. Nussbaum, A. Zauberman, R. Eren, I. Lubin, L. Neville, O. Ben-Moshe, A. Kischitzky, A. Litchi, I. Margalit, J. Gopher, S. Mounir, W. Cai, N. Daudi, A. Eid, O. Jurim, A. Czerniak, E. Galun, and S. Dagan. 2002. The hepatitis C virus (HCV)-Trimera mouse: a model for evaluation of agents against HCV. J. Infect. Dis. 185:153-161. [DOI] [PubMed] [Google Scholar]
- 20.Kishine, H., K. Sugiyama, M. Hijikata, N. Kato, H. Takahashi, T. Noshi, Y. Nio, M. Hosaka, Y. Miyanari, and K. Shimotohno. 2002. Subgenomic replicon derived from a cell line infected with the hepatitis C virus. Biochem. Biophys. Res. Commun. 293:993-999. [DOI] [PubMed] [Google Scholar]
- 21.Kneteman, N. M., A. J. Weiner, J. O'Connell, M. Collett, T. Gao, L. Aukerman, R. Kovelsky, Z. J. Ni, A. Hashash, J. Kline, B. Hsi, D. Schiller, D. Douglas, D. L. Tyrrell, and D. F. Mercer. 2006. Anti-HCV therapies in chimeric scid-Alb/uPA mice parallel outcomes in human clinical application. Hepatology 43:1346-1353. [DOI] [PubMed] [Google Scholar]
- 22.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 24.Lieber, M. R., J. E. Hesse, S. Lewis, G. C. Bosma, N. Rosenberg, K. Mizuuchi, M. J. Bosma, and M. Gellert. 1988. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell 55:7-16. [DOI] [PubMed] [Google Scholar]
- 25.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 26.Lindenbach, B. D., and C. M. Rice. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933-938. [DOI] [PubMed] [Google Scholar]
- 27.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 30.Mercer, D. F., D. E. Schiller, J. F. Elliott, D. N. Douglas, C. Hao, A. Rinfret, W. R. Addison, K. P. Fischer, T. A. Churchill, J. R. Lakey, D. L. Tyrrell, and N. M. Kneteman. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7:927-933. [DOI] [PubMed] [Google Scholar]
- 31.Murray, E. M., J. A. Grobler, E. J. Markel, M. F. Pagnoni, G. Paonessa, A. J. Simon, and O. A. Flores. 2003. Persistent replication of hepatitis C virus replicons expressing the β-lactamase reporter in subpopulations of highly permissive Huh7 cells. J. Virol. 77:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 33.Perni, R. B., S. J. Almquist, R. A. Byrn, G. Chandorkar, P. R. Chaturvedi, L. F. Courtney, C. J. Decker, K. Dinehart, C. A. Gates, S. L. Harbeson, A. Heiser, G. Kalkeri, E. Kolaczkowski, K. Lin, Y. P. Luong, B. G. Rao, W. P. Taylor, J. A. Thomson, R. D. Tung, Y. Wei, A. D. Kwong, and C. Lin. 2006. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 50:899-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 35.Rigel Pharmaceuticals, Inc. 22. November 2004. Poor bioavailability results in insignificant clinical effects for Rigel R803 in phase I/II HCV trial. Rigel Pharmaceuticals, Inc., San Francisco, Calif. [Press release.]
- 36.Sumpter, R., Jr., C. Wang, E. Foy, Y. M. Loo, and M. Gale, Jr. 2004. Viral evolution and interferon resistance of hepatitis C virus RNA replication in a cell culture model. J. Virol. 78:11591-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tedaldi, E. M., L. Chen, N. Markowitz, L. Kelly, and D. Abrams. 2005. Effect of IL-2 on hepatitis C virus RNA levels in patients co-infected with human immunodeficiency virus receiving HAART. J. Viral Hepat. 12:414-420. [DOI] [PubMed] [Google Scholar]
- 38.ViroPharma, Inc. 15. July 2003. ViroPharma announces results of hepatitis C phase 1b study. ViroPharma, Inc., Exton, Pa. [Press release.]
- 39.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, G. Y., M. Konishi, C. M. Walton, D. Olive, K. Hayashi, and C. H. Wu. 2005. A novel immunocompetent rat model of HCV infection and hepatitis. Gastroenterology 128:1416-1423. [DOI] [PubMed] [Google Scholar]
- 41.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]