Abstract
Women are at significant risk of human immunodeficiency virus (HIV) infection, with the cervicovaginal mucosa serving as a major portal for virus entry. Female-initiated preventatives, including topical microbicides, are urgently needed to help curtail the HIV/AIDS pandemic. Here we report on the development of a novel, live microbicide that employs a natural vaginal strain of Lactobacillus jensenii engineered to deliver the potent HIV inhibitor cyanovirin-N (CV-N). To facilitate efficient expression of CV-N by this bacterium, the L. jensenii 1153 genome was sequenced, allowing identification of native regulatory elements and sites for the chromosomal integration of heterologous genes. A CV-N expression cassette was optimized and shown to produce high levels of structurally intact CV-N when expressed in L. jensenii. Lactobacillus-derived CV-N was capable of inhibiting CCR5-tropic HIVBaL infectivity in vitro with a 50% inhibitory concentration of 0.3 nM. The CV-N expression cassette was stably integrated as a single copy into the bacterial chromosome and resolved from extraneous plasmid DNA without adversely affecting the bacterial phenotype. This bacterial strain was capable of colonizing the vagina and producing full-length CV-N when administered intravaginally to mice during estrus phase. The CV-N-producing Lactobacillus was genetically stable when propagated in vitro and in vivo. This work represents a major step towards the development of an inexpensive yet durable protein-based microbicide to block the heterosexual transmission of HIV in women.
Worldwide, over 40 million people are living with human immunodeficiency virus (HIV)/AIDS (33), and the predominant mode of HIV transmission is via heterosexual contact (36, 51). Women are increasingly at risk of HIV infection (1, 33) and now comprise over half of the new cases (33). Although condoms and mutual monogamy can prevent heterosexual HIV transmission, women are often prevented from employing these simple yet effective methods. While an effective HIV vaccine remains an important goal (7), the ability of HIV to mutate rapidly and evade the immune response (21) has made the development of a vaccine problematic. Topical microbicides that block HIV infection directly at mucosal surfaces represent an alternative preventative (44). For global use, microbicides need to be inexpensive to manufacture, stable, safe, and effective at reducing the transmission of HIV from an infected partner. To meet these challenges, we are developing a novel class of microbicide that uses live human vaginal Lactobacillus as a self-renewing delivery system for potent protein-based HIV inhibitors.
The vaginal mucosa of healthy women of childbearing age is populated with commensal bacteria typically dominated by H2O2-producing Lactobacillus species, which play a protective role in preventing urogenital infections (35). Depletion of vaginal lactobacilli is associated with establishment of opportunistic infections and increased risk of acquiring HIV and herpes simplex virus type 2 in women (10, 11, 37, 42). The principal Lactobacillus species isolated from the vaginal mucosa of healthy women include L. jensenii, L. crispatus, L. gasseri, and L. iners (2, 46, 54). L. jensenii 1153 was chosen for microbicide development based on its desirable growth and adherence properties, its association with reduced urogenital infections, and its suitability for genetic manipulation (21).
Cyanovirin-N (CV-N), isolated from the cyanobacterium Nostoc ellipsosporum (6), is an attractive HIV inhibitor because of its ability to potently inhibit virtually all clades of HIV in vitro (6, 13). CV-N also blocks vaginal HIV transmission in a nonhuman primate model (43) and exhibits a positive safety profile in both rabbits and macaques (6, 43). In addition, it is stable at low pH and resistant to chemical and thermal denaturation (6, 24). Mechanistically, CV-N blocks multiple steps leading to membrane fusion and virus entry (13). It impairs CD4-dependent and CD4-independent binding of gp120 to the target cells and blocks soluble CD4-induced binding of gp120 to coreceptors CXCR4 and CCR5 (13). The antiviral activity of CV-N is mediated through specific, high-affinity binding to the viral surface envelope glycoprotein gp120 and possibly also gp41 (38).
In a previous proof-of-concept study, we demonstrated that a human vaginal isolate of Lactobacillus could be engineered to secrete biologically active two-domain CD4, a prototypical HIV binding protein (8). In this report, we describe the development of a genetically stable strain of vaginal L. jensenii engineered to constitutively secrete high levels of structurally intact CV-N that potently inhibits both CCR5- and CXCR4-tropic HIV. We further show that this strain is capable of transiently colonizing the mouse vagina during estrus phase and producing full-length CV-N in vivo. Taken together, this work provides an important step forward in the development of a live vaginal microbicide for blocking mucosal transmission of HIV in women.
MATERIALS AND METHODS
Bacterial strains, culture, and transformation.
Naturally occurring isolates of L. jensenii, L. crispatus, and L. gasseri were obtained from vaginal swabs of healthy menarchial women and cultivated, as described previously (8). Shuttle plasmids were maintained and propagated in Escherichia coli, purified, and electroporated into lactobacilli (8). Transformed lactobacilli were routinely propagated on either MRS agar plates or in MRS broth containing 20 μg/ml erythromycin.
Sequencing of the L. jensenii 1153 genome.
To identify endogenous regulatory elements and chromosomal integration sites for increasing the efficiency of heterologous gene expression, the L. jensenii 1153 genome was sequenced using the whole-genome shotgun approach (14) at the Lawrence Berkeley National Laboratory. Genomic DNA was mechanically sheared using a HydroShear (GeneMachines, San Carlos, CA) to create 3- to 8-kb genomic libraries. Plasmid DNA was sequenced on an ABI PRISM 3700 automated DNA sequencer (Applied Biosystems, Foster City, CA) to provide threefold coverage of the L. jensenii 1153 genome. The sequence assembly was performed using Genome Assembler or CAP4 (Paracel, Inc.). Identification of open reading frames and prediction of protein function was made on the basis of BLASTP analysis against the nonredundant GenBank database.
Identification of strong L. jensenii 1153 promoters.
A shuttle vector was constructed, containing a promoterless green fluorescent protein (GFP) gene (50) downstream of an EcoRI restriction site and ribosomal binding site. L. jensenii genomic DNA was digested with restriction enzyme EcoRI and ligated into the EcoRI site of the GFP expression vector. Plasmids isolated from E. coli colonies that emitted the brightest green fluorescence upon exposure to UV light (302 to 312 nm) were electroporated into L. jensenii, and the resulting L. jensenii clones were screened for GFP expression by flow cytometry. For each clone, the fluorescence of 20,000 cells was measured using a FACScan system (Becton Dickinson, Franklin Lakes, NJ) and CellQuest software. The mean fluorescence intensity was calculated using FLOWJO software (Tree Star, Inc., Ashland, OR). The ability of the promoters to drive secreted CV-N expression in L. jensenii 1153 was also evaluated using the Lactococcus lactis P23 promoter for comparison. The expression level of soluble CV-N was quantified based on the band density of Coomassie blue R-250 stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels or Western blots in reference to a recombinant CV-N (P51G) protein standard (Laboratory of Chemical Physics, National Institutes of Health) at different concentrations.
Construction of CV-N expression vectors.
Lactobacilli are codon-biased organisms containing only about 36% G+C in their genomes (29). The nucleotide sequence of the CV-N gene was recoded by assembly PCR (41) to conform more closely to the optimal Lactobacillus codon usage (8). In addition, proline 51 was changed to glycine (P51G) by site-directed mutagenesis to stabilize CV-N in a properly folded monomeric form (24).
To express CV-N (P51G) in L. jensenii, the synthetic CV-N gene was cloned into the modular shuttle vector pOSEL175, a modified version of pOSEL144 (8). The expression cassette for secreted CV-N (P51G) contained a promoter, the signal sequence of the L. crispatus S-layer gene (CbsAss), from the ribosome binding site to the signal peptidase cleavage site (8) (Fig. 1A). The cassette also contained the CV-N (P51G) gene with a TAA stop codon at the 3′ end. A 4-amino-acid peptide (APVT), corresponding to the N terminus of the mature CbsA protein, was inserted downstream of the CbsA signal sequence to more closely resemble the native signal peptidase cleavage site of this protein. Site-directed mutagenesis was carried out using a QuickChange XL kit (Stratagene) and the following oligonucleotides: (5′-GTTTCAGCTGCTCCAGTTACTTTAGGTAAGTTTTC-3′ and 5′-GAAAACTTACCTAAAGTAACTGGAGCAGCTGAAAC-3′ [the codons of the tetrapeptide APVT are underlined]).
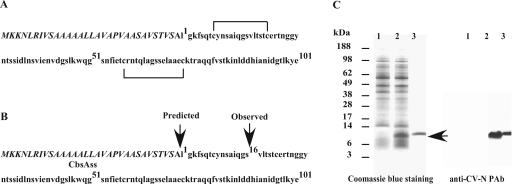
FIG. 1.
Proteolytic processing of L. jensenii-derived CV-N (P51G). (A) Schematic representation of CV-N (P51G) with the CbsA signal sequence (italicized). CV-N (P51G) possesses two disulfide bonds. (B) Proteolytic processing of Lactobacillus-derived CV-N (P51G). CV-N (P51G) is first synthesized as a CbsAss-bearing precursor protein. Following cleavage by a signal peptidase, the mature protein is released into the extracellular milieu. The predicted and observed cleavage sites are shown. (C) Production of proteolytically processed CV-N (P51G) by L. jensenii 1153. L. jensenii 1153 containing the empty expression vector (lane 1) and nonmodified CV-N (P51G) expression construct (lane 2) were cultured in Rogosa broth. Proteins from 300 μl of cell-free conditioned Rogosa media were separated by reducing SDS-PAGE, stained with Coomassie blue R-250, and compared to a CV-N (P51G) reference standard (NIH) (lane 3). Proteins in 5 μl of cell-free conditioned media were also probed with rabbit anti-CV-N polyclonal antibody (PAb). MALDI-TOF MS analysis revealed an average mass of 9,614 Da for the truncated CV-N (P51G).
Analysis of CV-N expression in L. jensenii.
Proteins in cell-free culture media were separated by electrophoresis using the 4 to 12% NuPAGE system (Invitrogen) (8), followed by Coomassie blue staining or Western blotting (8) using the rabbit anti-CV-N antibody OI5, developed in house. The ability of Lactobacillus-derived CV-N to interact with HIV-1 gp120 was analyzed using a gp120 capture enzyme-linked immunosorbent assay as described previously (8), except that the bound CV-N molecules were probed with antiserum against CV-N.
Purification and characterizations of Lactobacillus-derived CV-N.
The L. jensenii strains expressing CV-N were cultured in Rogosa SL broth (Difco) to stationary phase. Cell-free supernatants were dialyzed against 20 mM Bis-Tris, pH 5.4, at 4°C using a 3.5-kDa molecular-mass-cutoff dialysis membrane (Spectrum Laboratories) and passed sequentially over SP and Q Sepharose Fast Flow columns (Amersham). The CV-N-containing flowthrough fraction was dialyzed against 20 mM Tris, pH 8.8, and passed over an SP Fast Flow column. CV-N present in the flowthrough was then bound to Q Sepharose Fast Flow resin and eluted in a buffer containing 150 mM NaCl. The partially purified CV-N was then fractionated on a Superdex 75 column (Amersham) in phosphate-buffered saline (PBS). The amount of eluted CV-N was quantified by Coomassie blue staining and gp120 capture enzyme-linked immunosorbent assay using a CV-N reference standard for calibration (NIH).
N-terminal sequence analysis of CV-N was performed on an Applied Biosystems sequencer (Foster City, CA). Disulfide mapping was carried out by fast atom bombardment-mass spectrometry (FAB-MS) analysis (16) following tryptic digestion of purified Lactobacillus-derived APVT-CV-N (P51G) and the CV-N reference standard, with or without reduction of the disulfide bonds. The nominal molecular weight of CV-N was determined by linear matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (ABI 4700 TOF/TOF; Applied Biosystems) in a MALDI matrix of sinapinic acid in 50% acetonitrile-0.1% trifluoroacetic acid (Fluka, St. Louis, MO).
HIV-1 attachment inhibition assay.
The anti-HIV activity of Lactobacillus-derived CV-N was evaluated using the CCR5- and CXCR4-tropic HIV attachment inhibition assays (13) in collaboration with the Topical Microbicides Program at the Southern Research Institute, Frederick, MD. In this assay, monolayers of MAGI-R5-LTR-β-gal or HeLa-X4-LTR-β-gal cells were treated with Lactobacillus-derived CV-N (P51G) or a CV-N reference standard (NIH) for 30 min prior to addition of the cell-free virus HIV-1BaL or HIVIIIB. The cultures were incubated for 2 h with virus and then washed. Cells at 48 h postinfection were lysed to measure β-galactosidase activity. Cell viability was monitored on replicate plates using a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS)-based viability assay (Promega, Madison, WI).
Chromosomal integration of the CV-N expression cassette.
Derivatives of pUC18erm (18), which cannot replicate as extrachromosomal plasmids in L. jensenii, were used to integrate CV-N constructs into specific target sites within the L. jensenii chromosome by homologous recombination. To facilitate site-specific integration, genomic targeting sequences (∼2.8 kb) containing portions of the pox1 or pepO genes were cloned into pUC18erm. The targeting sequences were engineered to contain unique XbaI sites and in-frame stop codons to prevent formation of fusion proteins. The rpsU promoter expression cassette, PrpsU-APVT-CV-N (P51G), was inserted into the XbaI restriction site of the targeting sequence, and the resulting integration vector was transformed into L. jensenii 1153. The transformed bacteria were selected on MRS plates containing 3 μg/ml erythromycin and then screened by Western blotting for CV-N expression. Chromosomal DNA samples from putative integrants were also examined by PCR, with both integration site-specific and CV-N gene-specific primers, and by Southern blotting to ensure that the plasmid had integrated into the appropriate site. To resolve the single-crossover event, thereby removing the plasmid backbone (including the Ermr cassette) but not the CV-N expression cassette, the integrants were first grown without erythromycin selection in liquid MRS and then plated on nonselective MRS plates. Resolvants were identified by PCR as well as Western and Southern blot analyses. The bacterial strain containing the rpsU promoter-driven expression cassette integrated into the pox1 gene was designated L. jensenii 1153-1666. A CV-N expression cassette under the control of the ptsH promoter was also integrated into the pox1 gene and resolved. The resulting strain was designated L. jensenii 1153-2666. Another strain containing the rpsU promoter-driven expression cassette integrated into the pepO gene was designated L. jensenii 1153-3666.
Phenotypic assessment of CV-N expression strains.
To determine the rate of bacterial growth, a single colony was inoculated into MRS broth and grown at 37°C for different times, and absorbance at 600 nm was measured. d-Lactate production by each strain was determined using a standard d-lactate dehydrogenase-based assay (R-Biopharm, Darmstadt, Germany). Production of H2O2 by L. jensenii was detected using EM Quant Strips (Darmstadt, Germany).
Genetic and functional stability of a CV-N expression cassette in L. jensenii 1153-1666.
To evaluate the stability of the integrated and resolved CV-N expression cassette, individual colonies of L. jensenii 1153-1666 were grown in MRS medium and subcultured at 3-day intervals for a period of 2 weeks and then evaluated by Western blot analysis. Genomic DNA was isolated, and a DNA fragment containing portions of the pox1 gene and the complete CV-N expression cassette was generated by PCR using Pfu polymerase and the primers poxBamF (5′-GCACGGATCCCCACCTGGCATCAAG-3′) and poxBamR (5′-CTACGGATCCAGCAGCAGATATTGC-3′) (the codons of the BamHI sites are underlined). The PCR fragment was sequenced to confirm the integrity of the expression cassette.
In vivo experiments.
In vivo bacterial colonization was conducted in a murine model of bacterial colonization (34) at the Vaccine Research Center's AAALAC-approved facility at the NIH. The protocols and procedures were conducted under a protocol approved by the Animal Care and Use Committee. Sixteen female, outbred CD-1 mice, 4 to 8 weeks old (Charles River Laboratories, Raleigh, NC), were used in a typical experiment. In a mouse model of gonococcal genital tract infection, estradiol treatment was necessary to prolong colonization of Neisseria gonorrhoeae, a human-specific vaginal pathogen (17). To synchronize and prolong estrus phase, we implanted subcutaneous 21-day slow-release estradiol pellets in the animals (Innovation Research of America, Sarasota, FL). Vaginal cytology and evaluation confirmed that all the animals were in estrus phase 72 h after β-estradiol treatment. Mice in estrus phase were inoculated intravaginally with 5 × 107 CFU of L. jensenii 1153-1666 in 50 μl of PBS, or PBS alone, to evaluate colonization and CV-N expression in vivo. Vaginal washes were collected in approximately 50 μl of PBS at 48 h, 96 h, and 144 h postinoculation. Aliquots of the vaginal washes were plated on MRS agar for microbiological analysis, and Western blots were performed on pooled vaginal washes to detect CV-N expression. Colonies recovered from the MRS plates were grown in MRS broth, and Western blotting was carried out to compare protein expression and bacterial phenotype with the original strain. In addition, genomic DNA from these cultures was prepared to assess the stability of the integration site and CV-N expression cassette, as described above.
Bacterial colonization and vaginal tissue were evaluated histochemically. Briefly, animals were euthanized by CO2 asphyxiation and cervical dislocation. The lower reproductive tract was removed, fixed in 10% formalin, and embedded in paraffin. Sections were stained with either hematoxylin-eosin stain to determine the presence or absence of polymorphonuclear leukocytes or Brown-Hopps (B and H)-modified Gram stain to visualize Lactobacillus.
Cervicovaginal lavage (CVL) samples from adult female pigtailed macaques (Macaca nemestrina) were collected in an AAALAC-accredited animal facility, the regional Primate Research Center at the University of Washington. The procedures and protocols were approved by the Animal Care and Use Committee. In each experiment, two CVL samples (2 ml of saline per lavage) were collected from each of 6 monkeys. The first 2-ml CVL sample was used to culture Lactobacillus strains and evaluate CV-N expression.
Nucleotide sequence accession numbers.
The rpsU and ptsH promoter sequences of L. jensenii 1153 have been deposited in the GenBank database under accession numbers DQ812901 and DQ812902, respectively.
RESULTS
High-level plasmid-based expression of CV-N (P51G) in L. jensenii 1153.
To efficiently express CV-N in L. jensenii, we constructed a synthetic CV-N gene composed of Lactobacillus-preferred codons using assembly PCR (41). In addition, we replaced proline 51 with glycine (P51G) in the CV-N sequence, a mutation reported to improve the expression of properly folded, monomeric CV-N (24). Initially, we subcloned the CV-N (P51G) gene into the modular expression vector pOSEL175, immediately downstream of the Lactococcus lactis P23 promoter, and a signal sequence derived from the L. crispatus cbsA gene (8). However, only low levels of CV-N (<200 ng/ml) were secreted into the conditioned culture medium (Table 1).
TABLE 1.
Identification of strong endogenous promoters to drive efficient expression of heterologous proteins in L. jensenii 1153a
| Promoter | Expression of heterologous proteins
|
|
|---|---|---|
| Intracellular GFP (relative fluorescence intensity) | Extracellular CV-N (μg/ml in Rogosa broth) | |
| Vector | 4.52 ± 0.12 | ND |
| P23 | 24.60 ± 1.13 | 0.10 ± 0.05 |
| PptsH | 81.32 ± 3.93 | 4.41 ± 0.62 |
| PrpsU | 215.50 ± 16.26 | 5.02 ± 0.35 |
The ability of the promoters PrpsU and PptsH to drive expression of a heterologous protein in L. jensenii 1153 was examined using GFP and CV-N as reporter molecules. The lactococcal P23 promoter was used as a reference standard. GFP was measured by flow cytometric analysis. CV-N produced by engineered L. jensenii 1153 at a cell density approaching 109 CFU/ml and was quantified by the density of the protein bands on Coomassie blue R-250-stained SDS-PAGE or Western blot, in reference to a recombinant CV-N (P51G) protein reference standard (NIH) at different concentrations. The results (means ± standard deviations) from triplicate determinations in a single experiment are presented and were confirmed in three independent experiments. ND, not detected.
To improve the expression of CV-N in Lactobacillus, we employed a promoter-trapping strategy to identify native L. jensenii promoters more powerful than the lactococcal P23 promoter (8). Two DNA fragments were identified containing promoter elements that were significantly stronger than the P23 promoter. The promoter of the rpsU gene (encoding ribosomal small-subunit protein S21) drove GFP expression about 10-fold higher than P23, and the promoter of the ptsH gene (encoding the HPr protein of the phosphoenolpyruvate:sugar phosphotransferase system) was fourfold stronger. Replacement of P23 in the CV-N expression cassette with PrpsU or PptsH resulted in 50- or 40-fold higher levels of secreted CV-N (P51G) production, respectively (Table 1). The plasmid-based expression of secreted CV-N (P51G) reached 4 to 5 μg/ml (365 to 457 nM) using the rpsU promoter.
Proteolytic processing of L. jensenii-expressed CV-N (P51G).
Despite the good expression levels of CV-N described above, the protein was slightly smaller than the reference standard on Western blots. N-terminal amino acid sequencing of Lactobacillus-derived CV-N revealed that the protein was truncated 16 amino acids downstream of the predicted N terminus (Fig. 1). MALDI-TOF MS analysis revealed an average mass of 9,614 Da for truncated CV-N, consistent with the N-terminal sequencing results. Although the truncated CV-N protein could bind to recombinant HIV gp120, cleavage at the −16 position resulted in the loss of a cysteine residue required for formation of a critical disulfide bond that is necessary for anti-HIV activity (unpublished data).
Expression of structurally intact CV-N (P51G) in L. jensenii.
Proteins destined for secretion in gram-positive bacteria contain preferred amino acid sequences immediately downstream of the signal peptidase cleavage site (25). To address whether aberrant signal peptide processing contributed to truncation of the secreted CV-N species in L. jensenii, the sequence downstream of the predicted signal peptidase cleavage site in the expression cassette of pOSEL CV-N (P51G) was modified to include four extra amino acids (1APVT of mature CbsA, cloned from a proprietary strain) (Table 2). When the modified CV-N expression vector pOSEL APVT-CV-N (P51G) was introduced into L. jensenii 1153, full-length secreted APVT-CV-N was recovered in the culture medium, as determined by MALDI-TOF MS analysis (Table 2). In addition to APVT, the amino acid sequences APV and AP, but not A alone, also supported expression of full-length CV-N (P51G) (Table 2). Not surprisingly, the MS data also revealed that L. jensenii-expressed CV-N lacked N-linked glycosylation at Asn30. When expressed in eukaryotic hosts, glycosylation of CV-N at Asn30 CV-N is known to abolish its anti-HIV activity (24).
TABLE 2.
Characterizations of CV-N (P51G) secreted by L. jensenii 1153a
| Modification of signal sequence cleavage site | Observed mass/expected mass (Da) | N-terminal sequence |
|---|---|---|
| . . . SA-1CVN (P51G) | 9,614.85/11,060.70 | 16SVLTS . . . (truncated at −16) |
| . . . SA-APVT-1CVN (P51G) | 11,338.34/11,342.00 | APVT-1LGKF . . . (full length) |
| . . . SA-APV-1CVN (P51G) | 11,237.37/11,240.90 | APV-1LGKF . . . (full length) |
| . . . SA-AP-1CVN (P51G) | 11,144.32/11,141.80 | AP-1LGKF . . . (full length) |
| . . . SA-A-1CVN (P51G) | 9,614.85/11,060.70 | 16SVLTS . . . (truncated at −16) |
The Lactobacillus-derived proteins were semipurified and subjected to mass determination and N-terminal sequencing. Protein masses were compared with their predicted molecular weight when signal sequence cleavage sites were modified. The added-on modification sequences A-, AP-, APV-, and APVT- were derived from N-terminal sequences of mature protein native to CbsA signal sequence (CbsAss). The protein sequences from CbsAss are underlined. Plasmid-based expression of CV-N variants in full length has been achieved approaching 4 to 5 μg/ml (365 to 457 nM), based on Western analysis, relative to an NIH CV-N (P51G) reference standard. Superscripts indicate the amino acid position relative to the N terminus of full-length CV-N.
Biologically active CV-N requires proper disulfide bond formation (16). To determine whether the full-length APVT-CV-N (P51G) derived from L. jensenii adopted the correct disulfide-bonding pattern, the protein was purified and subjected to trypsin digestion, with or without reduction of the disulfide bonds. A CV-N (P51G) reference standard was treated identically for comparison. The tryptic peptides were analyzed by FAB-MS analysis (16), and the resulting mass spectra were consistent with correct disulfide bridging of the four cysteines in L. jensenii-derived APVT-CV-N (P51G) (data not shown). These results established the feasibility of expressing structurally intact CV-N in a vaginal Lactobacillus strain.
Full-length Lactobacillus-expressed CV-N (P51G) inhibits HIV infectivity in vitro.
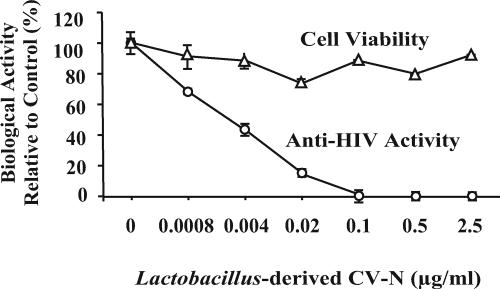
Sexually transmitted HIV-1 strains generally show a preference for the CCR5 coreceptor (R5) (4). The CCR5-tropic strains predominate during the early stages of HIV infection and are particularly relevant for HIV infections through the mucosa. To determine whether the L. jensenii-derived APVT-CV-N (P51G) was biologically active against a relevant CCR5-tropic HIV strain, the molecule was purified by ion exchange and size exclusion chromatography and tested in the MAGI-LTR-β-gal R5-tropic attachment assay (13). In general, a much higher multiplicity of infection is required for the attachment assays than for other types of HIV infectivity assays. As shown in Fig. 2, the full-length molecule potently and completely inhibited CCR5-tropic HIVBaL infection with a 50% inhibitory concentration (IC50) of 0.3 nM. No evidence of changes in cell viability was observed. In addition, the Lactobacillus-expressed molecule also inhibited CXCR4-tropic HIVIIIB in the HeLa-LTR-β-gal X4-tropic attachment assay with an IC50 of less than 3 nM (data not shown). Notably, the soluble Lactobacillus-derived APVT-CV-N (P51G) exhibited greater potency against HIV than CV-N displayed on the surface of an engineered oral commensal bacterium, Streptococcus gordonii (15). These results established that the full-length CV-N expressed in L. jensenii possessed full biological activity.
FIG. 2.
Biological activity of L. jensenii-derived APVT-CV-N (P51G). The anti-HIV activity was evaluated in a CCR5-tropic HIV-1BaL attachment inhibition assay. Cell viability was monitored on a replicate plate using the MTS dye reduction assay. The results (means ± standard deviations) from triplicate determinations in a single experiment are presented and were confirmed in three independent protein preparations.
Integration of the CV-N expression cassette into the L. jensenii 1153 chromosome.
To achieve stable expression of CV-N in Lactobacillus for use as a live vaginal microbicide, the APVT-CV-N (P51G) expression cassette was integrated in single copies into the L. jensenii 1153 genome by homologous recombination. Stable knockouts of either poxB in Lactobacillus plantarum or pepO in Lactobacillus helveticus have been shown not to affect the phenotypes of these bacteria (9, 23). Homologues of these genes, referred to as pox1 and pepO, were identified in the L. jensenii 1153 genome and used as integration sites for the optimized CV-N expression cassette via a single-crossover Campbell-type recombination event followed by resolution of the plasmid backbone.
The 1,881-bp putative pox1 coding sequence of L. jensenii is predicted to encode a 627-amino-acid polypeptide. The pox1 gene product of L. jensenii shows 79% identity with the pyruvate oxidase sequence of L. acidophilus NCFM (30). In addition, the pox1 gene appears to be a single transcriptional unit in L. jensenii, with a gene structure similar to that of the pox genes of L. johnsonii and L. plantarum (19, 48). In the integrated and resolved bacterial strain L. jensenii 1153-1666, the expression cassette PrpsU-APVT-CV-N (P51G) was inserted at nucleotides 1445 and 1450 of the pox1 coding sequence. The expression cassette PrpsU-APVT-CV-N (P51G) was in the opposite direction to the pox1 transcriptional unit to avoid the possible transcription interference by the native pox1 promoter.
The putative pepO gene of L. jensenii encodes a 614-amino-acid polypeptide. The PepO protein consists of two putative domains of peptidase family M13 and shares about ∼68% sequence identity with the endopeptidase of L. helveticus CNRZ32 (9). In the integrated and resolved bacterial strain L. jensenii 1153-3666, the expression cassette PrpsU-APVT-CV-N (P51G) was inserted between nucleotides 618 and 619 of the pepO coding sequence (1,842 bp) at the first N-terminal peptidase domain. The DNA sequence adjacent to the pepO gene of L. jensenii has a genome organization similar to that of the L. johnsonii and L. plantarum strains, which are predicted to function as single transcription units (19, 48).
Analysis of chromosomal DNA from strain L. jensenii 1153-1666 and L. jensenii 1153-3666 by PCR and sequencing confirmed that extraneous plasmid DNA, including the ermB gene, had been excised from the bacterial chromosome, and the nucleotide sequence of the CV-N expression cassette remained intact.
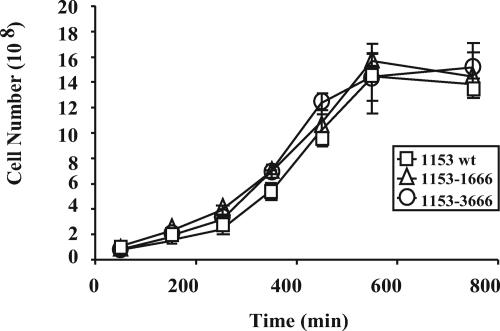
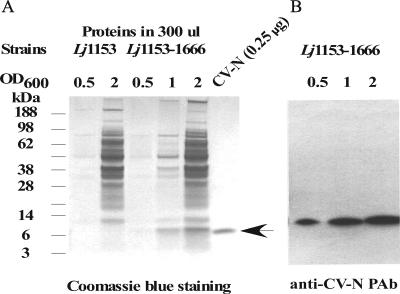
The phenotypes of the integrated and resolved bacterial strains were evaluated in vitro, focusing on rates of bacterial growth and production of lactic acid and hydrogen peroxide. As shown in Fig. 3, the growth rates of L. jensenii 1153-1666 (pox1::CV-N) and L. jensenii 1153-3666 (pepO::CV-N) resolvants were almost identical to the parental L. jensenii 1153 strain when cultured in MRS broth at 37°C. The three strains all produced about 4 g/liter of d-lactate in 24 h and produced similar amounts of hydrogen peroxide (data not shown). In addition, L. jensenii 1153-1666 secreted full-length CV-N into the culture medium at all bacterial growth phases, reaching 1.5 to 2 μg/ml (∼137 to 183 nM) of CV-N at Lactobacillus concentrations of 108 to 109 CFU/ml (Fig. 4). L. jensenii 1153-3666 secreted levels of full-length CV-N into the culture medium similar to those of L. jensenii 1153-1666 at all bacterial growth phases (data not shown). The expression levels of Lactobacillus-derived CV-N were also confirmed by Western blot analysis using several concentrations of the CV-N (P51G) standard in independent experiments (data not shown). These results suggested that insertion of the CV-N expression cassette into either the pox1 or pepO gene had no significant impact on several important characteristics of L. jensenii 1153. Furthermore, the recombinant CV-N protein was stably accumulated in conditioned Rogosa media when the CV-N-producing L. jensenii is cocultured with other members of human vaginal lactobacilli, such as L. gasseri and L. crispatus, or cocultured with human cervico-vaginal epithelium (data not shown).
FIG. 3.
Phenotypic assessment of CV-N expressing pox1 and pepO resolvant strains. Single colonies of the L. jensenii 1153-1666 (pox1::CV-N), L. jensenii 1153-1666 (pepO::CV-N), and parental strain (1153 wt) were inoculated in MRS and cultured at 37°C. Absorbance at 600 nm was performed at various time points to determine the rate of growth. The results (means ± standard deviations) from triplicate determinations in a single experiment are presented and were confirmed in three independent experiments.
FIG. 4.
Expression of full-length CV-N (P51G) by the stably integrated strain L. jensenii 1153-1666. L. jensenii 1153-1666 (Lj1153-1666) and the parental strain L. jensenii 1153 (Lj1153) were cultured in Rogosa broth to different bacterial densities. (A) Proteins from 300 μl of cell-free conditioned Rogosa media were separated by reducing SDS-PAGE and stained with Coomassie blue R-250. The CV-N proteins were quantified based on the band density on a Coomassie blue-stained SDS-PAGE gel in reference to a CV-N (P51G) reference standard (NIH) at different concentrations. (B) Proteins in 5 μl of cell-free conditioned media were also probed with rabbit anti-CV-N polyclonal antibody (PAb).
The CV-N expression by L. jensenii 1153-1666 remained constant over 2 weeks of continuous growth in MRS broth, with subculturing every 3 days. PCR and sequence analysis of genomic DNA indicated that no changes occurred in the expression cassette or the flanking DNA sequences during this time frame.
The pox1::CV-N resolvant expressed full-length CV-N when cultured in pigtailed macaque cervicovaginal lavage fluid.
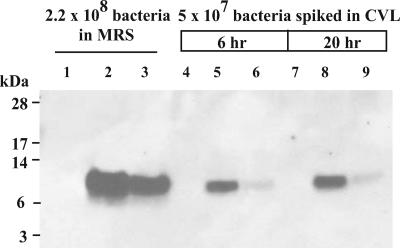
As a prelude to future in vivo CV-N expression studies in the nonhuman primate model of pigtailed macaques (Macaca nemestrina), we evaluated the ability of L. jensenii strain 1153-1666 and strain 1153-2666, a pox1::CV-N resolvant strain that employs a ptsH promoter, to produce CV-N in CVL fluid from pigtailed macaques. CVL fluid (400 μl) was inoculated with ∼5 × 107 bacteria and cultured at 37°C and 5% CO2 for 6 to 20 h, and CV-N expression was assessed by Western blot analysis. Although both strains expressed the 11-kDa full-length CV-N, L. jensenii 1153-1666, which uses the rpsU promoter, produced considerably more CV-N than L. jensenii 1153-2666, which contains the ptsH promoter, in cervicovaginal fluid (Fig. 5).
FIG. 5.
Expression of full-length CV-N (P51G) by L. jensenii strains cultured in pigtailed macaque CVL fluid. The following strains were used for the evaluation: L. jensenii 1153 (Lj1153, lanes 1, 4, and 7), L. jensenii 1153-1666 (lanes 2, 5, and 8), and L. jensenii 1153-2666 (lanes 3, 6, and 9). CVL fluid (400 μl) obtained from pigtailed macaques was inoculated with ∼5 × 107 bacteria and incubated at 37°C and 5% CO2 for 6 h (lanes 4 to 6) or 20 h (lanes 7 to 9). Proteins in cell-free supernatants (10 μl) were resolved by reducing SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane for immunodetection with anti-CV-N polyclonal antibodies.
Bacterial colonization and in vivo CV-N expression in a small-animal model.
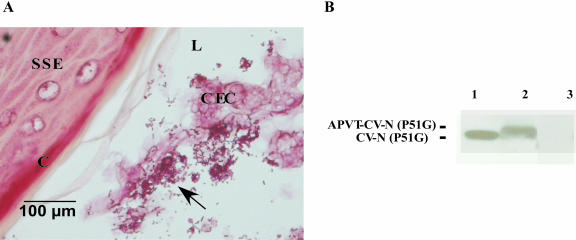
Mouse models have recently been used for preclinical safety testing of microbicides (40). To establish conditions for safety testing in the mouse and to build a foundation for future safety and efficacy studies in nonhuman primates, we established a mouse model to evaluate vaginal colonization and in vivo CV-N expression by different engineered Lactobacillus strains. Mice typically cycle through proestrus, estrus, metestrus, and diestrus phases every 4 days, and we determined that outbred CD-1 mice treated with β-estradiol to stay in prolonged estrus phase supported persistent vaginal colonization of Lactobacillus. Therefore, we implanted β-estradiol pellets in the mice 3 days prior to Lactobacillus inoculation in order to synchronize the animals and indefinitely prolong estrus phase. In the experiment shown in Fig. 6, 16 female mice that appeared to be in estrus by vaginal cytology were divided into two groups: eight animals were inoculated with L. jensenii 1153-1666, and eight were inoculated with the PBS vehicle. Two days following inoculation, vaginal washes were obtained for microbiological and CV-N analyses, and the reproductive tract was harvested for histological examination using a Brown and Hopps-modified Gram stain. Due to the small size of the female mouse reproductive tract, which naturally does not harbor Lactobacillus, only ∼104 CFU were recovered from each mouse following an intravaginal inoculation of 5 × 107 bacteria. Lactobacillus was not recovered from any of the mice in the PBS control group. The Brown and Hopps-modified Gram stain of vaginal tissue confirmed the presence of Lactobacillus, and the histopathological correlation was valid in 84% of the culture-positive animals. Lactobacilli, identified as gram-positive rods, were usually associated with the sloughed columnar epithelial cells, sometimes with the keratinized layer of epithelia, or were free in the vaginal lumen (Fig. 6A). No less than 10 ng/ml of soluble full-length CV-N was detected by Western analysis at day 2 in the pooled vaginal washes of the animals inoculated with L. jensenii 1153-1666, whereas none was found in the PBS control group (Fig. 6B). Full-length CV-N was still detected in the vaginal washes of the animals inoculated with L. jensenii 1153-1666 at day 4 and day 6 (data not shown). In addition, approximately 10 fields from each animal were examined histologically, and generally there did not seem to be evidence of changes in epithelial integrity or leukocytic infiltration of vaginal tissues during estrus phase, whether animals were inoculated with L. jensenii or PBS. An infiltration of polymorphonuclear leukocytes has been observed in late metestrus and is consistent with a previously published report that neutrophils infiltrate into the murine vagina postovulation (39).
FIG. 6.
Vaginal persistence of L. jensenii 1153-1666 and in vivo CV-N expression in the estrogenized CD1 mouse. (A) Brown and Hopps stain of Lactobacillus in the vagina 24 h postinoculation. Histology of the CD1 mouse vagina during estrus phase shows stratified squamous epithelium (SSE) with a cornified epithelial layer (C). Superficial columnar epithelial cells (CEC) slough into the vaginal lumen (L) and Lactobacillus, indicated by arrows, appear to associate with the sloughed columnar cells. (B) In vivo CV-N expression. Bacteria (∼5 × 107 CFU) or the PBS vehicle were administrated intravaginally to CD1 mice. Vaginal washes were collected in approximately 50 μl of PBS at 48 h postinoculation. The vaginal washes with total of 8 × 104 CFU bacteria were pooled from eight mice and trichloroacetic acid precipitated, and the expression of full-length CV-N was detected by Western blotting. Lane 1, CV-N (P51G) standard lacking APVT; lane 2, vaginal washes at 48 h postinoculation with L. jensenii 1153-1666; lane 3, vaginal washes at 48 h postinoculation with PBS control.
The stability of the CV-N expression cassette in the bacterial chromosome was examined in L. jensenii 1153-1666 recovered from inoculated animals. Sequence analysis of genomic DNA from 10 bacterial colonies did not reveal any mutations arising in the expression cassette or integration site during residence of the bacteria in the mouse vagina. These results further indicate that genetically engineered L. jensenii can stably express full-length CV-N under a variety of conditions and for a sustained period of time.
DISCUSSION
As the burden of HIV infection increasingly shifts towards women (32), numerous avenues are being pursued to develop effective vaginal microbicides to curtail heterosexual HIV transmission (8, 22, 44, 47). An effective topical microbicide should be able to inactivate HIV without irritating vaginal tissue or disturbing the natural microflora, as occurred with nonoxynol-9 (53). For global distribution, a topical microbicide should also be inexpensive to produce and stable without refrigeration. CV-N, as a conventional protein-based microbicide candidate, is expected to be costly to produce (12) and relatively unstable at ambient temperatures, making it an unrealistic product, particularly in economically disadvantaged regions. In addition, protein-based microbicides, as recently exemplified by a gel formulation of CV-N used in a macaque HIV vaginal transmission model (43), may also require administration of relatively high concentrations due to the relatively short half-life and/or rapid clearance of these molecules (43). Furthermore, conventional microbicide formulations typically require application at the time of potential exposure, which is not always practical. In this report, we have detailed the development of an innovative strategy that circumvents many of the deficiencies of conventional microbicides. It employs a live colonizing Lactobacillus from the protective vaginal microflora, which has been genetically enhanced to deliver an anti-HIV protein directly to the cervicovaginal mucosa, the major portal of virus entry in women. Since this microbicide is a bacterium rather than a purified protein, the costs associated with its manufacture and formulation are expected to be quite low compared to other biologics. We have formulated such bacteria as a vaginal suppository, which a woman can use discreetly on a regular basis to provide continuous protection.
The success of this novel Lactobacillus-based microbicide depends on whether modified lactobacilli can deliver sufficient quantities of CV-N to the vaginal mucosa to reduce heterosexual HIV transmission risk.
In a recent study, Pusch et al. (31) demonstrated that engineered lactic acid bacteria strains can secrete CV-N that inhibited HIV-1 infection in vitro. However, neither of the lactic acid bacteria strains used, Lactococcus lactis and L. plantarum, were human vaginal strains, nor would they be expected to colonize the human vagina. Without colonization, the levels of CV-N produced may not be sufficient to block HIV transmission.
Tsai et al. (43) reported that a single vaginal application of CV-N at a concentration of 5 mg/ml in a gel formulation could protect macaques (Macaca fascicularis) against vaginal challenge with a simian immunodeficiency virus (SIV)/HIV-1 chimera, SHIV89.6P. In this study, an unusually high challenge dose (5,000 50% tissue culture infectious doses) of virus was used to ensure that 100% of the animals became infected. In addition, progesterone treatment was employed to thin the vaginal epithelium to increase the infection rate. The high infection rate of this model does not reflect conditions of HIV transmission normally encountered by women (52). It should also be noted that the minimum dose for protection was not determined in this study. Given the low IC50 for CV-N in a human ectocervical explant model (43), it is likely that much lower doses will be effective, particularly in a more physiological challenge model in which animals are repeatedly exposed to low doses of virus (26).
In this report, we successfully achieved high-level expression of fully bioactive CV-N by a human vaginal Lactobacillus strain. The Lactobacillus-expressed CV-N dramatically decreased the infectivity of both CCR5-tropic HIVBaL and CXCR4-tropic HIVIIIB in vitro. When the optimized CV-N expression cassette was stably integrated in single copy within the L. jensenii chromosome, the resulting strains were capable of producing one-third to one-half the amount of CV-N produced by the plasmid-based system. Significantly, the strain L. jensenii 1153-1666 (pox1::CV-N resolvant) was able to secrete approximately 1.5 to 2 μg/ml (∼137 to 183 nM) of CV-N (Fig. 4) at bacterial concentrations similar to those (108 to 109 CFU) reported to exist within the normal vaginal microflora (5). These data demonstrate that physiological numbers of Lactobacillus are capable of expressing CV-N at concentrations approaching 100 times higher than the IC50 for inhibiting the most prevalent clades of HIV.
The endogenous promoters of the rpsU and ptsH genes drove high-level expression of biologically active CV-N in L. jensenii in vitro. The rpsU gene encodes a ribosomal small-subunit protein, S21, and the ptsH gene encodes the HPr protein of the phosphoenolpyruvate:sugar phosphotransferase system. The HPr protein is known to be highly expressed in a number of gram-positive bacteria (45), but less is known about the expression of S21 (49). The strains containing the rpsU promoter produced the highest levels of full-length CV-N (1.5 to 2.0 μg/ml), regardless of the signal sequence or integration site used. In comparison, strains utilizing the ptsH promoter produced at least twofold less CV-N. These differences in CV-N production were evident under a variety of experimental conditions, including growth in MRS or Rogosa broth, incubation in pigtailed macaque CVL fluid, coculture with reconstructed human cervicovaginal epithelium, and during vaginal colonization of CD1 mice. These results served to focus further development work on strains containing the rpsU promoter. However, it remains to be seen whether this promoter can drive the expression of CV-N to high enough levels in the cervicovaginal environment to block HIV transmission.
An important question is whether the genetic modifications made to L. jensenii will adversely affect its ability to colonize and compete with the microflora of the human vaginal mucosa. Bacterial colonization is a complex issue and is complicated further because L. jensenii is uniquely a member of the human vaginal microflora. To definitively answer this question, a human trial would have to be conducted comparing the engineered and parental strains. Nonetheless, the engineered L. jensenii strains described in this report were observed to retain several important phenotypic properties of the parental strain in vitro, including adherence to human vaginal epithelium, suggesting that they may be capable of vaginal colonization. We demonstrated that L. jensenii 1153-1666 could transiently colonize the mouse vagina during estrus phase and produce full-length CV-N in situ. This represents the first report of CV-N expression by a human commensal organism in vivo. While the reproductive tract physiology and vaginal microflora of the female mouse are clearly different from those of women, the estrogenized mouse model does provide a cost- and time-efficient means to compare the performance of engineered and parental Lactobacillus strains in an in vivo setting as well as a useful preclinical safety model. Others have shown that Streptococcus gordonii or Lactobacillus zeae strains engineered to produce single-chain antibodies, for example, can colonize the oral or vaginal mucosa of rats sufficiently well to prevent the development of dental caries and vaginal yeast infection (3, 20).
As a follow-up to the mouse model, we are planning to conduct similar studies in the pigtailed macaque, including vaginal colonization, safety, and in situ CV-N expression studies, culminating in an efficacy study using the siman-human immunodeficiency virus (a pathogenic HIV/SIV chimeric virus) vaginal challenge model (43). The pigtailed macaque has been extensively utilized as a model for human cervicovaginal physiology and response to sexually transmitted infections. The strength of the female pigtailed macaque is that the anatomy and physiology of the reproductive tract is similar to that of the human, including a Lactobacillus-dominant microflora (28) and a regular menstrual cycle. Unlike the mouse model, up to 108 Lactobacillus bacteria can colonize the vaginal mucosal surface of the pigtailed macaque (27).
Since CV-N is a bacterial protein, an important safety question is whether Lactobacillus-expressed CV-N can elicit a mucosal immune response strong enough to cause vaginal irritation, neutralize the anti-HIV activity, or impede the bacterial colonization of the vagina. In our short-term mouse colonization experiments, histological analysis did not reveal any evidence of changes in epithelial integrity or leukocytic infiltration of vaginal tissues. In addition, intravaginal administration of CV-N was reported to exhibit a positive safety profile in both rabbits and macaques (6, 43).
Given the potent anti-HIV activity demonstrated by Lactobacillus-derived CV-N and the high expression levels achieved by a vaginal Lactobacillus strain, the successful colonization of the vaginal mucosa by these bacteria should produce sufficient CV-N protein in the proximity of virus to significantly decrease the number of HIV particles and thus have a major impact on the frequency of heterosexual HIV transmission. Taken together, a natural vaginal isolate of Lactobacillus engineered to constitutively express secreted CV-N might represent the most practical and economically viable means of delivering this microbicide continuously to the vaginal mucosa. The findings reported in this paper represent an important step in the development of a much-needed female-initiated preventative against heterosexual transmission of HIV.
Acknowledgments
We thank John Lewicki for helpful discussions, Jan-Fang Cheng at the Lawrence Berkeley National Laboratory for directing genomic sequencing of L. jensenii 1153, Ken Frankel for bioinformatics support, Lewis Pannell at University of South Alabama for disulfide mapping, Carol Lackman-Smith and her colleagues at the Southern Research Institute for performing the HIV-1 neutralization assays, Dorothy Patton at the University of Washington for providing CVL samples of pigtailed macaques, Angela Gronenborn at the National Institute of Diabetes and Digestive and Kidney Diseases for providing a CV-N (P51G) reference standard, Laura Barrientos at the CDC for discussions on CV-N structure, Joy McFarlane and Saran Bao at VRC for assistance with all animal assays, and William Jewell at UC-Davis for linear MALDI-TOF analysis.
This work was supported in part by NIH Integrated Preclinical/Clinical Program for Topical Microbicides 1 grant U19 AI60615 and NIH Partnerships for Topical Microbicides grant U01 AI066708.
REFERENCES
- 1.al-Nozha, M., S. Ramia, A. al-Frayh, and M. Arif. 1990. Female to male: an inefficient mode of transmission of human immunodeficiency virus (HIV). J. Acquir. Immune Defic. Syndr. 3:193-194. [PubMed] [Google Scholar]
- 2.Antonio, M. A., S. E. Hawes, and S. L. Hillier. 1999. The identification of vaginal Lactobacillus species and the demographic and microbiologic characteristics of women colonized by these species. J. Infect. Dis. 180:1950-1956. [DOI] [PubMed] [Google Scholar]
- 3.Beninati, C., M. R. Oggioni, M. Boccanera, M. R. Spinosa, T. Maggi, S. Conti, W. Magliani, F. De Bernardis, G. Teti, A. Cassone, G. Pozzi, and L. Polonelli. 2000. Therapy of mucosal candidiasis by expression of an anti-idiotype in human commensal bacteria. Nat. Biotechnol. 18:1060-1064. [DOI] [PubMed] [Google Scholar]
- 4.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 5.Boskey, E. R., K. M. Telsch, K. J. Whaley, T. R. Moench, and R. A. Cone. 1999. Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect. Immun. 67:5170-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, M., K. Gustafson, J. McMahon, R. Shoemaker, B. O'Keefe, T. Mori, R. Gulakowski, L. Wu, M. Rivera, C. Laurencot, M. Currens, J. Cardellina, Jr., R. Buckheit, Jr., P. Nara, L. Pannell, R. Sowder, Jr., and L. Henderson. 1997. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: potential applications to microbicide development. Antimicrob. Agents Chemother. 41:1521-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2:706-713. [DOI] [PubMed] [Google Scholar]
- 8.Chang, T. L., C. H. Chang, D. A. Simpson, Q. Xu, P. K. Martin, L. A. Lagenaur, G. K. Schoolnik, D. D. Ho, S. L. Hillier, M. Holodniy, J. A. Lewicki, and P. P. Lee. 2003. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc. Natl. Acad. Sci. USA 100:11672-11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y. S., and J. L. Steele. 1998. Genetic characterization and physiological role of endopeptidase O from Lactobacillus helveticus CNRZ32. Appl. Environ. Microbiol. 64:3411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherpes, T. L., M. A. Melan, J. A. Kant, L. A. Cosentino, L. A. Meyn, and S. L. Hillier. 2005. Genital tract shedding of herpes simplex virus type 2 in women: effects of hormonal contraception, bacterial vaginosis, and vaginal group B streptococcus colonization. Clin. Infect. Dis. 40:1422-1428. [DOI] [PubMed] [Google Scholar]
- 11.Cohn, M. A., S. S. Frankel, S. Rugpao, M. A. Young, G. Willett, S. Tovanabutra, C. Khamboonruang, T. VanCott, L. Bhoopat, S. Barrick, C. Fox, T. C. Quinn, M. Vahey, K. E. Nelson, and D. Weissman. 2001. Chronic inflammation with increased human immunodeficiency virus (HIV) RNA expression in the vaginal epithelium of HIV-infected Thai women. J. Infect. Dis. 184:410-417. [DOI] [PubMed] [Google Scholar]
- 12.Colleluori, D. M., D. Tien, F. Kang, T. Pagliei, R. Kuss, T. McCormick, K. Watson, K. McFadden, I. Chaiken, R. W. Buckheit, Jr., and J. W. Romano. 2005. Expression, purification, and characterization of recombinant cyanovirin-N for vaginal anti-HIV microbicide development. Protein Expr. Purif. 39:229-236. [DOI] [PubMed] [Google Scholar]
- 13.Dey, B., D. L. Lerner, P. Lusso, M. R. Boyd, J. H. Elder, and E. A. Berger. 2000. Multiple antiviral activities of cyanovirin-N: blocking of human immunodeficiency virus type 1 gp120 interaction with CD4 and coreceptor and inhibition of diverse enveloped viruses. J. Virol. 74:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 15.Giomarelli, B., R. Provvedi, F. Meacci, T. Maggi, D. Medaglini, G. Pozzi, T. Mori, J. B. McMahon, R. Gardella, and M. R. Boyd. 2002. The microbicide cyanovirin-N expressed on the surface of commensal bacterium Streptococcus gordonii captures HIV-1. AIDS 16:1351-1356. [DOI] [PubMed] [Google Scholar]
- 16.Gustafson, K. R., R. C. Sowder, Jr., L. E. Henderson, J. H. Cardellina, Jr., J. B. McMahon, U. Rajamani, L. K. Pannell, and M. R. Boyd. 1997. Isolation, primary sequence determination, and disulfide bond structure of cyanovirin-N, an anti-HIV (human immunodeficiency virus) protein from the cyanobacterium Nostoc ellipsosporum. Biochem. Biophys. Res. Commun. 238:223-228. [DOI] [PubMed] [Google Scholar]
- 17.Jerse, A. E. 1999. Experimental gonococcal genital tract infection and opacity protein expression in estradiol-treated mice. Infect. Immun. 67:5699-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenney, T. J., and C. P. Moran, Jr. 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klaenhammer, T., E. Altermann, F. Arigoni, A. Bolotin, F. Breidt, J. Broadbent, R. Cano, S. Chaillou, J. Deutscher, M. Gasson, M. van de Guchte, J. Guzzo, A. Hartke, T. Hawkins, P. Hols, R. Hutkins, M. Kleerebezem, J. Kok, O. Kuipers, M. Lubbers, E. Maguin, L. McKay, D. Mills, A. Nauta, R. Overbeek, H. Pel, D. Pridmore, M. Saier, D. van Sinderen, A. Sorokin, J. Steele, D. O'Sullivan, W. de Vos, B. Weimer, M. Zagorec, and R. Siezen. 2002. Discovering lactic acid bacteria by genomics. Antonie Leeuwenhoek 82:29-58. [DOI] [PubMed] [Google Scholar]
- 20.Kruger, C., Y. Hu, Q. Pan, H. Marcotte, A. Hultberg, D. Delwar, P. J. Van Dalen, P. H. Pouwels, R. J. Leer, C. G. Kelly, C. Van Dollenweerd, J. K. Ma, and L. Hammarstrom. 2002. In situ delivery of passive immunity by lactobacilli producing single-chain antibodies. Nat. Biotechnol. 20:702-706. [DOI] [PubMed] [Google Scholar]
- 21.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 22.Lederman, M. M., R. S. Veazey, R. Offord, D. E. Mosier, J. Dufour, M. Mefford, M. Piatak, Jr., J. D. Lifson, J. R. Salkowitz, B. Rodriguez, A. Blauvelt, and O. Hartley. 2004. Prevention of vaginal SHIV transmission in rhesus macaques through inhibition of CCR5. Science 306:485-487. [DOI] [PubMed] [Google Scholar]
- 23.Lorquet, F., P. Goffin, L. Muscariello, J. B. Baudry, V. Ladero, M. Sacco, M. Kleerebezem, and P. Hols. 2004. Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J. Bacteriol. 186:3749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori, T., L. G. Barrientos, Z. Han, A. M. Gronenborn, J. A. Turpin, and M. R. Boyd. 2002. Functional homologs of cyanovirin-N amenable to mass production in prokaryotic and eukaryotic hosts. Protein Expr. Purif. 26:42-49. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 26.Otten, R. A., D. R. Adams, C. N. Kim, E. Jackson, J. K. Pullium, K. Lee, L. A. Grohskopf, M. Monsour, S. Butera, and T. M. Folks. 2005. Multiple vaginal exposures to low doses of R5 simian-human immunodefiviency virus: strategy to study HIV preclinical interventions in nonhuman primates. J. Infect. Dis. 191:164-173. [DOI] [PubMed] [Google Scholar]
- 27.Patton, D. L., Y. T. Cosgrove Sweeney, M. A. Antonio, L. K. Rabe, and S. L. Hillier. 2003. Lactobacillus crispatus capsules: single-use safety study in the Macaca nemestrina model. Sex. Transm. Dis. 30:568-570. [DOI] [PubMed] [Google Scholar]
- 28.Patton, D. L., Y. C. Sweeney, L. K. Rabe, and S. L. Hillier. 1996. The vaginal microflora of pig-tailed macaques and the effects of chlorhexidine and benzalkonium on this ecosystem. Sex. Transm. Dis. 23:489-493. [DOI] [PubMed] [Google Scholar]
- 29.Pouwels, P. H., and J. A. Leunissen. 1994. Divergence in codon usage of Lactobacillus species. Nucleic Acids Res. 22:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pusch, O., D. Boden, S. Hannify, F. Lee, L. D. Tucker, M. R. Boyd, J. M. Wells, and B. Ramratnam. 2005. Bioengineering lactic acid bacteria to secrete the HIV-1 virucide cyanovirin. J. Acquir. Immune Defic. Syndr. 40:512-520. [DOI] [PubMed] [Google Scholar]
- 32.Quinn, T. C. 2002. The global HIV/AIDS pandemic 2002: a status report. Hopkins HIV Rep. 14:12-15. [PubMed] [Google Scholar]
- 33.Quinn, T. C., and J. Overbaugh. 2005. HIV/AIDS in women: an expanding epidemic. Science 308:1582-1583. [DOI] [PubMed] [Google Scholar]
- 34.Rao, S., S. Hu, L. McHugh, K. Lueders, K. Henry, Q. Zhao, R. A. Fekete, S. Kar, S. Adhya, and D. H. Hamer. 2005. Toward a live microbial microbicide for HIV: commensal bacteria secreting an HIV fusion inhibitor peptide. Proc. Natl. Acad. Sci. USA 102:11993-11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redondo-Lopez, V., R. L. Cook, and J. D. Sobel. 1990. Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev. Infect. Dis. 12:856-872. [DOI] [PubMed] [Google Scholar]
- 36.Royce, R. A., A. Sena, W. Cates, Jr., and M. S. Cohen. 1997. Sexual transmission of HIV. N. Engl. J. Med. 336:1072-1078. [DOI] [PubMed] [Google Scholar]
- 37.Sha, B. E., M. R. Zariffard, Q. J. Wang, H. Y. Chen, J. Bremer, M. H. Cohen, and G. T. Spear. 2005. Female genital-tract HIV load correlates inversely with Lactobacillus species but positively with bacterial vaginosis and Mycoplasma hominis. J. Infect. Dis. 191:25-32. [DOI] [PubMed] [Google Scholar]
- 38.Shenoy, S. R., B. R. O'Keefe, A. J. Bolmstedt, L. K. Cartner, and M. R. Boyd. 2001. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J. Pharmacol. Exp. Ther. 297:704-710. [PubMed] [Google Scholar]
- 39.Sonoda, Y., N. Mukaida, J. B. Wang, M. Shimada-Hiratsuka, M. Naito, T. Kasahara, A. Harada, M. Inoue, and K. Matsushima. 1998. Physiologic regulation of postovulatory neutrophil migration into vagina in mice by a C-X-C chemokine(s). J. Immunol. 160:6159-6165. [PubMed] [Google Scholar]
- 40.Spencer, S. E., I. E. Valentin-Bon, K. Whaley, and A. E. Jerse. 2004. Inhibition of Neisseria gonorrhoeae genital tract infection by leading-candidate topical microbicides in a mouse model. J. Infect. Dis. 189:410-419. [DOI] [PubMed] [Google Scholar]
- 41.Stemmer, W. P., A. Crameri, K. D. Ha, T. M. Brennan, and H. L. Heyneker. 1995. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164:49-53. [DOI] [PubMed] [Google Scholar]
- 42.Taha, T. E., D. R. Hoover, G. A. Dallabetta, N. I. Kumwenda, L. A. Mtimavalye, L. P. Yang, G. N. Liomba, R. L. Broadhead, J. D. Chiphangwi, and P. G. Miotti. 1998. Bacterial vaginosis and disturbances of vaginal flora: association with increased acquisition of HIV. AIDS 12:1699-1706. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, C. C., P. Emau, Y. Jiang, M. B. Agy, R. J. Shattock, A. Schmidt, W. R. Morton, K. R. Gustafson, and M. R. Boyd. 2004. Cyanovirin-N inhibits AIDS virus infections in vaginal transmission models. AIDS Res. Hum. Retrovir. 20:11-18. [DOI] [PubMed] [Google Scholar]
- 44.Turpin, J. A. 2002. Considerations and development of topical microbicides to inhibit the sexual transmission of HIV. Expert Opin. Investig. Drugs 11:1077-1097. [DOI] [PubMed] [Google Scholar]
- 45.Vadeboncoeur, C., M. Frenette, and L. Lortie. 2000. Regulation of the pts operon in low G+C gram-positive bacteria. J. Mol. Microbiol. Biotechnol. 2:483-490. [PubMed] [Google Scholar]
- 46.Vasquez, A., T. Jakobsson, S. Ahrne, U. Forsum, and G. Molin. 2002. Vaginal Lactobacillus flora of healthy Swedish women. J. Clin. Microbiol. 40:2746-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veazey, R. S., R. J. Shattock, M. Pope, J. C. Kirijan, J. Jones, Q. Hu, T. Ketas, P. A. Marx, P. J. Klasse, D. R. Burton, and J. P. Moore. 2003. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 9:343-346. [DOI] [PubMed] [Google Scholar]
- 48.Ventura, M., C. Canchaya, D. Pridmore, B. Berger, and H. Brussow. 2003. Integration and distribution of Lactobacillus johnsonii prophages. J. Bacteriol. 185:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Versalovic, J., T. Koeuth, R. Britton, K. Geszvain, and J. R. Lupski. 1993. Conservation and evolution of the rpsU-dnaG-rpsD macromolecular synthesis operon in bacteria. Mol. Microbiol. 8:343-355. [DOI] [PubMed] [Google Scholar]
- 50.Waldo, G. S., B. M. Standish, J. Berendzen, and T. C. Terwilliger. 1999. Rapid protein folding assay using green fluorescent protein. Nat. Biotechnol. 17:691-695. [DOI] [PubMed] [Google Scholar]
- 51.Walker, P. R., M. Worobey, A. Rambaut, E. C. Holmes, and O. G. Pybus. 2003. Epidemiology: sexual transmission of HIV in Africa. Nature 422:679. [DOI] [PubMed] [Google Scholar]
- 52.Wawer, M. J., R. H. Gray, N. K. Sewankambo, D. Sewadda, and X. Li. 2005. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J. Infect. Dis. 191:1403-1409. [DOI] [PubMed] [Google Scholar]
- 53.Wilkinson, D., M. Tholandi, G. Ramjee, and G. W. Rutherford. 2002. Nonoxynol-9 spermicide for prevention of vaginally acquired HIV and other sexually transmitted infections: systematic review and meta-analysis of randomised controlled trials including more than 5000 women. Lancet Infect. Dis. 2:613-617. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565-2573. [DOI] [PubMed] [Google Scholar]