Abstract
Most methicillin-resistant Staphylococcus aureus (MRSA) isolates identified among blood isolates collected in Denmark between 1957 and 1970 belonged to either phage group III or the closely related 83A complex and had a PSTM antibiotype (resistance to penicillin [P], streptomycin [S], tetracycline [T], and methicillin [M]). Recently, some of these isolates were shown to have the same genetic backgrounds as contemporary epidemic MRSA isolates, and Danish methicillin-susceptible S. aureus (MSSA) isolates from the 1960s with a PST antibiotype were proposed to have been the recipients of the mecA gene in those lineages. In this study, we investigated the genetic backgrounds of isolates from the 83A complex that were fully susceptible or resistant to penicillin only in order to try to trace the evolutionary trajectory of contemporary MRSA lineages. We also studied MSSA and MRSA isolates from other phage groups in order to investigate if they had the potential to develop into contemporary MRSA clones. Most susceptible or penicillin-resistant isolates from phage group III or the 83A complex belonged to sequence type 8 (ST8) or ST5, while four isolates were ST254. STs 30, 45 and 25 were represented by MSSA isolates from other phage groups, which also included several singletons. Representatives of most of the current major epidemic MRSA lineages were identified among fully susceptible isolates collected in the 1960s, suggesting that these were MSSA lineages which carried genetic traits important for superior epidemicity before the acquisition of methicillin resistance.
Methicillin-resistant Staphylococcus aureus (MRSA) represents a major challenge to hospitals in all countries due to the emergence and spread of isolates with decreased susceptibilities to several antibiotic classes, in addition to methicillin and the other members of the β-lactam family. Molecular typing techniques applied to international collections of MRSA isolates have contributed to the understanding of the epidemiology and evolution of this infectious agent. In particular, data from multilocus sequence typing (MLST) in combination with staphylococcal chromosomal cassette mec (SCCmec) typing have indicated that methicillin resistance has emerged in the hospital setting as five main distinct genetic lineages and that most contemporary cases of MRSA disease are caused by a relatively small number of epidemic clones (15, 37).
A surveillance program operated by the Statens Serum Institut (Copenhagen, Denmark) produced a valuable collection of S. aureus isolates, which includes every strain recovered from patients with invasive disease in Denmark since 1957 to the present. The 5,304 isolates recovered between 1957 and 1973 are of particular interest because this period encompasses the years when resistance to the first antimicrobial agents emerged and spread among S. aureus isolates. Among these are the first MRSA isolates identified in Denmark in 1964, which followed closely the first British isolates collected in 1961. Analysis of these early MRSA isolates from the Danish collection by molecular typing techniques allowed us to identify the genetic backgrounds and the associated SCCmec types of these strains as representatives of what has become known as the archaic clone of MRSA, which was virtually identical to the very first isolates from the United Kingdom (11). Phage typing data and antibiotic susceptibility profiles indicated that most MRSA isolates collected between 1957 and 1970 belonged to either phage group III or the closely related 83A complex and, in addition to their resistance to methicillin (M), these isolates were also resistant to penicillin (P), streptomycin (S), and tetracycline (T) (PSTM antibiotype) (12). Recently, some of these isolates were shown to belong to the same genetic backgrounds as contemporary epidemic MRSA isolates by MLST and spa typing, and Danish methicillin-susceptible S. aureus (MSSA) isolates from the 1960s with a PST antibiotype were proposed to have been the recipients of the mecA gene in those lineages (11). Among the group III and 83A complex isolates, there were isolates that belonged to sequence type 250 (ST250), ST5, and ST30 (11).
The purpose of this study was to provide a more detailed portrait of the population structure of S. aureus before, during, and after the appearance of the first MRSA isolates in Denmark, on the basis of the characterization of additional strains from the collection of more than 5,000 isolates recovered during the 16-year period between 1957 and 1973. We characterized the genetic backgrounds of isolates from the 83A complex that were either fully susceptible or resistant to penicillin only in order to try to trace the evolutionary trajectory of the first MRSA lineages. The introduction of mecA into MSSA in the 1960s has been described within only one clonal complex (CC), namely, CC8, which led us to examine MRSA isolates from all other phage groups. Little is known about the prevalence of other CCs in MSSA at that time. MSSA isolates from other phage groups were studied in order to investigate whether these MSSA genetic backgrounds could be associated with the current MRSA epidemic clones.
MATERIALS AND METHODS
Bacterial isolates.
All S. aureus isolates in this study were clinical isolates from positive blood cultures collected in Denmark between 1957 and 1973 and preserved at the Statens Serum Institut. Ninety-four isolates were characterized by antimicrobial susceptibility testing, phage typing, and spa typing. Eighty-three isolates were selected for full characterization by MLST and SCCmec typing and for the presence of the Panton-Valentine leukocidin (PVL) genes.
Rationale for selection of bacterial isolates.
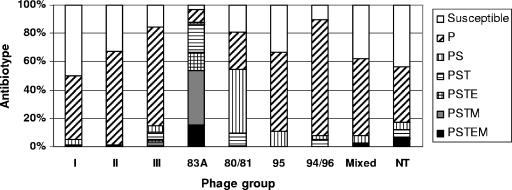
The majority of MRSA isolates identified among a total of 5,304 isolates collected in Denmark between 1957 and 1973 belonged to the 83A complex (95%), while most MSSA isolates, which were fully susceptible or resistant to penicillin only (94%), belonged to phage groups other than the 83A complex, including isolates which were nontypeable by phage typing (Fig. 1). Twenty-two MSSA isolates from the 83A complex and 16 MSSA isolates from the related phage group III were characterized by MLST and spa typing in order to assess whether the genetic backgrounds of these isolates were similar to those of MRSA isolates from the same phage groups which have been proposed to be the ancestors of contemporary epidemic MRSA clones. Since most MRSA isolates isolated from 1957 to 1973 belonged to the 83A complex, we also characterized 12 MRSA and 44 MSSA isolates from phage groups other than the 83A complex and group III that might correspond to contemporary successful clones. Among the 82 MSSA isolates studied, 56% caused hospital-acquired (HA) infections and 38% caused community-onset infections (the mode of acquisition was unknown for 6% of the isolates). Among the 12 MRSA isolates, 11 caused HA infections.
FIG. 1.
Distribution of S. aureus infection isolates collected between 1957 and 1973 in Denmark (n = 5,304) by the most prevalent antibiotypes and phage groups I (279 isolates), II (590 isolates), III (923 isolates), 83A complex (1,552 isolates), 80 complex (1,094 isolates), type 95 (9 isolates), 94,96 complex (39 isolates), mixed (362 isolates), and nontypeable (NT; 233 isolates).
Testing for susceptibility to penicillin, methicillin, streptomycin, erythromycin (E), and tetracycline was performed with Neosensitabs (Rosco, Taastrup, Denmark) on Danish blood agar (Statens Serum Institut) with a semiconfluent inoculum and overnight incubation at 35 to 36°C, according to the manufacturer's instructions and with the manufacturer's breakpoints (7, 43). Methicillin resistance was confirmed by mecA PCR.
Phage typing.
All study isolates were phage typed by the method of Blair and Williams (4) with the contemporary international set of typing phages and two Danish experimental phages (6, 26), which have been used since 1969 to differentiate among isolates of group III and the 83A complex.
MLST and spa typing.
MLST was performed as described previously (14), with the exception that primer arcCF2 (5′-CCT TTA TTT GAT TCA CCA GCG-3′) was used (11). MLST alleles and STs were identified by using the MLST database, available at http://www.mlst.net/. Analysis of the MLST STs was performed with eBURST, the enhanced version of the BURST (based upon related sequence types) algorithm, available at http://eburst.mlst.net/ (17). Molecular typing based on the sequence of the polymorphic region of protein A (spa typing) was performed as described previously (22, 45).
Detection of PVL genes.
Detection of PVL genes was performed by PCR, as described previously (29).
SCCmec typing.
SCCmec types were determined by a multiplex PCR strategy that derives a specific amplification pattern for each structural type, as described previously (35).
RESULTS
MSSA isolates from the 83A complex and phage group III.
ST8 was the predominant MLST profile among the MSSA isolates belonging to group III and the related 83A complex. It was present in four susceptible isolates and five penicillin-resistant isolates from the 83A complex (Table 1) and in seven mostly penicillin-resistant isolates from group III (Table 2). Eight isolates from these phage groups belonged to ST5, which was the second most represented genotype; six isolates belonged to ST1; three isolates belonged to ST250; and two isolates were ST50 (Tables 1 and 2). Interestingly, four isolates from group III had ST254 and are the first MSSA isolates with this genetic background identified (Table 2). Four new MLST profiles were identified among this set of isolates: ST569, which is a single-locus variant (SLV) of ST8 (Table 1); ST570, which is an SLV of ST5 (Table 2); ST568, which is an SLV of ST154 (Table 2); and ST138, which is an SLV of ST49 (Table 1).
TABLE 1.
Distribution of MSSA strains (n = 29) from the 83A complex, isolated from 1957 to 1973, by antimicrobial resistance, MLST, and spa types and presence of PVL
| Strain | Yr | Antibiogram | MLSTb
|
spa typing
|
PVL | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allelic profile | ST | CC | Kreiswirthc | Kreiswirth type | Ridomd | Ridom type | ||||
| E4344 | 1969 | Susceptible | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E4454 | 1970 | Susceptible | 3-3-1-1-4-4-3 | 8 | 8 | YHGGFMBQBLO | 59 | 11-19-12-12-21-17-34-24-34-22-25 | t211 | − |
| E4005 | 1969 | Susceptible | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBPBQBLO | 622 | 11-19-12-21-17-34-33-34-24-34-22-25 | t1187 | − |
| E4079 | 1969 | Susceptible | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E6321 | 1973 | Susceptible | 16-16-12-2-13-13-15 | 50 | 50 | ZDMJQDMO | 84 | 04-20-17-23-24-20-17-25 | t518 | − |
| E3410a | 1968 | Susceptible | 1-1-1-1-1-1-1 | 1 | 1 | UJFKBPE | 35 | 07-23-21-16-34-33-13 | t127 | |
| E3445a | 1968 | Susceptible | 1 | 1 | UJFKBPE | 35 | 07-23-21-16-34-33-13 | t127 | ||
| E691 | 1959 | Susceptible | 14-16-11-2-13-12-14 | 49 | 49 | ZDMMNNQMMMMMO | 677 | 04-20-17-17-31-31-24-17-17-17-17-17-25 | t127 | − |
| E5663 | 1972 | Susceptible | 14-16-11-2-47-12-14 | 138 | 49 | ZDMMNNQMMMMO | 623 | 04-20-17-17-31-31-24-17-17-17-17-25 | t208 | − |
| E676 | 1959 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E1570 | 1962 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E2129 | 1964 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E3008a | 1967 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | |
| E5589 | 1972 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E1044 | 1960 | P | 1-4-1-4-12-1-10 | 5 | 5 | TJMBMDMGMK | 2 | 26-23-17-34-17-20-17-12-17-16 | t002 | − |
| E1293 | 1961 | P | 1-4-1-4-12-1-10 | 5 | 5 | TJMBMDMGMK | 2 | 26-23-17-34-17-20-17-12-17-16 | t002 | − |
| E1436 | 1962 | P | 1-4-1-4-12-1-10 | 5 | 5 | TJMBMDMGMK | 2 | 26-23-17-34-17-20-17-12-17-16 | t002 | − |
| E2104a | 1964 | P | 1-4-1-4-12-1-10 | 5 | 5 | TJMBMDMGMK | 2 | 26-23-17-34-17-20-17-12-17-16 | t002 | |
| E6030 | 1972 | P | 1-4-1-4-12-1-10 | 5 | 5 | TJMBMDMGMK | 2 | 26-23-17-34-17-20-17-12-17-16 | t002 | − |
| E3001a | 1967 | P | 1-4-1-4-12-1-10 | 5 | 5 | TJMBMDMMK | 11 | 26-23-17-34-17-20-17-17-16 | t105 | |
| E2211 | 1965 | P | 1-1-1-1-1-1-1 | 1 | 1 | UJFKBPE | 35 | 07-23-21-16-34-33-13 | t127 | + |
| E2615a | 1966 | P | 1 | 1 | UJFKBPE | 35 | 07-23-21-16-34-33-13 | t127 | ||
| E1402 | 1962 | P | 1-1-1-1-1-1-1 | 1 | 1 | UJFKBP | 621 | 07-23-21-16-34-33-102 | t1186 | − |
| E228a | 1957 | PS | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E3733 | 1968 | PS | 3-3-1-1-4-4-16 | 250 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E2826 | 1966 | PS | 3-3-1-1-4-4-16 | 250 | 8 | YHGFBQBLO | 658 | 11-19-12-21-34-24-34-22-25 | t1188 | − |
| E4579 | 1970 | PS | 3-109-1-1-4-4-3 | 569 | 8 | YGFMBQBLO | 363 | 11-12-21-17-34-24-34-22-25 | t024 | − |
| E6674 | 1973 | PS | 1-4-1-4-12-1-10 | 5 | 5 | TMDMGMK | 47 | 26-17-20-17-12-17-16 | t045 | − |
| E229 | 1957 | PST | 3-3-1-1-4-4-16 | 250 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
TABLE 2.
Genetic backgrounds of MSSA strains (n = 16) from phage group III isolated between 1957 and 1964
| Strain | Yr | Antibiogram | MLST
|
spa typing
|
PVL | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allelic profile | ST | CC | Kreiswirtha | Kreiswirth type | Ridomb | Ridom type | ||||
| E230 | 1957 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E396 | 1958 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E402 | 1958 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E267 | 1957 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E201 | 1957 | ST | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E1264 | 1961 | P | 3-3-1-1-4-4-3 | 8 | 8 | YHGGFMBQBLO | 59 | 11-19-12-12-21-17-34-24-34-22-25 | t211 | − |
| E289 | 1957 | Susceptible | 3-3-1-1-4-4-3 | 8 | 8 | YHGGFMBQBQBLO | 461 | 11-19-12-12-21-17-34-24-34-24-34-22-25 | t1196 | − |
| E379 | 1958 | P | 3-32-1-1-4-4-3 | 254 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E219 | 1957 | PST | 3-32-1-1-4-4-3 | 254 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E203 | 1957 | Susceptible | 3-32-1-1-4-4-3 | 254 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E208 | 1957 | ST | 3-32-1-1-4-4-3 | 254 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E323 | 1958 | Susceptible | 1-4-1-4-12-1-10 | 5 | 5 | TJMBMDMGMK | 2 | 26-23-17-34-17-20-17-12-17-16 | t002 | − |
| E1603 | 1963 | P | 1-110-1-4-12-1-10 | 570 | 5 | TJMBMDMGMK | 2 | 26-23-17-34-17-20-17-12-17-16 | t002 | − |
| E247 | 1957 | P | 1-1-1-1-1-1-1 | 1 | 1 | UJFMEBKBPE | 426 | 07-23-21-17-13-34-16-34-33-13 | t273 | − |
| E268 | 1957 | P | 12-1-40-15-11-1-40 | 568 | 154 | UJFMBBBBPB | 427 | 07-23-21-17-34-34-34-34-33-34 | t1190 | + |
| E1114 | 1960 | P | 16-16-12-2-13-13-15 | 50 | 50 | ZDMJQDMO | 84 | 04-20-17-23-24-20-17-25 | t518 | − |
MSSA isolates from phage groups other than the 83A complex and phage group III.
The genetic backgrounds of MSSA isolates from phage groups other than the 83A complex and group III included ST30 (the most represented genotype present in 8 isolates from the 80 complex, 3 isolates from group I, and 3 nontypeable isolates), followed by ST25 (12 isolates from the 94,96 complex), ST45 (4 isolates from phage type 95), and ST121 (2 isolates from group II). STs 1, 8, 50, and 254, which were widely represented among MSSA isolates from group III and the 83A complex, were also present in a few isolates which were nontypeable or which had mixed phage types. Four new singletons, which were unique genotypes that cannot be identified with any clonal group in the MLST database, were identified in three susceptible or penicillin-resistant nontypeable isolates (STs 445 [n = 2] and 447) and in one susceptible isolate from group I (ST446) (Table 3).
TABLE 3.
Distribution of MSSA strains (n = 44) from phage groups other than 83A and III, isolated from 1957 to 1973, by phage group, antimicrobial resistance, MLST, and spa types and presence of PVL
| Strain | Yr | Phage group | Antibiogram | MLSTb
|
spa typing
|
PVL | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Allelic profile | ST | CC | Kreiswirthc | Kreiswirth type | Ridomd | Ridom type | |||||
| E216 | 1957 | NT | Susceptible | 1-61-1-31-12-4-10 | 445 | Singleton | YHMDMGMMMKK | 459 | 11-19-17-20-17-12-17-17-17-16-16 | t1191 | − |
| E227 | 1957 | NT | Susceptible | 1-61-1-31-12-4-10 | 445 | Singleton | YHMDMGMMMKK | 459 | 11-19-17-20-17-12-17-17-17-16-16 | t1191 | − |
| E3726 | 1968 | NT | Susceptible | 1-1-1-1-1-1-1 | 1 | 1 | UJGPLM | 125 | 07-23-12-33-22-17 | t156 | − |
| E3373 | 1967 | NT | P | 4-9-41-8-1-4-10 | 447 | Singleton | UG2MFBBLB | 414 | 07-06-17-21-34-34-22-34 | t164 | − |
| E253 | 1957 | NT | P | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQQ | 33 | 15-12-16-02-16-02-25-17-24-24 | t012 | + |
| E1410a | 1962 | NT | PS | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | - |
| E3245 | 1967 | NT | PST | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | + |
| E3175 | 1967 | NT | PSTE | 3-32-1-1-4-4-3 | 254 | 8 | YGFMBQBLO | 363 | 11-12-21-17-34-24-34-22-25 | t024 | − |
| E290 | 1957 | 80 | Susceptible | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | + |
| E367 | 1958 | 80 | P | 2-2-2-2-6-3-2 | 30 | 30 | WFGKAKAOMQQ | 158 | 15-21-12-16-02-16-02-25-17-24-24 | t238 | + |
| E200 | 1957 | 80 | PS | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | − |
| E217 | 1957 | 80 | PS | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | + |
| E610 | 1959 | 80 | PST | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | + |
| E3696 | 1968 | 80 | PST | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | − |
| E3742 | 1968 | 80 | PST | 2-2-2-2-6-3-2 | 30 | 30 | WGKAOMQ | 3 | 15-12-16-02-25-17-24 | t037 | + |
| E695 | 1959 | 80 | PSTE | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | + |
| E260 | 1957 | I | Susceptible | 18-62-6-2-38-50-5 | 446 | Singleton | ZMOKJBT2T2T2M | 430 | 04-17-25-16-23-34-50-50-50-17 | t1194 | + |
| E301 | 1957 | I | Susceptible | 2-2-2-2-6-3-2 | 30 | 30 | WGKAOMQQ | 234 | 15-12-16-02-25-17-24-24 | 275 | + |
| E266 | 1957 | I | P | 2-2-2-2-6-3-2 | 30 | 30 | W | 460 | 15 | t1192 | − |
| E2917 | 1966 | I | PS | 2-2-2-2-6-3-2 | 30 | 30 | WGKAKAOMQ | 43 | 15-12-16-02-16-02-25-17-24 | t021 | + |
| E231 | 1957 | Mixed | Susceptible | 16-16-12-2-13-13-15 | 50 | 50 | ZDMJQDMO | 84 | 04-20-17-23-24-20-17-25 | t518 | − |
| E220 | 1957 | Mixed | P | 1-1-1-1-1-1-1 | 1 | 1 | UJFMEBKBPE | 426 | 07-23-21-17-13-34-16-34-33-13 | t273 | + |
| E204 | 1957 | Mixed | PS | 3-32-1-1-4-4-3 | 254 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | − |
| E212 | 1957 | II | Susceptible | 6-5-6-2-7-14-5 | 121 | 121 | I2Z2EGMMJQ | 425 | 14-44-13-12-17-17-23-24 | t1193 | − |
| E269 | 1957 | II | P | 6-5-6-2-7-14-5 | 121 | 121 | I2Z2EGMMJZ2E | 428 | 14-44-13-12-17-17-23-44-13 | t1195 | + |
| E3812 | 1969 | 95 | Susceptible | 10-14-8-6-10-3-2 | 45 | 45 | A2AKBEMBKMBKB | 462 | 09-02-16-34-13-17-34-16-17-34-16-34 | t592 | − |
| E5655 | 1972 | 95 | Susceptible | 10-14-8-6-10-3-2 | 45 | 45 | XKAKBEMBKB | 73 | 08-16-02-16-34-13-17-34-16-34 | t015 | − |
| E6746 | 1973 | 95 | Susceptible | 10-14-8-6-10-3-2 | 45 | 45 | A2B | 187 | 09-34 | t362 | − |
| E5941 | 1972 | 95 | P | 10-14-8-6-10-3-2 | 45 | 45 | XKAKB | 278 | 08-16-02-16-34 | t230 | − |
| E5635 | 1972 | 95 | P | 45 | 45 | A2AKEEMBKB | 15 | 09-02-16-13-13-17-34-16-34 | t004 | ||
| E6020 | 1972 | 95 | P | 45 | 45 | A2AKBKB | 668 | 09-02-16-34-16-34 | t371 | ||
| E6261 | 1973 | 95 | P | 45 | 45 | XKB | 93 | 08-16-34 | t026 | ||
| E5318 | 1971 | 94,96 | Susceptible | 4-1-4-1-5-5-4 | 25 | 25 | ZGGGMM | 653 | 04-12-12-12-17-17 | t545 | − |
| E6566 | 1973 | 94,96 | Susceptible | 4-1-4-1-5-5-4 | 25 | 25 | ZFGU2DMGGM | 184 | 04-21-12-41-20-17-12-12-17 | t078 | − |
| E5223 | 1971 | 94,96 | Susceptible | 25 | 25 | ZFGU2DMGGGM | 548 | 04-21-12-41-20-17-12-12-12-17 | t258 | ||
| E4201 | 1969 | 94,96 | P | 4-1-4-1-5-5-4 | 25 | 25 | ZFGU2DMGGM | 184 | 04-21-12-41-20-17-12-12-17 | t078 | − |
| E4426 | 1970 | 94,96 | P | 25 | 25 | ZFGU2DMGGM | 184 | 04-21-12-41-20-17-12-12-17 | t078 | ||
| E5537 | 1972 | 94,96 | P | 25 | 25 | ZFGU2DMGGM | 184 | 04-21-12-41-20-17-12-12-17 | t078 | ||
| E4917 | 1971 | 94,96 | P | 25 | 25 | ZFGU2DMGM | 377 | 04-21-12-41-20-17-12-17 | t081 | ||
| E4520 | 1970 | 94,96 | PS | 25 | 25 | ZFGU2DMGGM | 184 | 04-21-12-41-20-17-12-12-17 | t078 | ||
| E6023 | 1972 | 94,96 | PS | 25 | 25 | ZFGU2DMGGM | 184 | 04-21-12-41-20-17-12-12-17 | t078 | ||
| E5331 | 1971 | 94,96 | PT | 25 | 25 | ZFGU2DMGGM | 184 | 04-21-12-41-20-17-12-12-17 | t078 | ||
| E5975 | 1972 | 94,96 | PST | 25 | 25 | ZFGMDMGGM | 618 | 04-21-12-17-20-17-12-12-17 | t349 | ||
| E6050 | 1972 | 94,96 | PST | 25 | 25 | ZFGMDMGGM | 618 | 04-21-12-17-20-17-12-12-17 | t349 | ||
MRSA isolates from phage groups other than the 83A complex and phage group III and nontypeable MRSA.
All isolates belonged to CC8. ST250 was the most common genotype among the nontypeable MRSA isolates. It was present in six of eight isolates collected between 1967 and 1970. STs 254 and 8 were each represented by a single nontypeable isolate. In addition, ST247 was the genetic background of all of the four isolates of mixed phage types. All isolates, including the ST247 isolates, were SCCmec type I (Table 4).
TABLE 4.
Genetic backgrounds of MRSA strains (n = 12) from phage groups other than 83A and III isolated between 1966 and 1970
| Strain | Yr | Phage group | Antibiogram | MLST
|
spa typing
|
SCCmec type | PVL | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allelic profile | ST | CC | Kreiswirtha | Kreiswirth type | Ridomb | Ridom type | ||||||
| E3717 | 1968 | NTc | PSTEM | 3-3-1-1-4-4-16 | 250 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | I | − |
| E4075 | 1969 | NT | PSTEM | 3-3-1-1-4-4-16 | 250 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | I | − |
| E4330 | 1969 | NT | PSTEM | 3-3-1-1-4-4-16 | 250 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | I | − |
| E3536 | 1968 | NT | PSTEM | 3-3-1-1-4-4-16 | 250 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | I | − |
| E4345 | 1969 | NT | PSTEM | 3-3-1-1-4-4-16 | 250 | 8 | YHGGFMBQBLO | 59 | 11-19-12-12-21-17-34-24-34-22-25 | t211 | I | − |
| E4676 | 1970 | NT | PSTEM | 3-3-1-1-4-4-16 | 250 | 8 | YHGGFMBQBLO | 59 | 11-19-12-12-21-17-34-24-34-22-25 | t211 | I | − |
| E3183 | 1967 | NT | PSTEM | 3-32-1-1-4-4-3 | 254 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | I | − |
| E4423 | 1970 | NT | PSTEM | 3-3-1-1-4-4-3 | 8 | 8 | YHGFMBQBLO | 1 | 11-19-12-21-17-34-24-34-22-25 | t008 | I | − |
| E2668 | 1966 | Mixed | PSTEM | 3-3-1-12-4-4-16 | 247 | 8 | YHFGFMBQBLO | 4 | 11-19-21-12-21-17-34-24-34-22-25 | t051 | I | − |
| E2779 | 1966 | Mixed | PSTEM | 3-3-1-12-4-4-16 | 247 | 8 | YHFGFMBQBLO | 4 | 11-19-21-12-21-17-34-24-34-22-25 | t051 | I | − |
| E2900 | 1966 | Mixed | PSTEM | 3-3-1-12-4-4-16 | 247 | 8 | YHFGFMBQBLO | 4 | 11-19-21-12-21-17-34-24-34-22-25 | t051 | I | − |
| E2940 | 1966 | Mixed | PSTEM | 3-3-1-12-4-4-16 | 247 | 8 | YHFGFMBQBLO | 4 | 11-19-21-12-21-17-34-24-34-22-25 | t051 | I | − |
Presence of PVL locus.
The presence of the PVL locus, a common virulence marker of community-acquired MRSA isolates, which included some of the genotypes observed in this study, was determined. The PVL genes were present in most isolates of ST30 (Table 3) and also in isolates belonging to STs 1 (Tables 1 and 3), 121 (Table 3), 446 (Table 3), and 568 (Table 2).
DISCUSSION
The emergence of pandemic clonal lineages of MRSA raised the question of whether these lineages possess features which are superior in terms of epidemicity to other genetic backgrounds. It has been suggested that the most widespread clones have been selected because they are good colonizers, in addition to being highly transmissible and highly virulent (30, 31). If this is the case, then, the MSSA ancestors of today's pandemic MRSA clones would already have been at an advantage in the pre-MRSA era and, therefore, would have been represented by isolates collected as far back in time as possible. In the absence of representative collections of pre-antibiotic-era S. aureus isolates, we studied the oldest isolates that have been collected and stored in Denmark by the Statens Serum Institut in order to try to identify the progenitors of today's pandemic clones.
Phage typing of the isolates, one of the few typing methods available in the 1960s, constituted the basis for the selection of the isolates together with the antimicrobial susceptibility patterns. As can be seen in the Results section, phage typing did not always group isolates in the same manner as more recent and powerful DNA typing approaches, as already documented by others (21, 52). In the collection examined in our work, there was a low typeability for phage typing, with 23% of the S. aureus isolates not belonging to recognized phage groups (the isolates were either nontypeable or of mixed phage types). These nontypeable and mixed-phage-type isolates belonged to multiple CCs. MSSA isolates from group III and the related 83A complex were more heterogeneous than MRSA isolates from these phage groups examined previously. While those isolates predominantly belonged to CC8 (25/45), like the contemporary MRSA isolates, 20 isolates were from five other CCs.
Major contemporary MRSA genetic backgrounds present in susceptible isolates from the 1960s.
Four of the five major contemporary epidemic genetic backgrounds of MRSA were already significantly represented among the Danish MSSA isolates from the 1960s: ST5, ST8, ST45, and ST30 (the predicted ancestral genotype of ST36) (Fig. 2). Sixty-five of the 94 isolates characterized in this work (69%) belonged to these genotypes. ST22 (the British epidemic MRSA type 15 [EMRSA-15] clone) is the only major contemporary epidemic clone not present in this collection. This may be due to a limitation imposed by the restricted geographic area of the sample or the early time period of our isolates rather than to the potential of this clone's genotype for geographical spread, which has been well documented (34, 37-39). We identified a new SLV of ST5, ST570, that differs from the ST5 profile in the arcC allele. ST5 is an old and globally spread lineage which is the predicted founder of CC5 (3, 8, 15, 18, 28, 40, 44, 48) and is the genetic background of most MRSA isolates with reduced susceptibility to vancomycin (vancomycin-intermediate S. aureus isolates) identified so far (23, 25, 32). The ST8 genetic background was proposed to represent the ancestor of the very first MRSA isolates and is the predicted founder of CC8 (15), which includes the Iberian clone, ST247 (11, 13), and the Brazilian clone, ST239 (36, 46), two of the most successful pandemic clones of MRSA. In this study, we identified a new genotype, ST569, which is another SLV of ST8, and the first MSSA isolates of the ST254 genetic background, which includes the epidemic strains ST254-MRSA-I and ST254-MRSA-IV (also known as the Hannover MRSA clone), identified in the United Kingdom and Germany (15), respectively. Only ST254 MRSA isolates have been characterized previously (41); and the identification of MSSA ST254 isolates provides an additional link in the evolution of CC8, which has been predicted to have existed in the evolution model of MRSA proposed by Robinson and Enright (41). ST45 MRSA, also known as the Berlin epidemic MRSA clone, was first observed in Germany in 1993 and was later found in several other European countries (49, 51). It has been shown that ST45 is a frequent genotype among contemporary MSSA isolates (50). The three fully susceptible ST45 isolates characterized in this study provide evidence that this genetic background was already represented among MSSA isolates collected in the 1960s.
FIG. 2.
Diagram of clonal groups defined for MSSA isolates with the eBURST algorithm. Clonal groups were defined by the less stringent approach: five or more shared alleles of the seven MLST loci. Single genotypes that do not correspond to any clonal group were identified as singletons. Each number represents an MLST ST. Blue numbers represent STs found in the present study, while black or white numbers represent STs which were already present in the database. The ST of the predicted ancestral genotype (predicted founder) is associated with a blue circle whose area indicates the prevalence of the ST in the entire MLST database. In parentheses, the number of isolates with a particular ST deposited in the database is followed by the number of isolates with that ST found in our study. Although STs 36 (EMRSA-16), 247 (Iberian clone), and 239 (Brazilian clone) have not been found among MSSA isolates in the present study, their numbers have been included in the diagram because of their epidemiological importance and relationship to STs in this work.
PVL-positive MSSA isolates from the 1960s belong to major community-acquired genotypes.
ST30 is the predicted ancestral genotype of CC30, whose most ubiquitous SLV is ST36, which corresponds to the British EMRSA-16 epidemic clone, which is another major contemporary genetic background (10, 15). Unlike ST36, ST30 MRSA has rarely been found among nosocomial isolates in Europe (1, 15, 35) but is very common in community isolates from Australia (9). Together with ST1, ST30 is among the six PVL-positive MRSA isolates which have been acquired in the community worldwide (47). Recently, it has been proposed that a PVL-positive branch of ST30 MSSA from the phage 80 complex has evolved into the community-acquired Southwest Pacific MRSA clone, while the hospital-acquired EMRSA-16 clone evolved from a PVL-negative ST30 lineage (42). In this study, we observed that both PVL-positive and PVL-negative MSSA ST30 isolates not only from the 80 complex but also from the related group I and from nontypeable isolates were present in the hospital setting in Denmark as early as the 1950s. Interestingly, 11 of 14 ST30 isolates were from HA infections. Moreover, the PVL genes, whose mobility is facilitated by carriage on bacteriophages, were also present in MSSA isolates from the ST 1, 121, and 568 genetic backgrounds and in the singleton ST446. This observation and the more recent identification of PVL-positive contemporary isolates also in diverse genetic backgrounds (24) support the notion that although the PVL toxin is widely present in MRSA isolates from the community, the PVL toxin is only one of a combination of genetic traits which confer a selective advantage to clones of S. aureus. ST121 is one of the genetic backgrounds displaying PVL-positive isolates both in the 1950s (this study) and today (24). Furthermore, only MSSA ST121 isolates have been identified so far (15, 24). It is possible that ST121 is one of the staphylococcal genetic backgrounds which has been designated nonpermissive of the mecA gene and its product (27) and, therefore, has never acquired and/or sustained the selective advantage of methicillin resistance, even though it possesses other genetic traits which have contributed to its persistence both in the hospital and in the community since the late 1950s. There may be a similar situation for ST25. ST25 MSSA has been present for over three decades in the hospital and in the community (although it is PVL negative) (15, 16; this study). ST25 is not a favorable genetic background for the constitutive expression of a plasmid-introduced mecA (27) but has recently been reported to be a highly transmissible clone associated with colonization and infection in the niche of AIDS patients with a history of drug use (20).
New genotypes observed among MSSA isolates from the 1960s.
While major contemporary MRSA lineages are represented among MSSA isolates from both phage group III and the 83A complex and phage groups other than group III and the 83A complex, two of the three singleton genotypes identified in this study (ST445 and ST447) were nontypeable by phage typing. These genotypes may be less fit than known lineages as well, since they have not been identified at all again since the time that these isolates were collected. It would be interesting to know in what way ST445, ST446, and ST447 differ from the five major genetic lineages of contemporary MRSA. The singleton MLST genotype in each of these isolates was associated with a new spa type (types 459, 430, and 414, respectively; Table 3). These new spa types only vaguely resemble more widespread types (http://spaserver.ridom.de). The other new spa types identified in this study appear to have originated by duplication and/or deletion of some repeats in more common types (e.g., spa types 621 to 658; Table 1). Another new MLST genotype identified among fully susceptible isolates from the 83A complex in this collection corresponds to ST138, an SLV of ST49. ST49 and ST50, also identified in this work, are the MLST profiles of only three other isolates in the MLST database (http://www.mlst.net). These isolates were collected in 1997 and 1998 in the United Kingdom, they were MSSA, and they were isolated from blood. Therefore, ST138, ST49, and ST50 are genotypes associated with susceptible isolates that were first identified in 1959 and that have still been isolated from blood in the recent past but have not spread globally.
MRSA clones in the 1960s and today.
Although the MSSA progeny of four of the five major contemporary MRSA clones were identified among isolates collected in the 1960s, only MRSA isolates belonging to the CC8 genetic background were present at that time. Therefore, it seems clear that the introduction of mecA into CC8 preceded the acquisition of methicillin resistance in the CC5, CC30, and CC45 backgrounds. Within CC8, only ST247 and ST250 had previously been identified among early MRSA isolates from phage group III and the 83A complex (11). Now, we report the presence of ST8 and ST254 isolates, besides ST247 and ST250 isolates, in mixed-phage-type and nontypeable isolates. We propose that these mixed-phage-type and nontypeable isolates have evolved out of group III and the 83A complex, in which most ST247 and ST250 isolates have been identified, and that ST8 and ST254 are variants of the CC8 background which appeared later in the MRSA epidemic. The early timing for the appearance of ST247 is corroborated by the identification of ST247 isolates with SCCmec type I. One such isolate is likely to be ancestral to contemporary ST247, which possess a cassette of type IA (not found anywhere else) that differs from cassette I by having pUB110 present and the hypervariable region deleted (36). Both ST247-SCCmec type I (19, 35) and ST8-SCCmec type I, identified for the first time in this study, may have evolved out of ST250 after its acquisition of methicillin resistance on the basis of the numbers of isolates with these backgrounds identified in the period from 1957 to 1973. This hypothesis contemplates only one exogenous acquisition of SCCmec type I in CC8 during this period. We propose a variation of the model originally proposed by Robinson and Enright (41) for the early evolution of MRSA within CC8 on the basis of the findings (STs and associated SCCmec types) for the earliest isolates identified and not on the basis of the prevalence of contemporary isolates with this genetic background (Fig. 3).
FIG. 3.
Model for the early evolution of MRSA in CC8. The scheme was based on the molecular typing of MRSA and MSSA isolates collected between 1957 and 1973 in Denmark. Arrows indicate the directions and changes between genotypes.
Evidence of the existence of more fit lineages of S. aureus.
Why are some isolates occasionally recovered from patients with invasive disease but not widespread as major MRSA clones? Several studies have shown that the genetic backgrounds of S. aureus isolates carried by healthy donors corresponded to the predominant CCs recovered from patients with invasive disease, the five major lineages of nosocomial MRSA: CC5, CC8, CC22, CC30, and CC45 (2, 16, 33). ST121 was among the STs recovered from the community in a Dutch study from 1997 to 2002 and was mostly associated with MSSA isolates causing nonbullous impetigo (33). Susceptible and penicillin-resistant isolates from ST121 in this study had also been recovered from blood in Denmark in 1957. Yet, ST121 is not dominant among the nasal carriage genotypes, contrary to the most successful clones, which appear to be apt both at causing disease and at colonizing the human host. Interaction of a pathogen with a host is a complex process, which may include searching for an entry site, targeting a place for multiplication in the host, and becoming persistent in the host or managing to reach the next host (5). Maintenance of an S. aureus clone in the original host and transmission to the next host are affected by a number of fitness factors which are broader in their scope than virulence factors and which include functions related to metabolic processes and other mechanisms affecting multiplication and survival. It is possible that STs 121, 25, 138, 49, and 50, in addition to corresponding to genotypes less favorable to the stable acquisition of the mecA gene, also possess characteristics which are less suitable to the persistence in the host and transfer between hosts.
In conclusion, MSSA representatives of most of the current major epidemic MRSA lineages (ST5, ST8, ST30, and ST45) were identified in the 1960s, suggesting that these were widespread lineages which carried genetic traits important for superior epidemicity, through their capacity to cause invasive disease and colonizing the host, well before the acquisition of antibiotic resistance.
Acknowledgments
We thank Alexander Tomasz for helpful discussions. We also thank Marilyn Chung from The Rockefeller University for collaboration with MLST and spa typing and S. Naidich and B. N. Kreiswirth for the assignment of new spa types.
This work was partially supported by project POCTI/SAU-ESP/57841/2004, and A. R. Gomes was supported by grants SFRH/BPD/9373/2002 and SFRH/BPD/26118/2005 from the Fundação para a Ciência e a Tecnologia, Lisbon, Portugal.
REFERENCES
- 1.Aires de Sousa, M., C. Bartzavali, I. Spiliopoulou, I. S. Sanches, M. I. Crisóstomo, and H. de Lencastre. 2003. Two international methicillin-resistant Staphylococcus aureus clones endemic in a university hospital in Patras, Greece. J. Clin. Microbiol. 41:2027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., T. Conceição, C. Simas, and H. de Lencastre. 2005. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 43:5150-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires de Sousa, M., H. de Lencastre, I. Santos Sanches, K. Kikuchi, K. Totsuka, and A. Tomasz. 2000. Similarity of antibiotic resistance patterns and molecular typing properties of methicillin-resistant Staphylococcus aureus isolates widely spread in hospitals in New York City and in a hospital in Tokyo, Japan. Microb. Drug Resist. 6:253-258. [DOI] [PubMed] [Google Scholar]
- 4.Blair, J. E., and R. E. O. Williams. 1961. Phage typing of staphylococci. Bull. W. H. O. 24:771-784. [PMC free article] [PubMed] [Google Scholar]
- 5.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bülow, P. 1968. A new epidemic phage type of Staphylococcus aureus. I. The experimental typing phage 6557′. Acta Pathol. Microbiol. Scand. 72:147-159. [DOI] [PubMed] [Google Scholar]
- 7.Casals, J. B., and O. G. Pedersen. 1972. Tablet sensitivity testing: a comparison of different methods. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. Immunol. 80:806-816. [DOI] [PubMed] [Google Scholar]
- 8.Chung, M., G. Dickinson, H. de Lencastre, and A. Tomasz. 2004. International clones of methicillin-resistant Staphylococcus aureus in two hospitals in Miami, Florida. J. Clin. Microbiol. 42:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coombs, G. W., G. R. Nimmo, J. M. Bell, F. Huygens, F. G. O'Brien, M. J. Malkowski, J. C. Pearson, A. J. Stephens, P. M. Giffard, and the Australian Group for Antibiotic Resistance. 2004. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42:4735-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, R. A., C. Conquest, C. Mallaghan, and R. R. Marples. 1995. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 29:87-106. [DOI] [PubMed] [Google Scholar]
- 11.Crisóstomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin- susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lencastre, H., M. Chung, and H. Westh. 2000. Archaic strains of methicillin-resistant Staphylococcus aureus: molecular and microbiological properties of isolates from the 1960s in Denmark. Microb. Drug Resist. 6:1-10. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez, M. A., H. de Lencastre, J. Liñares, and A. Tomasz. 1994. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J. Clin. Microbiol. 32:2081-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin- resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus. (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomes, A. R., I. S. Sanches, M. Aires de Sousa, E. Castañeda, and H. de Lencastre. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Colombian hospitals: dominance of a single unique multidrug-resistant clone. Microb. Drug Resist. 7:23-32. [DOI] [PubMed] [Google Scholar]
- 19.Gomes, A. R., S. Vinga, M. Zavolan, and H. de Lencastre. 2005. Analysis of the genetic variability of virulence-related loci in epidemic clones of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 49:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon, R. J., B. Quagliarello, C. Cespedes, M. Chung, H. de Lencastre, P. Vavagiakis, M. Miller, B. Zeller, and F. D. Lowy. 2005. A molecular epidemiological analysis of 2 Staphylococcus aureus clonal types colonizing and infecting patients with AIDS. Clin. Infect. Dis. 40:1028-1036. [DOI] [PubMed] [Google Scholar]
- 21.Grundmann, H., S. Hori, M. C. Enright, C. Webster, A. Tami, E. J. Feil, and T. Pitt. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40:4544-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harmsen, D., H. Claus, W. Witte, J. Rothganger, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, A., M. Ganner, S. McGuane, T. L. Pitt, B. D. Cookson, and A. M. Kearns. 2005. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 43:2384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howe, R. A., A. Monk, M. Wootton, T. R. Walsh, and M. C. Enright. 2004. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg. Infect. Dis. 10:855-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jessen, O., K. Rosendal, P. Bülow, V. Faber, and K. R. Eriksen. 1969. Changing staphylococci and staphylococcal infections. A ten-year study of bacteria and cases of bacteremia. N. Engl. J. Med. 281:627-635. [DOI] [PubMed] [Google Scholar]
- 27.Katayama, Y., D. A. Robinson, M. C. Enright, and H. F. Chambers. 2005. Genetic background affects stability of mecA in Staphylococcus aureus. J. Clin. Microbiol. 43:2380-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ko, K. S., J. Y. Lee, J. Y. Suh, W. S. Oh, K. R. Peck, N. Y. Lee, and J. H. Song. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 30.Lipsitch, M. 2001. Microbiology. Bacterial population genetics and disease. Science 292:59-60. [DOI] [PubMed] [Google Scholar]
- 31.Martinez, J. L., and F. Baquero. 2002. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 15:647-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melles, D. C., R. F. Gorkink, H. A. Boelens, S. V. Snijders, J. K. Peeters, M. J. Moorhouse, P. J. van der Spek, W. B. van Leeuwen, G. Simons, H. A. Verbrugh, and A. van Belkum. 2004. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Investig. 114:1732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, P. C., and J. A. Lindsay. 2002. Molecular characterisation of the dominant UK methicillin-resistant Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J. Med. Microbiol. 51:516-521. [DOI] [PubMed] [Google Scholar]
- 35.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 37.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 38.O'Neill, G. L., S. Murchan, A. Gil-Setas, and H. M. Aucken. 2001. Identification and characterization of phage variants of a strain of epidemic methicillin-resistant Staphylococcus aureus (EMRSA-15). J. Clin. Microbiol. 39:1540-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson, J. F., and S. Reith. 1993. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J. Hosp. Infect. 25:45-52. [DOI] [PubMed] [Google Scholar]
- 40.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 41.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson, D. A., A. M. Kearns, A. Holmes, D. Morrison, H. Grundmann, G. Edwards, F. G. O'Brien, F. C. Tenover, L. K. McDougal, A. B. Monk, and M. C. Enright. 2005. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet 365:1256-1258. [DOI] [PubMed] [Google Scholar]
- 43.Rosco. 2002. Neosensitabs® susceptibility testing. Users' guide, 13th ed. Rosco, Taastrup, Denmark.
- 44.Sá-Leão, R., I. Santos Sanches, D. Dias, I. Peres, R. M. Barros, and H. de Lencastre. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pediatric hospital in Portugal and in international samples: relics of a formerly widely disseminated strain? J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teixeira, L. A., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Velazquez-Meza, M. E., M. Aires de Sousa, G. Echaniz-Aviles, F. Solórzano-Santos, G. Miranda-Novales, J. Silva-Sanchez, and H. de Lencastre. 2004. Surveillance of methicillin-resistant Staphylococcus aureus in a pediatric hospital in Mexico City during a 7-year period (1997 to 2003): clonal evolution and impact of infection control. J. Clin. Microbiol. 42:3877-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wannet, W. J., E. Spalburg, M. E. Heck, G. N. Pluister, R. J. Willems, and A. J. de Neeling. 2004. Widespread dissemination in The Netherlands of the epidemic Berlin methicillin-resistant Staphylococcus aureus clone with low-level resistance to oxacillin. J. Clin. Microbiol. 42:3077-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witte, W., C. Braulke, C. Cuny, D. Heuck, and M. Kresken. 2001. Changing pattern of antibiotic resistance in methicillin-resistant Staphylococcus aureus from German hospitals. Infect. Control Hosp. Epidemiol. 22:683-686. [DOI] [PubMed] [Google Scholar]
- 51.Witte, W., M. Kresken, C. Braulke, and C. Cuny. 1997. Increasing incidence and widespread dissemination of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in central Europe, with special reference to German hospitals. Clin. Microbiol. Infect. 3:414-422. [DOI] [PubMed] [Google Scholar]
- 52.Zadoks, R. N., W. B. van Leeuwen, D. Kreft, L. K. Fox, H. W. Barkema, Y. H. Schukken, and A. van Belkum. 2002. Comparison of Staphylococcus aureus isolates from bovine and human skin, milking equipment, and bovine milk by phage typing, pulsed-field gel electrophoresis, and binary typing. J. Clin. Microbiol. 40:3894-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]